Abstract
Histone deacetylases mediate critical cellular functions but relatively little is known about mechanisms controlling their expression, including expression of HDAC4, a class II HDAC implicated in the modulation of cellular differentiation and viability. Endogenous HDAC4 mRNA, protein levels and promoter activity were all readily repressed by mithramycin, suggesting regulation by GC-rich DNA sequences. We validated consensus binding sites for Sp1/Sp3 transcription factors in the HDAC4 promoter through truncation studies and targeted mutagenesis. Specific and functional binding by Sp1/Sp3 at these sites was confirmed with chromatin immunoprecipitation (ChIP) and electromobility shift assays (EMSA). Cotransfection of either Sp1 or Sp3 with a reporter driven by the HDAC4 promoter led to high activities in SL2 insect cells (which lack endogenous Sp1/Sp3). In human cells, restored expression of Sp1 and Sp3 up-regulated HDAC4 protein levels, whereas levels were decreased by RNA-interference-mediated knockdown of either protein. Finally, variable levels of Sp1 were in concordance with that of HDAC4 in a number of human tissues and cancer cell lines. These studies together characterize for the first time the activity of the HDAC4 promoter, through which Sp1 and Sp3 modulates expression of HDAC4 and which may contribute to tissue or cell-line-specific expression of HDAC4.
INTRODUCTION
Histone deacetylases (HDACs) all share the ability to deacetylate specific lysines in the tail residues of the core histones, generally resulting in the compaction of chromatin, transcriptional repression, and silencing. There has been an explosion of interest in recent years in the HDACs, because of the panoply of critical cellular functions that have now been linked to HDACs (Grozinger and Schreiber, 2002; Verdin et al., 2003; Sengupta and Seto, 2004; Yang and Gregoire, 2005).
HDACs in general lack DNA binding activity, and appear to mediate their activities as part of large multiprotein complexes (such as the NuRD, Sin3, and CoREST complexes; Hassig et al., 1997; Laherty et al., 1997; Zhang et al., 1997, 1998), which include non-HDAC proteins and other unique polypeptides that together modulate histone deacetylase activity. An example of the regulation of HDAC activity mediated by protein-protein interactions relates to the SMRT/N-CoR complex. The silencing mediator of retinoid and thyroid receptor (SMRT) and nuclear receptor corepressor (N-CoR) are nuclear receptor corepressors that bind and enhance the HDAC activity of HDAC3 (Alland et al., 1997; Heinzel et al., 1997; Wen et al., 2000; Guenther et al., 2001; Zhang et al., 2002). In contrast to the effects on HDAC3, the deacetylase activity of the class II HDACs (HDAC4, 5, 7, and 9) are not enhanced by binding to the SMRT/N-CoR complex, suggesting that the class II HDACs may recruit enzymatically active HDAC3-SMRT/N-coR complexes for their functional effects (Fischle et al., 2002). Regulation of the expression of class II HDACs may therefore influence the overall function of such multiprotein complexes in regulating gene expression.
The role of class II HDACs such as HDAC4 in specific cellular and tissue functions is becoming clarified. HDAC4 has been found to regulate chondrocyte growth and differentiation during skeletogenesis through interactions with the runt-related transcription factor-2 (Runx2) (Vega et al., 2004a, 2004b). HDAC4 has also been found to mediate transcriptional repression through interactions with other DNA-binding factors, such as PLZF (promyelocytic leukemia zinc-finger) and BCL6, factors linked to hematopoietic cell differentiation, leukemogenesis, and inflammation (Lemercier et al., 2002; Chauchereau et al., 2004). HDAC4 regulates the activity of the myocyte enhancer factor-2 (MEF2) family of transcription factors, implicated in the cellular differentiation and proliferation of hematopoietic, nervous, and musculoskeletal systems (Miska et al., 1999; Wang et al., 1999; McKinsey et al., 2000a, 2000b, 2002; Chauchereau et al., 2004; Vega et al., 2004a, 2004b).
Perhaps reflecting the versatility of HDAC4 in regulating gene expression in diverse systems and tissues, the diversity of mechanisms regulating the activity of HDAC4 has been particularly striking. HDAC4 mRNA and protein are both highly unstable, with intracellular half-lives less than 8 h in HeLa cells (Liu et al., 2004). The interaction between HDAC4 and MEF2 is up-regulated by sumoylation of specific residues in HDAC4, which in turn leads to the sumoylation and repression of MEF2 (Wang et al., 1999; Gregoire and Yang, 2005). HDAC4 is phosphorylated at amino-terminal serine residues by the Ca2+/calmodulin-dependent kinase (CaMK) and protein kinase D, which appears to create docking sites for the 14-3-3 family of proteins, which results in its CRM1-dependent nuclear export (McKinsey et al., 2000a, 2000b, 2001; Wang et al., 2000; Zhou et al., 2000; Wang and Yang, 2001; Vega et al., 2004a, 2004b). HDAC4 is also regulated through caspase-mediated cleavage, which results in an nuclear localization signal (NLS)-containing amino-terminal fragment that translocates into the nucleus to effect transcriptional repression and decreases cell viability (Liu et al., 2004; Paroni et al., 2004).
Despite the diversity of pathways modulated by HDAC4 and mechanisms regulating the activity of HDAC4, surprisingly little is known about the mechanisms regulating its expression. We report here the identification of consensus binding sequences in the promoter of human HDAC4 for the Specificity Protein (SP) family of transcription factors. We find that both Sp1 and Sp3 bind to these sequences, and drive strong expression of a HDAC4 promoter-driven luciferase reporter. Expression of Sp1 and Sp3 in cells deficient for either transcription factor led to increased expression of HDAC4 protein. Consistent with direct roles in driving expression of HDAC4, knockdown of Sp1 and Sp3 led to reduced HDAC4 protein levels. Finally, a general concordance was found between Sp1 and HDAC4 protein expression in human cancer cell lines and normal human tissues. These results together strongly identify a role for the SP family of transcription factors in driving the expression of human HDAC4, an class II HDAC that may potentially modulate the activity of other transcription factors.
MATERIALS AND METHODS
Cell Culture, Reagents, and Treatments
All human cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA), and grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) or McCoy's media supplemented with 10% fetal calf serum (FCS). Drosophila melanogaster SL-2 cells were maintained in Schneider's insect medium supplemented with 10% FCS at 28°C. Mithramycin and trichostatin A (TSA) were from Sigma (St. Louis, MO). Mock-treated control cells were handled identically to drug-treated cells with the exception that medium alone was added.
Immunoblotting
Cell lysates were prepared via scraping on ice and pelleting at 4°C, followed by resuspension in Laemmli buffer and sonication. For immunoblotting, samples (50 μg/lane) were boiled for 5 min and separated via SDS-PAGE, and transferred to PVDF membranes. After transfer, the membranes were blocked with 5% nonfat milk in phosphate-buffered saline (PBS) and then probed with the indicated primary antibodies, followed by the appropriate secondary antibodies conjugated with horseradish peroxidase. Anti-HDAC2 antibodies were from Biomol (Plymouth Meeting, PA), and anti-HDAC6 were from Cell Signaling (Beverly, MA). Washes were performed with PBS with 0.1% Tween. Finally, after probing with primary and secondary antibodies, the membranes were exposed to film after enhanced chemiluminescence (ECL) (Amersham Biosciences, Piscataway, NJ). Densitometry of immunoblots was performed on images obtained under nonsaturated conditions and quantitated with NIH Image 1.54 software.
Reverse Transcriptase-PCR
Endogenous RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and assessed via RT-PCR. The Titan One Tube RT-PCR System (Roche, Indianapolis, IN) was used with the following primers: histone deacetylase 4 (HDAC4): 5′ CAA GAA CAA GGA GAA GGG CAA AG 3′ and 5′ GGA CTC TGG TCA AGG GAA CTG 3′; 53BP1: 5′ AGG TGG GTG TTC TTT GGC TTC C 3′ and 5′TTG GTG TTG AGG CTT GTG GTG ATA C 3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′ CAA CTT TGG TAT CGT GGA AGG ACT C 3′ and 5′ AGG GAT GAT GTT CTG GAG AGC C 3′. Reactions for all targeted mRNA were performed under similar conditions, with comparatively identical results.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay (ChIP) assays were performed via a commercially purchased chromatin immunoprecipitation kit (Upstate Biotechnology, Lake Placid, NY), using either anti-Sp1 (Upstate; 07-124), anti-Sp3 (Santa Cruz Biotechnology, Santa Cruz, CA; SC-644), or anti-NF-Y/CBF-B (Santa Cruz; SC-10779) antibodies. HeLa cells were first cross-linked for 10 min by adding formaldehyde directly to tissue culture medium to a final concentration of 1%. Cross-linked cells were then washed twice with cold PBS (with protease inhibitors), scraped, pelleted, resuspended in 200 μl SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0), and incubated for 10 min on ice. The lysates were then sonicated for five cycles of 30 s each, resting on ice for 1 min between cycles, on an Ultrasonic Processor W-385 sonicator (Plainview, NY) with settings of cycle time: 1 s, duty cycle: 20%, output power: 10%. After sonication, the samples were centrifuged and the supernatants diluted 10-fold in ChIP dilution buffer with protease inhibitors and precleared with 80 μl salmon sperm DNA/protein A Agarose-50% slurry for 30 min at 4°C. Cross-linked chromatin was incubated overnight with 3 μg Sp1, 3 μg Sp3, 3 μg NF-Y, or control IgG in a total volume of 1 ml at 4°C. Antibody-protein-DNA complexes were isolated by immunoprecipitation with 60 μl salmon sperm DNA/protein A. After extensive washing, pellets were eluted by freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3). Formaldehyde cross-linking was reversed by 5-12-h incubation at 65°C after adding 20 μl 5 M NaCl. Samples were purified through PCR purification kit columns (Qiagen, Chatsworth, CA) and used as a template in PCR. ChIP primers: 5′ TCC AGC AGC CAA TGA GGT CC 3′ and 5′ TTC TCC CCA CTC CAG CGT CG 3′ were used to amplify a 382-base pair fragment corresponding to the core HDAC4 promoter. Samples from at least three independent immunoprecipitations were analyzed.
