Abstract
Although circadian transcription of Period2 (Per2) is fundamental for the generation of circadian rhythm, the molecular mechanism remains unclear. Here we report that cell-autonomous circadian transcription of Per2 is driven by two transcriptional elements, one for rhythm generation and the other for phase control. The former contains the E-box-like sequence (CACGTT) that is sufficient and indispensable to drive oscillation, and indeed circadian transcription factors site-specifically bind to it. Furthermore, the nature of this atypical E-box is different from that of the classical circadian E-box. The current feedback loop model is based mainly on Period1. Our results provide not only compelling evidence in support of this model but also an explanation for a general basic mechanism to produce various patterns in the phase and amplitude of cell-autonomous circadian gene expression.
INTRODUCTION
In nearly all organisms, behavioral and physiological processes display ∼24 h rhythms that are controlled by circadian pacemakers (Pittendrigh, 1993). The circadian organization of physiology and behavior in mammals is governed by the suprachiasmatic nuclei (SCN), a defined pair of cell clusters in the anteroventral hypothalamus (Ralph et al., 1990). Circadian clocks can count time only approximately and must be adjusted every day by the photoperiod in order to be in harmony with the outside world (Menaker, 2003). Circadian oscillators also exist in most peripheral cells and even in cultured cells (Balsalobre et al., 1998; Yamazaki et al., 2000). It is thought that the phase of these peripheral time-keepers is reset by signals regulated by the SCN pacemaker (Akashi and Nishida, 2000; Schibler and Sassone-Corsi, 2002).
The molecular makeup of circadian clocks has been the subject of intense genetic and biochemical investigation in various organisms, including cyanobacteria, Neurospora, higher plants, Drosophila, and mammals (Dunlap, 1999; Kondo and Ishiura, 2000; Allada et al., 2001; Williams and Sehgal, 2001; Young and Kay, 2001; Reppert and Weaver, 2002). Over the last several years, orthologues of most Drosophila circadian clock genes have been cloned from mammals (Albrecht and Eichele, 2003; Lowrey and Takahashi, 2004). Although mPer2 was literally cloned as a secondary mammalian period gene (Albrecht et al., 1997; Takumi et al., 1998), gene-knockout analysis revealed that an mPer2 mutant displays a loss of circadian rhythmicity, revealing a prominent role for mPER2 in the mammalian clock (Zheng et al., 1999). Additionally familial advanced sleep phase syndrome has been attributed to a missense mutation in hPer2 (Toh et al., 2001). These studies demonstrate that a robust circadian fluctuation in Per2 transcription is an essential event for the generation of circadian rhythm.
Circadian oscillators appear to have been highly conserved throughout evolution and to involve transcription-translation negative feedback loops for the regulation of clock genes (Dunlap, 1999; Young and Kay, 2001). In mammals, in vitro studies have shown that the expression of Per1 (Period1) is driven by the CLOCK/BMAL1 transcription complex through an E-box enhancer and that PER proteins, together with CRY (Cryptochrome) proteins, serve to regulate the CLOCK/BMAL1 transcription complex negatively (Gekakis et al., 1998; Kume et al., 1999; Hida et al., 2000). This model has been thought to be applicable to other Per and Cry genes. However, several problems remain to be solved in the current model. First, there is a wide variation in the phase and amplitude of circadian accumulation of Per1, Per2, Per3, Cry1, and Cry2 mRNA levels in tissues and cultured cells (Yamamoto et al., 2004). The current feedback loop model in mammals cannot explain the mechanism to generate this variation. It is importantly to note that the phase and amplitude of circadian transcription cannot be verified by using an overexpression-based transient reporter assay. Second, although Per1, which has been analyzed in detail, serves as a foundation for the mammalian model, it has been reported recently that Per1 is not absolutely required for the generation and maintenance of circadian rhythms (Cermakian et al., 2001; Zheng et al., 2001). This issue remains controversial, therefore, because Bae et al. (2001) found that the loss of Per1 can result in arrhythmic mice. Thus, it is unclear whether the current model really reflects the core clock mechanism. Gene knockout studies of clock genes support the current model; for example, genetic deficiency of Bmal1 results in the down-regulation of Per expression (Vitaterna et al., 1999; Bunger et al., 2000; Bae et al., 2001). However, these studies cannot prove whether the regulation is direct or indirect. Overexpression-based transient reporter assay has shown that transcription of the Per2 gene, a key component for rhythm generation, is also up-regulated by coexpression of circadian transcription factors (BMAL1, CLOCK, and NPAS2), as shown for the Per1 gene (Travnickova-Bendova et al., 2002; Kaasik and Lee, 2004). Yet, although Per1 contains 5 classical circadian E-boxes (Hida et al., 2000; Yamamoto et al., 2004), no evolutionarily conserved E-box (the classical circadian E-box) has been identified in the upstream sequence of Per2. Therefore it remains unknown whether these circadian transcription factors directly activate the Per2 gene in a site-specific manner.
Herein we report the detailed analysis of the mechanism of a robust cell-autonomous circadian fluctuation in Per2 transcription, not by transient reporter assays but by monitoring transcriptional fluctuation of the luciferase reporter gene over several days. An ∼20-base pair region located near the transcription start site (TSS) was indispensable to drive cell-autonomous rhythmic transcription of Per2, whereas another region, located upstream from it, was shown to be responsible for phase control of cell-autonomous circadian transcription. An E-box-like sequence exists in the core region (the former region), and indeed circadian transcription factors activated Per2 transcription through site-specific binding to this element. The fact that this identified small region, indispensable for cell-autonomous rhythmic transcription of Per2, contains a functional E-box-like sequence is of great significance. Our results thus validate the current model by demonstrating that this model is actually applicable to cell-autonomous circadian fluctuation of Per2, a core component for the generation of circadian rhythm. We also show that this atypical E-box has properties different from those of the classical circadian E-box, which result in a pattern of circadian transcription different from that of Per1. Our results provide not only compelling evidence in support of the current feedback loop model in mammals but also an explanation for a general mechanism that generates wide variation in the expression patterns of circadian genes.
MATERIALS AND METHODS
Plasmid Construction
A bacterial artificial chromosome (BAC) clone containing the complete genomic sequence of the mouse Per2 (mPer2) gene was purchased from the BACPAC Resource Center (BPRC) at Children's Hospital Oakland Research Institute. The mPer2 promoter region was isolated and cloned in the pGL3-Basic vector (Promega, Madison, WI). The mPer2 region spans from -2811 to +110 (+1 is the putative TSS).
