Abstract
The recessive tall rice (Oryza sativa) mutant elongated uppermost internode (eui) is morphologically normal until its final internode elongates drastically at the heading stage. The stage-specific developmental effect of the eui mutation has been used in the breeding of hybrid rice to improve the performance of heading in male sterile cultivars. We found that the eui mutant accumulated exceptionally large amounts of biologically active gibberellins (GAs) in the uppermost internode. Map-based cloning revealed that the Eui gene encodes a previously uncharacterized cytochrome P450 monooxygenase, CYP714D1. Using heterologous expression in yeast, we found that EUI catalyzed 16α,17-epoxidation of non-13-hydroxylated GAs. Consistent with the tall and dwarfed phenotypes of the eui mutant and Eui-overexpressing transgenic plants, respectively, 16α,17-epoxidation reduced the biological activity of GA4 in rice, demonstrating that EUI functions as a GA-deactivating enzyme. Expression of Eui appeared tightly regulated during plant development, in agreement with the stage-specific eui phenotypes. These results indicate the existence of an unrecognized pathway for GA deactivation by EUI during the growth of wild-type internodes. The identification of Eui as a GA catabolism gene provides additional evidence that the GA metabolism pathway is a useful target for increasing the agronomic value of crops.
INTRODUCTION
Gibberellins (GAs) are a group of diterpenoid compounds, some of which act as growth-promoting hormones controlling such diverse processes as stem elongation, leaf expansion, seed germination, and flowering. The GA biosynthesis pathway has long been studied, and the majority of genes encoding enzymes in each step of the biosynthesis and catabolism pathways have been identified in the model plant species Arabidopsis thaliana and rice (Oryza sativa) (for reviews, see Hedden and Phillips, 2000; Olszewski et al., 2002; Sun and Gubler, 2004). In rice, some mutants with altered GA metabolism or signaling pathways have been studied in detail (Ueguchi-Tanaka et al., 2000; Ikeda et al., 2001; Sasaki et al., 2003; Sakamoto et al., 2004). Recently, GIBBERELLIN INSENSITIVE DWARF1 has been identified as a soluble GA receptor in rice (Ueguchi-Tanaka et al., 2005). Such GA-related mutants have been important not only to identify key components in the GA metabolism and signaling pathways but also to confer useful agronomic traits in cereals, some of which contributed to the success of the green revolution (Peng et al., 1999; Sasaki et al., 2002; Spielmeyer et al., 2002).
GAs are biosynthesized from geranylgeranyl diphosphate, a common C20 precursor for diterpenoids. Conversions of geranylgeranyl diphosphate into bioactive GAs, such as GA1 and GA4 (Figure 1), involve three classes of enzymes: plastid-localized terpene cyclases, membrane-bound cytochrome P450 monooxygenases (P450s), and soluble 2-oxoglutarate-dependent dioxygenases (2ODDs). In rice vegetative tissues, the early 13-hydroxylation pathway, leading to the formation of bioactive GA1, has been shown to occur predominantly (Figure 1), while GA4, the bioactive non-13-hydroxylated GA (13-H GA), accumulated to a high level in anthers (Kobayashi et al., 1988).
Figure 1.
The Major GA Biosynthesis and Catabolism Pathways in Plants.
Both GA1 and GA4 are biologically active GAs. Open circles, closed circles, and closed squares indicate reactions catalyzed by GA 20-oxidase, GA 3-oxidase, and GA 2-oxidase, respectively. The gene for GA 13-oxidase has not been identified. The structure of bioactive GA is also shown with carbon numbers. GGDP, geranylgeranyl diphosphate; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase.
Bioactive GA1 and GA4 and their immediate precursors GA20 and GA9, respectively, are deactivated by GA 2-oxidases (Figure 1), another class of 2ODDs (Thomas et al., 1999; Yamaguchi and Kamiya, 2000; Olszewski et al., 2002). Recently, a new class of 2ODD, including AtGA2ox7 and AtGA2ox8 of Arabidopsis, has been shown to catalyze 2-oxidation of C20-GAs (precursor GAs), including GA12 and GA53 (Schomburg et al., 2003; Lee and Zeevaart, 2005). In pea (Pisum sativum), a loss-of-function mutation in the PsGA2ox1 gene slender causes a tall phenotype (Lester et al., 1999). However, no loss-of-function mutant of the GA2ox gene family members has been recognized as a tall mutant in other plant species, including Arabidopsis and rice, likely due to functional redundancy among the family members (Hedden and Phillips, 2000; Sakamoto et al., 2004). Thus, it has yet to be clarified genetically whether GA 2-oxidation, the only well-characterized GA catabolism reaction so far, is commonly the major GA deactivation step(s) in plants or if any other catabolic route also plays a role in controlling bioactive GA levels.
Male sterile (MS) cultivars play a critical role in the production of hybrid rice seeds, an important strategy that has increased the rice yield significantly in agriculture (Li and Yuan, 2000). Despite the necessity of MS cultivars in the production of hybrid seeds, rice MS cultivars commonly have a defect in the growth of internodes (in particular, the uppermost internode), which causes an enclosure of the panicle in leaf sheaths at the heading stage. In agriculture, exogenous GA3 treatment has been used to stimulate panicle emergence in MS cultivars. However, the GA3 application also stimulates preharvest sprouting of grains and shortens the longevity of hybrid seeds because of the activity of this hormone to reduce seed dormancy and stimulate germination. The recessive tall rice mutant elongated uppermost internode (eui) (Rutger and Carnahan, 1981) exhibits a rapid and enhanced elongation of internodes, particularly the uppermost internode during the heading stage. Therefore, the eui mutation has been used to genetically improve the heading performance of MS cultivars (Shen and He, 1989; He and Shen, 1991, 1994; Yang et al., 2002). Introduction of the eui mutation has been a breakthrough technology in hybrid rice breeding, and rice varieties carrying the eui mutation have been officially released (Liu et al., 2004). Because the eui mutation is able to replace the effect of GA3 treatment in agriculture, it has been thought that the Eui gene might be related to GA metabolism or signaling. However, it has been unclear how EUI controls internode elongation at the molecular level.
Toward the identification of the Eui gene, we previously mapped this gene within a 98-kb region of chromosome 5 (Xu et al., 2004). In this work, we have isolated the Eui gene and studied its biological and biochemical functions. We show that eui plants accumulate extremely high levels of bioactive GAs in the uppermost internode at the heading stage. Map-based cloning reveals that the Eui gene encodes a previously uncharacterized P450, CYP714D1. Using heterologous expression in yeast, we demonstrate that EUI acts as a GA-deactivating enzyme through 16α,17-epoxidation of 13-H GAs. We discuss how EUI controls bioactive GA levels to modulate internode elongation in a tissue- and developmental stage–specific manner.
RESULTS
Characterization and GA Levels of the eui Mutant
During the seedling and tillering stages, eui plants were morphologically similar to wild-type plants (He and Shen, 1994). However, at the heading stage, the eui mutant exhibited an extremely elongated uppermost internode, with slightly elongated second and third internodes and panicle (Figures 2A and 2B). Because of the enhanced internode elongation, the stem exposed between the ear and the flag leaf sheath (panicle exsertion) is much longer in the eui mutant than in wild-type plants (Figure 2C). The enhanced internode elongation of the eui mutant was due to longitudinally increased cell lengths but not to an increase in the number of cells (Figure 2D). These observations suggested that the uppermost internode of the eui mutant might accumulate an excessive amount of biologically active GAs or exhibit an enhanced GA sensitivity.
Figure 2.
Phenotypic Characterization of the eui Mutant.
