Abstract
Tumor necrosis factor (TNF) is a cytokine that mediates many pathophysiologial processes, including angiogenesis. However, the molecular signaling involved in TNF-induced angiogenesis has not been determined. In this study, we examined the role of Etk/Bmx, an endothelial/epithelial tyrosine kinase involved in cell adhesion, migration, and survival in TNF-induced angiogenesis. We show that TNF activates Etk specifically through TNF receptor type 2 (TNFR2) as demonstrated by studies using a specific agonist to TNFR2 and TNFR2-deficient cells. Etk forms a preexisting complex with TNFR2 in a ligand-independent manner, and the association is through multiple domains (pleckstrin homology domain, TEC homology domain, and SH2 domain) of Etk and the C-terminal domain of TNFR2. The C-terminal 16-amino-acid residues of TNFR2 are critical for Etk association and activation, and this Etk-binding and activating motif in TNFR2 is not overlapped with the TNFR-associated factor type 2 (TRAF2)-binding sequence. Thus, TRAF2 is not involved in TNF-induced Etk activation, suggesting a novel mechanism for Etk activation by cytokine receptors. Moreover, a constitutively active form of Etk enhanced, whereas a dominant-negative Etk blocked, TNF-induced endothelial cell migration and tube formation. While most TNF actions have been attributed to TNFR1, our studies demonstrate that Etk is a TNFR2-specific kinase involved in TNF-induced angiogenic events.
Angiogenesis, a process of new blood vessel formation, is involved in many physiological and pathological settings, such as ischemia, atherosclerosis, and arthritis (7, 62). Although angiogenic factors such as VEGF and FGF promote angiogenesis in vitro and in vivo, inflammatory responses (as defined by the presence of infiltrated macrophages and proinflammatory cytokines) seem to be essential for angiogenesis (2, 28). Inflammatory angiogenesis is largely mediated by proinflammatory cytokines, such as tumor necrosis factor (TNF), which has been shown to stimulate angiogenesis in vitro, in vivo, and in disease settings (18, 27, 44, 48). However, the molecular signaling involved in TNF-induced angiogenesis has not been determined.
Vascular endothelial cells (EC) are among the principal physiological targets of TNF (37). In EC, as in other cell types, TNF elicits a broad spectrum of biological effects, including proliferation, differentiation, and apoptosis (31, 32, 49, 51). The nature of TNF effects depends on TNF concentration and the type and growth state of the target cells (17). These differences in TNF-induced response are due, in part, to the presence of two distinct TNF-specific plasma membrane-localized receptors, type 1 55-kDa TNFR (TNFR1) and type 2 75-kDa TNFR (TNFR2) (61). TNFR1 is expressed ubiquitously, whereas TNFR2 expression is tightly regulated and found predominantly on EC and hemopoietic cells (58). In many cell types, including EC, TNFR2 is primarily expressed on the cell surface, whereas TNFR1 is predominantly localized to the Golgi with little surface expression (6). The concept of ligand passing has been proposed in which plasma membrane TNFR2 binds TNF and efficiently passes the anchored ligand to TNFR1 (55). It has also been proposed that TNFR2 primarily responds to TNF expressed as an integral membrane protein on the cell surface, whereas TNFR1 primarily responds to soluble TNF (21, 22). However, recent evidence supports the idea that TNF utilizes TNFR1 and TNFR2 to trigger distinct signaling events and exert diverse biological functions in a context-dependent manner. TNFR1 is believed to mediate cell death, whereas TNFR2 serves to enhance TNFR1-induced cell death or to promote cell activation, migration, growth, or proliferation (9, 14, 15, 19, 35, 36, 45, 52, 56, 63). The role of TNFR2 in TNF signaling in EC is not clear. Studies utilizing receptor-specific neutralizing antibodies have shown that TNFR2 contributes to effects of lower concentrations of TNF (50). Furthermore, it has been shown that lower TNF concentrations stimulate proliferation and migration through TNFR2, whereas higher TNF concentrations inhibit both these responses through TNFR1 in EC and epithelial cells (23, 26).
TNFR1 and TNFR2, among members of the TNF receptor family, share a similar architecture with characteristic cysteine-rich motifs (52). Unlike the extracellular domains, the primary amino acid sequences of the cytoplasmic domains of TNFR1 and TNFR2 are unrelated. It is believed that the two receptors initiate distinct signal transduction pathways by interacting with different signaling proteins (12, 20, 29). A present model postulates that TNF binding triggers trimerization of TNFR1 and TNFR2, which recruit adaptor proteins and signaling molecules by their intracellular domains to form a receptor-signaling complex (4, 60). Many proteins have been shown to be recruited by TNFR1, including TRADD (TNFR-associated death-domain protein) (24, 25). TRADD functions as a platform adaptor that recruits TRAF2 (TNFR-associated factor), RIP (receptor-interacting protein kinase), and FADD (Fas-associated death-domain protein) to form TNFR1 signaling complex and to activate several distinct signaling cascades (11, 47, 53). TRAF2 acts as an assembly platform for the recruitment of additional factors, including the members of MAP kinase kinase kinase family, leading to activation of the JNK pathway (5).
In contrast, less is known regarding proteins recruited to TNFR2 and downstream signaling. Like TNFR1, TNFR2 can also recruit TRAF2 and utilize TRAF1 and the two cellular inhibitors of apoptotic proteins (cIAP1 and cIAP2) (46). However, the role of these factors in TNFR2-specific signaling has not been defined. So far no other signaling molecules have been shown to directly associate with TNFR2, and it is not known whether TRAF2-independent signaling pathways exist for TNFR2.
The endothelial/epithelial tyrosine kinase (Etk/Bmx), a member of the Btk nonreceptor tyrosine kinase family, has been implicated in cell adhesion, migration, proliferation, and survival (39, 40, 42, 54). Etk and three other members of this family, (Btk, Itk, and Tec) participate in signal transduction stimulated by growth factor receptors, cytokine receptors, G-protein-coupled receptors, antigen receptors, and integrins (10, 33, 41, 57). They share common structural domains, including a pleckstrin homology (PH) domain, a TEC homology (TH) domain, which has a PXXP motif (with exception of Etk), SH3 and SH2 domains, and a kinase domain. The mechanism(s) by which Btk family kinases are activated is not clear. It has been proposed that intramolecular interactions between the PXXP motif in the TH domain and the SH3 domain (with the exception of Etk) and between the PH domain and the kinase domain fold Btk family kinases into a closed inactive form. Btk activation occurs when stimuli disrupt these intramolecular interactions by promoting Btk translocation to the plasma membrane (10, 33, 38-42, 57).
Recently Etk has been implicated in the signaling of Tie-2 and VEGF receptors, two important receptor families essential for angiogenesis (41). In the present study we examined the role of Etk in TNF-induced angiogenesis. We show that TNF activates Etk through TNFR2 in a TRAF2-independent manner. TNFR2 associates with Etk through the C-terminal 16-amino-acid sequence of TNFR2 and multiple domains (the PH, TH, and SH2 domains) of Etk. Furthermore, we show that a constitutively active Etk mutant enhanced, whereas a dominant-negative Etk mutant blocked, TNF-induced angiogenesis by using in vitro models of EC migration and tube formation. Our data demonstrate that Etk is a critical mediator in TNF-induced angiogenic events through association with TNFR2.
MATERIALS AND METHODS
Plasmids.
Mammalian expression plasmids for the wild-type and the dominant-negative TRAF2 (WT-TRAF2 and DN-TRAF2, respectively) were provided by David Goeddel (Tularik); plasmids for Etk/Bmx wild type and mutants were provided by Dianqing Wu (University of Connecticut); and plasmids for TNFR1, TNFR2, and TNFR2 deletion mutants were provided by Jordan S. Pober (Yale University). The TNFR2 and Etk point mutants were generated by the Quickchange site-specific mutagenesis kit following the manufacturer's instructions (Stratagene). Etk-DK, -SK, -K, and -PTH were constructed into Flag-vector (Clontech). Etk-SK and Etk-DK were also cloned into an internal ribosome entry sequence vector coexpressing enhanced green fluorescence protein in a bicistronic mRNA (pIRES; Clontech).
Cells and cytokines.
Human umbilical vein EC (HUVEC) and bovine aorta endothelial cells (BAEC) were purchased from Clonetics (San Diego, Calif.). Lung endothelial cells isolated from TNFR-deficient mice (14, 35, 45) (The Jackson Laboratory) were isolated according to a procedure described previously (43). Minced tissue was digested with collagenase. Large tissue fragments were removed by filtration through a 100-m nylon screen. The filtrate was collected on a 20-m nylon screen, washed, and purified by Percoll gradient centrifugation. EC contained in the 3rd through 10th fractions from the top of the gradient (density, 1.00 to 1.050 g/ml) were collected, washed, and seeded into dishes containing EC growth medium. Contaminating non-EC were removed by mechanical weeding and by fluorescence-activated cell sorting with antibodies to PECAM-1 (CD31) to label the EC. The sorted cells were assessed for EC phenotype, including morphology and expression of von Willebrand Factor and PECAM-1. EC were used at passages 1 to 5. Human and murine recombinant TNF and polyclonal antibodies against TNFR1 and TNFR2 were from R&D Systems (Minneapolis, Minn.).
