Abstract
Among peroxins involved in peroxisome biogenesis, only Pex8p is predominantly intraperoxisomal at steady state. Pex8p is necessary for peroxisomal matrix protein import via the PTS1 and PTS2 pathways. It is proposed to bridge two peroxisomal membrane subcomplexes comprised of the docking (Pex13p, Pex14p, Pex17p) and RING (Pex2p, Pex10p, Pex12p) peroxins and is also implicated in cargo release of PTS1 proteins in the matrix. We show that Pichia pastoris Pex8p (PpPex8p) enters the peroxisome matrix using two redundant pathways in a Pex14p-dependent, but Pex2p-independent, manner, showing that the intact importomer and RING subcomplex are not required for its import. One pathway depends on the TPR motifs in Pex5p, the C-terminal PTS1 sequence (AKL) in PpPex8p, and the intraperoxisomal presence of this peroxin. The alternative pathway uses the PTS2 receptor, Pex7p, its accessory protein, Pex20p, and a putative PTS2 motif in PpPex8p, but does not require intraperoxisomal PpPex8p. Pex20p interaction with PpPex8p is independent of Pex7p, but the interaction of PpPex8p with Pex7p requires Pex20p. These data suggest a direct interaction between PpPex8p and Pex20p. Our studies shed light on the mechanism and evolution of the dual import pathways for PpPex8p.
INTRODUCTION
Protein import to the peroxisome matrix requires the coordinated action of ∼20 peroxins encoded by PEX genes (Lazarow, 2003). These peroxins are mostly localized in the cytosol (e.g., the cargo receptors Pex5p and Pex7p or the accessory protein, Pex20p) and the peroxisome membrane (e.g., components of the importomer). Of the peroxins in a given organism, only one, Pex8p, is known to be predominantly intraperoxisomal at steady state (Waterham et al., 1994; Liu et al., 1995; Rehling et al., 2000; Smith and Rachubinski, 2001). In contrast, the peroxisomal targeting signal (PTS) receptors, Pex5p and Pex7p, shuttle in and out of peroxisomes, transiently, although at steady state they are mostly localized to the cytosol (Dammai and Subramani, 2001; Nair et al., 2004).
Pex8p has been described as a central organizer of the importomer, a multisubunit complex of peroxisomal membrane and associated proteins, that is essential for matrix protein import into peroxisomes (Agne et al., 2003). Not surprisingly, Pex8p is necessary for peroxisomal matrix import of proteins carrying either a PTS1 or a PTS2 sequence (Waterham et al., 1994; Liu et al., 1995; Rehling et al., 2000; Smith and Rachubinski, 2001). In addition, it is also proposed to play a role in the release of PTS1 cargo within the peroxisomes, by interaction with cargo-loaded Pex5p (Wang et al., 2003). To perform these critical functions, Pex8p must first be translocated into the peroxisome matrix. How this is achieved is a mystery.
Pex8p was initially described in Hansenula polymorpha as a protein containing both putative PTS1 and PTS2 sequences that could be shown to serve as PTSs for heterologous passenger proteins (Waterham et al., 1994). The surprise however, was the observation that neither sequence was essential either for Pex8p targeting or function, leading to the conclusion that neither the PTS1 nor the putative PTS2 in HpPex8p was necessary for peroxisomal matrix targeting. Even the availability of a PTS2 sequence in HpPex8p or ScPex8p for binding to the PTS2-receptor, ScPex7p, could not be demonstrated (Rehling et al., 2000), raising a question about the in vivo functionality of this sequence. Pex8p was then analyzed in Pichia pastoris and Saccharomyces cerevisiae, where it was also shown to have a C-terminal PTS1 sequence (Liu et al., 1995; Rehling et al., 2000). However, Pex8p lacking the PTS1 was still targeted to peroxisomes and functional in matrix protein import in S. cerevisiae (Rehling et al., 2000). As with the work with H. polymorpha, these authors also concluded that Pex8p targeting to peroxisomes was independent of its PTS1 (Rehling et al., 2000). Pex8p in Yarrowia lipolytica has the C-terminal sequence GTL, which is not a PTS1 (Smith and Rachubinski, 2001). Collectively, these studies suggested the lack of involvement of a C-terminal tripeptide in Pex8p targeting to peroxisomes.
Studies in Y. lipolytica demonstrated that in the absence of Pex20p, an accessory protein for the PTS2 pathway, YlPex8p was associated with the organelle pellet (containing peroxisomes), suggesting the absence of a role for PTS2-pathway proteins in peroxisomal targeting of YlPex8p (Smith and Rachubinski, 2001). It was noted that an N-terminal fragment of ScPex8p (aa1–112) targeted a reporter protein to peroxisomes but the data were not shown, and neither the sequence nor the import pathway was identified (Rehling et al., 2000). Additionally, in the light of a newly defined consensus that predicts virtually all known PTS2 sequences, there is no consensus predicted PTS2 sequence in this fragment of ScPex8p (Petriv et al., 2004). These studies failed to implicate a role for the PTS2 pathway in Pex8p import into peroxisomes.
Thus, despite the acknowledged presence of PTS1 sequences in Pex8p from several yeasts and that of a putative PTS2 that is sufficient but not necessary for Pex8p targeting to peroxisomes in H. polymorpha, the evidence and conclusions in the literature suggest that neither the PTS1 nor the PTS2 sequence in Pex8p, nor PTS2-pathway proteins, such as Pex20p, are required for the peroxisomal matrix targeting of Pex8p.
Additional questions regarding the mechanism of Pex8p import into peroxisomes are raised by the proposed functions for Pex8p. If Pex8p is critical for assembly of the importomer, which is presumed to be required for all matrix protein import, how is Pex8p itself imported? Alternatively, if Pex8p is necessary for cargo release, how is Pex8p itself released inside peroxisomes?
We show, using PpPex8p, that the biological roles of the PTSs in Pex8p and of the proteins involved in these PTS-dependent import pathways may have been missed because of the existence of redundant import pathways. The entry of PpPex8p into peroxisomes by one of these pathways, and not the other, requires the presence of intraperoxisomal PpPex8p, providing an explanation for the evolution of the dual PpPex8p import pathways and for the coevolution of Pex8p and Pex20p-like proteins. Finally, we show that PpPex8p import into peroxisomes is independent of Pex2p, but requires Pex14p. These results shed light on the mechanism of entry of PpPex8p into peroxisomes and are relevant to its ability to function inside peroxisomes as a key component of matrix protein import pathways.
MATERIALS AND METHODS
Yeast Strains, Oligonucleotides, Plasmids, and Culture Conditions
The P. pastoris strains and plasmids used are listed in Table 1 and the oligonucleotides are in Table 2. Growth media components were as follows: rich medium YPD, 1% yeast extract, 2% peptone, 2% glucose; synthetic medium YNM, 0.67% yeast nitrogen base, 0.05% yeast extract, 0.5% (vol/vol) methanol; mineral oleate medium MMOT (Snyder et al., 1999), containing 0.2% (vol/vol) oleate and 0.02% (vol/vol) Tween-40.
Table 1.