HDAC4 Luciferase-Reporter Constructs
The HDAC4 promoter DNA containing a variety of Sp1/Sp3 binding sites was amplified from HeLa cell genomic DNA by Roche GC-rich PCR kit and primers A+B (composition of primers are listed in the next section). The amplified HDAC4 promoter segment was then digested with BglII and Hin-dIII and cloned into pGL3-basic luciferase reporter (Promega, Madison, WI) to generate (-317-+117) HDAC4-pGL3. Using (-317-+117) HDAC4-pGL3 construct as a template, a series of truncated HDAC4 promoter segments were amplified, digested, and cloned into pGL3-basic by using appropriate primers. The primers used for (-317-+7) HDAC4-pGL3 were A+C, for (-223-+7) HDAC4-pGL3 were D+C, for (-117-+7) HDAC4-pGL3 were E+C, and for (-103-+7) HDAC4-pGL3 were F+C. The construct “(-317-+7) HDAC4 mut Sp1/3,” in which the three contiguous and one proximal Sp1/Sp3 sites were mutated, was generated from (-317-+7) HDAC4-pGL3 via the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) by using primers G+H. All constructs were confirmed by direct DNA sequencing. For clarity, these constructs are listed in Figure 4 and discussed in the Results section by only the portion upstream of the transcription start site (because all constructs have in common the first seven nucleotides after the HDAC4 transcription start site as well as pGL3).
Figure 4.
The HDAC4 promoter activity depends on Sp1/Sp3 binding sequences. (A) Luciferase reporters designed to test the activity of different lengths of the HDAC4 promoter, including the effect of targeted mutations. The schematic shows the design of the pGL3 reporters to test the relative contribution of different portions of the HDAC4 promoter. The 317-base pair nucleotides upstream of the transcriptional initiation site of HDAC4, containing the seven potential Sp1/Sp3 binding sites indicated in Figure 4, were fused to the promoterless pGL3-Basic luciferase reporter to generate (-317) HDAC4. The four Sp1/Sp3 binding sites at the most 5′ end of the promoter region was mutated, resulting in (-317) mut Sp1/3 HDAC4, or removed, resulting in (-223) HDAC4. Further portions of the HDAC4 were removed, resulting in either (-177) HDAC4 or (-103) HDAC4, the latter lacking any of the potential Sp1/Sp3 binding sites indicated by the boxes in Figure 4. (B) The activity of the HDAC4 promoter is proportional to the number of intact Sp1/Sp3 binding sites. Each of the reporter constructs shown in A were transfected into individual plates of HeLa cells, and all plates were assayed for luciferase activity 48 h later after transfection. A set of plates was transfected with pGL3 alone as an additional control. In these experiments, β-galactosidase activity resulting from cotransfected CMV4-β-gal plasmid enabled normalization for transfection efficiency. All samples were performed in triplicate and the results were averaged and plotted as shown, with all results expressed as a percentage of the activity resulting from (-317) HDAC4 (which was set at 100%). Error bars, SE. The experiment was repeated three times with similar results. (C) The reporters described in A was transfected into PC3 cells, which were subsequently assayed in a manner that was otherwise identical to that described for B and with results calculated and plotted in a similar manner. This experiment was also repeated three times with similar results. (D) Either Sp1 or Sp3 can drive HDAC4 promoter activity in SL2 cells. SL2 insect cells were tested for reporter activity under different conditions. Individual plates were transfected with the promoterless pGL3 luciferase reporter alone or in combination with PacSp1 (which drives expression of Sp1 via an insect-specific promoter), in either case resulting in little activity (first and second bars). In contrast, transfection of (-317) HDAC4-pGL3 resulted in high levels of activity when Sp1 or Sp3 was coexpressed (respectively via PacSp1 or PacSp3, fourth and fifth bars). Cotransfection of (-317) HDAC4-PGL3 with the parental Pac vector resulted in minor activity only. In all plates, β-galactosidase activity resulting from cotransfected CMV4-β-gal plasmid enabled normalization for transfection efficiency. All samples were performed in triplicate and the results averaged and plotted as shown, with all results expressed as a percentage of the activity resulting from the HDAC4 promoter and PacSp3 ((-317) HDAC4 pGL3 + PacSp3, which was set at 100%). Error bars, SE. The experiment was repeated three times with similar results.
Primers used for construction of HDAC4-PGL3 Luciferase reporters were as follows: A) 5′ CAT AGA TCT GTG GGA GCA GAC GGG CTG TG 3′; B) 5′ CAT AAG CTT CAG GCT GGG AGG CTG TTC GG 3′; C) 5′ CAT AAG CTT CCA CAA CCT CCC CTC CTC ATT C 3′; D) 5′ CAT AGA TCT AAT TGA CGA GCT CTT CAT TAG 3′; E) 5′ CAT AGA TCT CAA GGG GAG GTG ACG CAA G 3′; F) 5′ CAT AGA TCT TGA CGG GCG TGC GGG GTG GCC TA 3′; G) 5′ CG CCC GGG TTG GTT GGT TGG GAG GAA GGG CCG AGC CGT 3′; and H) 5′ ACG GCT CGG CCC TTC CTC CCA ACC AAC CAA CCC GGG CG 3′.
Luciferase Assays in Mammalian Cells
For luciferase assays, 2.5 × 105 HeLa or 5 × 105 PC3 cells were plated in 60-mm plates. Luciferase reporter plasmids (2 μg for each) were transfected into HeLa or PC3 cells via 5 μl of Lipofectamine 2000 (Invitrogen). For all luciferase reporter assays, pRL-TK (Renilla Luciferase) was also cotransfected and measured to normalize transfection efficiency. After 48 h of transfection, cells were washed twice with PBS and lysed in situ with 300 μl passive lysis buffer with three freeze-thaw cycles. Luciferase activity of 20 μl of cell lysate was measured via the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Assays for all samples were performed in triplicate, and the results were averaged.
Luciferase Assay in Drosophila melanogaster SL2 Insect Cells
The day before transfection, 1 × 106 SL2 cells were plated in 60-mm plate and maintained at 28°C in Schneider's insect medium supplemented with 10% FCS. Drosophila-specific expression vector: Pac, PacSp1, or PacSp3 (each 0.8 μg) were cotransfected with 0.8 μg (-317-+7) HDAC4-pGL3 and 0.4 μg CMV4-β-gal into SL2 cells by 5 μl of Lipofectamine 2000 (Invitrogen). After 48 h of transfection, cells were washed twice with PBS and lysed by passive lysis buffer with three freeze-thaw cycles to accomplish complete lysis of cells. Luciferase activity of 20 μl of cell lysate was assessed via the Single-Luciferase reporter assay system (Promega). β-galactosidase activity of 20 μl of cell lysate was measured via spectrophotometry at 420 nm after incubating with 200 μl 5 mg/ml ONPG (Sigma; N-1127) in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, PH 7.2). Relative Luciferase activity was then normalized by β-gal activity. Assays for all samples were performed in triplicate, and the results were averaged.
Electrophoretic Mobility Shift Assay
These electrophoretic mobility shift assays (EMSAs) were performed with 1 × 106 HeLa cells grown in 10-cm cell culture plates. Nuclear proteins were extracted as described previously (Pore et al., 2004). The following oligonucleotides were synthesized and labeled with [γ-32P]ATP by T4 polynucleotide kinase: the sequences corresponding to -261 base pairs to -226 base pairs of human HDAC4 were 5′ GCG CCC GGG GCG GGC GGG CGG GAG GCG GGG CCG AG 3′ and 5′ CTC GGC CCC GCC TCC CGC CCG CCC GCC CCG GGC GC 3′ (corresponding to a 35-base pair section of the HDAC4 promoter) and 5′ GAT CGA TCG GGG CGG GGC GAT C 3′ and 5′ GAT CGC CCC GCC CCG ATC GAT C 3′ (the 22-base pair probe representing the canonical Sp1/Sp3 sequence). Unincorporated [γ-32P]ATP was removed by centrifugation through G-25 Sephadex column (Boehringer Mannheim, Indianapolis, IN) according to manufacturer's recommendations. The DNA-binding reaction was performed for 30 min at room temperature in a volume of 20 μl, containing 5 μg of nuclear protein extract, 2.5 mg/ml bovine serum albumin, 105 cpm, 0.1 mg/ml poly[dI:dC] (Sigma), 5 μl of 4× binding buffer (1× buffer: 10 mM Tris-Cl, pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, 10% [vol/vol] glycerol, 1 mM DTT) with or without 100-fold excess of unlabeled oligonucleotide competitor. Samples were subjected to electrophoresis on a native 5% polyacrylamide gel run in either 0.5× TBE (Tris-boric acid-EDTA) or 0.5× TGE (Tris-glycine-EDTA) for 3.5 h at 200 V.