Cell Culture, Transfection, and Reporter Assay
NIH3T3 cells were cultured and transfected as described previously (Akashi et al., 2002). For real-time PCR analyses, cells were immediately frozen in liquid nitrogen and stored at -80°C until processed for RNA. Cell lysates were used in the Dual Luciferase assay System (Promega) as described previously (Akashi and Takumi, 2005).
Real-Time Monitoring of Luciferase Activity in Living Cells
NIH3T3 cells were cultured, transfected with mPer2-luc and incubated for 24 h. Then the medium was exchanged for serum-rich medium (DMEM, supplemented with 50% serum). Two hours later this medium was replaced with normal culture medium. In the presence of 0.1 mM luciferin, light emission was measured and integrated for 1 min at intervals of 15 min with a photomultiplier tube (Hamamatsu Photonics, Hamamatsu, Japan) as described previously (Akashi and Takumi, 2005).
Data Analysis
Phase and period measurements were calculated as in previous studies (Abe et al., 2002; Yamazaki et al., 2002; Yoo et al., 2004). Data sets were detrended by subtracting the 24-h running average from the raw data. The maximum differences between the smoothed curves for each cycle (the peak and the trough) were used to calculate the amplitude of each cycle.
Animals
Mice were housed under a strict 12:12 h light/dark condition. Tissues were immediately frozen in liquid nitrogen and stored at -80°C until processed for RNA. All protocols of experiments using animals in this study were approved by the OBI (Osaka Bioscience Institute) Animal Research Committee.
Pulldown Experiment
Mouse liver extracts were prepared at 4-h intervals by homogenizing the tissue in ice-cold incubation buffer (Akashi et al., 2002), and then the extracts were incubated with an 80-base pair double-stranded biotinylated oligonucleotide that had been immobilized on streptavidin-Sepharose beads (Amersham, Piscataway, NJ). After having been washed with the incubation buffer, the resulting bound protein was subjected to immunoblot analysis. The designed sequences (response elements underlined) were the following: wild-type (mPer2, -105 to +15): 5′CTCAGGTTCCGCCCCGCCAGTATGCAAATGAGGTGGCACTCCGACCAATGGCGCGCGCAGGGGCGGGCTCAGCGCGCGCGGTCACGTTTTCCACTATGTGACAGCGGAGGGCGACGCGGC3′; mutant (mPer2, -105 to +15): 5′CTCAGGTTCCGCCCCGCCAGTATGCAAATGAGGTGGCACTCCGACCAATGGCGCGCGCAGGGGCGGGCTCAGCGCGCGCGGTCTTTCCACTATGTGACAGCGGAGGGCGACGCGGC3′; wild-type (mPer1, 3E-box): 5′GAAAGCTTTAGCCACGTGACAGTGAGGGGCACCCCTTAACGACACGTGGGCCCTCAATTGAGCACCCAAGTCCACGTGCAGGGATGTGTGGGGGCAGGGCCTGGCATTATGCAACCCGCCTCCCAGCCTC3′; and mutant (mPer1, 3E-box): 5′GAAAGCTTTAGCCGACAGTGAGGGGCACCCCTTAACGACGGGCCCTCAATTGAGCACCCAAGTCCGCAGGGATGTGTGGGGGCAGGGCCTGGCATTAACCCGCCTCCCAGCCTC3′.
Real-Time Quantitative RT-PCR
Quantification of relative RNA levels by the SYBER Green real-time PCR technology was done as described previously (Yamamoto et al., 2004). Briefly, DNase-treated total RNA (2.5 μg) was reverse-transcribed by using an oligo(dT) primer and Superscript reverse transcriptase (Invitrogen, Carlsbad, CA). The cDNA equivalent to 20 ng total RNA was PCR-amplified in an ABI PRISM 7900 HT sequence detection system (Applied Biosystems, Foster City, CA). Forward primers and reverse primers were as follows: Gapdh forward: 5′-CATCCACTGGTGCTGCCAAGGCTGT-3′; Gapdh reverse: 5′-ACAACCTGGTCCTCAGTGTAGCCCA-3′; mPer1 forward: 5′-CAGGCTAACCAGGAATATTACCAGC-3′; mPer1 reverse: 5′-CACAGCCACAGAGAAGGTGTCCTGG-3′; mPer2 forward: 5′-GGCTTCACCATGCCTGTTGT-3′; and mPer2 reverse: 5′-GGAGTTATTTCGGAGGCAAGTGT-3′.
The relative levels of each RNA were normalized to the corresponding Gapdh RNA levels. Relative RNA levels were then expressed as percentage of the maximal value obtained for each experiment.
RESULTS AND DISCUSSION
Cell-autonomous Circadian Transcription of Per2 Is Regulated by Two or More Transcriptional Regulatory Elements
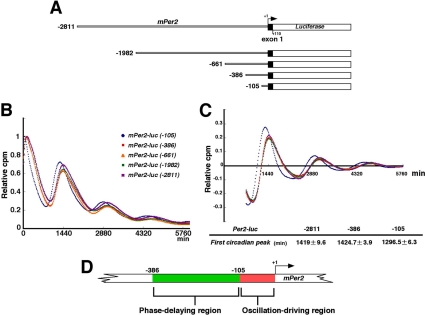
The core clock is a cell-autonomous system. Therefore, we should examine whether transcriptional regulatory elements function under a cell-autonomous condition. Our approach enables us to exclude internal environmental cues such as blood-borne factors and body temperature. To identify the transcriptional regulatory elements for cell-autonomous circadian transcription of Per2, we monitored transcriptional fluctuation of Per2 in real-time by using a deletion series of Per2 promoter-reporter constructs (Figure 1A). Note that exogenously transfected transgenes have no effect on endogenous cell-autonomous circadian oscillation and, therefore, that the deletion or mutation of constructs does not affect the endogenous pacemaker. NIH3T3 cells were transfected with the Per2-luc construct and then stimulated with a high concentration of serum. After the serum shock, in the presence of luciferin, light emission was measured and integrated for 1 min at intervals of 15 min. By using this in vitro luminescence reporter system, we can monitor cell-autonomous oscillators. Clearly, Per2-controlled fluctuations of luminescence resulted in a series of readily appreciable peaks and troughs, as exemplified in Figure 1. Almost the same phases and amplitudes were observed in the cells transfected with the constructs from -2811 to -386, whereas a phase advance (∼2 h earlier) was detected only when using the Per2 (-105) construct (Figure 1B). To better calculate phase differences, data sets were detrended, and the time of the first peak was used as a phase marker (Figure 1C). Again the Per2 (-105) construct showed a 2-h phase advance compared with the other constructs, as expected in Figure 1B. Thus, these data suggest that a phase delaying element is located between -386 and -106 base pairs upstream from the TSS(s) and that a rhythm-generating element exists between -105 and +1 base pairs upstream from the TSS(s) (Figure 1D). These results demonstrate that two or more transcriptional regulatory elements, i.e., a phase-delaying element and a rhythm-generating element located upstream from the TSS(s), are required for cell-autonomous circadian gene expression of Per2. Consistent with this cell-autonomous phenomenon, Yoo et al. (2005) has very recently reported that a 210-base pair fragment upstream from the TSS(s) drives Per2 circadian oscillation in vivo.