P. pastoris strains and plasmids used in this study
| Name | Genotype | Source |
|---|---|---|
| PPY12 | his4, arg4 | Gould et al. (1992) |
| JC121 | his4, arg4, pex8Δ::ARG4 | Liu et al. (1995) |
| PPY115 | his4, arg4, pex5Δ::ARG4 | McCollum et al. (1993) |
| SYE65 | his4, arg4, pex7Δ::ARG4 | Elgersma et al. (1998) |
| SEB3 | his4, arg4, pex20Δ::KanMx6 | Léon et al. (2006) |
| JC404 | his4, arg4, pex14Δ::ARG4 | Johnson et al. (2001) |
| JC214 | his4, arg4, pex2Δ::ARG4 | Waterham et al. (1994) |
| SLZ33 | his4, arg4, pex5Δ::ARG4, pex20Δ::KanMx6 | This study |
| SLZ56 | PPY12, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ58 | JC121, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ60 | PPY115, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ62 | SYE65, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ54 | SEB3, his4::pLZ119, arg4::pJCF235 | This study |
| SLZ64 | SLZ33, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ57 | PPY12, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ59 | JC121, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ61 | PPY115, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ63 | SYE65, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ55 | SEB3, his4::pLZ120, arg4::pJCF235 | This study |
| SLZ65 | JC404, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ67 | JC214, his4::pLZ119, arg4::pLZ127 | This study |
| SLZ66 | JC404, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ68 | JC214, his4::pLZ120, arg4::pLZ127 | This study |
| SLZ74 | PPY12, his4::pLZ125, arg4::pLZ127 | This study |
| SLZ75 | JC121, his4::pLZ125, arg4::pLZ127 | This study |
| SLZ76 | PPY115, his4::pLZ125, arg4::pLZ127 | This study |
| SLZ77 | SYE65, his4::pLZ125, arg4::pLZ127 | This study |
| SLZ78 | SEB3, his4::pLZ125, arg4::pLZ127 | This study |
| Plasmid | Properties |
|---|---|
| pLZ119 | pIB1-based with HIS4 PPEX8-GFP-PEX8 |
| pLZ120 | pIB1-based with HIS4 PPEX8-GFP-PEX8ΔAKL |
| pLZ125 | pIB1-based with HIS4 PPEX8-GFP-PEX8PTS2m |
| pLZ127 | pJC235 with ZeoR upstream of PPEX3-PEX3-mRFP |
| pJC235 | pIB1-derived with ARG4 PPEX3-PEX3-mRFP |
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| P8GBT-F | CGGAATTCATGTATAGATTGGGATCTCAG |
| P8hyb-R | ACGCGTCGACTTATAACTTTGCGGTTGATTG |
| P8hyb-R3 | ACGCGTCGACCTAGGTTGATTGGGCGTTTACATTCTC |
| P5GAD-F | CGGAATTCGATGTCGCTTATTGGCGGA |
| P5hyb-R | ACGCGTCGACCTAAAAGTCAAACATTTTTCTG |
| P5GAD-F1 | CGGAATTCCGATCCAGATGCCTATG |
| Y2H20.d | TCCGCCCGGGAATGTTTACTTCTAATGGCAG |
| Y2H20.r | TCCGGTCGACATTACCTTAGATCGGATAAAGG |
| 20–146.d | TCCGCCCGGGCAACCATATGCAACATAACAAAG |
| 20–146.r | TCCGGTCGACGTTGAACTCTTGAGTCCAAGC |
| 20–260.r | TCCGGTCGACCCACGTTGTTCATTTGGTTG |
| PPEX8-F | GGGGTACCGATCTGTGTTCCTAAG |
| P8-RSph | ACATGCATGCTTATAACTTTGCGGTTGATTG |
| P8ATG | GGACTAGTAGATCTCATGTATAGATTG |
| P8-R1Sph | ACATGCATGCTTAGGTTGATTGGGCGTTTACATTCTC |
| P8PTS2m-s | CCGTCATTTTCCAGAGAGGAGCTTTCCATTTTATTT |
| P8PTS2m-as | AAATAAAATGGAAAGCTCCTCTCTGGAAAATGACGG |
Yeast cells were grown at 30°C in rich medium (YPD) to 1 OD600/ml, washed with distilled H2O, and shifted either to synthetic methanol medium (YNM) for fluorescence microscopy, or to mineral oleate medium (MMOT) for biochemical experiments.
Generation of the Δpex5Δpex20 Mutant
To generate the Δpex5Δpex20 double deletion mutant (SLZ33), a linear DNA fragment containing the G418r gene flanked by the 5′ untranslated region (UTR) and 3′UTR of the PEX20 gene was amplified from pSEB47 (Léon et al., 2006) and introduced into the Δpex5 strain by electroporation to replace the PEX20 gene. The double mutant strain was confirmed by PCR and Western blot analysis of protein.
Subcellular Fractionation, Immunoprecipitation, and Protease Protection
Subcellular fractionation from oleate-induced yeast cells was performed as described previously (Faber et al., 1998). Immunoprecipitation of HA-tagged proteins was performed as follows. Cells (8 ODs) were broken with glass beads in 200 μl immunoprecipitate (IP) lysis buffer (50 mM HEPES-KOH, pH 7.5, 0.5 M NaCl, 0.5% NP-40, 10% glycerol, 1 mM EDTA, protease inhibitor cocktail), and centrifuged twice (14,000 × g, 10 min). Monoclonal anti-HA antibody (Covance, Madison, WI) was added to the supernatant (6 μl/ml lysate) and incubated overnight with the extract. GammaBind beads (Pharmacia, Piscataway, NJ) were added (25 μl) and incubated for 2 h. Beads were washed twice (1 ml) with the lysis buffer for 10 min, then three times (1 ml) with the wash buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA), and finally boiled in SDS loading buffer (50 μl). The equivalent of 0.2 ODs were loaded in the input (Inp) and unbound (Unb) lanes, whereas 1 OD equivalent was loaded in the IP lane.
Protease protection analysis was conducted with the P200 fraction isolated directly from the PNS in the absence of protease inhibitors. Two slightly different methods were used. In the first method, equal amounts (50 μg protein) of the P200 fraction were incubated with proteinase K (20 μg) in the absence (-) or presence (+) of 0.15% (vol/vol) Triton X-100, respectively, at 25°C for 0–30 min. In the second method, increasing amounts of proteinase K were used for 15 min. Reactions were terminated by precipitation with trichloroacetic acid (10%). The pelleted proteins were washed with cold acetone, dried by vacuum, resuspended in sample buffer, before equal volumes were subjected to SDS-PAGE, and immunoblotting.