(A) Wild-type ZS97 and eui-1 plants 10 d after heading. Bar = 10 cm.
(B) Culms of wild-type and eui-1 plants. Arrows indicate nodes. Bar = 10 cm.
(C) Panicle exsertion (heading performance) of wild-type and eui-1 plants. Bar = 10 cm.
(D) Longitudinal sections (×200) of the elongated regions of the uppermost internodes of wild-type and eui-1 plants.
To determine the specific cause of the eui phenotype, we measured endogenous GA levels in the uppermost internodes of both wild-type and eui plants using gas chromatography–mass spectrometry (GC-MS). In rice vegetative organs, the early 13-hydroxylation pathway has been thought to be the major route for GA biosynthesis, where GA1 functions as the active form (Kobayashi et al., 1988; Figure 1). GC-MS analysis showed that eui plants contained a much higher level of GA1 (24-fold) than did wild-type plants (Figure 3A; see Supplemental Table 1 online). More strikingly, GA4, the bioactive form in the non-13-hydroxylation pathway (Figure 1), was undetectable in wild-type plants but was present at an extremely high concentration in the eui mutant (Figure 3A; see Supplemental Table 1 online). These results indicated that the eui phenotype was caused by elevated levels of bioactive GAs.
Figure 3.
Endogenous GA Levels and GAox Gene Expression in eui and Wild-Type Plants.
(A) GA levels in the uppermost internodes of eui-2 (closed bar) and wild-type (open bar) plants. GA measurements were repeated using two other eui alleles (eui-1 and eui-3) with similar results (see Supplemental Table 1 online). Note that the results are shown in a discontinuous scale on the y axis, so that differences in the levels of the GAs other than GA4 can be seen clearly. GAs are displayed according to the order in the biosynthesis pathway (see Figure 1). Asterisks, undetectable GAs.
(B) Transcript levels of rice GA20ox2, GA3ox2, and GA2ox1 in the uppermost internodes of wild-type and eui-1 plants, as estimated by RT-PCR. Ubi-1, control for RT-PCR.
Some GA biosynthesis and catabolism genes have been shown to be regulated negatively and positively, respectively, by GA activity (for reviews, see Hedden, 1999; Olszewski et al., 2002). Figure 3B shows that the rice GA biosynthesis genes GA20ox2 and GA3ox2 were downregulated, while the rice GA catabolism gene GA2ox1 was upregulated in the eui mutant. These results are consistent with our finding that the eui mutant accumulated high levels of bioactive GAs (Figure 3A). Although the observed changes in gene expression would contribute to maintaining the homeostasis of bioactive GA levels (Olszewski et al., 2002), these regulatory mechanisms appear to be insufficient to fully circumvent the effect of the eui mutation.
Eui Encodes a P450
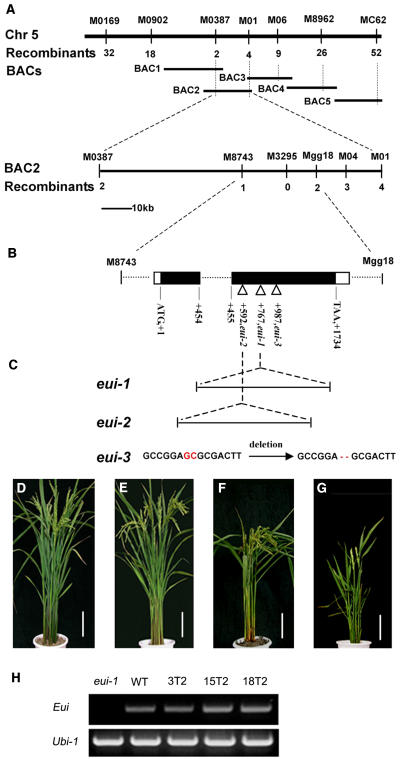
The Eui locus was previously mapped onto a 98-kb region of chromosome 5 (Xu et al., 2004). In this study, the gene was further narrowed down to a 24-kb genomic fragment containing only one annotated gene that exists as a single copy gene in the rice genome (Figures 4A and 4B). The identity of this annotated gene as Eui was initially confirmed by sequencing three independent eui alleles: two interrupted by either retrotransposon (eui-1) or a rice genomic fragment (eui-2) and one containing a two-nucleotide deletion (eui-3) in the coding region (Figure 4C). They are spontaneous or γ-ray–induced mutations (Rutger and Carnahan, 1981; Zhu et al., 2003). Sequence comparison between the isolated Eui cDNA and the published rice genomic DNA revealed a large intron (∼8 kb) that separates two exons (Figure 4C).
Figure 4.
Map-Based Cloning of Eui.
(A) Physical mapping of Eui. Eui was finally positioned on BAC2 (BAC OSJNBa0095J22) within a 24-kb region flanked by the markers M8743 and Mgg18 and cosegregated with the marker M3295. Numbers represent recombination events.
(B) Mutation positions of three eui alleles, eui-1 (307T), eui-2 (SKZ-T), and eui-3 (ZH11-T), in the Eui coding region that is separated by an intron.
(C) eui-1 and eui-2 mutations are attributed to insertions of a retrotransposon and a DNA fragment, respectively, while a two-nucleotide deletion was found in the eui-3 allele, in the Eui coding region.
(D) to (G) Genetic complementation of the eui mutant. Wild-type plant (ZS97) 10 d after heading (D). eui-1 plants complemented by one copy (3T2) (E), two copies (15T2) (F), and three copies (18T2) (G) of the wild-type Eui gene. Bars = 10 cm.
(H) Eui mRNA detected by RT-PCR in elongating internodes. Ubi-1 was used as a control. No Eui transcript was detected in the mutant. The RNA analysis was repeated three times with similar results.
The identity of the selected gene as Eui was further confirmed by a complementation test, in which a 13-kb wild-type genomic DNA fragment harboring the entire candidate gene rescued the eui-1 phenotype (Figures 4D and 4E). When multiple copies of the transgene were introduced, there was a reverse correlation between the length of the uppermost internode and the copy number and the transcript level of the transgene (Figures 4F to 4H), indicating that Eui controls the stem length in a dose-dependent manner.
A BLAST search revealed that the Eui gene encodes a putative P450 of 577 amino acids, with conserved amino acid sequences that may function as heme binding, oxygen binding and activation, and ERR triad domains (Schuler and Werck-Reichhart, 2003). P450s constitute a large superfamily in plants; 246 and 356 (putative) P450 genes have been identified in the genome of Arabidopsis and rice, respectively, and their phylogenetic relationships have been studied (Nelson et al., 2004). The EUI sequence belongs to the CYP714 family (Figure 5; see Supplemental Figure 1 online) and has been designated as CYP714D1 according to the Nelson nomenclature on the basis of the sequence identity of >40% with other family members (http://drnelson.utmem.edu/rice.color.sept12.html). No catalytic or biological function has been assigned to a CYP714 member.
Figure 5.
Phylogenetic Relationships among the CYP714 Family Members.
Amino acid sequences of the CYP714 family members were obtained from the Cytochrome P450 Homepage (http://drnelson.utmem.edu/cytochromeP450.html; see Supplemental Figure 1 online). The family includes CYP714A1 (accession number NM_122400) and CYP714A2 (NM_122399) of Arabidopsis, CYP714B1 (AP005099), CYP714C1 (AP008218), CYP714C2 (AK066943), CYP714C3 (AP008217), and CYP714D1/EUI (AK109526) of rice, and Lol-31-b (AF321861) and Lol-31-j (AF321862) of Lolium rigidum. The scale bar indicates the number of amino acid substitutions per site. Bootstrap values in percentage are shown at each branch point.