Knockdown of Sp1 or Sp3 via RNA Interference
Short interfering RNAs (siRNAs), designed to target human SP1 and SP3 mRNAs, were synthesized (Dharmacon, Lafayette, CO). The criteria for designing and methodology to introduce the siRNAs into cells were executed according to the company protocols. Briefly, 5 × 106 HeLa cells were plated the day before transfection in 10-cm plates. Oligofectamine (30 μl) and 60 μl of a 25 μM stock solution of siRNA were preincubated with 90 μl and 1050 μl of Opti-medium separately for 5 min and then mixed and vortexed gently and incubated at RT for an additional 20 min. The mixture was then evenly added to the plates. Forty-eight hours later, the cells were harvest by RIPA buffer. The forward strands of each duplex of siRNAs were as follows: Sp1: 5′NNA GCG CUU CA U GAG GAG UGA 3′; Sp3: 5′NNG CGG CAG GUG GAG CCU UCA CU 3′; and GFP: 5′GCA AGC TGA CCC TGA AGC TC 3′.
Immunohistochemistry
A microarray of human tissues taken from different organs (Zymed Max-Array, South San Francisco, CA) was assessed for expression of Sp1, HDAC4, and HDAC2 protein. After deparaffinization slides were immersed in 95°C 10 mM citric acid buffer (pH 6.0) for 15 min in a steamer, treated with hydrogen peroxide to block endogenous peroxides, and then blocked in 10% horse serum. The slides were then probed overnight with anti-Sp1 (Upstate Biotechnology), anti-HDAC4, or anti-HDAC2 (Biomol) antibodies in 4°C. A peroxidase labeled polymer (DakoCytomation Envision Plus Dual Link System Peroxidase, Carpinteria, CA) was applied at room temperature for 30 min. The slide was developed with Vector DAB Peroxidase Substrate Kit (SK-4100; Vector Laboratories, Burlingame, CA) and counterstained with hematoxylin.
RESULTS
Mithramycin Rapidly Leads to Decreased HDAC4 mRNA and Protein Levels and Decreased HDAC4 Promoter Activity
To begin to investigate mechanisms that might control expression of HDAC4 mRNA, we treated cells with mithramycin, a cell-permeable agent that binds to GC-rich DNA sequences and is frequently used to explore the sequence dependency of DNA-binding factors. The binding of mithramycin is thought to impede binding by GC-specific transcription factors by steric hindrance (Blume et al., 1991; Nehls et al., 1993). In an initial experiment, we found that treatment of HeLa cells with mithramycin rapidly led to decreased levels of HDAC4 mRNA and was almost undetectable by 16 h (Figure 1A). We therefore tested the effect of mithramycin on the cells by harvesting at shorter time points. This showed that the effect of mithramycin on HDAC4 mRNA was evident by 4 h and that HDAC4 mRNA expression was almost abolished by 8 h (Figure 1B).
Figure 1.

Mithramycin rapidly leads to decreased HDAC4 mRNA levels. (A) RT-PCR was performed on total mRNA isolated from HeLa cells harvested at the indicated times after exposure to 125 nM mithramycin. Reactions were performed in parallel under identical conditions using primer pairs targeting HDAC4, or as controls, 53BP1 and GADPH. The final products were separated via electrophoresis in ethidium bromide-labeled agarose and photographed under UV illumination. Each RT-PCR reaction yielded a single band as shown. (B) RT-PCR was performed as described for A, with the exception that the reactions were performed on total mRNA isolated from HeLa cells harvested at time points more immediately after mithramycin treatment, at the indicated times. (C) HeLa cells were mock-treated (“Control”) or treated with mithramycin at 125 nM for 8 or 16 h, or 250 nM for 8 h, and harvested and cell lysates were separated via SDS-PAGE, transferred to PVDF membranes, and immunoblotted for HDAC4 or alpha-tubulin (as a loading control). Both doses led to decreased HDAC4 protein. (D) Time course of mithramycin-mediated repression of HDAC4 protein and lack of effect on HDAC2 and HDAC6. Parallel plates of cells were mock-treated or treated with 125 nM mithramycin and harvested 4, 12, or 20 h later. The resultant cell lysates were immunoblotted for HDAC4, HDAC2, or HDAC6.
We then assessed the effect of mithramycin on levels of HDAC4 protein. Consistent with the decrease in levels of HDAC4 mRNA noted in Figure 1 and the previously reported instability of HDAC4 protein, which allows for changes of the levels of protein to be quickly evident (Liu et al., 2004), mithramycin treatment led to decreased HDAC4 protein levels (Figure 1C). A dose and time effect was detected: HDAC4 protein levels were visibly decreased after treatment with mithramycin at 125 nM for 8 h, and almost undetectable after 16 h. At 250 nM of mithramycin, 8 h of treatment was sufficient to render HDAC4 protein levels almost undetectable. In contrast to these effects on HDAC4, mithramycin treatment had no effect on HDAC2 and HDAC6 protein levels (Figure 1D). These results together indicated that binding of mithramycin to GC-rich DNA sequences leads to decreased levels of HDAC4 mRNA and protein, an effect that was not seen for all HDACs.
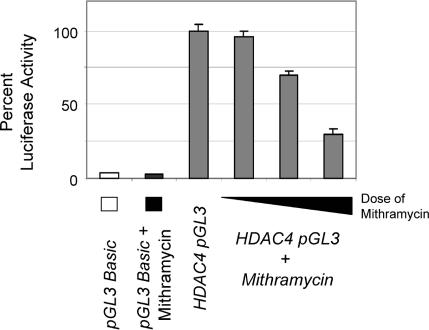
To assess whether the mithramycin directly inhibited HDAC4 mRNA transcription by affecting the promoter, we began by testing the ability of the sequences upstream of the transcription initiation site of HDAC4 to drive expression of a luciferase reporter. Analysis of this candidate HDAC4 promoter region revealed GC-rich sequences, which potentially might be targeted by mithramycin (and which are further addressed in Figure 3). The 317-base pair region upstream of the HDAC4 transcription start site was therefore cloned into pGL3-Basic (a promoterless expression vector that is commonly used as a reporter to assess the promoter activity of candidate sequences in driving luciferase expression). This sequence upstream of HDAC4 was indeed able to drive strong reporter expression (Figure 2; compare luciferase activity of HDAC4-pGL3 [third bar] to pGL3-Basic alone [first bar]). Furthermore, the addition of mithramycin led to a dose-dependent decrease in reporter activity driven by the putative HDAC4 promoter (the discrepancy between residual luciferase activity detectable here and the lack of HDAC4 protein shown in Figure 1 at the highest dose of mithramycin is likely attributable to the greater sensitivity of the luciferase assay vs. chemiluminescence). These observations together suggested that GC-rich sequences upstream of HDAC4 contained promoter activity, which could be blocked by mithramycin.
Figure 3.
The human HDAC4 promoter: binding by Sp1/Sp3. (A) Genomic sequences upstream of the beginning (indicated by arrow) of the mRNA sequence of HDAC4 are shown. Putative Sp1/Sp3 binding sites are boxed. Primers used for the Sp1/Sp3 chromatin immunoprecipitation experiment shown in B are indicated by the dotted arrows. CCAAT boxes are shown in bold. (HDAC4 mRNA sequence described by GenBank Accession number AB006626.2, GI: 6635126, clone ha061161/KIAA0288, submitted by O. Ohara (Kazusa DNA), as well as Entrez Nucleotide (NCBI), GI: 13259519) (B) Chromatin immunoprecipitation (ChIP) of Sp1 and Sp3 indicates binding to the HDAC4 promoter. Formaldehyde-treated HeLa cells were lysed, sonicated, and immunoprecipitated for Sp1 and Sp3, followed by PCR amplification with primers flanking the HDAC4 promoter (indicated in Figure 3A). The ChIP assays were performed with increasing amounts of purified immunoprecipitated DNA. Only the anti-Sp1 and Sp3 antibodies result in single bands consistent with the HDAC4 promoter and which were of the appropriate size. The assay was also performed with nonspecific immunoglobulin (Ig; Control Ig) under otherwise identical conditions as a negative control. As a positive control (Input), the assay was performed with the same primers on the genomic DNA on which the ChIP assays were performed. (C) EMSA assays of Sp1/Sp3 and the HDAC4 promoter sequences. Random oligonucleotides, or sequences corresponding to the Sp1/Sp3 consensus binding sequence or the human HDAC4 promoter were labeled with [γ-32P]ATP. These oligonucleotides were incubated with nuclear extracts from HeLa cells. The reactions with the canonical Sp1/Sp3 and the HDAC4 promoter resulted in protein:probe complexes, whereas the complexes were not seen with random oligonucleotides. The DNA-binding reaction was also performed in the absence (-) or presence (+) of 100-fold molar excess of unlabeled Sp1/Sp3 canonical HDAC4 or promoter oligonucleotides (as Unlabeled competitor), either of which diminished the protein:probe complexes seen in the absence of the competing sequences. The entire length of the gel is shown, including the free Sp1/Sp3 or HDAC4 promoter probes (Free Probe) at the bottom of the gel. The EMSA assay was repeated three times, including with TGE, with similar results each time. Faint bands below the protein:probe complexes are nonspecific bands that are not always seen when the EMSA was repeated.