Yeast Two-Hybrid Analysis
The Matchmaker two-hybrid protocol was followed (Clontech, Palo Alto, CA). Constructs were generated by ligating PCR-amplified full-length or truncated fragments in-frame and downstream of DNA encoding the DNA-binding (DB) or activation (AD) domains of GAL4 in plasmids pGBT9 and pGAD AH, respectively. To generate pGBT-PEX8 and pGBT-PEX8ΔAKL constructs, full-length PEX8 (Liu et al., 1995) and PEX8ΔAKL (Liu et al., 1995) were amplified with oligonucleotides P8GBT-F/P8hyb-R and P8GBT-F/P8hyb-R3, respectively, and ligated into EcoRI- and SalI-digested pGBT9 vector. pGAD-PEX5(1–576) and pGAD-PEX5(277–576) were generated by amplifying either the full-length or a fragment (277–576) of the PEX5 gene from plasmid pKSPas8 (Terlecky et al., 1995) with oligonucleotides P5GAD-F/P5hyb-R and P5GAD-F1/P5hyb-R, respectively, and ligated into EcoRI- and SalI-digested pGAD-GH vector. pGAD-PEX20, pGAD-PEX20(1–146), pGAD-PEX20(146–323), and pGAD-PEX20(146–260) were generated by amplifying either the full-length or the indicated fragments from genomic DNA with oligonucleotides Y2H20.d/Y2H20.r, Y2H20.d/20–146.r, 20–146.d/Y2H20.r, and 20–146.d/20–260.r, respectively (Léon et al., 2006) and ligated into the pGAD-GH vector digested with SmaI and SalI. pGBT-PEX8PTS2m was generated by site-directed mutagenesis with oligonucleotides P8PTS2m-s/P8PTS2m-as using pGBT-PEX8 as template, according to the Quick Change Site-Directed Mutagenesis instruction manual (Stratagene, La Jolla, CA). Interactions were tested by growth on plates lacking histidine but containing 50–100 mM 3-aminotriazole (3-AT) to eliminate background growth.
Fluorescence Microscopy
The fragment of PPex8-GFP-PEX8 was amplified with oligonucleotides PPex8-F/P8-RSph from plasmid pSS060 (a gift from Jay Sunga, Keck Graduate Institute, Claremont, CA) using PCR, and cloned into KpnI- and SphI-digested vector pIB1 to create pLZ119 (Table 1). GFP-PEX8ΔAKL was amplified with oligonucleotides P8ATG/P8-R1Sph from plasmid pSS060, and cloned into BglII- and SphI-digested plasmid pLZ119 to create pLZ120. Plasmids were linearized with SalI and inserted into the HIS locus of wild-type or pex mutant strains.
The construct pLZ127 containing PPEX3-PEX3-mRFP was modified from plasmid pJCF235 (a gift from Dr. J. C. Farré in this laboratory) by inserting a fragment containing the zeoR gene in the SmaI site in the upstream region of PEX3 promoter. The plasmid was linearized with NruI and inserted into the ARG4 locus of wild-type or pex mutant strains.
To mutate the putative PTS2 in PpPex8p, a construct pLZ125 was generated, which expresses GFP-Pex8p-PTS2m (i.e., GFP-Pex8p-K376E, I377E). A fragment was excised from pLZ119 (with BglII, SphI) and cloned into pUC18 (between the BamHI and SphI sites). Site-directed mutagenesis was performed with oligonucleotides P8PTS2m-s/P8PTS2m-as. The DNA fragment containing the mutations was then excised (with PstI and SphI) and cloned back into the digested pLZ119 (with PstI, SphI). The construct, pLZ125, was confirmed by sequencing.
Strains expressing both GFP-Pex8p and Pex3p-mRFP fusion proteins were induced in synthetic medium (YNM) for 4 h. After harvesting, cells were resuspended in SD medium and observed under a fluorescence microscope (Axioskop 2 Mot, Zeiss, Thornwood, NY). Images were captured using an AxioCam MR camera (Zeiss) and analyzed using AxioVision 4 software.
RESULTS
PpPex8p Targeting to the Peroxisome Matrix Occurs via Both the PTS1 and PTS2 Pathways
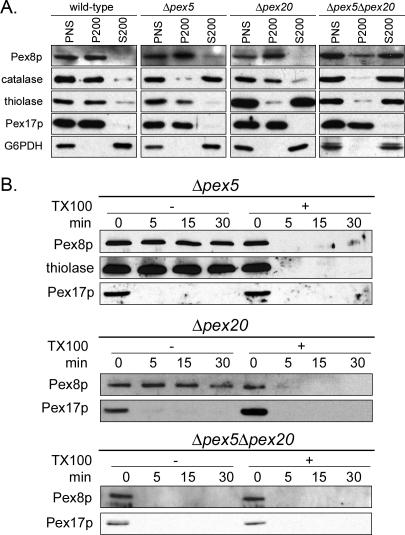
We analyzed the targeting of PpPex8p to the peroxisome matrix using subcellular fractionation and fluorescence microscopy (Figures 1 and 2). We first confirmed previous reports (Waterham et al., 1994; Liu et al., 1995; Rehling et al., 2000; Smith and Rachubinski, 2001) that Pex8p is predominantly a peroxisomal protein in wild-type (PPY12) cells, where PpPex8p was present solely in the organelle pellet (P200) fraction in differential centrifugation experiments (Figure 1A). The peroxisomal markers catalase, thiolase and Pex17p were predominantly in the pellet fraction, whereas the cytosolic marker, G6PDH, was in the S200 fraction, as expected. In both Δpex5 and Δpex20 cells, PpPex8p was predominantly in the organelle pellet (P200) fraction (Figure 1A). As expected, the Δpex5 mutant, defective specifically in the PTS1 import pathway, mislocalized catalase (a PTS1 protein) to the cytosol (S200 fraction), but not thiolase (a PTS2 protein), which was mostly in the organelle pellet (P200) fraction (Figure 1A; McCollum et al., 1993). Conversely, the Δpex20 mutant, compromised for the targeting of only PTS2 proteins, mislocalized thiolase to the cytosol, but not catalase (Figure 1A). However, in Δpex5Δpex20 cells, both catalase and thiolase were cytosolic, as expected; however, the PpPex8p was mostly cytosolic (S200 fraction), suggesting its mistargeting (Figure 1A). In these single and double mutants, the peroxisomal membrane and cytosolic markers, Pex17p and G6PDH, respectively, were in the pellet and supernatant fractions, as expected (Figure 1A).
Figure 1.
Targeting of PpPex8p to peroxisomes is impaired only in cells deficient in both PTS1 and PTS2 import pathways. (A) Oleate-grown wild-type (PPY12), Δpex5, Δpex20, and Δpex5Δpex20 P. pastoris cells were spheroplasted, lysed, and subjected to differential centrifugation. The postnuclear supernatants (PNS) were centrifuged at 200,000 × g. Equivalent fractions of PNS, 200,000 × g pellet (P200), and 200,000 × g supernatant (S200) were separated by SDS-PAGE and immunoblotted with antibodies to the indicated proteins. (B) Protease protection analysis of P200 fractions of oleate-grown Δpex5, Δpex20, and Δpex5Δpex20 cells. The organelle pellet was incubated at room temperature with 20 μg Proteinase K for the indicated times, in the presence (+) or absence (-) of Triton X-100. Proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
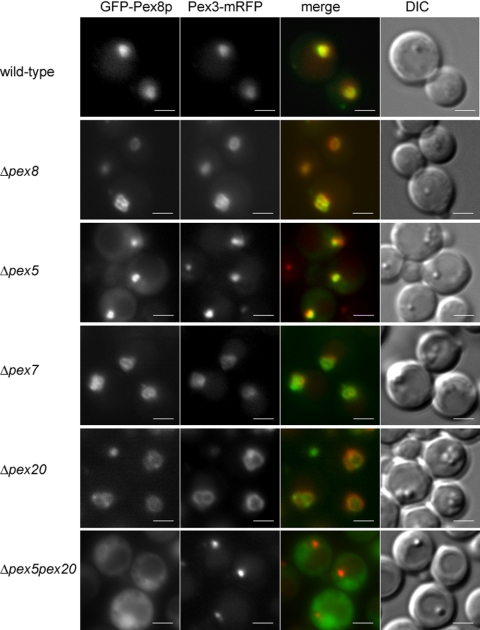
Figure 2.