Transfection.
Transfection of BAEC was performed with Lipofectamine 2000 according to the manufacturer's protocol (Gibco). Cells were cultured at 90% confluence in 6-well plates and were transfected with a total of 4 μg of plasmid constructs as indicated. Cells were harvested at 36 to 48 h posttransfection, and cell lysates were used for protein assays.
JNK kinase assay.
JNK assays were performed as described previously by using GST-c-Jun(1-80) fusion protein as a substrate (34). Briefly, a total of 400 μg of cell lysates was immunoprecipitated with 5 μg of antibody against JNK1 (Santa Cruz). The immunoprecipitates were mixed with 10 μg of GST-c-Jun(1-80) suspended in the kinase buffer (20 mM HEPES [pH 7.6], 20 mM MgCl2, 25 mM β-glycerophosphate, 100 μM sodium orthovanadate, 2 mM dithiothreitol, 20 μM ATP) containing 1 μl (10 μCi) of [γ-32P]ATP. The kinase assay was performed at 25°C for 30 min. The reaction was terminated by the addition of Laemmli sample buffer, and the products were resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis followed by protein transferring to a membrane (Immobilon P; Millipore, Milford, Mass.). Phosphorylated GST-c-Jun(1-80) was visualized by autoradiography. The membrane was further used for Western blotting with anti-JNK1.
Immunoprecipitation and immunoblotting.
After various treatments BAEC were washed twice with cold phosphate-buffered saline (PBS) and were harvested in a membrane lysis buffer (30 mM Tris [pH 8], 10 mM NaCl, 5 mM EDTA, 10 g of polyoxyethylene-8-lauryl ether/liter, 1 mM O-phenanthroline, 1 mM indoacetamide, 10 mM NaF, 5 mM orthovanadate, 10 mM sodium pyrophosphate). Cells were immediately frozen in liquid nitrogen. Cell lysates were then thawed on ice, scraped, sonicated, and centrifuged at 14,000 × g at 4°C for 15 min. Supernatants were used immediately for immunoblot or immunoprecipitation. For immunoprecipitation to analyze protein interaction in vivo, supernatant of cell lysates was diluted three times with a cold lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Triton X-100, 0.75% Brij 96, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 1 mM sodium pyrophosphate, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 2 mM phenylmethylsulfonyl fluoride, 1 mM EDTA). The lysates were then incubated with the first protein-specific antiserum (e.g., anti-Etk from Santa Cruz or anti-TNFR from R&D) for 2 h with 50 μl of GammaBind plus Sepharose. Immune complexes were collected after each immunoprecipitation by centrifugation at 13,000 × g for 10 min followed by three to five washes with lysis buffer. The immune complexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblot (Immobilon P) with the second protein (e.g., phosphotyrosine antibody; Upstate Biotechnology Inc., Lake Placid, N.Y.). The chemiluminescence was detected by using an ECL kit according to the instructions of the manufacturer (Amersham Life Science, Arlington Heights, Ill.). For detection of Flag-tagged proteins (Etk or TNFR2 mutants), anti-Flag M2 antibody (Sigma) was used for immunoblotting. For detection of T7-tagged proteins (Etk mutants), anti-T7 antibody (Novagen) was used for immunoblotting.
EC migration assay and image analysis.
BAEC were cultured in 0.5% fetal bovine serum (FBS) overnight and were subjected to wound injury with a yellow tip. Cells were washed with PBS once, and fresh medium (0.5% FBS) with or without TNF (1 ng/ml) was added. Cells were further cultured for the indicated times. The EC migration in culture was determined by measuring wound areas in cell monolayers. Three different images from each well along the wound were captured by a digital camera under a microscope (magnification, ×4). A hemocytometer (1 mm2/grid) was used as a standard. The wound area was measured (in square millimeters) and analyzed by using NIH Image 1.60.
EC tube formation assay in collagen gel culture.
Collagen gel culture was performed as follows. The ingredients 10× RPMI 1640 (Gibco), neutralizing buffer (260 mmol of NaHCO3/liter, 200 mmol of HEPES/liter, 50 mmol of NaOH/liter) and collagen gel (Vitrogen) were mixed at the ratio of 1:1:8 on ice and then were added to a 24-well plate (400 μl/well); the plate was then incubated at 37°C for 1 h. BAEC cells were seeded at 2 × 104/well, and the plate was incubated overnight in 5% CO2 at 37°C. The medium was removed and cells were gently washed with 1× PBS. The collagen gel mixture prepared as described above was added onto the cells, which had already attached to the first layer of collagen gel. The plate was incubated for 30 min at 37°C (while keeping the plate away from CO2). EC media were added at 500 μl/well, and the plates were incubated in 5% CO2 at 37°C. EC media were changed every 2 to 3 days, and tube formation was observed after 36 to 48 h. A branched EC network was visualized under a microscope. Images from three different areas in each well were captured by a digital camera under a microscope (magnification, ×4), and the area covered by branched cells was measured (in square millimeters) and analyzed by NIH Image 1.60.
RESULTS
TNF activates Etk through TNFR2.
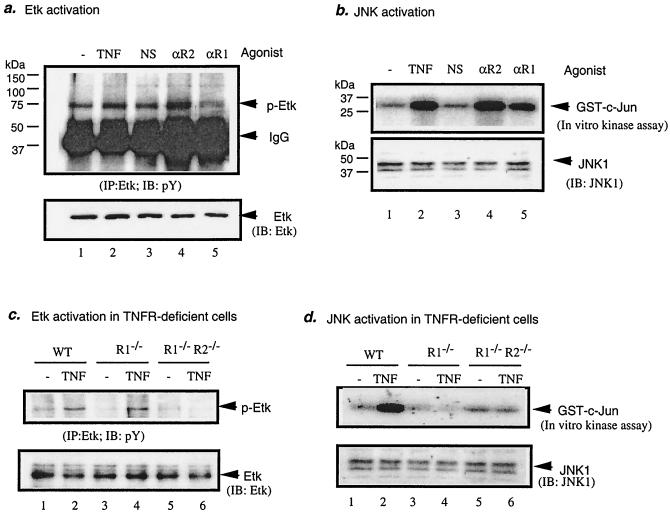
Recently it was observed that Etk activity was increased in human TNF transgenic mice, suggesting a role for Etk in TNF signaling (64 and W. Min, unpublished data). To determine if TNF activates Etk, HUVEC were treated with TNF (1 ng/ml for 15 min) or an antibody against TNFR (5 μg/ml for 15 min). TNFR1 antibody and TNFR2 antibody specifically recognize the extracellular domain of the receptor and can function as receptor agonists (15, 19, 56). Etk activation was determined by measuring tyrosine phosphorylation of Etk. Results show that TNF and TNFR2 agonist activated Etk. However, TNFR1 agonist did not activate Etk, suggesting that TNF activates Etk through TNFR2 (Fig. 1a). As a control, JNK activity was also measured by an in vitro kinase assay. As expected, TNF, TNFR1 agonist, and TNFR2 agonist all activated JNK (Fig. 1b). Similar results were obtained with BAEC (data not shown). To define further the role of TNFR2 in Etk activation, TNF-induced Etk activation in EC from mice deficient in TNFR1 or both TNFR1 and TNFR2 was examined. Results show that TNFR1 deficiency had no effect on Etk activation compared to that of wild-type cells. However, double deficiency (TNFR1−/−R2−/−) (14, 35, 45) resulted in diminished Etk activation in response to TNF (Fig. 1c). In contrast, JNK activation by TNF was blunted in both cell types compared to that of the wild type (Fig. 1d), indicating that TNFR1 is required for JNK but not for Etk activation by TNF. Taken together these data demonstrate that TNF activates Etk specifically through TNFR2 in EC.
FIG. 1.