Figure 1.
A phase-delaying element and a rhythm-generating element are required for robust cell-autonomous circadian gene expression of Per2. (A) Schematic representation of deletion mutants of the mPer2 promoter. +1 corresponds to the transcription start site. (B) Transcriptional oscillation of mPer2 was monitored by using the cell culture-based luminescent reporter assay. NIH3T3 cells were transfected with the mPer2-luc construct and then stimulated with a high concentration of serum. After the serum shock, in the presence of luciferin, light emission was measured and integrated for 1 min at intervals of 15 min (vertical scale: relative cpm; horizontal scale: 1440 min = 1 d). Peak values of the curves were set to 1. A representative result of three independent experiments is shown. (C) The signals obtained in B were detrended. The time of the first peak was calculated as a phase marker (mean ± SEM; n = 4). (D) Schematic representation of the results obtained from the deletion analysis.
An ∼20-Base Pair Region Located near the TSS(s) Was Indispensable to Drive Cell-autonomous Rhythmic Transcription of Per2
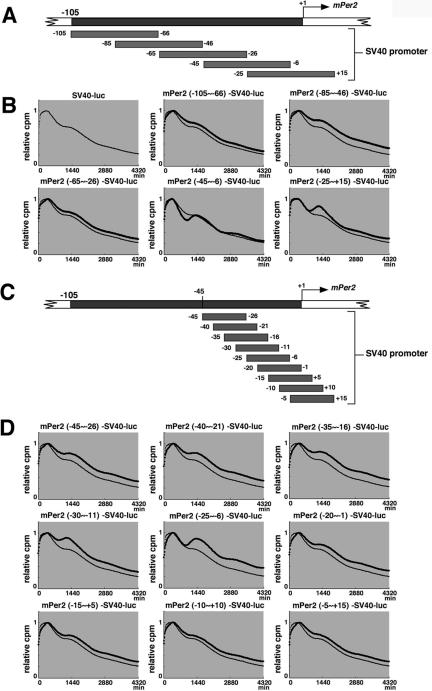
To identify transcription factors required to drive cell-autonomous rhythmic transcription of Per2, we exactly narrowed the rhythm-generating region by monitoring in real-time Per2-controlled fluctuations of luminescence in cells transfected with another set of constructs, in which a fragment from Per2 (-105) was inserted into the upstream of the SV40 promoter (Figure 2, A and C). When 40-base pair fragments overlapping by 20 base pairs were used (Figure 2A), the Per2 (-45 to -6)-SV40-luc construct and the Per2 (-25 to +15)-SV40-luc construct clearly showed Per2-controlled fluctuations of luminescence as compared with the other constructs (Figure 2B). The detrended data suggests that the SV40 promoter activity exhibits not a drastic but significant circadian fluctuation (Supplementary Figure 1A), indicating that this promoter contains some circadian enhancer elements. The detrended bioluminescence data sets make clear that only the Per2 (-45 to -6)-SV40-luc and Per2 (-25 to +15)-SV40-luc construct exhibit a higher amplitude of oscillation than the SV40 promoter. Next, to compare the magnitude of fluctuation in these two constructs, the amplitude in each cycle was calculated by subtracting the value of the trough from that of the peak (Supplementary Figure 1B). The higher amplitude of oscillation was maintained over the cycles in Per2 (-45 to -6)-SV40-luc. The Per2 (-45 to -6)-SV40-luc/Per2 (-25 to +15)-SV40-luc amplitude ratio was obtained by dividing the Per2 (-45 to -6)-SV40-luc wave amplitude by the Per2 (-25 to +15)-SV40-luc wave amplitude in each cycle (Supplementary Figure 1C), demonstrating that the amplitude of Per2 (-45 to -6)-SV40-luc gradually grew to be twofold higher than that of Per2 (-25 to +15)-SV40-luc, as cycle number increases. Taken together, the region ranging from -45 to +15 was shown to possess the ability to drive circadian oscillation of transcription.
Figure 2.
Identification of the region responsible for driving cell-autonomous circadian oscillation of Per2 transcription. (A) Schematic representation of a set of constructs in which a 40-base pair fragment from Per2 (-105 to +15) was inserted into the upstream of the SV40 promoter attached to the luciferase gene. These fragments overlap by 20 base pairs. (B) Transcriptional oscillation of mPer2-SV40-luc was monitored. NIH3T3 cells were transfected and then stimulated with a high concentration of serum. After the serum shock, light emission was measured and integrated for 1 min at intervals of 15 min (vertical scale: relative cpm; horizontal scale: 1440 min = 1 d). Peak values of the curves were set to 1. A representative result of three independent experiments is shown. For accurate comparison, thin lines show the curve for “SV40-luc.” (C) Schematic representation of another set of constructs, in which a 20-base pair fragment from Per2 (-45 to +15) was inserted into the upstream of the SV40 promoter attached to the luciferase gene. These fragments overlap by 15 base pairs. (D) Transcriptional oscillation of mPer2-SV40-luc was monitored. Peak values of the curves were set to 1 (horizontal scale: 1440 min = 1 d). A representative result of three independent experiments is shown. For accurate comparison, thin lines show the curve for “SV40-luc.”
Next, to further define this region, we used 20-base pair fragments overlapping by 15 base pairs (Figure 2C). The results show that the Per2 (-25 to -6) fragment possessed the most potent ability to oscillate SV40 promoter activity (Figure 2D). The detrended data highlighted the Per2 (-25 to -6) and Per2 (-30 to -11) fragments' ability to drive oscillation (Supplementary Figure 1D). The Per2 (-25 to -6)-SV40-luc maintained a higher amplitude in each cycle than that of Per2 (-30 to -11)-SV40-luc (Supplementary Figure 1E). Additionally, the Per2 (-25 to -6)-SV40-luc/Per2 (-30 to -11)-SV40-luc amplitude ratio in each cycle demonstrated that the amplitude of Per2 (-25 to -6)-SV40-luc was also considered, being twofold higher over cycles than that of Per2 (-30 to -11)-SV40-luc (Supplementary Figure 1F). The merge of Per2 (-25 to -6)-SV40-luc (Supplementary Figure 1D) and Per2 (-45 to -6)-SV40-luc (Supplementary Figure 1A) illustrated almost the same pattern of oscillation, confirming that the Per2 (-25 to -6) region is necessary and sufficient to drive Per2 (-45 to -6)-SV40-luc oscillation (Supplementary Figure 1G). This region (-25 to -6), therefore, contains the core element for rhythmic transcription of Per2.