Cytosolic mislocalization of PpPex8p in cells deficient in both PTS1 and PTS2 import pathways. Wild-type (PPY12) and pex mutant strains expressing both functional GFP-Pex8p and Pex3p-mRFP fusion proteins were grown in synthetic medium (YNM) for 4 h and visualized by fluorescence microscopy and Nomarski optics. Scale bar, 2 μm.
Further evidence for the intraperoxisomal location of PpPex8p in the Δpex5 and Δpex20 cells came from protease-protection experiments performed with the P200 organelle pellet fraction of oleate-grown cells (Figure 1B). PpPex8p was resistant to added protease in both mutants, whereas a peroxisomal membrane protein, Pex17p, was susceptible. However, upon addition of detergent and protease, the PpPex8p was degraded, showing that sufficient protease had been added. The matrix protein, thiolase, was protected from protease action by the peroxisome membrane in the Δpex5 cells. In the Δpex5Δpex20 cells, however, the PpPex8p associated with the organelle pellet (Figure 1A) was completely susceptible to protease, even in the absence of added detergent (Figure 1B). These experiments demonstrate that PpPex8p is still targeted to the peroxisome matrix in cells affected for only the PTS1 or PTS2 import pathways, but importantly, the protein does not reach its normal intraperoxisomal destination in cells lacking both import pathways.
In addition to differential centrifugation and protease protection, we also used fluorescence microscopy to examine the targeting of GFP-Pex8p to peroxisomes. This fusion protein was targeted to peroxisomes in wild-type and Δpex8 cells by virtue of its colocalization with the peroxisomal membrane marker, Pex3p-mRFP (Figure 2). It was also fully functional in its ability to complement the mutant strain for growth on methanol (unpublished data). GFP-Pex8p was also targeted to punctate structures in methanol-grown Δpex5, Δpex7, and Δpex20 cells (Figure 2). However, as suggested by the data in Figure 1, GFP-Pex8p was cytosolic and failed to colocalize with peroxisome remnants, in Δpex5 Δpex20 cells (Figure 2).
One Pathway for PpPex8p Import Requires its PTS1 and the Second Pathway Requires Both Pex7p and Pex20p
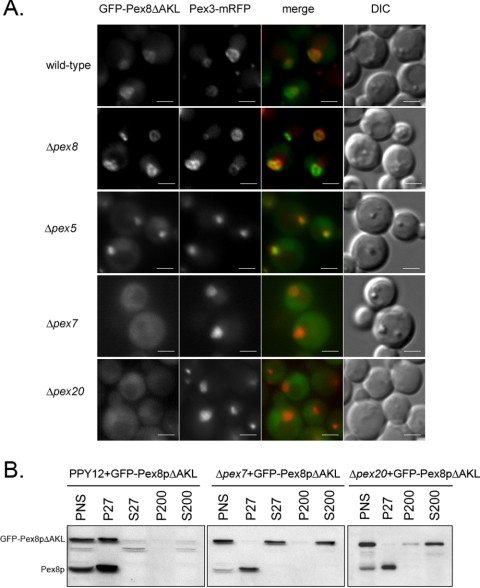
The presence of a C-terminal PTS1 (AKL) in PpPex8p made it an obvious basis for its peroxisomal targeting by the Pex5p-dependent pathway. However, previous reports have concluded that the PTS1 sequence plays no role in the targeting or function of Pex8p in other organisms (Waterham et al., 1994; Rehling et al., 2000). When we analyzed the targeting of GFP-Pex8pΔAKL, which lacked the last three amino acids of PpPex8p, we found that the fusion protein was peroxisomal (Figure 3A, wild-type and Δpex8 panels) and functional in its ability to complement the Δpex8 strain (unpublished data). Because the peroxisomal targeting of PpPex8p relies on two independent and redundant pathways (Figures 1 and 2), we reasoned that the targeting role of the PTS1 sequence might have been masked by presence of another pathway relying on the PTS2-targeting machinery. Indeed, the importance of the PTS1 of PpPex8p for its targeting via the Pex5p-dependent pathway was apparent in Δpex20 cells, in which the GFP-Pex8p fusion (containing the PTS1) was peroxisomal (Figure 2), but the GFP-Pex8pΔAKL fusion was not (Figure 3A). This result clearly shows that in the absence of Pex20p, a component of the PTS2 import pathway, the peroxisomal targeting of PpPex8p was completely dependent on its PTS1.
Figure 3.
GFP-Pex8pΔAKL targeting to peroxisomes is dependent on both Pex7p and Pex20p. (A) GFP-Pex8pΔAKL and Pex3p-mRFP were expressed in wild-type and pex mutant strains. Cells grown in synthetic medium (YNM) for 4 h were visualized by fluorescence microscopy and Nomarski optics. (B) Differential centrifugation analysis of peroxisomal targeting of the GFP-Pex8pΔAKL fusion in wild-type, Δpex7, and Δpex20 cells using the anti-Pex8p antibody.
Interestingly, when the Pex5p-dependent targeting of PpPex8p was inactivated by deletion of its PTS1 sequence, GFP-Pex8pΔAKL was still targeted to peroxisomes in Δpex5 and Δpex8 cells, but was cytosolic in Δpex7 and Δpex20 cells (Figure 3A). Additionally, differential centrifugation experiments confirmed that although GFP-Pex8pΔAKL was in the organelle pellet (P27) fraction in wild-type (PPY12) cells, it was cytosolic (S27 and/or S200 fractions) in both Δpex7 and Δpex20 cells (Figure 3B), whereas endogenous PpPex8p was correctly targeted to peroxisomes, and was in the P27 fraction. Thus, the alternative targeting pathway for PpPex8p requires both Pex7p, the PTS2 receptor, and its accessory protein, Pex20p.
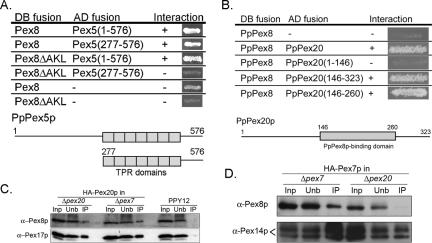
The PTS1 in PpPex8p Interacts with Pex5p
If the C-terminal AKL sequence of PpPex8p functions as a PTS1 sequence in vivo, it should interact with the tetratricopeptide repeat (TPR) domains of Pex5p, the PTS1 receptor (Terlecky et al., 1995). Both full-length Pex5p and its C-terminal TPR domains interacted with full-length PpPex8p, but the TPR domains of Pex5p failed to interact with Pex8pΔAKL, consistent with the binding of the C-terminal AKL of PpPex8p to the TPR motifs in Pex5p (Figure 4A). These results are similar to those described for ScPex8p (Rehling et al., 2000). The Pex8pΔAKL also interacted with full-length Pex5p because the latter protein has a second binding site for PpPex8p that is independent of the PTS1. This additional binding site in Pex5p is not relevant for PpPex8p targeting (Figure 3A), but is a useful control showing that the Pex8pΔAKL construct was capable of protein-protein interactions.
Figure 4.