TNF activates Etk specifically through TNFR2. (a) TNFR2 (R2) agonist, but not TNFR1 (R1) agonist, activates Etk in EC. BAEC were treated with human TNF (1 ng/ml), anti-TNFR1 (αR1; 5 μg/ml), anti-TNFR2 TNFR1 (αR2; 5 μg/ml), or a normal goat serum (5 μg/ml; NS; TNFR1) for 15 min or were left untreated. Etk activation was detected by immunoprecipitation with anti-Etk followed by Western blot with antiphosphotyrosine (pY). Total Etk was detected by Western blot with anti-Etk. IgG, immunoglobulin G; IB, immunoblot.(b) JNK activation was determined by an in vitro kinase assay with GST-c-Jun as a substrate. JNK1 protein in the immunoprecipitates was determined by Western blot with anti-JNK1. (c and d) TNF fails to activate Etk in TNFR2-deficient cells. Lung EC from wild type (WT), TNFR1−/−, or TNFR1−/−TNFR2−/− mice were treated with murine TNF at 1 ng/ml for 15 min, and Etk (c) and JNK (d) activation was determined.
Etk binds to TNFR2 but not TNFR1.
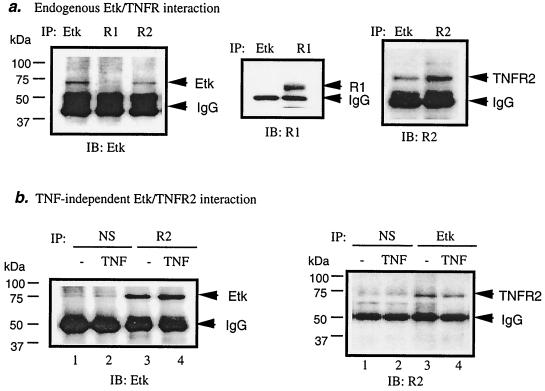
To define the mechanisms by which TNFR2 activates Etk, we examined association of endogenous Etk and TNFR2 in EC (both HUVEC and BAEC). Interaction of Etk with TNFR1 and TNFR2 was determined by coimmunoprecipitation assay. The ability of an antibody against either Etk or TNFR2 to precipitate a complex of Etk and TNFR2 suggests that Etk and TNFR2 interact in vivo (Fig. 2a). In contrast, Etk and TNFR1 failed to be coimmunoprecipitated by either anti-Etk or anti-TNFR1 (Fig. 2a), indicating that Etk and TNFR1 do not interact. To determine if association of Etk with TNFR2 is TNF-dependent, HUVEC were treated with TNF. Association of Etk with TNFR2 was examined by coimmunoprecipitation assay, as described above, with anti-TNFR2 or anti-Etk (Fig. 2b). Results showed that TNF did not enhance the association of Etk with TNFR2, indicating that Etk interacts with TNFR2 in a ligand-independent fashion (Fig. 2b). Similar results for TNFR2-Etk interaction were obtained with BAEC. These results indicate that HUVEC and BAEC share a common pathway for TNF-induced Etk activation.
FIG. 2.
Association of TNFR2 with Etk in EC is ligand independent. BAEC were either treated with TNF (1 ng/ml for 15 min) or were left untreated. An antibody against either Etk, TNFR1, or TNFR2 was used to examine its ability to precipitate a complex of Etk and TNFRs. A total of 40 μg of cell lysate and 6 μg of antibody was used for each immunoprecipitation (IP). (a) Etk associates with TNFR2 but not with TNFR1 in EC. Anti-Etk was used as a positive control for immunoprecipitation. (b) Association of Etk with TNFR2 is TNF independent in EC. Normal goat sera (NS) were used as controls. IgG, immunoglobulin G; IB, immunoblot; R1, TNFR1; R2, TNFR2.
Multiple domains (PH, TH, and SH2) of Etk are critical for association with TNFR2.
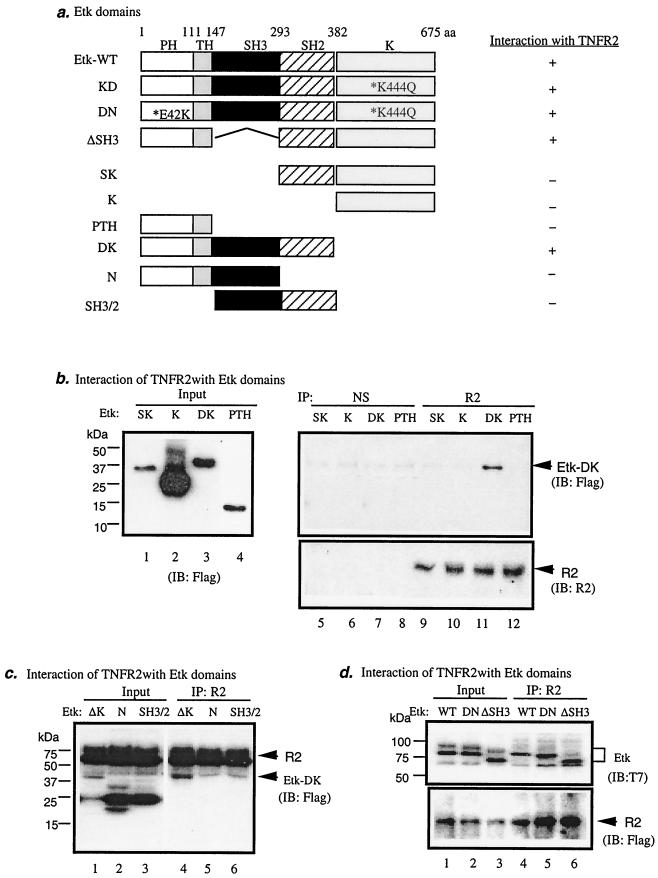
Etk contains several structural domains (PH, TH, SH3, SH2, and kinase domains; Fig. 3a). To map the domain of Etk responsible for interaction with TNFR2, we generated Flag-tagged expression constructs encoding different Etk structural domains, as shown in Fig. 3a. Etk-SK contains the SH2 domain and the kinase domain, Etk-K contains the kinase domain only, Etk-PTH contains the PH and TH domains, and Etk-DK has a deletion of the kinase domain. Interaction of Etk proteins with endogenous TNFR2 was performed by coimmunoprecipitation with anti-TNFR2 followed by Western blot with anti-Flag. Results showed that Etk-DK, but not Etk-SK, -K, or -PTH, interacts with TNFR2 (Fig. 3b). These data suggest that TNFR2-interacting sequence is located in the N-terminal (from PH to SH2) domains of Etk.
FIG. 3.
Multiple domains (the PH, TH, and SH2 domains) of Etk are responsible for association with TNFR2 in EC. (a) Schematic diagram of Etk domains and expression constructs. The kinase-dead Etk (Etk-KD) has a mutation at the kinase domain (K444Q). The dominant-negative Etk (Etk-DN) has mutations at the kinase domain (K444Q) and the phospholipid-binding PH domain (E42K). Etk-ΔSH3 has a deletion of the SH3 domain. Etk-SK contains the SH2 and the kinase domains. Etk-K contains the kinase domain only. Etk-PTH contains the PH and TH domains. Etk-DK has a deletion of the kinase domain. Etk-N contains the PTH and SH3 domains. Etk-SH3/2 contains the SH3 and SH2 domains. A summary of interaction with TNFR2 is shown on the right. Etk-WT, wild-type Etk; aa, amino acid. (b) Mapping the Etk domain interacting with TNFR2. Flag-tagged Etk-SK, Etk-K, Etk-PTH, and Etk-DK were transfected in EC, and expression was determined by anti-Flag. Interaction of Etk domains with TNFR2 was examined by immunoprecipitation with TNFR2 or normal serum (NS) followed by Western blot with anti-Flag as indicated. TNFR2 protein in the immunoprecipitate was detected by Western blot with anti-TNFR2. (c) Mapping the TNFR2-interacting domain in the N-terminal portion of Etk. Flag-tagged TNFR2 was cotransfected in EC with Etk-DK, Etk-N, or Etk-SH3/2. Expression was determined by anti-Flag (lanes 1 to 3). Interaction of Etk domains with TNFR2 was determined by immunoprecipitation with TNFR2 followed by Western blot with anti-Flag (lanes 4 to 6). (d) Determining the role of the SH3 domain in Etk for TNFR2 association. T7-tagged Etk-WT, DN, and Etk-ΔSH3 were cotransfected with Flag-TNFR2 in EC, and interaction of Etk with TNFR2 was examined by immunoprecipitation with TNFR2 followed by Western blot with anti-T7. TNFR2 was detected by Western blot with anti-Flag. IP, immunoprecipitation; IB, immunoblot; R2, TNFR2.
To further map the interacting domain in Etk, we used Etk-DK to generate expression constructs for Etk-N (the PTH and SH3 domains) and Etk-SH3/2 (the SH3 and SH2 domains). BAEC were cotransfected with Flag-tagged Etk mutants and TNFR2 constructs, and interaction of Etk proteins with TNFR2 was performed by coimmunoprecipitation with anti-TNFR2 followed by Western blot with anti-Flag. Results showed that Etk-DK, but not Etk-N or Etk-SH3/2, associated with TNFR2 (Fig. 3c). These data suggest that either all domains (PTH, SH3, and SH2) or a combination of the PTH and SH2 domains are required for TNFR2 association.