Figure 2.
HDAC4 promoter activity is repressed by mithramycin. HDAC4 promoter sequence was cloned to the promoterless pGL3-Basic luciferase reporter to generate (-317) HDAC4-pGL3. The PGL3-basic and (-317) HDAC4-pGL3 were transfected into HeLa cells. Twenty-four hours after transfection, cells were mock-treated or treated with 62.5, 125, or 250 nM mithramycin. Another 24 h after treatment, all plates were harvested and the resultant luciferase activity was measured. Equal amounts of DNA (2 μg) was transfected for each sample. The luciferase activity of cells transfected with HDAC4 pGL3 and unexposed to mithramycin was set to 100% (third bar), and the activities of all other samples were calculated and plotted as a percentage of this value. To control for transfection efficiency, the relative luciferase activity of each sample was also normalized to the activity of a cotransfected Renilla luciferase plasmid (pRL-TK, 0.1 μg). All assays were performed in triplicate, and the results were averaged. Error bars, SE. This experiment was repeated three times, with similar results.
The Transcription Factor Specificity Protein 1 (Sp1) Binds to HDAC4 Promoter In Vivo and In Vitro
Analysis of the GC-rich sequences of the putative HDAC4 promoter revealed a number of sites that appeared to be consistent with the consensus binding sequence for the Specificity Protein (Sp1/Sp3) family of transcription factors (GGCGGG or GGGCGG; indicated by the boxes in Figure 3A). To determine whether Sp1 or Sp3 in fact binds to this area, we performed chromatin immunoprecipitation (ChIP) of the HDAC4 promoter with anti-Sp1 and anti-Sp3 antibodies. PCR (PCR) amplification of the immunoprecipitated DNA (using primers indicated by dotted line in Figure 3A) resulted in single bands of a size consistent with the region of the HDAC4 promoter being amplified (377 base pairs), the identity of which was confirmed by direct DNA sequencing (Figure 3B, and unpublished data).
Within the region of the HDAC4 promoter that was successfully immunoprecipitated with Sp1 and Sp3, we identified seven potential binding sites for Sp1/Sp3 (as indicated by the boxes in Figure 3A, and also diagrammed in Figure 4A). One particular 22-base pair sequence was especially interesting in that it contained three contiguous Sp1/Sp3 binding sites ((-253) GG CGG GCG GGC GGG (-239)) in addition to being separated by a single adenine (A) from another potential Sp1/Sp3 binding site ((-237) GGCGGG). As an additional test of specific binding of Sp1/Sp3 to this region of the HDAC4 promoter, we performed EMSAs using an unrelated nucleotide sequence, a synthesized portion of the HDAC4 promoter or a canonical Sp1/Sp3 sequence as the radiolabeled probes. The assay was performed in HeLa cell lysates, resulting in protein:probe complexes with the Sp1/Sp3 binding (“Canonical Sp1/Sp3”) or HDAC4 promoter sequences that were readily detectable via autoradiography, complexes that did not appear with an unrelated nucleotide sequence (Random nucleotides; Figure 3C). For both the canonical Sp1/Sp3 and HDAC4 promoter probes, addition of either the unlabeled Sp1/Sp3 or HDAC4 promoter sequence (as Unlabeled competitor) resulted in diminished complex formation, supporting the specificity of the interaction between Sp1 and the HDAC4 promoter. The EMSA assay was repeated using 0.5× TGE instead of 0.5× TBE as the running buffer, with similar results (unpublished data).
Sp1/Sp3 Binding Sequences Proportionally Contribute to HDAC4 Promoter Activity
Whereas the experiments performed strongly supported the specific binding of Sp1/Sp3 to the HDAC4 promoter, we wanted to assess the functional consequence of the binding and to assess the relative contribution of specific sequences to expression. We amplified different portions of the HDAC4 promoter (see Materials and Methods) and fused each of these to the pGL3-Basic luciferase reporter (Figure 4A). As an additional step to assess the contribution of Sp1 sites within (-253) GG CGG GCG GGC GGG GAG GCG GG (-232) to promoter activity independent of promoter length, we mutated each of the three contiguous and the next adjacent Sp1 site (to (-253) GG TTG GTT GGT TGG GAG GAA GG (-232). Each of these constructs was then expressed in HeLa cells, along with the promoterless pGL3-Basic parental vector alone as a negative control, and the resultant luciferase activity was measured (Figure 4B). The entire promoter sequence containing all seven Sp1 sites (“(-317) HDAC4”) resulted in the highest level of expression. Mutation of the four contiguous and adjacent Sp1 sites (“(-317) mut sp1 HDAC4”) resulted in a large decrease in activity. Interestingly, further decreases in the length of the promoter sequences (i.e., “(-223), (-177), or (-103) HDAC4”), resulting in progressively fewer Sp1 binding sites, resulted in progressively decreased expression of the reporter as well. To further broaden the applicability of these results, we repeated the experiments in PC3 cells, with similar results (Figure 4C). These results together suggested that for both HeLa and PC3 cells, additional Sp1 binding sites positively contribute to driving HDAC4 expression.
Either Sp1 or Sp3 Protein Increases HDAC4 Promoter Activity
The GC-rich boxes identified in the HDAC4 promoter could be potentially bound by either Sp1 or Sp3, both of which are often ubiquitously expressed in mammalian cells (Kingsley and Winoto, 1992; Azizkhan et al., 1993). Whole cell lysates of mammalian cells often contain significant amounts of both transcription factors. Studies on the relative activities of specific Sp family members are therefore often investigated in insect cell lines such as SL2, which is devoid of endogenous human Sp family members. (Yamada et al., 2000; Koutsodontis et al., 2005). We cotransfected into SL2 cells, (-317) HDAC4-pGL3, along with either PacSp1 or PacSp3 (insect expression vectors encoding human Sp1 or Sp3). Either Sp1 or Sp3 drove strong expression of the (-317) HDAC4-pGL3 reporter, suggesting that either transcription factor alone can positively drive HDAC4 expression (Figure 4D). In comparison, the three controls showed comparatively little activity. Interestingly, the HDAC4 promoter-fused reporter resulted in a low level (<10%) of activity in the absence of human Sp1 or Sp3 (possibly due to other endogenous transcription factors in the insect cells that have a minor influence on HDAC4 promoter activity).
Either Sp1 or Sp3 Can Modulate HDAC4 Protein Levels
The reporter assays described above indicate that both Sp1 and Sp3 can drive HDAC4 expression. To better understand possible functional consequences of this pathway, we investigated the relationship between Sp1 and Sp3 expression and HDAC4 protein levels. This was performed via two complementary approaches: 1) by overexpressing Sp1 and Sp3 via mammalian expression vectors (CMV4-Sp1 and CMV4-Sp3), and 2) by knocking down Sp1 and Sp3 via RNA interference (RNAi). U2OS cells express low endogenous levels of either Sp1 or Sp3 (Figure 5A), which allowed us to express graduated levels of Sp1 or Sp3 protein. Indeed, transfecting increasing amounts of the expression vector CMV4-Sp1 resulted in correspondingly greater levels of Sp1 protein. This in turn was associated with correspondingly increased levels of HDAC4 protein, whereas levels of alpha-tubulin remained unchanged (Figure 5B). Transfection of CMV4-Sp3 also led to increased HDAC4 protein levels, whereas alpha-tubulin remained unchanged as well (Figure 5C). These results, together with the reporter assays shown in Figures 4 and 5, are consistent with the ability of Sp1 and Sp3 to modulate HDAC4 protein levels by driving the transcription of HDAC4 mRNA.
Figure 5.

Overexpression of Sp1 or Sp3 increases HDAC4 protein levels. (A) U2OS cells express low levels of Sp1 and Sp3 protein. Asynchronous HeLa, PC3, or U2OS cells in exponential growth phase were trypsinized and pelleted, and whole cell lysates were prepared. These were then separated on SDS-PAGE gels, transferred to PVDF, and immunoblotted for Sp1, Sp3, or alpha-tubulin (as loading control). (B) Individual plates of U2OS cells were nontransfected or transfected with CMV4 vector (6 μg) alone, or with 2, 4, or 6 μg of CMV4-Sp1. The total amount of DNA transfected into all cells was adjusted to 6 μg by the addition of parental CMV4 empty vector. Twenty-four hours after transfection, all cells were harvested and the individual cell lysates were immunoblotted for Sp1, HDAC4, or alpha-tubulin (loading control). The transfection of increasing amounts of Sp1 expression vector led to increased levels of Sp1 protein, which also led to increasing levels of HDAC4 protein. In contrast, alpha-tubulin protein levels remained unaffected. (C) Cells were prepared and treated in a manner similar to B except that CMV4-Sp3 (instead of CMV4-Sp1) was transfected. Transfection of increasing amounts of Sp3 expression vector led to increased levels of Sp3, as well as HDAC4 protein.