Protein-protein interactions of PpPex8p with Pex5p, Pex20p, and Pex7p. (A) Interaction of the PTS1 of PpPex8p with TPR domains of Pex5p. Numbers refer to amino acid residues from Pex5p. Diagram shows domain map of Pex5p. Boxes represent TPR domains. (B) PpPex8p interaction with Pex20p. The domain of Pex20p that interacts with PpPex8p is shown. AH109 (S. cerevisiae) cells cotransformed with genes encoding Gal4p DNA-binding domain (DB, first column) and activation-domain (AD, second column) fusion proteins were tested for activation of HIS3 gene, which results in the growth (+) on medium without histidine (third column). (C) Immunoprecipitation of HA-Pex20p from wild type, Δpex7, and Δpex20 strains followed by immunoblotting with antibodies to indicated proteins. (D) Immunoprecipitation of HA-tagged Pex7p from Δpex7 or Δpex20 cells using a monoclonal HA antibody. The immunoprecipitates were immunoblotted as shown.
PpPex8p Interacts with Pex20p Independently of Pex7p, and Interaction between PpPex8p and Pex7p Requires Pex20p
Pex20p facilitates the targeting of PpPex8p by an interaction between these two proteins that was detectable both by the yeast two-hybrid method and by coimmunoprecipitation (Figure 4, B and C). A two-hybrid interaction was observed both with full-length Pex20p and the truncated Pex20p(146–323) and Pex20p(146–260) constructs (Figure 4B). The interaction of Pex20p with PpPex8p may be direct, or at least independent of the presence of Pex7p, because the amount of Pex20p that coimmunoprecipitated with PpPex8p was unchanged in the presence or absence of Pex7p (Figure 4C). Mapping of the Pex20p-binding region in PpPex8p failed because most of the PpPex8p fragments used did not interact with Pex20p (unpublished data). However, as shown later, a mutation of the putative PTS2 sequence of PpPex8p abolished its interaction with Pex20p. These data are consistent with a direct interaction between PpPex8p and Pex20p analogous to that described in Y. lipolytica (Smith and Rachubinski, 2001).
In view of the Pex7p-independent interaction between PpPex8p and Pex20p, the dependence of the PTS2 import pathway for PpPex8p on Pex7p was surprising and warranted further investigation of interactions between these proteins. Pex7p and PpPex8p could be coimmunoprecipitated (Figure 4D), but their interaction was strictly dependent on the presence of Pex20p. No interaction was detected between Pex7p and Pex8p in the yeast two-hybrid system (unpublished data) or in previous studies with their counterparts in S. cerevisiae (Rehling et al., 2000). These results suggest that there might be a complex containing PpPex8p-Pex20p-Pex7p that is essential for the proper delivery of PpPex8p into the peroxisomal matrix. Analogous to this is the observation that thiolase translocation also requires both Pex20p and Pex7p (Léon et al., 2006).
Mutation of a Putative PTS2 Sequence in PpPex8p Abolishes Its Import via the Pex7p- and Pex20p-dependent Pathway and Makes Its Import Pex5p-dependent
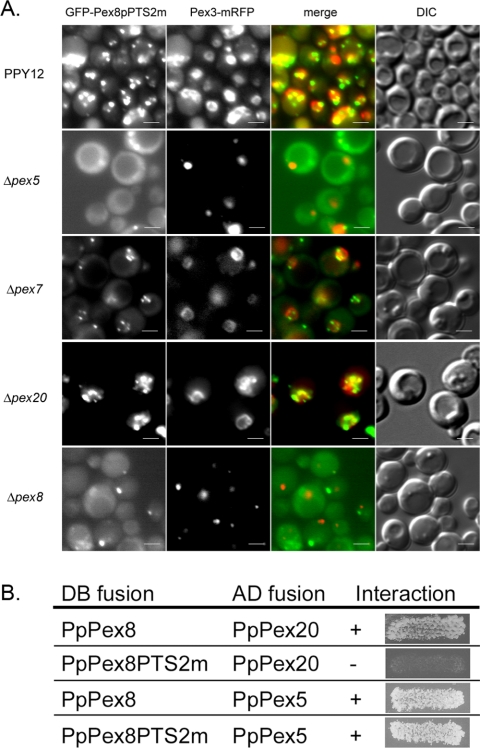
Despite our inability to detect a direct interaction between Pex7p and Pex8p, we were intrigued by the presence of a putative PTS2 sequence, KILSILFNL, between amino acids 376–384 in PpPex8p. This fits the newly defined PTS2 consensus (Petriv et al., 2004). This sequence was mutated to EELSILFNL (PTS2m, with the K376E I377E mutations) in GFP-Pex8p-PTS2m, and the effect of these mutations on the targeting of this fusion protein was analyzed. It localized to punctate structures in wild-type (PPY12) cells and colocalized with the peroxisomal marker, Pex3p-mRFP (Figure 5A). This peroxisomal targeting of GFP-Pex8p-PTS2m in wild-type cells was most likely mediated by the PTS1 on the protein because in the absence of Pex5p, the fusion protein was cytosolic (Figure 5A), a result that was confirmed further by the cytosolic location of GFP-Pex8pΔAKL-PTS2m in wild-type cells (Supplementary Figure S1). Not surprisingly, GFP-Pex8p-PTS2m colocalized with peroxisomes in Δpex7 and Δpex20 cells because it uses the PTS1 pathway (Figure 5A). These results demonstrate that the PTS2-motif on PpPex8p is necessary for its peroxisomal targeting via the Pex7p and Pex20p-dependent pathway, when the PTS1 import pathway for PpPex8p is inactivated either by deletion of its C-terminal AKL sequence (Supplementary Figure S1) or by the absence of Pex5p in the cells (Figure 5A).
Figure 5.
Peroxisomal targeting of GFP-Pex8p-PTS2m and protein-protein interactions of Pex8p-PTS2m with Pex5p and Pex20p. (A) Targeting of GFP-Pex8p-PTS2m in wild-type and pex mutants of P. pastoris. The localization of the PpPex8p fusion and its colocalization with Pex3p-mRFP was monitored by fluorescence microscopy and Nomarski optics. (B) Interactions between Pex8p-PTS2m and Pex5p or Pex20p were detected by yeast two-hybrid analysis.
Interestingly, mutation of the putative PTS2 motif in PpPex8p (PpPex8-PTS2m) abolished its interaction with Pex20p (Figure 5B). As a control for the proper expression and stability of this construct, it exhibited a robust interaction with Pex5p (via its PTS1), as expected. This is the first clear evidence that the PTS2 motif on PpPex8p is necessary for its peroxisomal import in the context of the full-length protein. As noted earlier, previous studies had only demonstrated that protein fragments containing the putative PTS2 of HpPex8p and ScPex8p were sufficient for peroxisomal targeting of a passenger protein, but the key necessity test could not be demonstrated (Waterham et al., 1994; Rehling et al., 2000).
Dependence of the Two PpPex8p Import Pathways on Intraperoxisomal Pex8p
GFP-Pex8pΔAKL, which uses the PTS2 import pathway for PpPex8p targeting, is peroxisomal in both wild-type and in Δpex8 cells (Figure 3A). In fact, this fusion complements the Δpex8 cells for Pex8p function and the cells have normal peroxisomes. This important result shows that the entry of Pex8p into peroxisomes using its PTS2-dependent pathway does not require the prior presence of PpPex8p within peroxisomes. However, GFP-Pex8p-PTS2m, which can only be imported into peroxisomes via its PTS1 signal, was only targeted to peroxisomes of cells that already have intraperoxisomal Pex8p (wild-type, Δpex7, and Δpex20 cells in Figure 5A), but not into peroxisomes of Δpex8 cells, as judged by the lack of colocalization of the GFP fluorescence with Pex3p-mRFP (Figure 5A). Therefore, the PTS2 pathway for PpPex8p import into peroxisomes does not require the prior presence of functional intraperoxisomal PpPex8p, but the PTS1 pathway does.