To define the role of the SH3 domain in TNFR2 binding, we generated Etk-ΔSH3 (deletion of the SH3 domain) from Etk-WT. BAEC were transfected with various T7-tagged Etk expression constructs (Etk-WT, -DN, and -ΔSH3) and Flag-TNFR2, and interaction of Etk proteins with TNFR2 was performed by coimmunoprecipitation with anti-TNFR2 followed by Western blot with anti-T7. The kinase-dead Etk (Etk-KD) has a mutation at the kinase domain (K444Q). The dominant-negative form (Etk-DN) has mutations at both the kinase domain (K444Q) and the phospholipid-binding PH domain (E42K) (Fig. 3a). Results showed that Etk-DN and Etk-ΔSH3 bound to TNFR2 as well as Etk-WT did (Fig. 3d), indicating that the Etk kinase activity and phospholipid-binding activity are not required for interaction with TNFR2. The SH3 domain is not required for TNFR association. Taken together, we conclude that the combination of multiple domains (the PH, TH, and SH2 domains) in Etk is critical for Etk-TNFR2 complex formation.
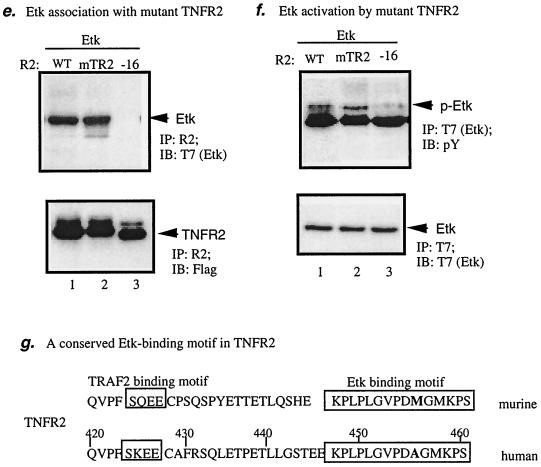
A non-TRAF2-binding motif at the C-terminal domain (16-amino-acid residues) in TNFR2 is critical for Etk association and activation.
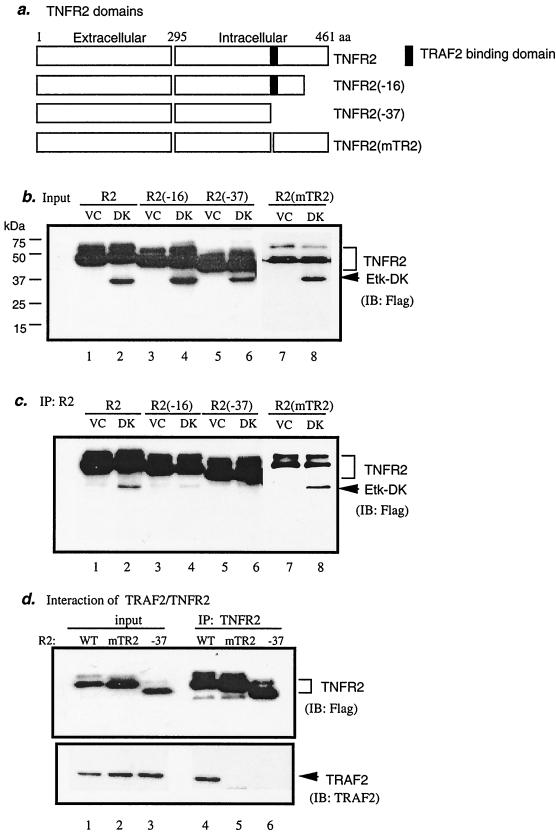
To map the residues in the intracellular domain of TNFR2 important for Etk binding, Flag-TNFR2 mutants containing truncated C termini were used (16). TNFR2(−16) and TNFR2(−37) are mutants lacking 16- and 37-amino-acid residues at the C terminus, respectively (Fig. 4a). We also constructed a TNFR2 mutant in which the TRAF2-binding motif SKEE has been changed to AKAA (mTR2; Fig. 4g). EC were transfected with Etk-DK and TNFR2 expression constructs (Fig. 4b). Interaction of Etk-DK with TNFR2 proteins was performed as described in Materials and Methods. Results showed that the full-length TNFR2-wild type (WT) and the TRAF2-binding motif mutant (mTR2), but not TNFR2(−16) or TNFR2(−37), interacted with Etk-DK (Fig. 4c). A similar result was observed for Etk-WT (see Fig. 4e). Consistent with previous reports (47), TNFR2-WT, but not TNFR2(−37) (deletion of the TRAF2-binding motif) or TNFR2(mTR2) (mutations at the TRAF2-binding motif SKEE), bound to TRAF2 (Fig. 4d). These data suggest that Etk associates with TNFR2 through a non-TRAF2-binding motif.
FIG. 4.
The C-terminal 16-amino-acid residues in TNFR2 are critical for association and activation of Etk. (a) Schematic diagram of TNFR2 domains and the deletion mutants at the C terminus. TNFR2(−16) and TNFR2(−37) are mutants lacking 16- and 37-amino-acid residues at the C terminus, respectively. The black box indicates the TRAF2-binding domain. TNFR2(mTR2) is a mutant containing mutations at the TRAF2-binding motif. All TNFR2 expression constructs are Flag-tagged at the N termini. (b) Flag-tagged Etk-DK or the VC was cotransfected in EC with TNFR2 expression constructs, and expression was determined by anti-Flag. (c) Interaction of Etk-DK with full-length TNFR2 and TNFR2(mTR2) but not with TNFR2(−16) or TNFR2(−37). Interaction was determined by immunoprecipitation with anti-TNFR2 (recognizes the extracellular domain of TNFR2) followed by Western blot with anti-Flag. (d) Interaction of TRAF2 with TNFR2-WT but not with TNFR2(mTR2) or TNFR2(−37). Flag-tag TNFR2-WT, TNFR2(mTR2), or TNFR2(−37) was cotransfected in EC with a TRAF2 expression construct. Association of TRAF2 with TNFR2 or TNFR2(−37) was determined by immunoprecipitation with anti-TNFR2 followed by Western blot with anti-TRAF2. (e) Association of Etk with TNFR2 mutants. BAEC were transfected with T7-tag Etk-WT and Flag-TNFR2 constructs (WT, −16, or mTR2) as indicated. Association of Etk with TNFR2 proteins was determined by immunoprecipitation with anti-TNFR2 followed by Western blot with anti-T7. TNFR2 protein was determined by Western blot with anti-Flag. (f) Activation of Etk by TNFR2 mutants. Etk activation was determined by immunoprecipitation with anti-T7 followed by Western blot with anti-pY. Total Etk was determined by Western blot with anit-T7. (g) Schematic diagram of Etk-binding/activating motif and TRAF2-binding motif in the C-terminal intracellular domain of human and murine TNFR2. The Etk-binding/activating motif is highly conserved between the human and murine TNFR2 and differs in only one residue (in bold). IP, immunoprecipitation; IB, immunoblot; R2, TNFR2; aa, amino acid.
To determine if association of TNFR2 with Etk is required for Etk activation, we examined Etk activation by TNFR2 mutants. BAEC were cotransfected with Etk with TNFR2-WT, TNFR2(mTR2), or TNFR2(−16). Association of Etk with TNFR2 was determined by immunoprecipitation with anti-Flag (for TNFR2) followed by Western blot with anti-T7 (for Etk). Activation of Etk by TNFR2 was determined by Etk tyrosine phosphorylation. As expected, wild-type TNFR2 bound to and activated Etk (lane 1 in Fig. 4e and f). TNFR2(mTR2) (TRAF2-binding deficient) still retains the ability to bind and activate Etk (lane 2 in Fig. 4e and f), further supporting the hypothesis that TNFR2 activates Etk in a TRAF2-independent manner. In contrast, TNFR2(−16) was unable to associate and activate Etk (lane 3 in Fig. 4e and f), suggesting that association of Etk with TNFR2 is required for Etk activation. These data define the C-terminal 16 residues (amino acids 445 to 461 in human TNFR2) as an Etk-binding/activating motif. Interestingly, this Etk-binding/activating motif is highly conserved between the human and murine TNFR2, and only one residue differs (M456 in human and A456 in murine molecules) (Fig. 4g). However, the sequences between the TRAF2-binding motif and the Etk-binding/activating motif are quite variable. These results warrant further mutagenesis analysis to define the critical residues in TNFR2 sequence for Etk binding and activation.