To further establish the link between Sp1 and Sp3 and HDAC4 protein levels, we assessed the effects of Sp1 and Sp3 knockdown via RNAi. Treatment of HeLa cells (which express high levels of both Sp1 and Sp3 protein) with siRNA against either Sp1 or Sp3 led to greater than 90% decreases in levels of the respective targeted proteins, compared with control siRNA targeting green fluorescent protein (GFP). Knockdown of either Sp1 or Sp3 was in turn associated with marked decreases in HDAC4 protein levels (Figure 6A). As an additional test that the effects of siRNA targeted against Sp1 or Sp3 on HDAC4 protein levels was due to effects on transcription driven by the HDAC4 promoter, we also assessed for HDAC4 mRNA expression. SiRNA-mediated knockdown of Sp1 and Sp3 led to decreased HDAC4 mRNA (Figure 6B), which is therefore consistent with the effects on HDAC4 protein. The results of overexpression and knockdown of Sp1 and Sp3 together provide additional support that either Sp1 or Sp3 can modulate intracellular HDAC4 protein levels through effects on the HDAC4 promoter.
Figure 6.

Knockdown of either Sp1 or Sp3 results in decreased HDAC4 protein levels. (A) Individual plates of HeLa cells were transfected with control siRNA targeting GFP (control) or siRNA targeting either Sp1 or Sp3. Forty-eight hours after transfection, all cells were harvested and cell lysates immunoblotted for the proteins indicated. SiRNA targeting either Sp1 or Sp3 led to decreased levels of its target and also resulted in decreased HDAC4 protein levels. In contrast, siRNA targeting GFP had no effect on HDAC4. (B) Knockdown of Sp1 or Sp3 via siRNA leads to decreased HDAC4 mRNA levels. HeLa cells were treated as in A, but harvested for total mRNA, which was then assessed via RT-PCR with primers specific for HDAC4 or GAPDH.
Expression of Sp1, HDAC4, and HDAC2 Protein in Human Tissues
Given the link established between Sp1 and expression of HDAC4, we wanted to determine if levels or expression patterns of the respective proteins correlated in different human tissues and if Sp1 correlated with a different HDAC protein. We therefore assessed the expression of Sp1, HDAC4, and HDAC2 protein in a panel of human tissues taken from different organs. In the cerebral cortex, strong expression of all three proteins was readily detected in pyramidal neurons (cells with larger nuclei, designated by white arrowheads in the first column of Figure 7A). As expected, HDAC4 was expressed in both the nucleus and cytoplasm of these cells, consistent with the presence of both NLS and cytoplasmic retention signals (CRS) in the protein (Wang et al., 2001), whereas as a transcription factor Sp1 was exclusively nuclear. In contrast to expression of all three proteins in neurons, strong expression of Sp1 and HDAC4 but not HDAC2 was detectable in the testis, which was predominantly manifested by the spermatogenic germ cells (designated by stippled arrowheads in the second column of Figure 7A) in the lumen of each seminiferous tubule. Expression of Sp1 and HDAC4 but not HDAC2 was also seen in the prostate, which was especially conspicuous in the cells of the prostatic glands (glandular layer designated by the black arrowheads in the third column of Figure 7A). Expression of Sp1 and HDAC4 was also detected in other tissues such as cardiac muscle, breast, pancreas, and ovary (unpublished data), whereas a number of tissues such as thyroid showed neither protein nor HDAC2 (fourth column of Figure 7A).
Figure 7.
Concordance between Sp1 and HDAC4 protein expression in human tissues. (A) Immunohistochemical analysis of Sp1, HDAC4, and HDAC2 protein expression was performed on a human tissue array containing sections from the respective organs. Sections from the cerebral cortex, testis, prostate, and thyroid tissues are shown. Pyramidal neurons in the cerebral cortex (select neurons indicated by white arrowheads in the first column of figures) show expression of Sp1, HDAC4, and HDAC2 protein. Protein expression is indicated by the brown signal elicited by the detection system against the background blue hematoxylin staining. As a transcription factor, Sp1 localizes to the nucleus, whereas HDAC4 is found in both the nucleus and cytoplasm. Strong expression of Sp1 and HDAC4 protein but not HDAC2 was found in the testis, manifested conspicuously by the developing spermatozoa (select cells indicated by stippled arrowheads in the second column). Strong expression of Sp1 and HDAC4 protein but not HDAC2 was also detected in the prostate, as detected in the cells of the prostatic glands (designated by black arrowheads in the third column). The expression of neither Sp1, HDAC4, and HDAC2 in contrast was not detected in the thyroid gland (fourth column). (B) Expression of Sp1 and HDAC4 protein in the layers of the epidermis. Immunohistochemical analysis for Sp1, HDAC4, and HDAC2 protein was performed as in A. Specific layers of the epidermis show Sp1 and HDAC4 protein expression, including cells of stratum basale (indicated by the black arrowheads), and especially strong in the stratum spinosum (stippled arrowheads). Sp1 and HDAC4 protein in contrast is weaker in the stratum granulosum (white arrowheads) and undetectable in the stratum corneum lying at the very surface (and which consists of cornified dead cells prone to flaking). In contrast to the concordance of Sp1 and HDAC4 protein expression and the differences of expression patterns in different strata, HDAC2 expression appears more variable, with many cells showing little to no HDAC2, but occasional cells of all layers showing HDAC2 protein.
The strong expression of Sp1 and HDAC4 in the maturing spermatocytes shown in Figure 7A suggested a potential role for these proteins in differentiating cells. This possibility seemed to be reflected in the epidermis as well. In the epidermis, the continuous maturation of keratinocytes can be easily distinguished in specific cellular layers of the tissue. The surface of the skin (the stratum corneum) consists of cornified dead cells, whereas the layer immediately below (the stratum granulosum) consists of keratinizing cells that are elongated and stain darker for hematoxylin (designated by the white arrowheads in Figure 7B). Cells below this layer show bigger nuclei and compose the stratum spinosum (designated by stippled arrowheads). Finally the stratum basale, lying immediately above the basement membrane, is composed of columnar cells (designated by black arrowheads) with nuclei smaller than the cells of the stratum spinosum. Interestingly, although almost all cells throughout the epidermis showed expression of both Sp1 and HDAC4 protein, differences in the expression levels appeared to be detectable. The cells of the stratum spinosum (stippled arrowheads) showed strongest expression of both Sp1 and HDAC4. Although the cells of the stratum basale (black arrowheads) also showed Sp1 protein, here the levels of HDAC4 seemed slightly weaker, possibly reflecting the beginning of (Sp1-driven?) HDAC4 expression. Sp1 protein expression levels were weakest in the stratum granulosum (white arrowheads), and in these cells HDAC4 protein expression seemed diminished as well. Finally, the stratum corneum facing the surface showed neither Sp1 nor HDAC4 protein. In contrast to the expression of Sp1 and HDAC4 protein throughout the epidermis, the expression pattern of HDAC2 protein seemed more random. Occasional cells of the stratum granulosum, spinosum, and basale showed HDAC2 protein, but many cells of all three layers did not. These results together indicate that there is cellular- and tissue-specific expression of Sp1 and HDAC4, which did not coincide with that of HDAC2. Although Sp1 is sometimes referred as an “ubiquitous protein,” differences in protein expression levels were detectable between different tissues and sometimes within the cell types of the tissue.
Correlations between Sp1 Protein and HDAC4 mRNA and Protein Levels in Additional Cell Lines
To further broaden the potential applicability of our findings of cell-specific differences in expression levels of Sp1 and HDAC4, we also assessed levels of the respective proteins in different human cancer cell lines (Figure 8A). Our experiments in human cancer lines as well as insect cells together suggest that Sp1 and Sp3 can drive HDAC4 promoter activity and influence HDAC4 mRNA and protein levels. In assessing additional human cancer cell lines, HeLa (derived from cervical cancer), U373 (glioblastoma), and HCT116 (colorectal cancer) cells showed the highest levels of Sp1 protein, whereas SKBR3 (breast cancer) showed the lowest levels. OVCAR (ovarian cancer) and T98G (glioblastoma) cells showed intermediate levels of Sp1 protein. There appeared to be a correlation between levels of Sp1 and HDAC4 protein expression levels. For example, the high Sp1-expressing HeLa and U373 cells also expressed high levels of HDAC4 protein, whereas the low Sp1-expressing SKBR3 and HT29 cells likewise expressed little HDAC4, and OVCAR and NHA (astrocytoma-derived) cells expressed intermediate levels of both Sp1 and HDAC4 proteins. In contrast to the link between Sp1 and HDAC4 levels, there was no correlation between Sp1 and HDAC2 protein levels, and levels of alpha-tubulin protein were nearly equivalent between all cell lines. Interestingly, there was variation in the degree of correlation of Sp1 and HDAC4 between cell lines. For example, T98G cells expressed intermediate levels of Sp1 but high levels of HDAC4; in contrast, HCT116 cells expressed high Sp1 but intermediate levels of HDAC4. Finally, levels of HDAC4 protein tended to match that of its mRNA in the different cell lines (Figure 8B). Cell lines with high HDAC4 protein levels also showed high mRNA levels, whereas cell lines with low HDAC4 mRNA levels showed low levels of HDAC4 (with the exception of HT29 cells, which showed intermediate levels of HDAC4 mRNA but little HDAC4 protein). These results suggest that HDAC4 protein levels closely follow levels of the mRNA, supporting the notion that promoter-driven control of transcription of HDAC4 mRNA by Sp transcription factors can potentially modulate HDAC4 protein expression. Furthermore, these results also suggest that although Sp1 is sometimes described as an “ubiquitous protein,” differences in protein levels were detectable between different cell lines and indeed, possibly throughout embryogenesis (Supplemental Figure 1).
Figure 8.