Translocation of PpPex8p into Peroxisomes Is Pex14p-dependent and Pex2p-independent
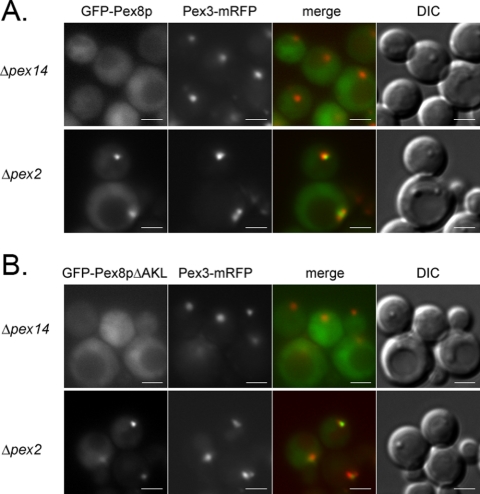
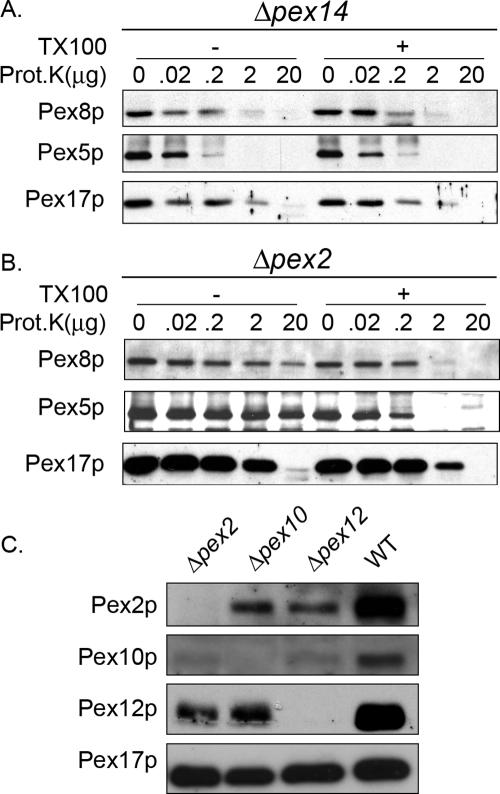
Two distinct multiprotein subcomplexes comprised of the docking and RING peroxins exist in the peroxisomal membrane and these also assemble into a joint complex called the importomer (Hazra et al., 2002; Agne et al., 2003). However, each of these subcomplexes has been referred to as the translocon in the peroxisome membrane, leading to uncertainty regarding the precise roles of these subcomplexes (Chang et al., 1999; Sacksteder and Gould, 2000; Holroyd and Erdmann, 2001; Purdue and Lazarow, 2001). We used the intraperoxisomal targeting of PpPex8p to ask whether a key component of one or both of these subcomplexes is necessary for its transport into the matrix. In cells lacking a central component, Pex14p, of the docking subcomplex, targeting of both GFP-Pex8p and GFP-Pex8pΔAKL to peroxisomes was abolished (Figure 6, A and B). Furthermore, the organelle-associated PpPex8p was completely protease sensitive, as was Pex5p (Figure 7A; see lane with 20 μg proteinase K). This protease sensitivity of PpPex8p and Pex5p in Δpex14 cells was true whether the analysis was performed with 20 μg proteinase K over a period of 30 min (Supplementary Figure S2) or when varying amounts of proteinase K were used for 15 min (Figure 7A; see lane with 20 μg proteinase K). In Δpex2 cells, lacking a key component of the RING subcomplex, in marked contrast, both GFP-Pex8p and GFP-Pex8pΔAKL were targeted to punctate peroxisome remnants that contained Pex3p-mRFP (Figure 6, A and B). In this mutant, both PpPex8p and Pex5p were protease resistant (Figure 7B; see lane with 20 μg proteinase K), under conditions where Pex17p was protease sensitive. In cells lacking Pex2p, the other components of the RING subcomplex, Pex10p and Pex12p, are either absent or present in reduced amounts (Figure 7C), making it likely that little, if any, of the complete RING subcomplex is stable in Δpex2 cells (Hazra et al., 2002). These data demonstrate that the targeting of PpPex8p requires an intact docking, but not a complete RING, subcomplex. Pex5p was also protease protected in Δpex2 cells, strongly supporting the view that Pex14p, but not Pex2p, is necessary for the peroxisomal entry of PpPex8p and Pex5p.
Figure 6.
Location of GFP-Pex8p and GFP-Pex8pΔAKL in Δpex14 and Δpex2 cells. GFP-Pex8p (A) and GFP-Pex8pΔAKL (B) were detected by fluorescence microscopy in cells grown in YNM. Peroxisome remnants were identified using Pex3p-mRFP. Scale bar, 2 μm.
Figure 7.
Pex14p, but not Pex2p, is required for the peroxisomal entry of PpPex8p. Protease protection analysis of the P200 fraction (obtained directly from the PNS) of oleate-grown cells. Δpex14 (A) and Δpex2 (B) strains. The P200 fraction was incubated at room temperature with the indicated amounts of Proteinase K for 15 min, in the presence (+) or absence (-) of Triton X-100. Samples were subjected to SDS-PAGE and immunoblotting. (C) Cell lysates of oleate-grown wild-type, Δpex2, Δpex10, and Δpex12 cells were immunoblotted with the indicated antibodies.
DISCUSSION
Two Redundant Pathways for PpPex8p Targeting to the Peroxisome Matrix
Although previous studies had noted that Pex8p of H. polymorpha, P. pastoris, and S. cerevisiae have a conserved C-terminal PTS1, as well as a putative PTS2 in HpPex8p, deletion of either sequence in Pex8p from several yeast species did not affect either its peroxisomal targeting or its function in peroxisome biogenesis (Waterham et al., 1994; Rehling et al., 2000). Similarly, studies with Y. lipolytica concluded that Pex8p-HA targeting to peroxisomes was unaffected in Δpex20 cells (Smith and Rachubinski, 2001). These studies suggested that neither the PTS1 or PTS2 of Pex8p, nor Pex20p, were involved in the targeting of Pex8p to peroxisomes (Waterham et al., 1994; Rehling et al., 2000; Smith and Rachubinski, 2001). However, our data show that in Δpex7 and in Δpex20 cells, both Pex5p and the PTS1 of PpPex8p are critical for its peroxisomal targeting (Figures 2, 3A, 5A, and 8). In cells lacking the PTS1 on PpPex8p, its targeting is dependent on both Pex20p and Pex7p (Figure 3A). Conversely, in cells lacking the PTS2 motif in PpPex8p, its targeting requires Pex5p and the PTS1 in PpPex8p (Figures 5 and 8 and Supplementary Figure S1). Both Pex7p and Pex20p have been described to bind PTS2 motifs in vitro (Rehling et al., 1996; Otzen et al., 2005).