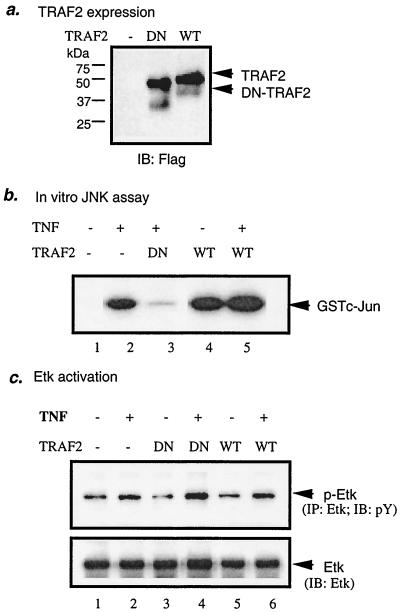
TRAF2 is not involved in Etk activation by TNF.
TRAF2 is an adaptor protein that was shown to be recruited by both TNFR1 and TNFR2 and that couples TNF signaling to NF-κB and JNK activation (46, 47). The N-terminal domain of TRAF2, containing the ring and zinc fingers, is critical for its activity. A dominant-negative TRAF2 (DN-TARF2) contains the TRAF domain but lacks the functional N terminus and presumably acts by competing with endogenous TRAF2 for binding to receptors or receptor complexes (47). To determine the role of TRAF2 in Etk activation by TNF and/or TNFR2, we examined effects of TRAF2 on TNF-induced Etk activation. EC were transfected with an expression construct for WT-TRAF2 or DN-TRAF2 (Fig. 5a). Consistent with previous observations (34), TRAF2 expression alone activated JNK in EC, whereas DN-TRAF2 blocked TNF-induced JNK activation (Fig. 5b). In contrast, expression of WT-TRAF2 did not activate Etk and DN-TRAF2 had no effect on TNF-induced Etk activation (Fig. 5c). These data indicate that Etk activation by TNF is TRAF2 independent.
FIG. 5.
TRAF2 is not involved in TNF-induced Etk activation in EC. BAEC were transfected with WT-TRAF2 or DN-TRAF2. Expression of TRAF2 was determined by Western blot with anti-Flag (a). Twenty-four hours posttransfection, cells were treated with human TNF (1 ng/ml for 15 min) or were left untreated. Etk (b) and JNK (c) activation were determined as described in the legend to Fig. 1. IP, immunoprecipitation; IB, immunoblot.
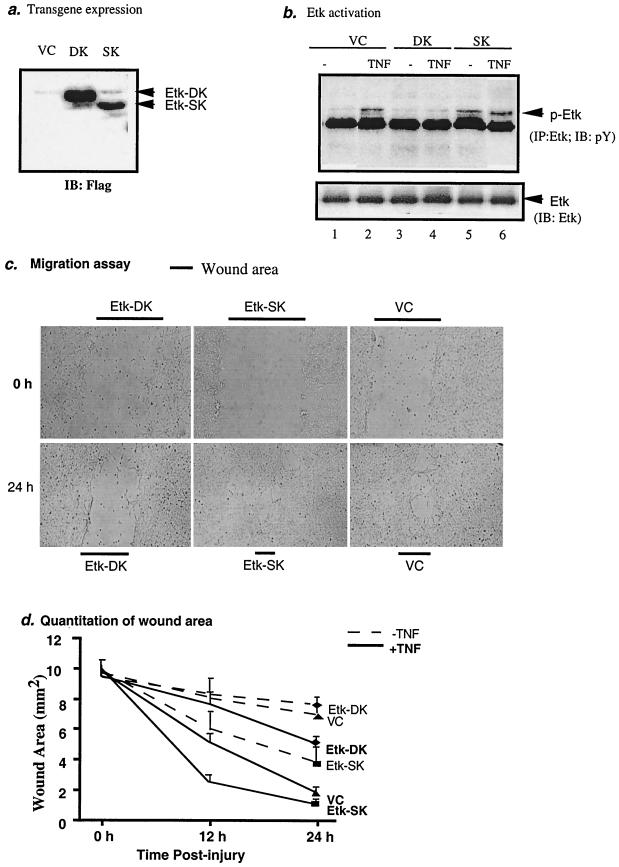
A critical role of Etk in TNF-induced angiogenesis.
Etk has been implicated in cell migration and tumor metastases (3, 10). To examine the biological consequence of Etk activation by TNF in EC, we reasoned that Etk might be involved in TNF-induced angiogenesis. To test this hypothesis, we employed in vitro angiogenesis models to determine effects of Etk on TNF-induced EC migration and tube formation, two critical steps in the process of angiogenesis. First we generated two Etk mutants: Etk-SK contains the SH2 domain and the kinase domain, and Etk-DK contains a deletion of the kinase domain (see Fig. 3a). Etk-SK and Etk-DK were coexpressed with an enhanced green fluorescent protein in a bicistronic mRNA, linked by an internal ribosome entry sequence. BAEC were transfected with Etk-SK and Etk-DK by using Lipofectamine 2000. Transfection efficiency was determined by fluorescence microscopy and usually reached 90%. This high transfection efficiency allowed us to examine effects of the transgene on endogenous Etk activity (30) (Fig. 5). Expression of Etk-DK and Etk-SK was determined by Western blot anti-Flag (Fig. 6a). It has been proposed that an intramolecular interaction between the PH domain and the kinase domain folds Etk into a closed inactive form. Thus, the PH domain is an inhibitory domain and deletion of the PH domain (Etk-SK) should render Etk active. It is conceivable that overexpressed Etk-SK binds to the PH domain of endogenous Etk to release the inhibitory PH domain from Etk, leading to Etk activation. In contrast, overexpressed Etk-DK (containing the inhibitory PH domain) should keep endogenous Etk in an inactive form. We examined the effect of Etk-SK and Etk-DK on TNF-induced endogenous Etk phosphorylation. Results showed that Etk-SK increased both basal and TNF-induced phosphorylation of endogenous Etk, suggesting that Etk-SK is a constitutively active form (Fig. 6b and c). In contrast, Etk-DK blocked TNF-induced activation of endogenous Etk (Fig. 6b), indicating that Etk-DK is a dominant-negative form. As expected, we did not observe phosphorylation of Etk-SK (lacking Y40 in the PH domain) or Etk-DK (lacking the kinase domain) (data not shown).
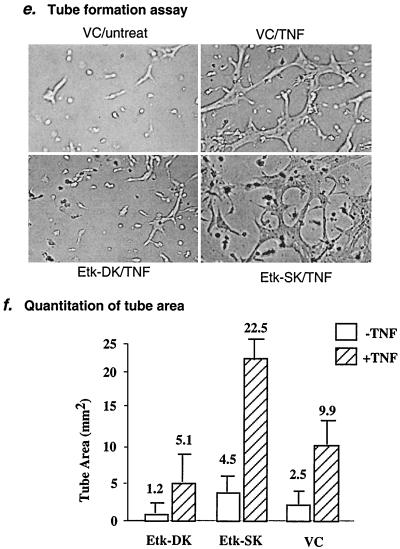
FIG. 6.
A critical role of Etk in TNF-induced angiogenesis. BAEC in a 6-well plate were transfected with VC, Etk-SK, or Etk-DK, which coexpresses an enhanced green fluorescent protein. Twenty-four hours postinfection, transfection efficiency and transgene expression were determined. Cells were split into a 24-well plate to reach 100% confluency for migration and tube formation assays (see Materials and Methods). (a) Expression of Etk-SK and Etk-DK was determined by Western blot with anti-Flag. (b) Etk-SK functions as a constitutive active form, whereas Etk-DK functions as a dominant-negative form of Etk. VC, Etk-SK, and Etk-DK-expressing cells were treated with TNF (1 ng/ml for 15 min), and activation of endogenous Etk was determined by immunoprecipitation with anti-Etk followed by Western blot with anti-phosphotyrosine. (c) Etk-SK promotes, while Etk-DK inhibits, TNF-induced EC migration. Transfected BAEC were subjected to injury, and wound healing was measured in the absence or presence of TNF (1 ng/ml) at 0, 12, and 24 h postinjury. Zero and 24 h in the presence of TNF are shown. Triplets for each sample were performed. Experiments were repeated two times. (d) Quantitation of EC migration. Image capture and analyses were performed as described in Materials and Methods. Data presented are means ± standard errors of the means of the triplicate in one experiment. Similar results from two other experiments were obtained. (e) Etk-SK promotes, while Etk-DK inhibits, TNF-induced EC tube formation. Transfected cells were seeded in collagen gels as described in Materials and Methods in the presence or absence of TNF (1 ng/ml), and tubes were visualized on day 5. Image capture and analyses were performed as described in Materials and Methods. Triplets for each sample were performed. Experiments were repeated one time. (f) Quantitation of EC tube formation. Three images from each well were captured, and total tube areas were measured in each field (30 mm2). Data presented are means ± standard errors of the means of the two triplicates from two independent experiments. IP, immunoprecipitation; IB, immunoblot.