Concordance between Sp1 and HDAC4 protein levels, and the correlation between HDAC4 mRNA and protein. (A) The following cell lines were grown under identical conditions: HeLa (cervical cancer), U373 (glioma), OVCAR (ovarian), T98G (glioma), HCT116 (colorectal), NHA (astrocytic), SKBR3 (breast), and HT29 (colorectal). Cells were harvested in exponential phase, and the resultant lysates separated via SDS-PAGE, transferred to nitrocellulose, and immunoblotted for Sp1, HDAC4, HDAC2, and alpha-tubulin (loading control). (B) The respective cell lines shown in A were grown and harvested under identical conditions, but processed for total mRNA. RT-PCR was performed using primers specific for HDAC4 and GAPDH (as assay control), and the resultant products were separated via agarose.
Do Additional Mechanisms Influence HDAC4 Promoter Activity?
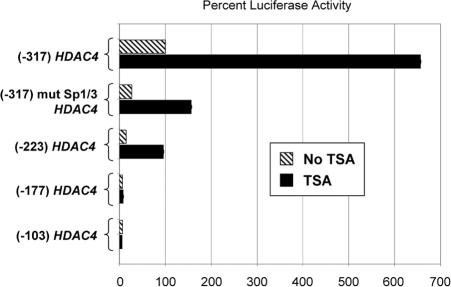
We have described here the influence of Sp1 and Sp3 on HDAC4 promoter activity. There are likely additional mechanisms that also control the activity of this promoter, such as nucleosomal structure and other transcription factors. The influence of chromatin structure on gene expression is often probed with TSA, a general histone deacetylase inhibitor that leads to the hyperacetylation of core histones that comprise the nucleosome, which results in the expansion of chromatin (Yoshida et al., 1995) and which can result in both up- as well as down-regulation of expression of certain genes (Glaser et al., 2003). To assess whether TSA might influence the promoter activity of HDAC4, we transfected the reporters for the HDAC4 promoter used in the experiments described in Figure 4, B and C, followed by mock treatment or treatment with TSA (Figure 9). As before, in the absence of TSA, (-317) HDAC4 led to the highest levels of expression, whereas expression was progressively decreased in the reporters composed of smaller portions of the HDAC4 promoter. Exposure of the cells to TSA had a number of interesting and impressive effects (Supplemental Figure 1). The promoter activity of (-317) HDAC4, (-317) mut Sp1/Sp3 HDAC4, or (-223) HDAC4 was further increased, respectively, about seven-, five-, and fivefold by TSA. The promoter activity of (-177) HDAC4 and (-103) HDAC4 remained comparatively low, and the effects of TSA for these were negligible. These results together indicate that HDAC4 promoter activity is increased by TSA (in a manner dependent on promoter length), potentially suggesting that the activity of the promoter may also be influenced by nucleosomal structure. However, the possible contribution of effects on nonhistone proteins by TSA cannot be excluded.
Figure 9.
HDAC4 promoter activity is enhanced by the histone deacetylase inhibitor trichostatin A (TSA). Cells were transfected with the luciferase reporters described in Figure 4A, representing different lengths of the HDAC4 promoter as well as with targeted mutations. As previously described, these pGL3 luciferase reporters consist of progressively shorter portions of the HDAC4 promoter, which contain respectively fewer Sp1/Sp3 binding sites. The (-317) mut Sp1/3 HDAC4 reporter is identical to (-317) HDAC4 with the exception that the four most upstream Sp1/Sp3 binding sites are mutated. Twenty-four hours after transfection, all cells were mock-treated or treated with 2 μM TSA, and after another 24 h the resultant luciferase activity was assayed in all cells. All samples were performed in triplicate and the results were averaged and plotted as shown, with all results expressed as before as a percentage of the activity resulting from (-317) HDAC4, which was set at 100%. TSA substantially increased the activities of (-317) HDAC4, (-317) mut Sp1/3 HDAC4, and (-223) HDAC4. In all plates, normalization for transfection efficiency was performed by measuring β-galactosidase activity resulting from a CMV4-β-gal plasmid that was cotransfected. The experiment was repeated three times with similar results.
It is also likely that other transcription factors and chromatin remodeling factors (such as P300/CBP and HDAC1) bind to the HDAC4 promoter and influence activity (Doetzlhofer et al., 1999; Zhang and Dufau, 2003; Varshochi et al., 2005). In support of this notion, we also note the presence of CCAAT boxes in the HDAC4 promoter (shown in bold in Figure 3). These often represent potential binding sites for the NF-Y transcription factor, which also influence promoter activity (Maity and de Crombrugghe, 1998; Mantovani, 1999; Matuoka and Yu Chen, 1999). We performed ChIP assays for NF-Y and indeed found evidence consistent with the binding of NF-Y to the HDAC4 promoter (Figure 10). Whether or not NF-Y positively drives HDAC4 promoter activity, similar or contributing to that of to the activity of Sp1/Sp3, is currently being investigated but will likely underscore the complexity of mechanisms controlling the HDAC4 promoter.
Figure 10.
Binding of the NF-Y transcription factor to the HDAC4 promoter. Chromatin immunoprecipitation (ChIP) indicates binding of NF-Y to the HDAC4 promoter. Formaldehyde-treated HeLa cells were lysed, sonicated, and immunoprecipitated for NF-Y, followed by PCR-amplification with primers flanking the HDAC4 promoter (identical to that used for the assays shown in Figure 3). The assays were performed on increasing amounts of purified immunoprecipitated DNA. The assay was also performed with nonspecific Ig (Control Ig) under otherwise identical conditions as a negative control. As a positive control (Input), the assay was performed with the same primers on the genomic DNA on which the ChIP assays were performed.
DISCUSSION
We have described here the identification of regions containing Sp1 and Sp3 consensus binding sites in the human HDAC4 promoter, confirmed the binding of Sp1 and Sp3 to these sequences, and found that this in turn drove HDAC4 promoter activity. The role of Sp1 and Sp3 in controlling HDAC4 expression was further solidified by restoration experiments and by knockdown of Sp1 or Sp3 by RNAi, which in turn led to decreased HDAC4 mRNA and protein levels. In linking the expression of HDAC4 to the activity of a specific family of transcription factors in human cells, we believe this may be the first description of mechanism(s) controlling the expression of a class II HDAC.
Sp1 and Sp3 are zinc finger proteins that belong to the Specificity Factor (Sp) family of transcriptional factors, which regulates the transcription of tissue-specific, viral and inducible genes by binding and acting through the GC boxes (GGGCGG; for a recent review see Li et al., 2004). The Sp family members are characterized by a motif of three conserved Cys2His2 zinc fingers, which form the DNA-binding domain (Suske, 1999). Sp1 and Sp3 share more than 90% sequence homology in the DNA-binding domain and bind to the same GC-rich DNA domains, which have been hypothesized to lead to either activation or repression of gene activity based on the promoter context or cellular background (Majello et al., 1997; Bouwman and Philipsen, 2002; Ammanamanchi et al., 2003; Yu et al., 2003).
The novelty of this work also includes what we believe to be the first survey of relative Sp1 and HDAC4 protein expression levels in human tissues and cancer cell lines, which showed general concordance between the two proteins. These results appear to be consistent with previous studies of relative HDAC4 mRNA levels in different human tissues, which found high levels in the brain, skeletal muscle, heart, and testis (Fischle et al., 1999; Grozinger et al., 1999; Wang et al., 1999). Other investigations have identified roles of Sp1 and other Sp family proteins in driving promoter activity in different human tissues or tissue-specific promoters, including that of neurons, the testis, keratinocytes, the prostate, and the pancreas (McClure et al., 1999; Zhang et al., 1999; Shi et al., 2001; Kaufman et al., 2002; Wilkerson et al., 2002; Naso et al., 2003; Abdelrahim et al., 2004; Tang et al., 2004; Benfante et al., 2005; Shin et al., 2005; Wang and Bannon, 2005). Interestingly, these investigations are supported by our findings of high Sp1 levels in the cortical brain, the testis, the prostate, epidermal skin, and pancreas. Because high levels of HDAC4 protein were also identified in these tissues, the potential role(s) of HDAC4 in influencing intracellular processes in those tissues and the degree to which these roles are modulated by Sp1/Sp3 may be intriguing to pursue. Although HDAC4 protein expression generally matched that previously found for the mRNA, such as the strong expression of both HDAC4 protein and mRNA in the brain, we also noted individual differences (e.g., strong HDAC4 protein but weaker mRNA expression in the prostate), which may reflect increased protein stability or decreased protein degradation in these tissue.
Changes in expression levels of Sp1 over time or in response to stimuli that result in tissue-wide changes have been previously noted. During murine development, Sp1 is expressed at varying levels in the tissues of different organs, which can change depending on the age of the embryo. For example, Sp1 mRNA levels in the heart are increased four-fold at day 30 from levels at day 15, whereas levels in other tissues such as the thymus may vary up to 100-fold (Saffer et al., 1991). Sp1 has also been shown to drive expression of genes implicated in muscle atrophy, such as atrophy associated with exposure to dexamethasone. Dexamethasone treatment of muscle cells leads to Sp1-mediated upregulation of the expression of ubiquitin C (Marinovic et al., 2000; Price, 2003), which becomes covalently linked to and leads to accelerated proteasome-mediated proteolysis of muscle proteins and which then likely contributes to muscle atrophy (Wing et al., 1995; Solomon et al., 1998; Lecker et al., 1999; Marinovic et al., 2000). In contrast to these observations, continuous skeletal contraction led to decreased Sp1 mRNA levels, potentially to help maintain muscle mass (Irrcher and Hood, 2004). Interestingly, HDAC4 has been associated with repression of the myogenic MEF2 transcription factor (Miska et al., 2001), and HDAC4 has been found to be substantially up-regulated during muscle atrophy (Table 1 in Supplemental Data section of Bodine et al., 2001), which might be consistent with parallel up-regulation by Sp1.