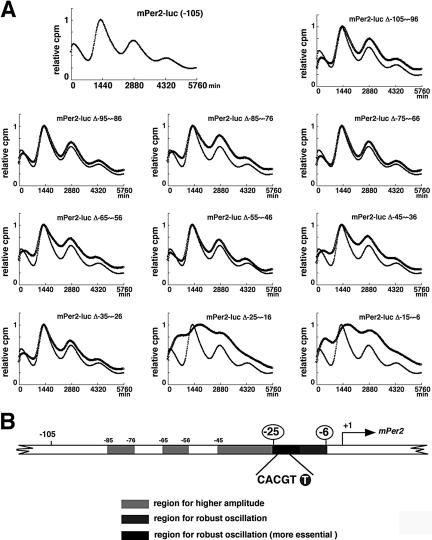
We also constructed mutants lacking 10 base pairs in the -105 to +6 region, and monitored transcriptional fluctuation of Per2 in real-time (Figure 3A). The amplitude of circadian transcription of Per2 was strongly diminished in mPer2-luc Δ (-25 to -16) and mPer2-luc Δ (-15 to -6). Furthermore, both of these two constructs exhibited a phase delay, illustrating that both 10-base pair regions contribute to maintain the original phase. Other regions were required for amplification, whereas these two regions were essential for driving rhythmic transcription of Per2. After removal of baseline changes (Supplementary Figure 2A), the period was obtained from regression analysis of a circadian marker (trough) in Supplementary Figure 2B. As compared with wild-type Per2-luc, mPer2-luc Δ (-95 to -86), mPer2-luc Δ (-85 to -76), mPer2-luc Δ (-75 to -66), mPer2-luc Δ (-65 to -56), mPer2-luc Δ (-55 to -46), and mPer2-luc Δ (-45 to -36) showed a 20-40-min shorter period. Notably, the -75 to -46 region seemed to largely contribute to the determination of period length. Basically, exogenously transfected transgenes have no effect on endogenous cell-autonomous circadian oscillation. Therefore, these slight changes in period lengths may be attributable to desynchronization of reporter expression from the endogenous oscillator and elicitation of transcriptional noise, because these mutations resulted in loss of robust transcriptional oscillation. The amplitude of mPer2-luc Δ (-85 to -76), mPer2-luc Δ (65 to -56), mPer2-luc Δ (-45 to -36), or mPer2-luc Δ (-35 to -26) was significantly smaller over cycles than that of wild-type Per2-luc (Supplementary Figure 2C), and deletion of -15 to -6 or -25 to -16 caused a remarkable inhibition of circadian fluctuation, as shown in Figure 3A. The others showed only a slight decrease of the initial amplitude. To compare the damping pattern in the constructs, the amplitude in the initial cycle was set to 100 (Supplementary Figure 2D). Although mPer2-luc Δ (-25 to -16) showed a slightly irregular damping pattern, the others had almost the same damping pattern as that of wild-type Per2-luc. Taken together, the -85 to -76, -65 to -56, -45 to -36, and -35 to -26 regions work to enhance the amplitude of transcriptional oscillation, whereas the -15 to -6 and -25 to -16 regions are most essential to drive circadian oscillation. There are no regions that obviously affect the period length and damping rate.
Figure 3.
Mapping of the rhythm-generating element by using deletion mutants of Per2. (A) Transcriptional oscillation of a series of mPer2-luc 10-base pair deletion constructs was monitored in real time. NIH3T3 cells were transfected and then stimulated with a high concentration of serum. Peak values of the curves were set to 1 (vertical scale: relative cpm; horizontal scale: 1440 min = 1 d). A representative result of three independent experiments is shown. For accurate comparison, thin lines show the curve for “mPer2-luc (-105).” (B) Schematic representation of the results obtained from the 10-base pair deletion analysis, taken together with those in Figure 2. Sequence inspection reveals an E-box-like sequence (CACGTT) in the region essential for circadian transcription of Per2.
To rule out the possibility that these mutations disrupt the core promoter activity, we examined the basal transcriptional activity of these constructs by conducting reporter experiments. These mutations did not interfere with the core promoter activity, because these constructs did not show severe attenuation of the transcriptional activity found for the full-length construct (Supplementary Figure 3). These results are consistent with those in Figure 2, B and D. Taken together, our data indicate that the -25 to -6 region upstream from the TSS is indispensable for robust cell-autonomous circadian gene expression of Per2. Sequence inspection revealed an E-box-like sequence (CACGTT, instead of the known clock E-box sequence CACGTG) in this region (Figure 3B).
Endogenous Circadian Transcription Factors Bind Site-specifically to the Per2 E-Box-like Sequence
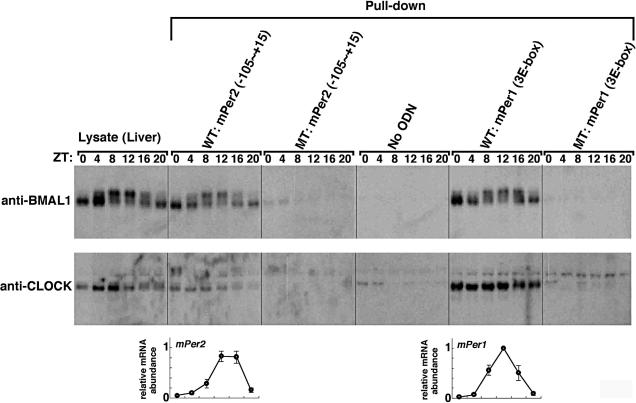
To examine whether circadian transcription factors indeed bind to the Per2 E-box-like sequence, we investigated the binding of endogenous BMAL1 and CLOCK to Per2 (-105 to +15) double-stranded DNA fragments immobilized on streptavidin beads (Figure 4). After having been entrained to LD (12-h light/12-h dark) cycles, BALB/c mice were transferred to DD (constant dark). Liver lysates were prepared at 4-h intervals and immunoblotted with anti-BMAL1 or anti-CLOCK antibody. The shifted bands correspond to phosphorylated BMAL1, as reported recently (Lee et al., 2001; Kondratov et al., 2003; Tamaru et al., 2003). The phosphorylation levels peaked at ZT (Zeitgeber Time) 8 to ZT12. By pulldown experiments using the Per2 promoter fragment immobilized to beads, protein precipitation of BMAL1 and phosphorylated BMAL1 was observed. The peak of phosphorylated BMAL1 bound to the Per2 promoter correlated with that of Per2 mRNA expression in the liver (bottom panel). This correlation indicates that phosphorylated BMAL1 activated transcription of the Per2 gene, consistent with the report that formation of the CLOCK/BMAL1 complex is followed by their codependent phosphorylation (Kondratov et al., 2003). When we used the Per2 promoter fragment with a mutated E-box-like sequence, precipitated BMAL1 protein and phosphorylated BMAL1 were almost completely undetectable. Thus, BMAL1 and phosphorylated BMAL1 were confirmed to specifically recognize and bind to the Per2 E-box-like sequence. We also confirmed that the BMAL1-CLOCK-mediated transcription of the Per2 gene was dependent on this E-box-like sequence by performing traditional luciferase assays (unpublished data). As a positive control, when a DNA fragment containing three different Per1 E-boxes was used for pulldown assays, a similar pattern of BMAL1 binding was detected. Mutation of these Per1 E-boxes completely inhibited BMAL1 binding. As observed in the Per2 pulldown assays, the peak of phosphorylated BMAL1 bound to the Per1 E-boxes correlated with that of Per1 mRNA expression (bottom panel). Our results indicate that the Per2 E-box-like sequence, as well as the classical circadian E-box, binds to endogenous circadian transcription factors and that transcriptional activation of Per2 and Per1 correlates with phosphorylation of BMAL1.