Next we examined effects of Etk-DK or Etk-SK expression on EC migration induced by wounding. EC were cultured in a medium with 0.5% FBS for 24 h followed by a wound healing assay in the presence of various concentrations of TNF (0, 1, 5, and 10 ng/ml). Consistent with previous reports on TNF-induced angiogenesis (18, 27, 44, 48), we found that TNF at a low concentration (1 ng/ml) stimulated EC migration to heal the wound (Fig. 6c). However, TNF at high concentrations (5 and 10 ng/ml) inhibited EC migration (data not shown). The effects of Etk-SK and Etk-DK expression on TNF-induced EC migration rates were determined by measuring the wound area (in square millimeters/field) in the cell monolayer at various time points after wounding (0, 12, and 24 h). Results showed that Etk-SK expression stimulated basal (−TNF) and TNF-induced (+TNF) EC migration (Fig. 6c and d). In contrast, Etk-DK significantly delayed TNF-induced EC wound healing compared to that for vector control (VC)-infected cells (Fig. 6c and d). These data indicate that Etk is involved in TNF-induced EC migration.
Finally, we examined the effects of Etk-DK or Etk-SK expression on TNF-induced EC tube formation. EC were cultured in 0.5% FBS medium in a three-dimensional collagen gel. TNF had a dose effect on EC tube formation similar to that observed in the EC migration assay (data not shown). TNF at 1 ng/ml stimulated tube formation as indicated by the EC network structure (Fig. 6e). The effects of Etk-SK and Etk-DK expression on TNF-induced tube formation were determined by measuring areas covered by the branched cell network (in square millimeters/field) on day 5 after addition of the second layer of collagen gel. Results showed that expression of Etk-DK reduced, whereas Etk-SK enhanced, TNF-induced tube formation (Fig. 6e and f). These data demonstrate that Etk is a critical mediator in TNF-induced EC tube formation.
DISCUSSION
In this study we show that TNF activates Etk specifically through TNFR2, as demonstrated by studies using a specific agonist to TNFR2, overexpression of TNFR2, and TNFR2-deficient cells. Etk binds to a non-TRAF2-binding motif in TNFR2 intracellular domains and is activated by TNF in a TRAF2-independent manner. Moreover, we show that Etk is a critical mediator in TNF-induced angiogenesis by in vitro models of EC migration and EC tube formation. Our findings demonstrate that Etk is the first kinase to associate specifically with TNFR2 (but not with TNFR1).
EC are among the restricted cell types that express TNFR2 (6). The role of TNFR2 in TNF signaling in EC is not clear. Here we show that TNF through TNFR2, but not TNFR1, activates an EC-expressed kinase, Etk. This is supported by several lines of evidence. First, an agonist to TNFR2, but not to TNFR1, activates Etk in EC. Second, overexpression of TNFR2, but not TNFR1, in EC activates Etk. Third, TNF fails to activate Etk in TNFR2-deficient cells, but Etk activation by TNF is still present in TNFR1-deficient cells.
It is generally accepted that TNF utilizes TNFR1 and TNFR2 to trigger distinct signaling and to exert diverse biological functions in a context-dependent manner. TNFR1 is believed to mediate cell death, whereas TNFR2 serves to promote cell activation, migration, growth, or proliferation (9, 36, 47, 52, 55, 56, 63). In T cells, TNF through TNFR2 promotes T-cell proliferation at an early stage of activation (36). It has been shown that TNFR2, but not TNFR1, is critical in TNF-induced proliferation of oligodendrocyte progenitors and remyelination (1). However, the mechanisms by which TNF through TNFR2 exerts these functions have not been defined. The reason, in part, is that specific proteins recruited by TNFR2 have not been identified. TRAF2 can be recruited to both TNFR1 and TNFR2 signaling complexes (46, 47) and is the only signaling molecule found to associate with TNFR2. Several factors, including cellular inhibitor of apoptosis and caveolin-1, are recruited to TNFR2 through TRAF2 (16, 46). Thus, TRAF2 and TRAF2-associated factors have been implicated in TNFR2-induced antiapoptotic or apoptotic responses.
In contrast to these findings, we show that TNF-induced activation of Etk through TNFR2 is independent of TRAF2. The interactions between Etk and TNFR2 are dependent on the N-terminal portion (the PH, TH, and SH3 domains) of Etk and the last 16 amino acids at the C-terminal intracellular domain of TNFR2. This Etk-binding sequence is distinct from the TRAF2-binding motif. Specifically, the TNFR2(−16) deletion mutant lacking the last 16 residues retains TRAF2 binding but fails to associate with and activate Etk. Also, WT-TRAF2 does not activate Etk, and DN-TRAF2 does not block TNF-induced Etk activation.
An interaction between the Btk family and the TNFR family has been demonstrated previously (59). Similar to Etk-TNFR2 interaction, Btk interacts with Fas, a member of the TNFR family involved in apoptosis in a ligand-independent manner (59). However, the association of Btk and Fas is dependent on the PH and kinase domains but not on the SH3 domain of Btk. This interaction disrupts the association of FADD with Fas, leading to inhibited Fas-induced apoptosis. Consistent with our finding, Btk does not interact with TNFR1 signaling complex molecules such as TRADD, FADD, and FLICE (59).
The mechanism by which Etk is activated is not clear. On the basis of data from Etk activation by focal adhesion kinase (FAK), it has been proposed that integrin-induced binding of the PH domain of Etk to the FERM domain of FAK leads to conformational change of the PH domain and phosphorylation of Y40 concomitant with the membrane translocation of Etk (10). This process resembles the effects of phospholipid binding to the PH domains of Btk family kinases in response to growth factors or cytokines. Membrane targeting of Etk will open up the closed conformation of inactive Etk and will allow the kinase to be phosphorylated by Src family kinases at the highly conserved tyrosine residue Y566 in the catalytic domain, which is originally masked by the PH domain. Tyrosine phosphorylation activates the Etk kinase, leading to autophosphorylation and activation. We show that, unlike what is shown by this two-step model, Etk forms a preexisting complex with TNFR2 located in the cytoplasm membrane. This association is independent of the phospholipid-binding PH domain of Etk, as the phospholipid-binding deficient mutant (Etk-DN) still binds to TNFR2. This membrane-bound Etk should still be in a closed inactive form. TNFR2 has no kinase activity, and it is not clear how Etk is activated in response to TNF. It has been shown that TNFR superfamily can form preassembled trimers, and ligand induces a conformation change (8). One possibility for Etk activation by TNFR2 is that TNF-induced TNFR2 conformational change triggers a change in Etk conformation to open up the closed conformation (Fig. 7). Alternatively, TNF-induced recruitment of additional factor(s) (e.g., a kinase) to a TNFR2/Etk complex, which phosphorylates Etk to open up the closed conformation, resembling phosphorylation of Y40 in the PH domain by FAK (10). The second step of Etk activation in response to TNF has not been determined. The tyrosine phosphorylation of the kinase-inactive Etk (Etk-DK) is undetectable in response to TNF/TNFR2, indicating that Etk activation induced by TNF/TNFR2 is largely due to autokinase activity. This is similar to Etk activation by interleukin-6 (40).
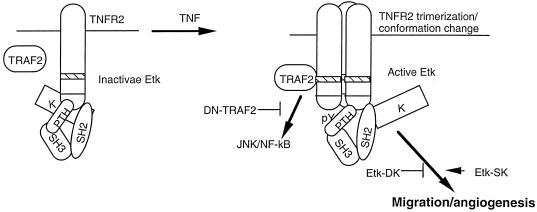
FIG. 7.
A model for Etk activation by TNF/TNFR2 in EC angiogenesis. Etk forms a preexisting complex with TNFR2, and Etk remains in a closed inactive form. TNF induces trimerization or conformational change of TNFR2, leading to an open conformation of Etk followed by Etk activation (tyrosine phosphorylation) and EC angiogenesis.
Our data demonstrate that Etk plays an important role in TNF-induced EC migration and tube formation by in vitro models, suggesting that Etk is a critical mediator in TNF-induced angiogenesis. The roles of Etk in cell activation, proliferation, and migration have been demonstrated in other cell types, including tumor cell lines and epithelial cells. Recently it has been shown that expression of Etk is much higher in metastatic carcinoma cells than in nonmetastatic carcinoma cells (10). Mechanistic studies suggest that interactions of Etk and FAK synergistically promote migratory potential of carcinoma cells. Etk is highly expressed in cultured EC (HUVEC and BAEC) (10 and our data) and in vivo endothelium (13). However, the role of Etk in angiogenesis has not been elucidated. Recent studies show that although Etk-deficient mice have no obvious defect in vasculogenesis, Etk is activated by EC growth factor receptor tyrosine kinases VEGFR1 and Tie-2. These data led the authors to postulate that Etk has a redundant function downstream of receptors for angiogenic factor (41). Angiogenesis under pathological conditions (e.g., inflammation) has not been examined with mice. Our data demonstrate that Etk mediates TNF-induced EC migration and tube formation in vitro, implying that Etk may play a critical role in inflammatory angiogenesis, such as occurs with ischemia, atherosclerosis, and rheumatoid arthritis. Our studies suggest that Etk might be a target for treatment of the inflammatory angiogenesis-dependent disease.