Regulation of the activities of Sp family members of transcription factors have also been found to include posttranslational mechanisms as well as protein-to-protein interactions. Posttranslational modifications such as phosphorylation, glycosylation, or sumoylation have been reported to influence the activity of Sp1 (Jang and Steinert, 2002; Sun et al., 2002; Zhang and Dufau, 2002; Chu and Ferro, 2005). The ratio of Sp1 to Sp3 in certain cellular contexts may determine the degree of activation (Wong et al., 2003; Pang et al., 2004). Sp1 has been reported to associate with chromatin remodeling factors (such as p300/CBP or PCAF) and DNA-binding proteins (Chapman and Perkins, 2000; Xiao et al., 2000; Jang and Steinert, 2002; Suzuki et al., 2003; Zhang and Dufau, 2003; Li et al., 2004; Huang et al., 2005; Varshochi et al., 2005).
Although we found a general correlation between Sp1 and HDAC4 protein levels in tissues and cell lines, posttranslational mechanisms may account for individual variations in the degree of correlation. For example, a portion of Sp1 in T98G cells migrates slower than the Sp1 in OVCAR cells (compare lanes 4 and 3 in Figure 8) possibly suggesting the presence of a greater proportion of “activated” phosphorylated Sp1, which may in turn contribute to increased expression of HDAC4 mRNA and protein. Nonetheless, a threshold level of Sp1 may be required for HDAC4 expression, as suggested in our RNAi experiments, in which reduction of Sp1 led to reduced HDAC4 levels. We noted that SKBR3 cells expressed the lowest levels of Sp1, and HDAC4 in this cell line was undetectable. Finally, it is also likely that that factors specific to HDAC4 may influence the ultimate levels of the protein, such as translational efficiency, the relative stabilities of HDAC4 mRNA and protein in different cell lines, and the presence of mechanisms that lead to mRNA and protein degradation (Liu et al., 2004).
The mechanisms we describe here that control the promoter activity of HDAC4 are reminiscent of that previously described for HDAC1, a class I HDAC (Schuettengruber et al., 2003). Similar to what we have found for HDAC4, the activation of HDAC1 expression by Sp1 was dependent on specific sequences in the promoter, which was increased by TSA. However, unlike HDAC4, HDAC1 has not been previously implicated in muscle differentiation or development, but was linked instead to adipocyte differentiation (Wiper-Bergeron et al., 2003). To our knowledge, the relative expression patterns of HDAC1 in different human cell lines have not been reported. Nonetheless, these findings together therefore raise the possibility that the Sp family of transcription factors have different target genes in different tissues, but share similarities in the mechanisms of activation.
Supplementary Material
Acknowledgments
We are grateful to the other members of the Kao Laboratory past and present for indispensable and expert assistance, especially Shary Parker, Katie Murphy, Jessica Liao, Andrew Boethy, and Geoffrey Geiger. We thank Dr. Jon Horowitz for the generous gifts of pPac, pPacSP1, and pPacSP3 and for helpful comments. We thank Mark Patrick for excellent administrative support. G.D.K. was supported by funds from the University of Pennsylvania Research Foundation, the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (Advanced Career Research Award), and the National Institutes of Health (CA107956). A.M. was supported by the National Institutes of Health (CA093638).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0775) on November 9, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abdelrahim, M., Smith, R., 3rd, Burghardt, R., and Safe, S. (2004). Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res, 64, 6740-6749. [DOI] [PubMed] [Google Scholar]
- Alland, L., Muhle, R., Hou, H., Jr., Potes, J., Chin, L., Schreiber-Agus, N., and DePinho, R. A. (1997). Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49-55. [DOI] [PubMed] [Google Scholar]
- Ammanamanchi, S., Freeman, J. W., and Brattain, M. G. (2003). Acetylated sp3 is a transcriptional activator. J. Biol. Chem. 278, 35775-35780. [DOI] [PubMed] [Google Scholar]
- Azizkhan, J. C., Jensen, D. E., Pierce, A. J., and Wade, M. (1993). Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit. Rev. Eukaryot. Gene Expr. 3, 229-254. [PubMed] [Google Scholar]
- Benfante, R., Antonini, R. A., Vaccari, M., Flora, A., Chen, F., Clementi, F., and Fornasari, D. (2005). The expression of the human neuronal alpha3 Na+,K+-ATPase subunit gene is regulated by the activity of the Sp1 and NF-Y transcription factors. Biochem. J. 386, 63-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume, S. W., Snyder, R. C., Ray, R., Thomas, S., Koller, C. A., and Miller, D. M. (1991). Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J. Clin. Invest. 88, 1613-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine, S.C. et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704-1708. [DOI] [PubMed] [Google Scholar]
- Bouwman, P., and Philipsen, S. (2002). Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195, 27-38. [DOI] [PubMed] [Google Scholar]
- Chapman, N. R., and Perkins, N. D. (2000). Inhibition of the RelA(p65) NF-kappaB subunit by Egr-1. J. Biol. Chem. 275, 4719-4725. [DOI] [PubMed] [Google Scholar]
- Chauchereau, A., Mathieu, M., de Saintignon, J., Ferreira, R., Pritchard, L. L., Mishal, Z., Dejean, A., and Harel-Bellan, A. (2004). HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF. Oncogene 23, 8777-8784. [DOI] [PubMed] [Google Scholar]
- Chu, S., and Ferro, T. J. (2005). Sp1, regulation of gene expression by phosphorylation. Gene 348, 1-11. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer, A., Rotheneder, H., Lagger, G., Koranda, M., Kurtev, V., Brosch, G., Wintersberger, E., and Seiser, C. (1999). Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19, 5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Dequiedt, F., Hendzel, M. J., Guenther, M. G., Lazar, M.A., Voelter, W., and Verdin, E. (2002). Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45-57. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Emiliani, S., Hendzel, M. J., Nagase, T., Nomura, N., Voelter, W., and Verdin, E. (1999). A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274, 11713-11720. [DOI] [PubMed] [Google Scholar]
- Glaser, K. B., Staver, M. J., Waring, J. F., Stender, J., Ulrich, R. G., and Davidsen, S. K. (2003). Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2, 151-163. [PubMed] [Google Scholar]
- Gregoire, S., and Yang, X. J. (2005). Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25, 2273-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger, C. M., Hassig, C. A., and Schreiber, S. L. (1999). Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96, 4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger, C. M., and Schreiber, S. L. (2002). Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9, 3-16. [DOI] [PubMed] [Google Scholar]
- Guenther, M. G., Barak, O., and Lazar, M. A. (2001). The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig, C. A., Fleischer, T. C., Billin, A. N., Schreiber, S. L., and Ayer, D. E. (1997). Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89, 341-347. [DOI] [PubMed] [Google Scholar]
- Heinzel, T. et al. (1997). A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43-48. [DOI] [PubMed] [Google Scholar]
- Huang, W., Zhao, S., Ammanamanchi, S., Brattain, M., Venkatasubbarao, K., and Freeman, J. W. (2005). Trichostatin A induces transforming growth factor beta type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1.NF-Y complex. J. Biol. Chem. 280, 10047-10054. [DOI] [PubMed] [Google Scholar]
- Irrcher, I., and Hood, D. A. (2004). Regulation of Egr-1, SRF, and Sp1 mRNA expression in contracting skeletal muscle cells. J. Appl. Physiol. 97, 2207-2213. [DOI] [PubMed] [Google Scholar]
- Jang, S. I., and Steinert, P. M. (2002). Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J. Biol. Chem. 277, 42268-42279. [DOI] [PubMed] [Google Scholar]
- Kaufman, C. K., Sinha, S., Bolotin, D., Fan, J., and Fuchs, E. (2002). Dissection of a complex enhancer element: maintenance of keratinocyte specificity but loss of differentiation specificity. Mol. Cell. Biol. 22, 4293-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley, C., and Winoto, A. (1992). Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 12, 4251-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsodontis, G., Vasilaki, E., Chou, W. C., Papakosta, P., and Kardassis, D. (2005). Physical and functional interactions between members of the tumor suppressor p53 and the Sp families of transcription factors: importance for the regulation of genes involved in cell cycle arrest and apoptosis. Biochem. J. 389, 443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty, C. D., Yang, W. M., Sun, J. M., Davie, J. R., Seto, E., and Eisenman, R. N. (1997). Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89, 349-356. [DOI] [PubMed] [Google Scholar]
- Lecker, S. H., Solomon, V., Price, S. R., Kwon, Y. T., Mitch, W. E., and Goldberg, A. L. (1999). Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J. Clin. Invest. 104, 1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier, C., Brocard, M. P., Puvion-Dutilleul, F., Kao, H. Y., Albagli, O., and Khochbin, S. (2002). Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 277, 22045-22052. [DOI] [PubMed] [Google Scholar]
- Li, L., He, S., Sun, J. M., and Davie, J. R. (2004). Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 82, 460-471. [DOI] [PubMed] [Google Scholar]
- Liu, F., Dowling, M., Yang, X. J., and Kao, G. D. (2004). Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279, 34537-34546. [DOI] [PubMed] [Google Scholar]
- Maity, S. N., and de Crombrugghe, B. (1998). Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23, 174-178. [DOI] [PubMed] [Google Scholar]