Figure 4.
Temporal patterns of the site-specific binding of endogenous circadian transcription factors to the Per2 E-box-like sequence. Mouse liver extracts were harvested at 4-h intervals and then subjected to immunoblot analysis with anti-BMAL1 antibody (top panels) or anti-CLOCK antibody (middle panels). Mouse liver extracts were incubated with a double-stranded biotinylated oligonucleotide including the consensus-predicted Per2 E-box-like sequence (CACGTT) or three different Per1 E-boxes (wild-type, WT; mutant, MT), which was immobilized on streptavidin-Sepharose beads. The negative control samples were treated with the beads without an oligonucleotide (No ODN). The resulting precipitates were subjected to immunoblot analysis with anti-BMAL1 antibody (top panels) or anti-CLOCK antibody (middle panels). Temporal expression patterns of the mPer2 and mPer1 genes in mouse liver were assayed by real-time quantitative RT-PCR (bottom panels). Each value represents the average of three independent RT-PCR experiments. The relative levels were normalized to the corresponding Gapdh RNA levels. Peak values of the mPer2 and mPer1 curves were set to 1.
The Per2 E-Box-like Sequence Has Functional Characteristics Different from Those of the Classical Circadian E-Box
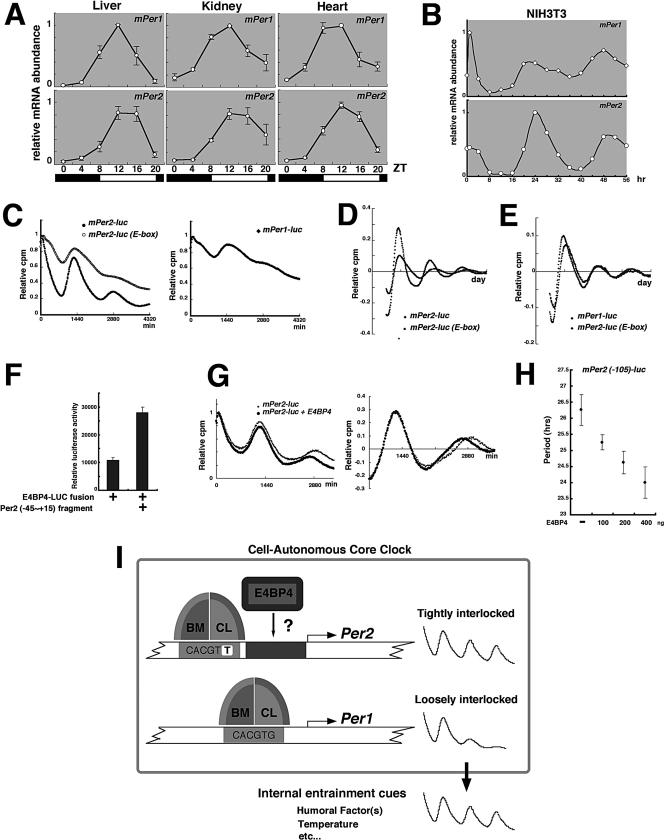
Both Per2 and Per1 mRNA expression showed circadian oscillations with high amplitude in peripheral tissues (Figure 5A), whereas the amplitude of Per1 mRNA rhythms was significantly lower than that of Per2 mRNA rhythms in serum-stimulated NIH3T3 cells (Figure 5B). This observation was reproducible when Rat-1 fibroblasts were used (unpublished data; Balsalobre et al., 1998). A very recent report demonstrated that in vitro cultured fibroblasts harbor self-sustained and cell-autonomous circadian clocks similar to those operative in SCN neurons (Nagoshi et al., 2004). Therefore, the high amplitude of Per1 mRNA oscillation in peripheral tissues largely depends on the extracellular environment such as blood-borne factors and body temperature, which changes cyclically around the clock, rather than on the cell autonomous core clock. In support of this idea, in mPer1::Luc transgenic animals, peripheral organs fail to express persistent circadian rhythms in reporter gene activity (Yamazaki et al., 2000). In contrast, in mPer2-Luciferase knockin mice, peripheral tissues in explant cultures show robust and self-sustained circadian rhythms (Yoo et al., 2004). On the other hand, gene knockout studies have indicated that mPer1-deficient mice display a persistent circadian rhythm (Bae et al., 2001; Cermakian et al., 2001; Zheng et al., 2001), whereas mice deficient for mPer2 have no circadian rhythms in locomotor activity (Zheng et al., 1999; Bae et al., 2001; Zheng et al., 2001). Consistent with these behavioral phenotypes, disruption of mPer2 results in reduced levels of clock gene expression in the SCN; and in contrast, mice homozygous for the targeted mPer1 allele have unaltered SCN gene expression rhythms (Bae et al., 2001). Taken together, these reports also indicate that the Per1 gene might not be tightly incorporated into the cell-autonomous core clock mechanism. However, this issue remains controversial, because the loss of Per1 can result in arrhythmic mice, in prolonged constant dark conditions (Bae et al., 2001). Also, as for the above in vivo experiments, the difference between transgenics and knockins should be recognized; the latter are in their native context and have significantly more potential cis-acting sequences surrounding them. If there are important elements that are distant from the TSS, they might be lost in the Per1 transgenics.
Figure 5.