Acknowledgments
This work was supported by a grant from NIH 1R01HL65978-01 to WM.
We thank David Goeddel for TRAF2 constructs, Dianqing Wu for Etk/Bmx constructs, and Jordan S. Pober for TNFR1 and TNFR2 constructs. We thank Bradford C. Berk for discussions and critical reading of the manuscript.
REFERENCES
- 1.Arnett, H. A., J. Mason, M. Marino, K. Suzuki, G. K. Matsushima, and J. P. Ting. 2001. TNFα promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 4:1116-1122. [DOI] [PubMed] [Google Scholar]
- 2.Arras, M., W. D. Ito, D. Scholz, B. Winkler, J. Schaper, and W. Schaper. 1998. Monocyte activation in angiogenesis and collateral growth in the rabbit hindlimb. J. Clin. Investig. 101:40-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagheri-Yarmand, R., M. Mandal, A. H. Taludker, R. A. Wang, R. K. Vadlamudi, H. J. Kung, and R. Kumar. 2001. Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J. Biol. Chem. 276:29403-29409. [DOI] [PubMed] [Google Scholar]
- 4.Baud, V., and M. Karin. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11:372-377. [DOI] [PubMed] [Google Scholar]
- 5.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley, J. R., S. Thiru, and J. S. Pober. 1995. Disparate localization of 55-kd and 75-kd tumor necrosis factor receptors in human endothelial cells. Am. J. Pathol. 146:27-32. [PMC free article] [PubMed] [Google Scholar]
- 7.Carmeliet, P., and R. K. Jain. 2000. Angiogenesis in cancer and other diseases. Nature 407:249-257. [DOI] [PubMed] [Google Scholar]
- 8.Chan, F. K., H. J. Chun, L. Zheng, R. M. Siegel, K. L. Bui, and M. J. Lenardo. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351-2354. [DOI] [PubMed] [Google Scholar]
- 9.Chan, F. K., and M. J. Lenardo. 2000. A crucial role for p80 TNF-R2 in amplifying p60 TNF-R1 apoptosis signals in T lymphocytes. Eur J. Immunol. 30:652-660. [DOI] [PubMed] [Google Scholar]
- 10.Chen, R., O. Kim, M. Li, X. Xiong, J. L. Guan, H. J. Kung, H. Chen, Y. Shimizu, and Y. Qiu. 2001. Regulation of the PH-domain-containing tyrosine kinase Etk by focal adhesion kinase through the FERM domain. Nat. Cell Biol. 3:439-444. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan, A. M., K. O'Rourke, M. Tewari, and V. M. Dixit. 1995. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81:505-512. [DOI] [PubMed] [Google Scholar]
- 12.Dembic, Z., H. Loetscher, U. Gubler, Y. C. Pan, H. W. Lahm, R. Gentz, M. Brockhaus, and W. Lesslauer. 1990. Two human TNF receptors have similar extracellular, but distinct intracellular, domain sequences. Cytokine 2:231-237. [DOI] [PubMed] [Google Scholar]
- 13.Ekman, N., A. Lymboussaki, I. Vastrik, K. Sarvas, A. Kaipainen, and K. Alitalo. 1997. Bmx tyrosine kinase is specifically expressed in the endocardium and the endothelium of large arteries. Circulation 96:1729-1732. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, S. L., F. J. de Sauvage, K. Kikly, K. Carver-Moore, S. Pitts-Meek, N. Gillett, K. C. Sheehan, R. D. Schreiber, D. V. Goeddel, and M. W. Moore. 1994. Decreased sensitivity to tumour-necrosis factor but normal T-cell development in TNF receptor-2-deficient mice. Nature 372:560-563. [DOI] [PubMed] [Google Scholar]
- 15.Espevik, T., M. Brockhaus, H. Loetscher, U. Nonstad, and R. Shalaby. 1990. Characterization of binding and biological effects of monoclonal antibodies against a human tumor necrosis factor receptor. J. Exp. Med. 171:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, X., M. L. Gaeta, L. A. Madge, J. H. Yang, J. R. Bradley, and J. S. Pober. 2001. Caveolin-1 associates with TRAF2 to form a complex that is recruited to tumor necrosis factor receptors. J. Biol. Chem. 276:8341-8349. [DOI] [PubMed] [Google Scholar]
- 17.Fiers, W., R. Beyaert, E. Boone, S. Cornelis, W. Declercq, E. Decoster, G. Denecker, B. Depuydt, D. De Valck, G. De Wilde, V. Goossens, J. Grooten, G. Haegeman, K. Heyninck, L. Penning, S. Plaisance, K. Vancompernolle, W. Van Criekinge, P. Vandenabeele, W. Vanden Berghe, M. Van de Craen, V. Vandevoorde, and D. Vercammen. 1995. TNF-induced intracellular signaling leading to gene induction or to cytotoxicity by necrosis or by apoptosis. J. Inflamm. 47:67-75. [PubMed] [Google Scholar]
- 18.Frater-Schroder, M., W. Risau, R. Hallmann, P. Gautschi, and P. Bohlen. 1987. Tumor necrosis factor type alpha, a potent inhibitor of endothelial cell growth in vitro, is angiogenic in vivo. Proc. Natl. Acad. Sci. USA 84:5277-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehr, G., R. Gentz, M. Brockhaus, H. Loetscher, and W. Lesslauer. 1992. Both tumor necrosis factor receptor types mediate proliferative signals in human mononuclear cell activation. J. Immunol. 149:911-917. [PubMed] [Google Scholar]
- 20.Goodwin, R. G., D. Anderson, R. Jerzy, T. Davis, C. I. Brannan, N. G. Copeland, N. A. Jenkins, and C. A. Smith. 1991. Molecular cloning and expression of the type 1 and type 2 murine receptors for tumor necrosis factor. Mol. Cell. Biol. 11:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grell, M., E. Douni, H. Wajant, M. Lohden, M. Clauss, B. Maxeiner, S. Georgopoulos, W. Lesslauer, G. Kollias, K. Pfizenmaier, et al. 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83:793-802. [DOI] [PubMed] [Google Scholar]
- 22.Grell, M., H. Wajant, G. Zimmermann, and P. Scheurich. 1998. The type 1 receptor (CD120a) is the high-affinity receptor for soluble tumor necrosis factor. Proc. Natl. Acad. Sci. USA 95:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heba, G., T. Krzeminski, M. Porc, J. Grzyb, A. Ratajska, and A. Dembinska-Kiec. 2001. The time course of tumor necrosis factor-alpha, inducible nitric oxide synthase and vascular endothelial growth factor expression in an experimental model of chronic myocardial infarction in rats. J. Vasc. Res. 38:288-300. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, H., J. Xiong, and D. V. Goeddel. 1995. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 81:495-504. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y., J. D. Woronicz, W. Liu, and D. V. Goeddel. 1999. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 283:543-546. (Erratum, 283:1852.) [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, G. C., and D. B. Polk. 1997. Tumor necrosis factor alpha regulates proliferation in a mouse intestinal cell line. Gastroenterology 112:1231-1240. [DOI] [PubMed] [Google Scholar]
- 27.Koch, A. E. 1998. Review: angiogenesis: implications for rheumatoid arthritis. Arthritis Rheum. 41:951-962. [DOI] [PubMed] [Google Scholar]
- 28.Leibovich, S. J., P. J. Polverini, H. M. Shepard, D. M. Wiseman, V. Shively, and N. Nuseir. 1987. Macrophage-induced angiogenesis is mediated by tumour necrosis factor-alpha. Nature 329:630-632. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, M., L. A. Tartaglia, A. Lee, G. L. Bennett, G. C. Rice, G. H. W. Wong, E. Y. Chen, and D. V. Goeddel. 1991. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc. Natl. Acad. Sci. USA 88:2830-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Y., and W. Min. 2002. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 90:1259-1266. [DOI] [PubMed] [Google Scholar]
- 31.Loetscher, H., Y. E. Pan, H. W. Lahm, R. Gentz, M. Brockhaus, H. Tabuchi, and W. Lesslauer. 1990. Molecular cloning and expression of the human 55kd tumor necrosis factor receptor. Cell 61:351-359. [DOI] [PubMed] [Google Scholar]
- 32.Madge, L. A., and J. S. Pober. 2001. Tnf signaling in vascular endothelial cells. Exp. Mol. Pathol. 70:317-325. [DOI] [PubMed] [Google Scholar]