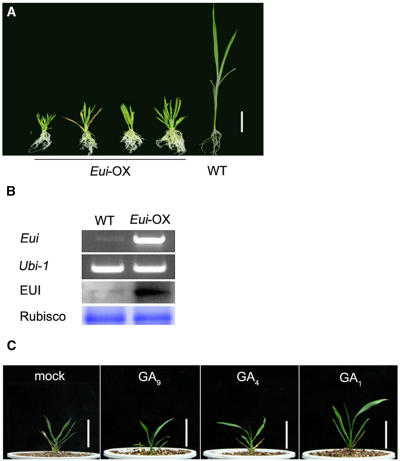
Overexpression of Eui Causes Severe Dwarfism
To further investigate the role of EUI in plant development, we generated transgenic rice plants that overexpress the Eui cDNA (Eui-OX) driven by the constitutively active cauliflower mosaic virus 35S promoter. The Eui-OX plants were extremely dwarfed and failed to produce seeds (Figure 6A). Overexpression of the Eui gene in transgenic lines was confirmed by RT-PCR and immunoblot analysis (Figure 6B). Treatment with 10 μM GA1 promoted the growth of dwarfed Eui-OX plants (Figure 6C). GA4 and GA9, both of which are 13-H GAs (Figure 1), were not as effective as GA1 in rescuing the dwarfed Eui-OX phenotype, in comparison with GA1 (Figure 6C), while GA1 and GA4 were nearly equally active in promoting the growth of eui seedlings when applied exogenously (data not shown).
Figure 6.
Overexpression of Eui (Eui-OX) in Transgenic Rice.
(A) Two-month-old Eui-OX plants and 2-week-old wild-type plant. Bar = 5 cm.
(B) Confirmation of transgene expression in Eui-OX plants. A representative RT-PCR analysis (Eui) and immunoblot detection by an anti-EUI antibody (EUI) are shown. Ubi-1, control for RT-PCR; Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase used as loading control for immunoblot analysis.
(C) Effect of different GAs on the growth of 1-month-old Eui-OX plants. Plants were treated with 10 μM GA solution for 1 week. Bars = 5 cm.
EUI Catalyzes 16α,17-Epoxidation of 13-H GAs
Given the morphological and biochemical features of the eui mutants and Eui-OX transgenic plants and the predicted protein identity of the Eui gene, we suspected that EUI might be a novel GA-deactivating enzyme. To directly test our hypothesis, we examined the enzyme activity by incubating a microsomal fraction of EUI-expressing yeast with various GAs as putative substrates. Because GA4 was the most abundant GA accumulated in the eui mutant (Figure 3A), we initially incubated [17-14C]GA4 with recombinant EUI. Analysis on silica-gel thin layer chromatography revealed that [17-14C]GA4 was converted to a polar product by the EUI-containing microsomal proteins (Figure 7A), whereas no conversion occurred when a microsomal preparation from the control yeast carrying the empty vector was used (data not shown). Using nonlabeled GA4 as a substrate, the product of EUI was identified as 16,17-dihydro-16α,17-dihydroxy GA4 (16α,17-[OH]2-GA4) by full-scan GC-MS, based on its retention time on GC and the mass spectrum (Table 1, Figure 7B).
Figure 7.
Enzymatic Function of EUI and Biological Activity of EUI Products.
(A) [14C]GA4 was incubated with a yeast microsomal preparation containing EUI. Metabolites were separated by silica-gel thin layer chromatography. Control, yeast microsome without EUI. Position of authentic GA4 is shown by a black arrow. The white arrow indicates the EUI product.
(B) Reactions catalyzed by recombinant EUI in vitro. GA12, GA9, and GA4 were converted to 16α,17-epoxy derivatives by EUI (Table 2). The epoxides were chemically converted to 16,17-dihydro-16α,17-dihydroxy GAs when the products were purified in the presence of 1% acetic acid (Table 1).
(C) Biological activity of EUI products. One (gray bar) or 10 (black bar) ng of GA samples was applied to seedlings of Tan-ginbozu. Data are means ± se (n = 12). Mock, treatment without GA; (OH)2-GA4, synthetic 16,17-dihydro-16α,17-dihydroxy GA4; +EUI, GA4 incubated with EUI-containing yeast microsome (16α,17-epoxy GA4); –EUI, unmetabolized GA4 after incubation with the control yeast microsome. Asterisks indicate the amount of microsome-treated GA4 (or its metabolite) applied to the plant estimated from the amount used as a substrate, based on the assumption that the recovery is 100%.
Table 1.
GC-MS Profiles of GA Metabolites by Recombinant EUIa
| Sample | Retention Time on GC in min (KRI)b | Characteristic Ions, m/z (Relative Intensity)c |
|---|---|---|
| GA4 metabolited | 10.85 (2927) | 581 [M+-15] (2), 493 (100), 299 (3), 217 (14), 181 (7), 147 (30), 129 (13), 73 (95) |
| Authentic 16α,17-[OH]2 GA4d | 10.87 (2929) | 581 [M+-15] (2), 493 (100), 299 (4), 217 (9), 181 (10), 147 (25), 129 (12), 73 (90) |
| Authentic 16β,17-[OH]2 GA4d | 10.58 (2895) | 581 [M+-15] (2), 493 (100), 299 (4), 217 (6), 181 (9), 147 (21), 129 (10), 73 (87) |
| GA9 metabolited | 9.77 (2792) | 493 [M+-15] (2), 405 (100), 345 (2), 301 (4), 217 (10), 183 (14), 147 (29), 73 (76) |
| Authentic 16α,17-[OH]2 GA9d | 9.80 (2793) | 493 [M+-15] (2), 405 (100), 345 (2), 301 (4), 217 (10), 183 (15), 147 (25), 73 (77) |
| Authentic 16β,17-[OH]2 GA9d | 9.60 (2769) | 493 [M+-15] (3), 405 (100), 345 (2), 301 (4), 217 (6), 183 (13), 147 (21), 73 (77) |
| GA12 metabolited | 9.80 (2797) | 523 [M+-15] (5), 506 (7), 448 (6), 435 (60), 416 (10), 375 (22), 239 (20), 147 (28), 73 (100) |
| Authentic 16α,17-[OH]2 GA12d | 9.80 (2796) | 523 [M+-15] (3), 506 (4), 448 (5), 435 (60), 416 (6), 375 (26), 239 (17), 147 (30), 73 (100) |
Metabolites were purified in the presence of 1% acetic acid.
Kovats retention index.
m/z, mass-to-charge ratio.
Methyl ester-trimethylsilyl (TMSi) ether derivative.
Because P450s are known to function as monooxygenases (Schuler and Werck-Reichhart, 2003), 16α,17-[OH]2-GA4 is unlikely to be the direct product by EUI activity from GA4. We therefore speculated that a 16α,17-epoxy GA was initially produced and then hydrated to form the diol during the purification in the presence of acetic acid, which was included in the initial experiment for a better separation of the products on a reverse-phase column. We prepared 16,17-epoxy GA4 chemically (as methyl ester) and tested this possibility. When acetic acid was omitted during purification, the product of GA4 by EUI activity was identified to be 16α,17-epoxy GA4 by GC-MS (Table 2). From these results, we can conclude that EUI is a GA 16α,17-epoxidase and that 16α,17-epoxy GA4 is converted to 16α,17-[OH]2-GA4 in vitro in an acidic condition (Figure 7B).
Table 2.