- Majello, B., De Luca, P., and Lania, L. (1997). Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 272, 4021-4026. [DOI] [PubMed] [Google Scholar]
- Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15-27. [DOI] [PubMed] [Google Scholar]
- Marinovic, A. C., Mitch, W. E., and Price, S. R. (2000). Tools for evaluating ubiquitin (UbC) gene expression: characterization of the rat UbC promoter and use of an unique 3′ mRNA sequence. Biochem. Biophys. Res. Commun. 274, 537-541. [DOI] [PubMed] [Google Scholar]
- Matuoka, K., and Yu Chen, K. (1999). Nuclear factor Y (NF-Y) and cellular senescence. Exp. Cell Res. 253, 365-371. [DOI] [PubMed] [Google Scholar]
- McClure, R. F., Heppelmann, C. J., and Paya, C. V. (1999). Constitutive Fas ligand gene transcription in Sertoli cells is regulated by Sp1. J. Biol. Chem. 274, 7756-7762. [DOI] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., Lu, J., and Olson, E. N. (2000a). Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2000b). Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97, 14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2001). Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2, a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40-47. [DOI] [PubMed] [Google Scholar]
- Miska, E. A., Karlsson, C., Langley, E., Nielsen, S. J., Pines, J., and Kouzarides, T. (1999). HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18, 5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska, E. A., Langley, E., Wolf, D., Karlsson, C., Pines, J., and Kouzarides, T. (2001). Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29, 3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naso, M., Uitto, J., and Klement, J. F. (2003). Transcriptional control of the mouse Col7a1 gene in keratinocytes: basal and transforming growth factor-beta regulated expression. J. Invest. Dermatol. 121, 1469-1478. [DOI] [PubMed] [Google Scholar]
- Nehls, M. C., Brenner, D. A., Gruss, H. J., Dierbach, H., Mertelsmann, R., and Herrmann, F. (1993). Mithramycin selectively inhibits collagen-alpha 1(I) gene expression in human fibroblast. J. Clin. Invest. 92, 2916-2921. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pang, R. T., Lee, L. T., Ng, S. S., Yung, W. H., and Chow, B. K. (2004). CpG methylation and transcription factors Sp1 and Sp3 regulate the expression of the human secretin receptor gene. Mol. Endocrinol. 18, 471-483. [DOI] [PubMed] [Google Scholar]
- Paroni, G., Mizzau, M., Henderson, C., Del Sal, G., Schneider, C., and Brancolini, C. (2004). Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell 15, 2804-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pore et al. (2004). Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol. Biol. Cell 15, 4841-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, S. R. (2003). Increased transcription of ubiquitin-proteasome system components: molecular responses associated with muscle atrophy. Int. J. Biochem. Cell Biol. 35, 617-628. [DOI] [PubMed] [Google Scholar]
- Saffer, J. D., Jackson, S. P., and Annarella, M. B. (1991). Developmental expression of Sp1 in the mouse. Mol. Cell. Biol. 11, 2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber, B., Simboeck, E., Khier, H., and Seiser, C. (2003). Autoregulation of mouse histone deacetylase 1 expression. Mol. Cell. Biol. 23, 6993-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, N., and Seto, E. (2004). Regulation of histone deacetylase activities. J. Cell. Biochem. 93, 57-67. [DOI] [PubMed] [Google Scholar]
- Shi, Q., Le, X., Abbruzzese, J. L., Peng, Z., Qian, C. N., Tang, H., Xiong, Q., Wang, B., Li, X. C., and Xie, K. (2001). Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 61, 4143-4154. [PubMed] [Google Scholar]
- Shin, T., Sumiyoshi, H., Matsuo, N., Satoh, F., Nomura, Y., Mimata, H., and Yoshioka, H. (2005). Sp1 and Sp3 transcription factors upregulate the proximal promoter of the human prostate-specific antigen gene in prostate cancer cells. Arch. Biochem. Biophys. 435, 291-302. [DOI] [PubMed] [Google Scholar]
- Solomon, V., Baracos, V., Sarraf, P., and Goldberg, A. L. (1998). Rates of ubiquitin conjugation increase when muscles atrophy, largely through activation of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 95, 12602-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. M., Chen, H. Y., Moniwa, M., Litchfield, D. W., Seto, E., and Davie, J. R. (2002). The transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2. J. Biol. Chem. 277, 35783-35786. [DOI] [PubMed] [Google Scholar]
- Suske, G. (1999). The Sp-family of transcription factors. Gene 238, 291-300. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Muto, S., Miyamoto, S., Aizawa, K., Horikoshi, M., and Nagai, R. (2003). Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J. Biol. Chem. 278, 28758-28764. [DOI] [PubMed] [Google Scholar]
- Tang, S., Bhatia, B., Zhou, J., Maldonado, C. J., Chandra, D., Kim, E., Fischer, S. M., Butler, A. P., Friedman, S.L., and Tang, D. G. (2004). Evidence that Sp1 positively and Sp3 negatively regulate and androgen does not directly regulate functional tumor suppressor 15-lipoxygenase 2 (15-LOX2) gene expression in normal human prostate epithelial cells. Oncogene 23, 6942-6953. [DOI] [PubMed] [Google Scholar]
- Varshochi, R., Halim, F., Sunters, A., Alao, J. P., Madureira, P. A., Hart, S. M., Ali, S., Vigushin, D. M., Coombes, R. C., and Lam, E. W. (2005). ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor alpha from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J. Biol. Chem. 280, 3185-3196. [DOI] [PubMed] [Google Scholar]
- Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., and McKinsey, T. A. (2004a). Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, R. B. et al. (2004b). Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555-566. [DOI] [PubMed] [Google Scholar]
- Verdin, E., Dequiedt, F., and Kasler, H. G. (2003). Class II histone deacetylases: versatile regulators. Trends Genet. 19, 286-293. [DOI] [PubMed] [Google Scholar]
- Wang, A. H., Bertos, N. R., Vezmar, M., Pelletier, N., Crosato, M., Heng, H. H., Th'ng, J., Han, J., and Yang, X. J. (1999). HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19, 7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. H., Kruhlak, M. J., Wu, J., Bertos, N. R., Vezmar, M., Posner, B. I., Bazett-Jones, D. P., and Yang, X. J. (2000). Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20, 6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. H., and Yang, X. J. (2001). Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21, 5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Bannon, M. J. (2005). Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J. Neurochem. 93, 474-482. [DOI] [PubMed] [Google Scholar]
- Wen, Y. D., Perissi, V., Staszewski, L. M., Yang, W. M., Krones, A., Glass, C. K., Rosenfeld, M. G., and Seto, E. (2000). The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97, 7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson, D. C., Wolfe, S. A., and Grimes, S. R. (2002). Sp1 and Sp3 activate the testis-specific histone H1t promoter through the H1t/GC-box. J. Cell. Biochem. 86, 716-725. [DOI] [PubMed] [Google Scholar]
- Wing, S. S., Haas, A. L., and Goldberg, A. L. (1995). Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochem. J. 307(Pt 3), 639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiper-Bergeron, N., Wu, D., Pope, L., Schild-Poulter, C., and Hache, R. J. (2003). Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 22, 2135-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, W. K., Chen, K., and Shih, J. C. (2003). Decreased methylation and transcription repressor Sp3 up-regulated human monoamine oxidase (MAO) B expression during Caco-2 differentiation. J. Biol. Chem. 278, 36227-36235. [DOI] [PubMed] [Google Scholar]
- Xiao, H., Hasegawa, T., and Isobe, K. (2000). p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275, 1371-1376. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Tanaka, T., Miyamoto, K., and Noguchi, T. (2000). Sp family members and nuclear factor-Y cooperatively stimulate transcription from the rat pyruvate kinase M gene distal promoter region via their direct interactions. J. Biol. Chem. 275, 18129-18137. [DOI] [PubMed] [Google Scholar]
- Yang, X. J., and Gregoire, S. (2005). Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25, 2873-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M., Horinouchi, S., and Beppu, T. (1995). Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17, 423-430. [DOI] [PubMed] [Google Scholar]
- Yu, B., Datta, P. K., and Bagchi, S. (2003). Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 31, 5368-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Kalkum, M., Chait, B. T., and Roeder, R. G. (2002). The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611-623. [DOI] [PubMed] [Google Scholar]
- Zhang, L. P., Stroud, J., Eddy, C. A., Walter, C. A., and McCarrey, J. R. (1999). Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biol. Reprod. 60, 1329-1337. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Dufau, M. L. (2002). Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277, 33431-33438. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Dufau, M. L. (2003). Dual mechanisms of regulation of transcription of luteinizing hormone receptor gene by nuclear orphan receptors and histone deacetylase complexes. J. Steroid Biochem. Mol. Biol. 85, 401-414. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Iratni, R., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (1997). Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89, 357-364. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S., and Reinberg, D. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279-289. [DOI] [PubMed] [Google Scholar]
- Zhou, X., Richon, V. M., Wang, A. H., Yang, X. J., Rifkind, R. A., and Marks, P. A. (2000). Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl. Acad. Sci. USA 97, 14329-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.