A potential mechanism by which the cell-autonomous core clock generates more overt circadian oscillations in Per2 transcription than in Per1 transcription. (A and B) Temporal expression patterns of the mPer1 and mPer2 genes, in mouse peripheral tissues (A) and in serum-stimulated NIH3T3 cells (B) were assayed by real-time quantitative RT-PCR. Mice were kept in a 12-h light:12-h dark cycle (LD, lights on 8 a.m.; lights off 8 p.m.) for 2 wk to establish entrainment. Three animals were killed at the times given on the abscissas of the diagrams. Each value represents the average of three independent RT-PCR experiments. The relative levels of each RNA were normalized to the corresponding Gapdh RNA levels. Peak values of the mPer1 and mPer2 curves were set to 1. (C) Transcriptional oscillation of mPer2-luc, mPer2-luc (E-box), and mPer1-luc was monitored in real time. NIH3T3 cells were transfected and then stimulated with a high concentration of serum. Peak values of the curves were set to 1 (vertical scale: relative cpm; horizontal scale: 1440 min = 1 d). A representative result of three independent experiments is shown. (D and E) The signals obtained in C were detrended. (F) COS7 cells were transfected with the E4BP4-luciferase fusion expression vector. Cell extracts were incubated with the double-stranded biotinylated Per2 (-45 to +15) oligonucleotide, including the consensus-predicted E4BP4 response elements, which was immobilized on streptavidin-Sepharose beads. The negative control samples were treated with streptavidin-Sepharose beads without an oligonucleotide. The resulting precipitates were subjected to luciferase assays. Data represent the mean ± SEM of triplicate samples. (G) Transcriptional oscillation of Per2 (-105)-luc was monitored, in the presence or absence of E4BP4 (left). The signals obtained were detrended (right). (H) With increasing dose of the E4BP4 expression plasmid, transcriptional oscillation of Per2 (-105)-luc was monitored. The periods were obtained from analysis of a circadian marker. Data represent the mean ± SEM of triplicate samples. (I) Schematic model representing BMAL1:CLOCK-mediated control of cell-autonomous Per1 and Per2 oscillation. A combination of the E-box-like sequence and a cooperating element increases the amplitude of rhythmic transcription of Per2, and consequently the amplitude of Per2 mRNA rhythms is significantly higher than that of Per1 mRNA rhythms. The 1-base pairs difference (CACGTT) from the classical circadian E-box may be indispensable for this combination. The cell-autonomous core clock generates overt circadian oscillations in Per2 transcription, whereas the high amplitude of Per1 oscillation in vivo largely depends on the extracellular environment, which changes cyclically around the clock, rather than on the core clock. Thus, the Per1 gene might not be tightly incorporated into the cell-autonomous core clock mechanism.
Per1 is highly sensitive to various extracellular stimuli in vitro (Akashi and Nishida, 2000; Balsalobre et al., 2000); and also in vivo, circadian changes in the extracellular environment can readily induce expression of the Per1 gene. Thus, the dramatic circadian changes in Per1 mRNA accumulation in peripheral tissues (Figure 5A) may be attributed to this mechanism. Increased expression of the Per1 gene in response to changes in the extracellular environment may be functional in the entrainment of peripheral oscillators. In fact, in peripheral tissues of mPer1-deficient mice, the phase of clock gene expression is not only delayed, but the peak of expression is broadened (Cermakian et al., 2001)
A Mechanism by Which the Per2 E-Box-like Sequence Generates a Higher Amplitude of Circadian Gene Expression than Does the Classical Circadian E-Box
As shown in Figure 3A, even when the region including the E-box-like sequence was deleted, the promoter activity still fluctuated in a circadian manner. These data suggest that the neighboring -15 to -6 region also contains another element that regulates circadian transcription of Per2 by cooperating with the E-box-like sequence. This -15 to -6 region contains a consensus E4BP4 binding site (8/10 base pairs match), suggesting that DBP and E4BP4 may cooperate with BMAL1-CLOCK/PER-CRY to drive robust circadian gene expression of Per2. In Figure 5C, we substituted the Per2 E-box-like sequence (CACGTT) with the classical circadian E-box (CACGTG) by site-directed mutagenesis and monitored in real-time its transcriptional fluctuation. This substitution did not markedly affect either basal promoter activity or BMAL1-CLOCK-induced transactivation (unpublished data). Interestingly, this 1-base pair substitution resulted in small amplitude of circadian gene expression of Per2 (Figure 5C, left panel), as observed in Per1-luc (Figure 5C, right panel). The detrended bioluminescence data sets made it clear that compared with Per2-luc, Per2-luc (E-box) showed a small amplitude, and additionally that its period time-dependently became longer (Figure 5D). Interestingly, Per2-luc (E-box) had a very similar pattern of amplitude, period, and damping rate to those of Per1-luc (Figure 5E). Thus, our data suggest that the Per2 E-box-like sequence generates high amplitude of circadian gene expression through cooperation between these two distinct elements (the -25 to -16 region and the -15 to -6 region) and that the 1-base pair difference (CACGTT) is indispensable for this cooperation.
To examine whether E4BP4 controls transcriptional oscillation of Per2, we studied the binding of E4BP4 to Per2 (-45 to +15). To confirm that the E4BP4-LUCIFERASE fusion protein binds to the Per2 fragment, we pulled down the fusion protein by using biotinylated double-strand Per2 (-45 to +15) containing the putative E4BP4 consensus (Figure 5F). In the presence of this Per2 fragment, the luciferase activity in the precipitate was enhanced, demonstrating the E4BP4 binding to this fragment. Next, in order to examine the role of E4BP4 protein in cell-autonomous transcriptional oscillation of Per2, we monitored the Per2 (-105)-luc activity in real time. As shown in Figure 5G, coexpression of E4BP4 resulted in a gradual reduction of the basal transcriptional activity (left) and a shortened period length of circadian transcription (right, detrended data). The expression of E4BP4 shortened the period in a dose-dependent manner (Figure 5H). These results indicate that E4BP4 may be a transcriptional repressor in Per2 transcription and control the period length of Per2 oscillation. As shown in Figures 3B and 5C, Per2 Δ (-15 to -6)-luc and Per2 (E-box)-luc did not show deep troughs in circadian transcription, as wild-type Per2-luc did, and therefore, we speculate that E4BP4 may be required for the trough formation in circadian transcription of Per2.