GC-MS Profiles of GA Metabolites by Recombinant EUIa
| Sample | Retention Time on GC in min (KRI) | Characteristic Ions, m/z (Relative Intensity) |
|---|---|---|
| GA4 metaboliteb | 9.07 (2709) | 434 [M+] (5), 305 (15), 249 (32), 245 (23), 241 (37), 217 (22), 183 (23), 129 (42), 73 (100) |
| Authentic 16α,17-epoxy GA4b | 9.05 (2707) | 434 [M+] (6), 305 (15), 249 (34), 245 (25), 241 (42), 217 (24), 183 (26), 129 (46), 73 (100) |
| Authentic 16β,17-epoxy GA4b | 9.00 (2700) | 434 [M+] (5), 305 (16), 249 (28), 245 (23), 241 (40), 217 (20), 183 (21), 129 (38), 73 (100) |
| GA9 metabolitec | 7.97 (2524) | 346 [M+] (2), 286 (8), 270 (8), 242 (100), 225 (19), 185 (37), 183 (32), 159 (21), 129 (20) |
| Authentic 16α,17-epoxy GA9c | 7.97 (2524) | 346 [M+] (2), 286 (7), 270 (7), 242 (100), 225 (18), 185 (37), 183 (32), 159 (18), 129 (18) |
| Authentic 16β,17-epoxy GA9c | 7.90 (2512) | 346 [M+] (2), 286 (8), 270 (6), 242 (100), 225 (20), 185 (38), 183 (39), 159 (20), 129 (20) |
| GA12 metabolitec | 8.22 (2556) | 376 [M+] (1), 344 (100), 316 (38), 301 (38), 285 (29), 257 (77), 241 (43), 227 (40), 199 (63) |
| Authentic 16α,17-epoxy GA12c | 8.22 (2556) | 376 [M+] (2), 344 (100), 316 (38), 301 (36), 285 (25), 257 (76), 241 (35), 227 (40), 199 (59) |
Metabolites were purified in the absence of acetic acid.
Methyl ester-TMSi ether derivative.
Methyl ester derivative. M+, molecular ion.
To clarify whether EUI uses other GAs and/or GA precursors as substrates, we incubated ent-kaurenoic acid, GA12, GA9, and GAs in the early 13-hydroxylation pathway (GA53, GA20, and GA1; Figure 1) with recombinant EUI. GC-MS analysis of the reaction products showed that GA12 and GA9 were metabolized by EUI into 16α,17-epoxy GA12 and 16α,17-epoxy GA9, respectively (Figure 7B, Table 2). As was the case with 16α,17-epoxy GA4, the 16α,17-epoxides of GA12 and GA9 were converted to corresponding 16α,17-[OH]2-GAs in the presence of acetic acid (Figure 7B, Table 1). When GA1, GA20, GA53, and ent-kaurenoic acid were tested as substrates, the majority of the added substrates remained unmetabolized as in the control (no EUI) reaction mixture, which is in contrast with the case for GA4, GA9, and GA12, all of which were nearly undetectable after the incubation with EUI (see Supplemental Figure 2 online). These results indicate that recombinant EUI can catalyze 16α,17-epoxidation of both bioactive and precursor GAs and that 13-H GAs (GA4, GA9, and GA12), rather than their 13-hydroxylated forms (GA1, GA20, and GA53), are preferred substrates for EUI in vitro.
Identification of 16α,17-[OH]2-GAs in Rice
It was previously reported that a fraction of isotopically labeled GA4 applied to rice seedlings was converted to 16α,17-[OH]2-GA4 (Kobayashi et al., 1993). The 16α,17-[OH]2-GA4 was also identified as an aglycon of an endogenous GA glucoside in rice anthers (Hasegawa et al., 1994). To address whether EUI functions as GA 16α,17-epoxidase in planta, we analyzed endogenous 16α,17-epoxy GAs and their hydrated products in Eui-OX and wild-type plants. Unlike the case with yeast microsome preparations, we found it technically difficult to purify 16α,17-epoxy GAs from plant extracts without being exposed to any acidic condition due to much larger amounts of impurities. In addition, it is possible that 16α,17-[OH]2-GAs are formed from the corresponding 16α,17-epoxy GAs in planta, and it may not be possible to identify the epoxides in plant extracts. We therefore focused on the hydrated products (16α,17-[OH]2-GAs) purified under acidic conditions in the following experiments. GC-MS and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses showed that levels of 16α,17-[OH]2-GA12 and 16α,17-[OH]2-GA9 were 18- and 12-fold greater in Eui-OX plants than in the wild type (Table 3). In addition, the levels of all GAs that we measured (including GA12, GA9, and metabolites of GA1; Figure 1) were decreased in Eui-OX plants in comparison with the wild type (see Supplemental Table 2 online). These results support the idea that EUI functions as GA 16α,17-epoxidase in planta. The 16α,17-[OH]2-GA4 was not detected in Eui-OX and wild-type plants due to its low abundance, possibly because the substrate GA4 was not present at high concentrations. Because GA12 is a biosynthetic precursor for both 13-H and 13-hydroxylated GAs (13-OH GAs) (Figure 1), the depletion of the GA12 pool by EUI activity would result in a reduction in the levels of bioactive GAs, which is consistent with the observed dwarfism of Eui-OX plants (Figure 6A). Because of the necessity of exogenous GA treatment to maintain the viability of Eui-OX plants, we could not determine the endogenous level of GA1 in the Eui-OX plants in this experiment (see Supplemental Methods online). The occurrence of 16α,17-[OH]2-GA4 and -GA9 in the uppermost internode of wild-type plants was also suggested by GC-MS analysis (see Supplemental Methods online).
Table 3.
Levels of 16α,17-Dihydroxy GAs in Eui-OX and Wild-Type Plants
| Sample | Ions Used for Quantification, m/z (Relative Intensity)a | Relative Levelb |
|---|---|---|
| Wild-type | Endogenous: 365 [M-H]− (100), 347 (8), 321 (8) | 1 |
| 16α,17-[OH]2 GA12c | Internal standard (2H2): 367 [M-H] (100), 349 (8), 323 (7) | |
| Eui-OX | Endogenous: 365 [M-H]− (100), 347 (10), 321 (7) | 18 |
| 16α,17-[OH]2 GA12c | Internal standard (2H2): 367 [M-H] (100), 349 (8), 323 (7) | |
| Wild-type | Endogenous: 493 [M+-15] (5), 405 (100), 345 (4) | 1 |
| 16α,17-[OH]2 GA9d | Internal standard (2H3): 496 [M+-15] (3), 408 (100), 348 (2) | |
| Eui-OX | Endogenous: 493 [M+-15] (3), 405 (100), 345 (3) | 12 |
| 16α,17-[OH]2 GA9d | Internal standard (2H3): 496 [M+-15] (3), 408 (100), 348 (2) |
Only representative ions are shown. The intensity of the most abundant ion was set as 100.
The values for wild-type samples were arbitrarily set as 1.
Determined without derivatization on LC-MS/MS.
Determined as methyl ester-TMSi ether derivatives on GC-selected ion monitoring. The 16α,17-[OH]2 GA4 could not be quantified due to its low abundance in both samples and comigration of impurities.
GA 16α,17-Epoxidation Is a Deactivation Reaction
The tall and dwarfed phenotypes of the eui mutants (Figure 2A) and the Eui-OX transgenic plants (Figure 6A) suggest that EUI plays a role in GA deactivation. To verify this premise, we examined the biological activity of 16α,17-epoxy GA4 and 16α,17-[OH]2-GA4 using seedlings of Tan-ginbozu, a GA-deficient semidwarf cultivar with reduced KO activity (Itoh et al., 2004). Because synthetic 16α,17-epoxy GA4 was available only as a methyl ester derivative, we used 16α,17-epoxy GA4 prepared enzymatically for bioassay. Figure 7C shows that treatment with EUI significantly reduced the biological activity of GA4 and that the activity of synthetic 16α,17-[OH]2-GA4 was much lower than that of GA4. These results demonstrate that EUI functions as a GA-deactivating enzyme in rice.