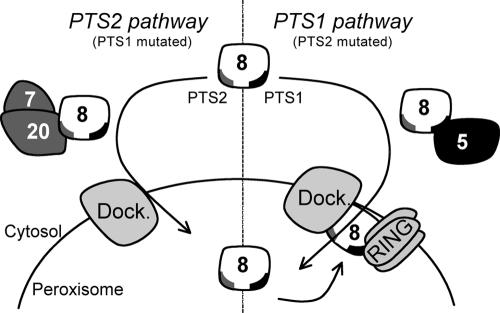
Figure 8.
Working model for the targeting of P. pastoris Pex8p to the peroxisomal matrix. PpPex8p is targeted to peroxisomes by two independent pathways, involving either its PTS1 or its PTS2 (Figures 1, 2, 3 and 5). The PTS1 targeting pathway requires the PTS1 signal in PpPex8p, which can interact with the TPR repeats of Pex5p and therefore suggests a cargo/receptor-like interaction (Figure 4). Targeting by the PTS2 pathway involves the auxiliary protein, Pex20p, and the PTS2 receptor, Pex7p (Figure 3A). An intact PTS2-like motif in PpPex8p is required for interaction with Pex20p, but PpPex8p interacts with Pex7p only in the presence of Pex20p (Figures 4 and 5). Deletion of the PTS1 tripeptide does not affect the targeting of PpPex8p by the PTS2 pathway (Figure 3A). Furthermore, peroxisomal import of PpPex8p via the PTS2 pathway is independent of the RING subcomplex (Figure 6B), as well as intraperoxisomal PpPex8p (Figure 3A), which bridges the docking and RING subcomplexes in the importomer (Agne et al., 2003). Mutations in the PTS2 sequence do not affect PpPex8p targeting as long as its PTS1 sequence, intraperoxisomal PpPex8p and Pex5p are present, suggesting the requirement of the full importomer for PTS1-dependent import of PpPex8p, as would be expected for other PTS1 cargo. Dock denotes the docking subcomplex and RING represents the RING subcomplex.
We also uncovered new functions for Pex20p and Pex7p in PpPex8p translocation into peroxisomes. PpPex8p likely interacts with Pex20p directly (Figure 4, B and D; Smith and Rachubinski, 2001), and Pex20p interacts with Pex7p (Léon et al., 2006), but no stable interaction was detected between Pex7p and PpPex8p directly (Figure 4D; Rehling et al., 2000). Our working hypothesis is that Pex8p/Pex20p behave as a receptor/cargo pair whose entry into peroxisomes depends on Pex7p. Pex20p enters the peroxisome matrix (Léon et al., 2006) and so does Pex7p during its extended shuttle mechanism of cycling between the cytosol and the peroxisome matrix (Nair et al., 2004).
Evolution of the Dependence of Peroxisomal Matrix Protein Import on Pex8p and Coevolution of Pex8p and Pex20p-like Proteins
The redundant import pathways for Pex8p (summarized in Figure 8) raise the important question regarding why this protein evolved dual pathways. If PpPex8p enters peroxisomes using the PTS1 and PTS2 import pathways, does it behave exactly like other cargo in requiring the prior presence of intraperoxisomal PpPex8p for its own import? If so, how did PpPex8p first enter peroxisomes? Our data demonstrate that GFP-Pex8pΔAKL, which uses the PTS2 import pathway for PpPex8p targeting, is peroxisomal in both wild-type and in Δpex8 cells (Figure 3A). However, GFP-Pex8p-PTS2m, which can only be imported into peroxisomes via its PTS1 signal, is only targeted to peroxisomes of cells that already have functional intraperoxisomal PpPex8p (wild-type, Δpex7, and Δpex20 cells in Figure 5A), but not into peroxisomes of Δpex8 cells (Figure 5A). Therefore, the PTS2 pathway for PpPex8p import into peroxisomes does not appear to require the prior presence of functional intraperoxisomal Pex8p, but the PTS1 pathway does (Figure 8). One concern with respect to this interpretation is whether the targeting and/or function of GFP-Pex8p-PTS2m may have been compromised. This fusion is targeted to peroxisomes (using its PTS1 sequence) in wild-type, Δpex7, and Δpex20 cells, and it also interacts with Pex5p (Figure 5B), but it is impossible at present to address its ability to support import. The dependence of the PTS1-import pathway for PpPex8p on intraperoxisomal PpPex8p is reminiscent of the behavior of other PTS1 cargoes. However, the lack of reliance of the import of PpPex8p, via the PTS2 pathway, on intraperoxisomal PpPex8p is surprising and suggests that PpPex8p does not behave like other PTS2 cargoes in this respect, as well as the fact that it does not interact with Pex7p directly. Interestingly, Pex20p, an essential component of the PTS2 import pathway, also does not require intraperoxisomal Pex8p for its accumulation within peroxisomes (Smith and Rachubinski, 2001).
The use of two redundant peroxisomal import pathways for PpPex8p may have aided the evolution of a system in which PpPex8p import became dependent on the prior presence of PpPex8p in the matrix. During evolution, Pex8p may have needed Pex20p and Pex7p to first enter peroxisomes without requiring Pex8p to be present inside the organelle. It may have then evolved a redundant pathway, either as a backup or for an increased efficiency of Pex8p import. YlPex8p does not have a C-terminal PTS1, but it interacts directly with Pex20p (Smith and Rachubinski, 2001), suggesting that, like PpPex8p, it might also use the PTS2 pathway for peroxisomal targeting. However, apparently contradicting this, Yl-Pex8P-HA was found in the organelle pellet fraction in Y. lipolytica pex20Δ cells (Smith and Rachubinski, 2001). One explanation for this result is that protease protection analysis was not done to confirm that YlPex8p was located in the peroxisome matrix, leaving open the possibility that it was on the cytosolic side of the peroxisomal membrane. A more likely alternative is that YlPex8p may also have an alternative redundant targeting pathway that is not dependent on the presence of a C-terminal PTS1 in YlPex8p. There is precedence for the existence of such PTS1-independent, but Pex5p-dependent, peroxisomal matrix import pathways (Klein et al., 2002; Gunkel et al., 2004). The existence and nature of such an alternative Pex20p-independent peroxisomal import pathway for YlPex8p remains to be confirmed.
Supporting the idea of a conserved Pex20p-dependent peroxisomal import pathway for Pex8p targeting to the peroxisomes matrix is the interesting fact that Pex8p is found only in fungi, but every organism with the PEX8 gene also has a PEX20-like gene. These two sets of genes appear to have coevolved. Mammals dispensed with the PEX20 gene by incorporating its PTS2-pathway function into an alternative exon in the PEX5 gene (Dodt et al., 2001). As a consequence, organisms such as plants and mammals that use the long (Pex5L) isoform of Pex5 to perform Pex20p-like functions, also lack a PEX8 gene. The presence of multiple targeting pathways may also allow organisms to increase the efficiency of peroxisomal targeting in certain metabolic states as illustrated by the dependence of peroxisomal targeting of ScPex8p on its PTS1 sequence in basal, but not in, proliferated peroxisomes (Wang et al., 2004).