- 33.Mao, J., W. Xie, H. Yuan, M. I. Simon, H. Mano, and D. Wu. 1998. Tec/Bmx non-receptor tyrosine kinases are involved in regulation of Rho and serum response factor by Galpha12/13. EMBO J. 17:5638-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min, W., and J. S. Pober. 1997. TNF initiates E-selectin transcription in human endothelial cells through parallel TRAF-NF-kappa B and TRAF-RAC/CDC42-JNK-c-Jun/ATF2 pathways. J. Immunol 159:3508-3518. [PubMed] [Google Scholar]
- 35.Pfeffer, K., T. Matsuyama, T. M. Kundig, A. Wakeham, K. Kishihara, A. Shahinian, K. Wiegmann, P. S. Ohashi, M. Kronke, and T. W. Mak. 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73:457-467. [DOI] [PubMed] [Google Scholar]
- 36.Pimentel-Muinos, F. X., and B. Seed. 1999. Regulated commitment of TNF receptor signaling: a molecular switch for death or activation. Immunity 11:783-793. [DOI] [PubMed] [Google Scholar]
- 37.Pober, J. S., and R. S. Cotran. 1990. Cytokines and endothelial cell biology. Physiol. Rev. 70:427-451. [DOI] [PubMed] [Google Scholar]
- 38.Pursglove, S. E., T. D. Mulhern, J. P. Mackay, M. G. Hinds, and G. W. Booker. 2001. The solution structure and intramolecular associations of the Tec kinase SH3 domain. J. Biol. Chem. 29:29. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, Y., and H. J. Kung. 2000. Signaling network of the Btk family kinases. Oncogene 19:5651-5661. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, Y., D. Robinson, T. G. Pretlow, and H. J. Kung. 1998. Etk/Bmx, a tyrosine kinase with a pleckstrin-homology domain, is an effector of phosphatidylinositol 3′-kinase and is involved in interleukin 6-induced neuroendocrine differentiation of prostate cancer cells. Proc. Natl. Acad. Sci. USA 95:3644-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajantie, I., N. Ekman, K. Iljin, E. Arighi, Y. Gunji, J. Kaukonen, A. Palotie, M. Dewerchin, P. Carmeliet, and K. Alitalo. 2001. Bmx tyrosine kinase has a redundant function downstream of angiopoietin and vascular endothelial growth factor receptors in arterial endothelium. Mol. Cell. Biol. 21:4647-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rawlings, D. J., and O. N. Witte. 1995. The Btk subfamily of cytoplasmic tyrosine kinases: structure, regulation and function. Semin. Immunol. 7:237-246. [DOI] [PubMed] [Google Scholar]
- 43.Redmond, E. M., J. P. Cullen, P. A. Cahill, J. V. Sitzmann, S. Stefansson, D. A. Lawrence, and S. S. Okada. 2001. Endothelial cells inhibit flow-induced smooth muscle cell migration: role of plasminogen activator inhibitor-1. Circulation 103:597-603. [DOI] [PubMed] [Google Scholar]
- 44.Ristimaki, A., K. Narko, B. Enholm, V. Joukov, and K. Alitalo. 1998. Proinflammatory cytokines regulate expression of the lymphatic endothelial mitogen vascular endothelial growth factor-C. J. Biol. Chem. 273:8413-8418. [DOI] [PubMed] [Google Scholar]
- 45.Rothe, J., W. Lesslauer, H. Lotscher, Y. Lang, P. Koebel, F. Kontgen, A. Althage, R. Zinkernagel, M. Steinmetz, and H. Bluethmann. 1993. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature 364:798-802. [DOI] [PubMed] [Google Scholar]
- 46.Rothe, M., M. G. Pan, W. J. Henzel, T. M. Ayres, and D. V. Goeddel. 1995. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 83:1243-1252. [DOI] [PubMed] [Google Scholar]
- 47.Rothe, M., S. C. Wong, W. J. Henzel, and D. V. Goeddel. 1994. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell 78:681-692. [DOI] [PubMed] [Google Scholar]
- 48.Sarma, V., F. W. Wolf, R. M. Marks, T. B. Shows, and V. M. Dixit. 1992. Cloning of a novel tumor necrosis factor-alpha-inducible primary response gene that is differentially expressed in development and capillary tube-like formation in vitro. J. Immunol. 148:3302-3312. [PubMed] [Google Scholar]
- 49.Schall, T. J., M. Lewis, K. J. Koller, A. Lee, G. C. Rice, G. H. W. Wong, T. Gatanaga, G. A. Granger, R. Lentz, H. R. J. Kohr, and D. V. Goeddel. 1990. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 61:361-370. [DOI] [PubMed] [Google Scholar]
- 50.Slowik, M. R., L. G. DeLuca, W. Fiers, and J. S. Pober. 1993. Tumor necrosis factor activates human endothelial cells through the p55 tumor necrosis factor receptor but the p75 receptor contributes to activation at low tumor necrosis factor concentration. Am. J. Pathol. 143:1724-1730. [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, C. A., T. Davis, D. Anderson, L. Solam, M. P. Beckmann, R. Jerzy, S. K. Dower, D. Cosman, and R. G. Goodwin. 1990. A receptor for tumor necrosis factor defines an unusual family of cellular and viral proteins. Science 248:1019-1022. [DOI] [PubMed] [Google Scholar]
- 52.Smith, C. A., T. Farrah, and R. G. Goodwin. 1994. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell 76:959-962. [DOI] [PubMed] [Google Scholar]
- 53.Stanger, B. Z., P. Leder, T. H. Lee, E. Kim, and B. Seed. 1995. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 81:513-523. [DOI] [PubMed] [Google Scholar]
- 54.Tamagnone, L., I. Lahtinen, T. Mustonen, K. Virtaneva, F. Francis, F. Muscatelli, R. Alitalo, C. I. Smith, C. Larsson, and K. Alitalo. 1994. BMX, a novel nonreceptor tyrosine kinase gene of the BTK/ITK/TEC/TXK family located in chromosome Xp22.2. Oncogene 9:3683-3688. [PubMed] [Google Scholar]
- 55.Tartaglia, L. A., D. Pennica, and D. V. Goeddel. 1993. Ligand passing: the 75-kDa tumor necrosis factor (TNF) receptor recruits TNF for signaling by the 55-kDa TNF receptor. J. Biol. Chem. 268:18542-18548. [PubMed] [Google Scholar]
- 56.Tartaglia, L. A., R. F. Weber, I. S. Figari, C. Reynolds, J. M. A. Palladino, and D. V. Goeddel. 1991. The two different receptors for tumor necrosis factor mediate distinct cellular responses. Proc. Natl. Acad. Sci. USA 88:9292-9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai, Y. T., Y. H. Su, S. S. Fang, T. N. Huang, Y. Qiu, Y. S. Jou, H. M. Shih, H. J. Kung, and R. H. Chen. 2000. Etk, a Btk family tyrosine kinase, mediates cellular transformation by linking Src to STAT3 activation. Mol. Cell. Biol. 20:2043-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandenabeele, P., W. Declercq, R. Beyaert, and W. Fiers. 1995. Tumor necrosis factor receptors - structure and function. Trends Cell Biol. 5:392-399. [DOI] [PubMed] [Google Scholar]
- 59.Vassilev, A., Z. Ozer, C. Navara, S. Mahajan, and F. M. Uckun. 1999. Bruton's tyrosine kinase as an inhibitor of the Fas/CD95 death-inducing signaling complex. J. Biol. Chem. 274:1646-1656. [DOI] [PubMed] [Google Scholar]
- 60.Wajant, H., M. Grell, and P. Scheurich. 1999. TNF receptor associated factors in cytokine signaling. Cytokine Growth Factor Rev. 10:15-26. [DOI] [PubMed] [Google Scholar]
- 61.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 62.Walsh, D. A. 1999. Angiogenesis and arthritis. Rheumatology (Oxford) 38:103-112. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, T., M. Grell, B. Hessabi, S. Bourteele, G. Muller, P. Scheurich, and H. Wajant. 1997. Enhancement of TNF receptor p60-mediated cytotoxicity by TNF receptor p80: requirement of the TNF receptor-associated factor-2 binding site. J. Immunol. 158:2398-2404. [PubMed] [Google Scholar]
- 64.Yin, G., W. Liu, P. An, P. Li, I. Ding, V. Planelles, E. M. Schwarz, and W. Min. 2002. Endostatin gene transfer inhibits joint angiogenesis and pannus formation in inflammatory arthritis. Mol. Ther. 5:547-554. [DOI] [PubMed] [Google Scholar]