EUI Protein Is Anchored on the Endoplasmic Reticulum
Plant P450s are usually anchored on the cytoplasimc surface of the endoplasmic reticulum (ER) and are occasionally associated with the plastids (Schuler and Werck-Reichhart, 2003). We first estimated subcellular localization of EUI protein by instantly expressing a chimeric EUI–green fluorescent protein (EUI-GFP) in onion epidermic cells and showed that the EUI-GFP fusion protein was targeted to the cytoplasm (Figures 8A and 8B). A more accurate assay by immunogold detection using an anti-EUI antibody revealed that EUI protein was localized on the cytoplasmic surface of the ER (Figures 8C and 8D). A similar ER-localized pattern was also found for P450 CYP80B1 and CYP2B1 (Alcantara et al., 2005; Lengler et al., 2005).
Figure 8.
Subcellular Localization of EUI Protein.
(A) and (B) Cytoplasm localization of the EUI-GFP fusion protein (A), with GFP alone as a control (B). N, nucleus. Bar = 100 μm.
(C) and (D) Immunogold detection of the EUI protein on the cytoplasmic surface of the ER in a wild-type (C) and eui-1 (D) cell (×50,000) in the uppermost internode. Arrows indicate gold particles. CW, cell wall.
Developmental Regulation of the Eui Gene
To assess developmental regulation of Eui expression, β-glucuronidase (GUS) activity was examined histochemically in transgenic plants carrying an Eui promoter–GUS reporter gene fusion. Eui-GUS expression was observed with a pattern of strong tissue and development specificity; GUS activity was detected mainly in rapidly elongating or dividing tissues, including the shoot apical meristem, the divisional (intercalary meristem) and elongating zones of internodes, nodes of an elongating stem, and panicle (Figures 9B to 9I and 9L) but not in young seedlings, roots, and the flag leaf (Figures 9A, 9J, and 9K). Before heading, the highest Eui-GUS expression was detected in the divisional and node areas of internodes and young panicles (Figure 9B, 9F, and 9G). During the heading stage, the highest Eui-GUS expression was detected in the flowering spikelets, the divisional zone, and the node of the uppermost internode (Figures 9C, 9E, 9H, and 9I).
Figure 9.
Developmental Regulation of Eui Expression.
(A) No GUS staining in young seedlings 1 week after germination.
(B) and (C) GUS activity in young panicle and internodes 4 weeks (B) and 1 week (C) before heading.
(D) and (E) GUS activity in young spikelets (D) and flowering spikelets (E).
(F) and (G) GUS activity in the shoot apical meristem and the divisional zone and node region of internodes during the tillering (F) and the elongating/bolting (G) stages.
(H) and (I) GUS activity in the divisional zone and node region of the young uppermost internode 1 week before heading (H) and the newly elongated uppermost internodes (I) at the heading stage.
(J) and (K) No GUS staining in roots and flag leaves.
(L) Schematic representation of the rice stem. The three regions are shown in the uppermost internode.
(M) Eui transcript detected by RT-PCR DNA gel blot hybridization in different tissues, using ubi-1 as control. LS, leaf of seedling; SS, stem of seedling; RS, root of seedling; YP, young panicle 2 weeks before heading as in (C); SB, shoot during the bolting stage as in (G); LH, flag leaf during heading stage; PH, flowering panicle during the heading stage as in (E); IL, the lower part of the uppermost internode, including the divisional, node, and elongating regions, as in (H) and (I); IU, the upper elongated part of the uppermost internode; SI, second internode during the heading stage as in (I). Independent experiments showed similar results. Arrows indicate the uppermost internode in (B), (C), (H), and (I). Bars = 1 cm.
To examine whether Eui-GUS expression correlates with Eui transcript abundance, we performed analysis of the Eui transcript using RNA samples from different tissues/organs (Figure 9M). Our results showed that flowering panicles accumulated the Eui transcript at the highest level, followed by unflowered panicles and the lower part (including the elongating and divisional zones and the node) of the uppermost internode at the heading stage. These data are largely in agreement with the pattern of Eui-GUS expression (Figures 9A to 9K) and with the developmental stage–specific eui mutant phenotypes (Figure 2). Previous work has indicated that the rice GA biosynthesis genes GA20ox2 and GA3ox2 are also predominantly expressed in the divisional zone of elongating internodes and flowers (Kaneko et al., 2003). Therefore, our results from Eui-GUS expression and Eui transcript analysis suggest that the sites of GA deactivation by EUI and the synthesis of bioactive GAs may partly overlap at the heading stage.
DISCUSSION
EUI Is a New GA-Deactivating Enzyme
In this article, we have demonstrated that the tall recessive rice mutant eui contains large amounts of bioactive GAs in the uppermost internode and that the Eui gene encodes a P450 that deactivates GAs through GA 16α,17-epoxidation. The only well-characterized GA deactivation reaction has been GA 2-oxidation (Thomas et al., 1999; Olszewski et al., 2002), which is catalyzed by soluble 2ODDs. Because some 2β-hydroxylated GAs, such as GA8, GA29, GA34, and GA51, were detectable in the uppermost internode (Figure 3A; see Supplemental Table 1 online), GA 2-oxidation may also take place in this tissue. However, the extremely large amounts of GA1 and GA4 found in the eui mutants illustrate that GA 2-oxidase activity is clearly insufficient to deactivate a fraction of GA pools that are normally metabolized by EUI in wild-type plants. These observations suggest that EUI is a major GA catabolism enzyme in internodes of rice at the heading stage.
Recombinant EUI produced in yeast was able to catalyze 16α,17-epoxidation of multiple GA substrates, including precursor GAs (GA12 and GA9) and bioactive GA4 (Figure 7A). The accumulation of 16α,17-[OH]2-GA12 at a high level in Eui-OX transgenic rice suggests that EUI is capable of reducing the pool size of an early precursor, GA12, in planta (Table 3). Increased levels of GA12 in the eui mutants relative to wild-type plants in the uppermost internode (see Supplemental Table 1 online) also support this possibility. Because both KAO and EUI are located on the ER membrane (Figure 8; Helliwell et al., 2001b), GA12 produced by KAO might be used efficiently as a substrate by EUI in the cell. Recent work has shown that a new type of 2ODD catalyzes 2-oxidation of precursor GAs (C20-GAs), such as GA12 and GA53, in Arabidopsis and spinach (Spinacia oleracea) (Schomburg et al., 2003; Lee and Zeevaart, 2005). In spinach, one of such GA 2-oxidase, SoGA2ox3, is regulated by photoperiod, suggesting that catabolism of precursor GAs may play a key regulatory role in altering bioactive GA levels in some cases. Our data suggest that at least part of EUI activity is responsible for decreasing the pool size of precursor GAs. The promiscuous nature of EUI in terms of the recognition of ring A modifications will allow this enzyme to use multiple substrates in the GA metabolism pathway, and this feature may contribute to altering the levels of bioactive GAs drastically (Figure 3A).
Our results using recombinant EUI indicated that 13-hydroxylation greatly affects the substrate preference of this enzyme; 13-H GAs were more efficiently metabolized by EUI than 13-OH GAs in vitro (see Supplemental Figure 2 online). In addition, GA1 was more effective in rescuing the dwarfed Eui-OX phenotype than GA4 (Figure 6C), suggesting that GA4 might be deactivated more efficiently than GA1 by EUI in planta as well. At the moment, the biological role of GA 13-hydroxylation is not clear. Identification of genes encoding GA 13-hydroxylase would facilitate our understanding on the biological meaning of the substrate preference of EUI.