CONCLUSIONS
The current feedback loop model has been based mainly on Per1. A publication that appeared after this article was submitted demonstrated that a 210-base pair region including the E-box-like sequence is sufficient for Per2 oscillation in vivo (Yoo et al., 2005). Consistent with this report, our data indicate that the -25 to -6 region upstream from the TSS, including the same E-box-like sequence, is indispensable for cell-autonomous circadian gene expression of Per2. Not only the E-box described by Yoo et al. (2005) and Ueda et al. (2005) but also the 10-base pair nucleotides next to the E-box are necessary for this oscillatory regulation. Furthermore, our biochemical data show that endogenous BMAL1 and CLOCK site-specifically bind to the E-box in a phosphorylation- and time-dependent manner (Figure 4), which is novel observation. Importantly, our experiments were mainly performed on the cell-based real-time assay, because the core clock is a cell-autonomous system. To investigate if transcriptional oscillation is driven in the core clock system, we need to examine whether transcriptional regulatory elements function under a cell-autonomous condition. Our approach enables us to exclude internal environmental cues such as blood-borne factors and body temperature. Thus, these two reports verified that the current feedback loop model reflects a core clock mechanism.
The current molecular model for the mammalian circadian clock has been established mainly on the basis of data obtained by conventional reporter assays on the Per1 promoter, implying that several issues remain to be clarified. First, it is difficult to interpret whether the experimental system based on ectopic overexpression reflects physiological phenomena. The E-box consensus sequence, which consists of only six-base pair nucleotides, could appear at some frequency in the promoter region of many genes, leading to the possibility that overexpressed BMAL1 and CLOCK non-specifically bind to E-box and E-box-like sequences and activate transcription (Munoz et al., 2002). Second, these transient assays are not suitable to monitor temporal changes, and therefore the obtained data do not reflect mechanisms of circadian rhythm generation. Mutant mice also could not clarify the role of BMAL1 and CLOCK in Per1 oscillation, because of the possibility of indirect effects. Thus, to date there is no compelling evidence that BMAL1 and CLOCK regulate circadian transcription of Per1 and that E-boxes are indispensable for circadian fluctuation of Per1 transcription. Third, the BMAL1:CLOCK regulation might be specific for Per1 transcription, because detailed analyses have not been performed on the involvement of BMAL1 and CLOCK in circadian transcription of other clock genes. So far at least we could not exclude the possibility that other as yet unknown components have pivotal roles in circadian transcription of other clock genes. In this report, by the real-time monitoring of bioluminescence in cultured cells, we demonstrated that the BMAL1 and CLOCK binding region is included in only the 20-base pair region essential for circadian transcription of Per2, an indispensable gene for the mammalian circadian clock. Our monitoring system was performed under the condition that the endogenous cell-autonomous circadian pacemaker was indeed operating, without the use of overexpression.
Our results are summarized in Figure 5I. Next to the Per2 E-box-like sequence, there is another region that generates more robust cell-autonomous oscillation in transcription of Per2. A combination of the E-box-like sequence and this cooperating element increases the amplitude of cell-autonomous rhythmic transcription of Per2, and consequently the amplitude of Per2 mRNA rhythms is significantly higher than that of Per1 mRNA rhythms. The 1-base pair difference (CACGTT) from the classical circadian E-box is indispensable for this combination. This discovery suggests that the atypical E-box (CACGTT) might be more functional for circadian gene expression rather than the classical E-box (CACGTG) that has been believed to be as the BMAL1: CLOCK binding consensus. However, we still need to note that the reduction in clock-gene amplitude may not necessarily indicate that the pacemaker amplitude is reduced. Future studies will define the precise relationship between the amplitude of clock gene oscillation and that of pacemaker function. The cell-autonomous core clock generates overt circadian oscillations in Per2 transcription, whereas the high amplitude of Per1 oscillation in vivo largely depends on the extracellular environment, which changes cyclically around the clock, rather than on the core clock. This increased expression of the Per1 gene in response to circadian changes of the extracellular environment may be functional in the entrainment of peripheral oscillators.
Among genes that are expressed in a circadian manner, there are clock genes that are central to the timing mechanism and output genes that directly or indirectly mediate physiology under circadian control. The phases and amplitudes in the circadian expression of these genes are different from each other (Panda et al., 2002; Storch et al., 2002). Nevertheless, it is thought that a few enhancer elements, such as E-box, RORE, and DBPE, generate a broad range of phases and amplitudes in circadian gene expression (Yamamoto et al., 2004). Our results provide an explanation for why a small number of elements generate various patterns of circadian gene expression. We showed that two or more transcriptional regulatory elements, such as a phase-delaying element and a rhythm-generating element, are required for robust circadian gene expression of Per2, illustrating that even when the same rhythm-generating element regulates several different genes, combinations with other enhancer elements can advance or delay the phase of these circadian gene expressions. The copy number and various combinations of elements would create unlimited patterns in phases. On the other hand, we showed that the Per2 E-box-like element might generate high amplitude of circadian gene expression by cooperating with a neighboring element. This result demonstrates that synergistic cooperation of several elements may generate more dynamic oscillation in circadian gene expression. In fact, we found that 2 ROR response elements synergistically function in circadian transcription of Bmal1 (Akashi and Takumi, 2005), illustrating that cooperation of elements can enhance the amplitude. The molecular mechanism by which circadian expression of clock and clock-related genes shows a variety of phases and amplitudes will be more clearly revealed by further detailed analyses of the regulatory mechanism for circadian transcription of each known clock gene.