A Negative Regulation Mechanism by EUI
Our data demonstrate that EUI activity can alter bioactive GA levels drastically in the uppermost internode at the heading stage. Tissue- and stage-specific occurrences of the Eui expression (Figure 9) and the eui mutant phenotypes (Figure 2) suggest that EUI regulation of bioactive GA levels operates in restricted developmental processes in rice. At the heading stage, the operation of the EUI deactivation mechanism will allow a rapid and drastic change in bioactive GA content, thereby helping the control of the timing and speed of the uppermost internode elongation and panicle emergence under various environmental conditions.
Recent studies have shown that DELLA proteins function as negative regulators of the GA signaling pathway and that GA signal is transmitted via removing these negative regulators (for a review, see Sun and Gubler, 2004). Our data illustrate that a negative regulation mechanism also exists in the GA metabolism pathway; the active GA levels can be elevated effectively by removing the deactivation reaction by EUI (Figure 3) because there is a large flux of the synthesis of inactive GAs at the basal state. The occurrence of such regulatory components in both GA metabolism and signaling pathways would contribute to flexibly modulating the GA-mediated signal both negatively and positively during plant development and in response to environmental cues.
EUI Is a Member of the CYP714 Family
A number of P450s have been shown to be involved in the biosynthesis and/or catabolism of plant hormones (Schuler and Werck-Reichhart, 2003; Nelson et al., 2004), including brassinosteroids, abscisic acid, cytokinins, and jasmonic acid. In the GA biosynthesis pathway, KO and KAO are P450s that belong to the CYP701A and CYP88A subfamilies, respectively (Helliwell et al., 1999, 2001a; Nelson et al., 2004). EUI is the only P450 identified so far in GA catabolism. P450s share the same CYP number when the sequence identity is >40%; they are further grouped into subfamilies (indicated by a letter) based on the sequence identity of >55% (Schuler and Werck-Reichhart, 2003). In comparison with hormone-related P450 sequences highly conserved across species, CYP714 family members are more divergent. For example, rice and Arabidopsis KO and KAO belong to the same subfamilies, whereas Arabidopsis does not have the CYP714D/EUI subfamily; the Arabidopsis P450 closest to CYP714D1/EUI in sequence is CYP714A1 (Figure 5). We have incubated microsome preparations from recombinant yeast expressing Arabidopsis CYP714A1 with a range of GA substrates under the same experimental conditions as used for CYP714D1/EUI, but GA 16α,17-epoxidase activity has not been detected so far (data not shown).
No 16α,17-epoxy GA has been identified before as an endogenous compound in plants, although it was speculated that 16α,17-[OH]2-GAs might be formed by hydration of the 16,17-epoxides (Santes et al., 1995). In fact, 16α,17-epoxy GAs were easily hydrated to form 16α,17-[OH]2-GAs during the routine GA analysis procedure involving acidic conditions (Figure 7B). In rice, 16α,17-[OH]2-GA4 has previously been identified as a metabolite of exogenously applied GA4 to seedlings (Kobayashi et al., 1993) and as an aglycon of a GA glucoside in anthers (Hasegawa et al., 1994). The 16α,17-[OH]2-GAs were also found in extracts of other plant species, including Pisum sativum fruits (Santes et al., 1995), Lupinus albus seeds (Gaskin et al., 1992), developing Malus domestica seeds (Hedden et al., 1993), sporophytes of a tree fern (Cibotium glaucum) (Yamane et al., 1988), seedlings of Prunus avium (Blake et al., 2000), and capsules of Populus trichocarpa (Pearce et al., 2002). These 16(α),17-[OH]2-GAs might have been produced via 16(α),17-epoxy GAs by EUI-related enzymes, suggesting that 16α,17-epoxidation might participate in GA deactivation in a variety of plant species. Functional characterization of these relatively divergent members in the CYP714 family is important to elucidate whether GA 16α,17-epoxidation is a common GA deactivation reaction in other plant species and if other CYP714s in rice also play a role in the GA metabolism pathway.
METHODS
Characterization of Mutants
Plants of wild-type (ZS97) and eui-1 were grown in the experimental field (Xu et al., 2004). Elongating regions of the uppermost internode of the eui-1 and wild-type plants (ZS97) were sliced into 8- to 10-μm sections and were observed under bright field with a microscope (Leica DMLB).
Analyses of Endogenous GAs
For GA measurements, elongating uppermost internodes (∼30 g fresh weight per analysis) were harvested and lyophilized. Quantitative analysis of GAs shown in Figure 3A and Supplemental Table 1 online was performed using 2H-labeled GAs as internal standards by GC-MS (selected ion monitoring SIM mode) as described before (Gawronska et al., 1995).
We prepared 2H-labeled 16α,17-[OH]2-GA12, -GA9, and -GA4 by incubating [17,17-2H2]-GA12, [2,2,6-2H3]-GA9, and [2,2,6-2H3]-GA4, respectively (1 μg each), with recombinant EUI (see below) for quantitative analysis in vegetative tissues of Eui-OX and wild-type samples. The 2H-labeled GA diols produced by recombinant EUI were purified on a Bond Elut C18 column (ODS; Varian) in the presence of 1% acetic acid (see below) and then purified through reverse-phase HPLC. Because the 2H-labeled substrates were available only in a limited amount, we were unable to measure the absolute amount of 2H-labeled 16α,17-[OH]2-GAs after purification. Therefore, we used these 2H-labeled 16α,17-[OH]2-GAs as internal standards to compare the relative levels in wild-type and Eui-OX samples (Table 3). Assuming that the conversion and recovery of the enzymatic preparations of internal standards are 100%, the amounts of endogenous 16α,17-[OH]2-GAs in the wild-type sample in Table 3 are calculated to be ∼1 ng/g dry weight. The 16α,17-[OH]2-GA12 and -GA9 were purified according to the method for normal GAs (Gawronska et al., 1995). Quantification of 16α,17-[OH]2-GA9 was performed by GC-MS. The 16α,17-[OH]2-GA12 was identified and quantified on LC-ESI-MS/MS (Q-Tof Premier equipped with Acquity Ultra Performance LC; Waters) using a reverse-phase column (Acquity UPLC BEH-C18; Waters). We used LC-ESI-MS/MS for the analysis of this particular compound because major fragment ions of [16,17-2H2]16α,17-[OH]2-GA12 (as a methyl ester-TMSi ether derivative) on GC-MS do not contain 2H-labels. Unlike other GAs, 16α,17-[OH]2-GA4 was found to be present in the aqueous phase during solvent partitioning due to its high polarity. The 16α,17-[OH]2-GA4 in the aqueous phase was extracted with n-butanol and purified through a Bond Elut C18 column, Bond Elut DEA column (ODS; Varian), and reverse-phase HPLC before GC-MS analysis.
Cloning of Eui, Mutant Allele Assay, and Complementation
Eui was finally placed within a 24-kb region between the markers M01 and M0387. The Nipponbare BAC OSJNBa0095J22 bearing Eui (Xu et al., 2004) was used to make subclones with the vector pCAMBIA1300 to produce a plasmid pCEUI-B3 with the entire 13-kb genomic DNA, including the coding region and the 5′ promoter region of EUI. This plasmid and the empty vector pCAMBIA1300 (accession number AF234269) were introduced into the eui-1 mutant plants (307T) by Agrobacterium tumefaciens–mediated transformation. More than 55 independent transgenic lines were produced, which were assayed in the second (T1) or third (T2) generations with 12 to 24 sibling plants. The transgene was detected by DNA gel blotting. The same Eui regions in three eui alleles, eui-1 (Xu et al., 2004), eui-2 (SKZ-T), and eui-3 (ZH11-T) (Zhu et al., 2003), were amplified with Pyrobest Tag DNA polymerase (Takara) and were sequenced by a 3700 DNA sequencer (Applied Biosystems) to identify mutations.
Phylogenetic Analysis
The BLAST search program (http://www.ncbi.nlm.nih.gov/BLAST/) was used to look for protein sequences homologous to EUI. Amino acid sequences of the CYP714 family members were obtained from the Cytochrome P450 Homepage (http://drnelson.utmem.edu/cytochromeP450.html). The obtained CYP714 sequences were aligned using MEGA version 3.1 software (Kumar et al., 2004; see Supplemental Figure 1 online), and the neighbor-joining tree (Figure 5) was generated with the Poisson correction method using the same software. Bootstrap replication (500 replications) was used for a statistical support for the nodes in the phylogenetic tree.
Overexpression of Eui
A plasmid, pCD1900 containing the 1.9-kb Eui full-length cDNA inserted into the overexpression vector 35S-C1301 (provided by Pamela Ronald, University of California, Davis, CA), was transformed into the wild-type cultivar TP309 to generate >70 independent Eui-OX plants.
Promoter Activity
The Eui promoter region, 2.5 kb upstream of the coding region, was fused to the GUS reporter gene with the nopaline synthase terminator and cloned into pCAMBIA1300 (Eui-GUS). The Eui-GUS construct was introduced into TP309 to generate 16 independent transgenic lines. Histochemical assay for GUS activity in transgenic plants was performed as described (Jefferson et al., 1987).
Heterologous Expression in Yeast and Identification of the EUI Products
The full-length coding region of Eui was ligated into the BamHI and KpnI sites of pYeDP60 that was transformed into yeast strain WAT11 engineered to coproduce the Arabidopsis thaliana NADPH P450 reductase-1 (Pompon et al., 1996). Transformed colonies were inoculated into 10 mL of SGI medium (7 g/L yeast nitrogen base, 1 g/L bactocasamino acid, 20 g/L glucose, and 20 mg/L tryptophan) and were grown at 30°C for 1 d. The culture was then inoculated into 500 mL of SLI medium (7 g/L yeast nitrogen base, 1 g/L bactocasamino acid, 20 g/L galactose, and 20 mg/L tryptophan) and grown at 28°C until the cell density reached 5 × 107 cells mL−1. To prepare microsomal proteins, collected yeast cells were suspended in 10 mL of 0.1 M potassium phosphate buffer, pH 7.6, and passed through a French press (25,000 psi). The broken cells were centrifuged at 10,000g for 15 min, and the resulting supernatant was centrifuged at 100,000g for 1 h. The microsomal fraction was suspended in 0.1 M potassium phosphate buffer, pH 7.6. Substrate GA (30 μM) was incubated with 150 μg (in a 50-μL volume) of microsomal protein in the presence of 500 μM NADPH at 25°C for 4 h. The reaction mixture was loaded onto a Bond Elut C18 column in 1% acetic acid or water. The products were eluted with 80% methanol and further analyzed on silica-gel thin layer chromatography (number 5533; Merck) or on full-scan GC-MS as described previously (Yamaguchi et al., 1998).
GA Bioactivity Assay
The microdrop bioassay was performed using the GA-deficient cultivar Tan-ginbozu (Itoh et al., 2004) as described previously (Ishii and Nishijima, 1995). Seeds were germinated at 30°C, planted in a vial filled with 0.8% (w/v) agar, and then incubated at 30°C under continuous light (85 μmol m−2 s−1) for 3 d. GA samples were applied in a 1-μL 50% acetone solution onto the region between the first and second leaves. The length of the second leaf sheath was measured 2 d after the GA treatment.
Preparation of 16,17-Epoxy GAs
The 16,17-epoxy GA9 and GA4 derivatives were prepared by treatment of the respective parent GA methyl esters with 3-chloroperoxybenzoic acid as described by Gaskin et al. (1992) for a mixture of GA4/GA7 methyl esters. The lower retention factor products on silica-gel thin layer chromatography were shown to be the 16α epimers in each case. Similarly, 16,17-epoxy GA12 derivatives were prepared by treating GA12 with 3-chloroperoxybenzoic acid.
Subcellular Localization and Protein Gel Blot Analyses of EUI Protein
EUI-GFP fusion was made by in-frame fusion of the 1.9-kb full-length Eui cDNA with GFP (accession number U87973). The fusion gene was inserted into the vector 35S-C1301. Transient expression of the EUI-GFP fusion and GFP alone (as a control) in onion epidermal cells was performed as previously described (Collings et al., 2000) using a helium biolistic device (Bio-Rad PDS-1000). The samples were observed with a confocal laser microscope (Zeiss LSM510). EUI protein was further immunogold detected in wild-type cells as described (Alcantara et al., 2005). Protein gel blotting of EUI protein in wild-type and transgenic plants was performed with the SuperSignal West chemiluminescence kit according to the manufacturer's protocol (Pierce Chemical).
RNA Preparation and Transcript Analysis
Total RNA was prepared from the elongating uppermost internodes of eui-1 and wild-type plants and different tissues of wild-type plants using TRIzol reagent according to the manufacturer's protocol (Gibco BRL). The Eui transcripts were detected by RT-PCR using the primers EuiF1 and EuiR1 EuiF1 (5′-CGGGACTTCGAGAAGGACG-3′ and 5′-GATGCTGAAGATGACGCTGGT-3′) that amplify a 549-bp fragment of the 3′-end of the Eui cDNA, using the following PCR conditions: 94°C for 4 min, followed by 35 cycles at 94°C for 20 s, 64°C for 40 s, and 72°C for 30 s and an elongation step at 72°C for 10 min. The rice (Oryza sativa) ubiquitin (Ubi-1) cDNA served as an internal control (Kachroo et al., 2003). Because the Eui cDNA is GC rich, we could not detect the transcript with either RNA gel blot, real-time PCR, or in situ hybridization after many attempts. The RT-PCR products of different tissues of wild-type plants were subjected to DNA gel blot analysis to confirm the Eui expression levels revealed by the GUS fusion activity in different tissues. Briefly, RT-PCR was performed for 30 cycles in a 25-μL reaction mixture using the above conditions. Each 18 μL of PCR product and 6 μL of the diluted (20×) Ubi-1 control PCR products were loaded on 0.8% agarose gels for electrophoresis and then blotted to Hybond N+ membranes (Amersham). The Eui and Ubi-1 cDNA fragments were labeled with [α-32P]dCTP using a random primer labeling kit (Takara) for hybridization and autoradiography. RT-PCR was also performed to determine the transcript levels of the other rice GA metabolism genes, GA20ox2 (Sd1), GA3ox2 (D18), and GA2ox1, using the same primers as reported (Sakamoto et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Eui genomic DNA (AY987039), cDNA and protein (AY987040), inserted fragment (DQ004852), and inserted retrotransposon (DQ004853).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Endogenous GA Levels in the Uppermost Internodes of eui and Wild-type Plants.
Supplemental Table 2. Levels of Endogenous GAs in Eui-OX and Wild-Type Plants.
Supplemental Figure 1. Sequence Alignments of EUI-Related P450s (Members of the CYP714 Family) Used for the Generation of Phylogenetic Tree Shown in Figure 5.
Supplemental Figure 2. Substrate Preference of EUI.
Supplemental Methods.