Supplementary Material
Acknowledgments
We thank Setsuko Tsuboi, Yukari Shima, and Daniel Trcka for their expert technical assistance, as well as Takuro Yamamoto, Yasukazu Nakahata, Atsuko Takano, Mayumi Yoshida, and Pabel Delgado for help, reagents, and discussions. We also thank Eisuke Nishida and Keiji Tanaka for general support. We are grateful to Teruya Tamaru for providing the anti-BMAL1 antibody and to Shin Yamazaki for helpful suggestions on data analysis. This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science, and Technology. The support of fellowships from the Japan Society for the Promotion of Science (M.A.) is also acknowledged.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0396) on November 9, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abe, M., Herzog, E. D., Yamazaki, S., Straume, M., Tei, H., Sakaki, Y., Menaker, M., and Block, G. D. (2002). Circadian rhythms in isolated brain regions. J. Neurosci. 22, 350-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, M., and Nishida, E. (2000). Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645-649. [PMC free article] [PubMed] [Google Scholar]
- Akashi, M., and Takumi, T. (2005). The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12, 441-448. [DOI] [PubMed] [Google Scholar]
- Akashi, M., Tsuchiya, Y., Yoshino, T., and Nishida, E. (2002). Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol. Cell. Biol. 22, 1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, U., and Eichele, G. (2003). The mammalian circadian clock. Curr. Opin. Genet. Dev. 13, 271-277. [DOI] [PubMed] [Google Scholar]
- Albrecht, U., Sun, Z. S., Eichele, G., and Lee, C. C. (1997). A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055-1064. [DOI] [PubMed] [Google Scholar]
- Allada, R., Emery, P., Takahashi, J. S., and Rosbash, M. (2001). Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci. 24, 1091-1119. [DOI] [PubMed] [Google Scholar]
- Bae, K., Jin, X., Maywood, E. S., Hastings, M. H., Reppert, S. M., and Weaver, D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525-536. [DOI] [PubMed] [Google Scholar]
- Balsalobre, A., Damiola, F., and Schibler, U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929-937. [DOI] [PubMed] [Google Scholar]
- Balsalobre, A., Marcacci, L., and Schibler, U. (2000). Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 10, 1291-1294. [DOI] [PubMed] [Google Scholar]
- Bunger, M. K., Wilsbacher, L. D., Moran, S. M., Clendenin, C., Radcliffe, L. A., Hogenesch, J. B., Simon, M. C., Takahashi, J. S., and Bradfield, C. A. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian, N., Monaco, L., Pando, M. P., Dierich, A., and Sassone-Corsi, P. (2001). Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 20, 3967-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C. (1999). Molecular bases for circadian clocks. Cell 96, 271-290. [DOI] [PubMed] [Google Scholar]
- Gekakis, N., Staknis, D., Nguyen, H. B., Davis, F. C., Wilsbacher, L. D., King, D. P., Takahashi, J. S., and Weitz, C. J. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564-1569. [DOI] [PubMed] [Google Scholar]
- Hida, A., Koike, N., Hirose, M., Hattori, M., Sakaki, Y., and Tei, H. (2000). The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics 65, 224-233. [DOI] [PubMed] [Google Scholar]
- Kaasik, K., and Lee, C. C. (2004). Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467-471. [DOI] [PubMed] [Google Scholar]
- Kondo, T., and Ishiura, M. (2000). The circadian clock of cyanobacteria. Bioessays 22, 10-15. [DOI] [PubMed] [Google Scholar]
- Kondratov, R. V., Chernov, M. V., Kondratova, A. A., Gorbacheva, V. Y., Gudkov, A. V., and Antoch, M. P. (2003). BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 17, 1921-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume, K., Zylka, M. J., Sriram, S., Shearman, L. P., Weaver, D. R., Jin, X., Maywood, E. S., Hastings, M. H., and Reppert, S. M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193-205. [DOI] [PubMed] [Google Scholar]
- Lee, C., Etchegaray, J. P., Cagampang, F. R., Loudon, A. S., and Reppert, S. M. (2001). Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855-867. [DOI] [PubMed] [Google Scholar]
- Lowrey, P. L., and Takahashi, J. S. (2004). Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5, 407-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menaker, M. (2003). Circadian rhythms. Circadian photoreception. Science 299, 213-214. [DOI] [PubMed] [Google Scholar]
- Munoz, E., Brewer, M., and Baler, R. (2002). Circadian transcription. Thinking outside the E-Box. J. Biol. Chem. 277, 36009-36017. [DOI] [PubMed] [Google Scholar]
- Nagoshi, E., Saini, C., Bauer, C., Laroche, T., Naef, F., and Schibler, U. (2004). Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693-705. [DOI] [PubMed] [Google Scholar]
- Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S., and Hogenesch, J. B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307-320. [DOI] [PubMed] [Google Scholar]
- Pittendrigh, C. S. (1993). Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16-54. [DOI] [PubMed] [Google Scholar]
- Ralph, M. R., Foster, R. G., Davis, F. C., and Menaker, M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975-978. [DOI] [PubMed] [Google Scholar]
- Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935-941. [DOI] [PubMed] [Google Scholar]
- Schibler, U., and Sassone-Corsi, P. (2002). A web of circadian pacemakers. Cell 111, 919-922. [DOI] [PubMed] [Google Scholar]
- Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H., and Weitz, C. J. (2002). Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78-83. [DOI] [PubMed] [Google Scholar]
- Takumi, T., Matsubara, C., Shigeyoshi, Y., Taguchi, K., Yagita, K., Maebayashi, Y., Sakakida, Y., Okumura, K., Takashima, N., and Okamura, H. (1998). A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells 3, 167-176. [DOI] [PubMed] [Google Scholar]
- Tamaru, T., Isojima, Y., van der Horst, G. T., Takei, K., Nagai, K., and Takamatsu, K. (2003). Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells 8, 973-983. [DOI] [PubMed] [Google Scholar]
- Toh, K. L., Jones, C. R., He, Y., Eide, E. J., Hinz, W. A., Virshup, D. M., Ptacek, L. J., and Fu, Y. H. (2001). An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040-1043. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova, Z., Cermakian, N., Reppert, S. M., and Sassone-Corsi, P. (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. USA 99, 7728-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H. R., Hayashi, S., Chen, W., Sano, M., Machida, M., Shigeyoshi, Y., Iino, M., and Hashimoto, S. (2005). System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37, 187-192. [DOI] [PubMed] [Google Scholar]
- Vitaterna, M. H., Selby, C. P., Todo, T., Niwa, H., Thompson, C., Fruechte, E. M., Hitomi, K., Thresher, R. J., Ishikawa, T., Miyazaki, J., Takahashi, J. S., and Sancar, A. (1999). Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 96, 12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. A., and Sehgal, A. (2001). Molecular components of the circadian system in Drosophila. Annu. Rev. Physiol. 63, 729-755. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Nakahata, Y., Soma, H., Akashi, M., Mamine, T., and Takumi, T. (2004). Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, S., Numano, R., Abe, M., Hida, A., Takahashi, R., Ueda, M., Block, G. D., Sakaki, Y., Menaker, M., and Tei, H. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682-685. [DOI] [PubMed] [Google Scholar]
- Yamazaki, S., Straume, M., Tei, H., Sakaki, Y., Menaker, M., and Block, G. D. (2002). Effects of aging on central and peripheral mammalian clocks. Proc. Natl. Acad. Sci. USA 99, 10801-10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S. H., Ko, C. H., Lowrey, P. L., Buhr, E. D., Song, E. J., Chang, S., Yoo, O. J., Yamazaki, S., Lee, C., and Takahashi, J. S. (2005). A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 102, 2608-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S. H. et al. (2004). PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101, 5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M. W., and Kay, S. A. (2001). Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702-715. [DOI] [PubMed] [Google Scholar]
- Zheng, B. et al. (2001). Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683-694. [DOI] [PubMed] [Google Scholar]
- Zheng, B., Larkin, D. W., Albrecht, U., Sun, Z. S., Sage, M., Eichele, G., Lee, C. C., and Bradley, A. (1999). The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169-173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.