Supplementary Material
Acknowledgments
We acknowledge Jiayang Li for suggestions and support of the study and Jianming Li for critical reading of the manuscript. We thank Jianjun Wang, Linyou Wang, and Zongxiu Sun for help with rice transformation and growth. We also thank Yaoguang Liu for the construction of transformation subclones, Rod Wing and Bin Han for providing BAC clones, the National Institute of Agrobiological Sciences (Ibaraki, Japan) for the full-length cDNA, and Pamela Ronald for the vector 35S-C1301. This work was supported by grants from the Ministry of Science and Technology of China (2002AA2Z1003), the National Natural Science Foundation of China (30270136 and 30421001), and the Shanghai Municipal Science and Technology Commission (03DJ14016) to Z.H. This work was also supported in part by a Grant-in-Aid for Scientific Research (17770048) to S.Y. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. T.N. is supported by the Special Postdoctoral Program from RIKEN.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zuhua He (zhhe@sibs.ac.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038455.
References
- Alcantara, J., Bird, D.A., Franceschi, V.R., and Facchini, P.J. (2005). Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol. 138 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, P.S., Browning, G., Benjamin, L.J., and Mander, L.N. (2000). Gibberellins in seedlings and flowering trees of Prunus avium L. Phytochemistry 53 519–528. [DOI] [PubMed] [Google Scholar]
- Collings, D.A., Carter, C.N., Rink, J.C., Scott, A.C., Wyatt, S.E., and Allen, N.S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12 2425–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin, P., Hoad, G.V., MacMillan, J., Makinson, I.K., and Readman, J.E. (1992). Gibberellins A82 and A83 in seed of Lupinus albus. Phytochemistry 31 1869–1877. [Google Scholar]
- Gawronska, H., Yang, Y.Y., Furukawa, K., Kendrick, R.E., Takahashi, N., and Kamiya, Y. (1995). Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol. 36 1361–1367. [Google Scholar]
- Hasegawa, M., Nakajima, M., Takeda, K., Yamaguchi, I., and Murofushi, N. (1994). A novel gibberellin glucoside, 16α,17-dihydroxy-16,17-dihydro gibberellin A4-17-O-β-D-glucopyranoside, from rice anthers. Phytochemistry 37 629–634. [Google Scholar]
- He, Z., and Shen, Z. (1991). Inheritance of panicle exsertion and improvement of male sterile line in rice. Chinese J. Rice Sci. 5 1–6. [Google Scholar]
- He, Z., and Shen, Z. (1994). Sensitivity of elongated internode gene to GA3 and improvement of MS line in rice. Acta Agron. Sinica 20 161–167. [Google Scholar]
- Hedden, P. (1999). Regulation of gibberellin biosynthesis. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Amsterdam: Elsevier Science), pp. 161–188.
- Hedden, P., Hoad, G.V., Gaskin, P., Lewis, M.J., Green, J.R., Furber, M., and Mander, L.N. (1993). Kaurenoids and gibberellins, including the newly characterized gibberellin A88, in developing apple seeds. Phytochemistry 32 231–237. [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Chandler, P.M., Poole, A., Dennis, E.S., and Peacock, W.J. (2001. a). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Poole, A., Peacock, W.J., and Dennis, E.S. (1999). Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 119 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Sullivan, J.A., Mould, R.M., Gray, J.C., Peacock, W.J., and Dennis, E.S. (2001. b). A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 28 201–208. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T., and Nishijima, T. (1995). Inhibition of gibberellin-induced elongation growth of rice by feruloyl oligosaccharides. Plant Cell Physiol. 36 1447–1451. [PubMed] [Google Scholar]
- Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., Ashikari, M., Ichihara, S., and Matsuoka, M. (2004). A rice semi-dwarf gene, Tan-ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54 533–547. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.V. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., He, Z., Patkar, R., Zhu, Q., Zhong, J., Li, D., Ronald, P., Lamb, C., and Chattoo, B.B. (2003). Induction of H2O2 in transgenic rice leads to cell death and enhanced resistance to both bacterial and fungal pathogens. Transgenic Res. 12 577–586. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., Itoh, H., Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35 104–115. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M., Gaskin, P., Spray, C.R., Suzuki, Y., Phinney, B.O., and MacMillan, J. (1993). Metabolism and biological activity of gibberellin A4 in vegetative shoots of Zea mays, Oryza sativa, and Arabidopsis thaliana. Plant Physiol. 102 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., Yamaguchi, I., Murofushi, N., Ota, Y., and Takahashi, N. (1988). Fluctuation and localization of endogenous gibberellins in rice. Agric. Biol. Chem. 52 1189–1194. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lee, D.J., and Zeevaart, J.A.D. (2005). Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 138 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengler, J., Holzmuller, H., Salmons, B., Gunzburg, W.H., and Renner, M. (2005). FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1-reporter fusion protein. Anal. Biochem. 343 116–124. [DOI] [PubMed] [Google Scholar]
- Lester, D.R., Ross, J.J., Smith, J.J., Elliott, R.C., and Reid, J.B. (1999). Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 19 65–73. [DOI] [PubMed] [Google Scholar]
- Li, J., and Yuan, L. (2000). Hybrid rice: Genetics, breeding and seed production. Plant Breed. Rev. 17 15–158. [Google Scholar]
- Liu, L., van Zanten, L., Shu, Q., and Maluszynski, M. (2004). Officially released mutant varieties in China. Mutation Breeding Review 14 1–64. [Google Scholar]
- Nelson, D.R., Schuler, M.A., Paquette, S.M., Werck-Reichhart, D., and Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, D.W., Hutt, O.E., Rood, S.B., and Mander, L.N. (2002). Gibberellins in shoots and developing capsules of Populus species. Phytochemistry 59 679–687. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Pompon, D., Louerat, B., Bronine, A., and Urban, P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272 51–64. [DOI] [PubMed] [Google Scholar]
- Rutger, J.N., and Carnahan, H.L. (1981). A fourth genetic element to facilitate hybrid cereal production. A recessive tall in rice. Crop Sci. 21 373–376. [Google Scholar]
- Sakamoto, T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santes, C.M., Hedden, P., Gaskin, P., and Garcia-Martinez, J. (1995). Gibberellins and related compounds in young fruits of pea and their relationship to fruit-set. Phytochemistry 40 1347–1355. [Google Scholar]
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416 701–702. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M., Bizzell, C.M., Lee, D.J., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54 629–667. [DOI] [PubMed] [Google Scholar]
- Shen, Z., and He, Z. (1989). Interaction between eui gene and WAMS cytoplasm of rice and improvement of panicle exsertion of MS line. SABRAO J. 6 753–756. [Google Scholar]
- Spielmeyer, W., Ellis, M.H., and Chandler, P.M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Zhu, Y., Zhou, H., Li, Q., Sun, Z., Liu, Y., Lin, H., and He, Z. (2004). Identification of a 98-kb DNA segment containing the rice Eui gene controlling uppermost internode elongation, and construction of a TAC transgene sublibrary. Mol. Genet. Genomics 272 149–155. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41 251–257. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-P. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, H., Fujioka, S., Spray, C.R., Phinney, B.O., MacMillan, J., Gaskin, P., and Takahashi, N. (1988). Endogenous gibberellins from sporophytes of two tree ferns, Cibotium glaucum and Dicksonia antarctica. Plant Physiol. 86 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R., Zhang, S., Huang, R., Yang, S., and Zhang, Q. (2002). Breeding technology of eui hybrids of rice. Scientia Agricultura Sinica 35 233–237. [Google Scholar]
- Zhu, X., Chen, H., and Min, S. (2003). Allelism analysis and comparison of plant height, panicle and internode lengths in tall-culm mutants of rice (Oryza sativa L.). Acta Agron. Sinica 29 591–594. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.