Pex2p and the Intact Importomer Are Not Necessary for Entry of PpPex8p into Peroxisomes
The Pex14p-dependent, but Pex2p-independent, mechanism of entry of PpPex8p into peroxisomes provides a simple answer to another chicken-and-egg paradox. How does Pex8p enter peroxisomes under conditions wherein the assembly of the importomer itself might be severely compromised because of the absence of a key component or by the absence of pre-existing, intraperoxisomal Pex8p, which is supposed to organize the importomer (Agne et al., 2003)? Pex2p is a key component of the RING subcomplex and the importomer (Hazra et al., 2002; Agne et al., 2003). In the absence of Pex2p, this (Figure 7C) and previous studies (Hazra et al., 2002; Agne et al., 2003) show that Pex10p and Pex12p are either absent or unstable, making it unlikely that the RING subcomplex or the complete importomer are necessary for PpPex8p import into peroxisomes. Previous reports have suggested that the RING subcomplex is the translocon in the peroxisome membrane (Chang et al., 1999; Sacksteder and Gould, 2000; Holroyd and Erdmann, 2001; Purdue and Lazarow, 2001), but this clearly could not apply to PpPex8p or Pex5p. We cannot address unambiguously whether one or both import pathways for PpPex8p is independent of the complete importomer. However, it seems more likely that the PTS2, and perhaps not the PTS1, import pathway for PpPex8p is the one that is independent of the RING subcomplex (Figure 8). Support for this view comes from our finding that peroxisomal import of PpPex8p via the PTS2 pathway, and of Pex20p (Smith and Rachubinski, 2001), does not need intraperoxisomal PpPex8p (Figure 3A), which is necessary to bridge the docking and RING subcomplexes. Additionally, the import of Pex8pΔAKL (Figure 6B) and of PpPex20p into peroxisomes (Léon et al., 2006) does not require the RING subcomplex. PTS1-dependent import of PpPex8p has similarities to other PTS1 cargo and likely requires the full importomer.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DK41737 and DK59844 to S.S. We thank Dr. R. Tsien (UCSD) for the mRFP construct.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0758) on November 30, 2005.
Abbreviations used: 3-AT, 3-aminotriazole; G6PDH, glucose-6-phosphate dehydrogenase; PTS, peroxisomal targeting signal; TPR, tetratricopeptide repeat; UTR, untranslated region.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Agne, B., Meindl, N. M., Niederhoff, K., Einwachter, H., Rehling, P., Sickmann, A., Meyer, H. E., Girzalsky, W., and Kunau, W. H. (2003). Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635-646. [DOI] [PubMed] [Google Scholar]
- Chang, C. C., Warren, D. S., Sacksteder, K. A., and Gould, S. J. (1999). PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J. Cell Biol. 147, 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai, V., and Subramani, S. (2001). The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105, 187-196. [DOI] [PubMed] [Google Scholar]
- Dodt, G., Warren, D., Becker, E., Rehling, P., and Gould, S. J. (2001). Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J. Biol. Chem. 276, 41769-41781. [DOI] [PubMed] [Google Scholar]
- Elgersma, Y., Elgersma-Hooisma, M., Wenzel, T., McCaffery, J. M., Farquhar, M. G., and Subramani, S. (1998). A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J. Cell Biol. 140, 807-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, K. N., Heyman, J. A., and Subramani, S. (1998). Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 18, 936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. J., McCollum, D., Spong, A. P., Heyman, J. A., and Subramani, S. (1992). Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast 8, 613-628. [DOI] [PubMed] [Google Scholar]
- Gunkel, K., van Dijk, R., Veenhuis, M., and van der Klei, I. J. (2004). Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery. Mol. Biol. Cell 15, 1347-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra, P. P., Suriapranata, I., Snyder, W. B., and Subramani, S. (2002). Peroxisome remnants in pex3Δ cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3, 560-574. [DOI] [PubMed] [Google Scholar]
- Holroyd, C., and Erdmann, R. (2001). Protein translocation machineries of peroxisomes. FEBS Lett. 501, 6-10. [DOI] [PubMed] [Google Scholar]
- Johnson, M. A., Snyder, W. B., Cereghino, J. L., Veenhuis, M., Subramani, S., and Cregg, J. M. (2001). Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast 18, 621-641. [DOI] [PubMed] [Google Scholar]
- Klein, A. T., van den Berg, M., Bottger, G., Tabak, H. F., and Distel, B. (2002). Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277, 25011-25019. [DOI] [PubMed] [Google Scholar]
- Lazarow, P. B. (2003). Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 15, 489-497. [DOI] [PubMed] [Google Scholar]
- Léon, S., Zhang, L., McDonald, W. H., Yates, J., III, Cregg, J. M., and Subramani, S. (2006). Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Liu, H., Tan, X., Russell, K. A., Veenhuis, M., and Cregg, J. M. (1995). PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J. Biol. Chem. 270, 10940-10951. [DOI] [PubMed] [Google Scholar]
- McCollum, D., Monosov, E., and Subramani, S. (1993). The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J. Cell Biol. 121, 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, D. M., Purdue, P. E., and Lazarow, P. B. (2004). Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 167, 599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen, M., Wang, D., Lunenborg, M. G., and van der Klei, I. J. (2005). Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2). J. Cell Sci. 118, 3409-3418. [DOI] [PubMed] [Google Scholar]
- Petriv, O. I., Tang, L., Titorenko, V. I., and Rachubinski, R. A. (2004). A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 341, 119-134. [DOI] [PubMed] [Google Scholar]
- Purdue, P. E., and Lazarow, P. B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17, 701-752. [DOI] [PubMed] [Google Scholar]
- Rehling, P., Marzioch, M., Niesen, F., Wittke, E., Veenhuis, M., and Kunau, W. H. (1996). The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 15, 2901-2913. [PMC free article] [PubMed] [Google Scholar]
- Rehling, P., Skaletz-Rorowski, A., Girzalsky, W., Voorn-Brouwer, T., Franse, M. M., Distel, B., Veenhuis, M., Kunau, W. H., and Erdmann, R. (2000). Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor Pex5p. J. Biol. Chem. 275, 3593-3602. [DOI] [PubMed] [Google Scholar]
- Sacksteder, K. A., and Gould, S. J. (2000). The genetics of peroxisome biogenesis. Annu. Rev. Genet. 34, 623-652. [DOI] [PubMed] [Google Scholar]
- Smith, J. J., and Rachubinski, R. A. (2001). A role for the peroxin Pex8p in Pex20p-dependent thiolase import into peroxisomes of the yeast Yarrowia lipolytica. J. Biol. Chem. 276, 1618-1625. [DOI] [PubMed] [Google Scholar]
- Snyder, W. B., Faber, K. N., Wenzel, T. J., Koller, A., Luers, G. H., Rangell, L., Keller, G. A., and Subramani, S. (1999). Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell 10, 1745-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky, S. R., Nuttley, W. M., McCollum, D., Sock, E., and Subramani, S. (1995). The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 14, 3627-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Visser, N. V., Veenhuis, M., and van der Klei, I. J. (2003). Physical interactions of the peroxisomal targeting signal 1 receptor Pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 278, 43340-43345. [DOI] [PubMed] [Google Scholar]
- Wang, X., McMahon, M. A., Shelton, S. N., Nampaisansuk, M., Ballard, J. L., and Goodman, J. M. (2004). Multiple targeting modules on peroxisomal proteins are not redundant: discrete functions of targeting signals within Pmp47 and Pex8p. Mol. Biol. Cell 15, 1702-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham, H. R., Titorenko, V. I., Haima, P., Cregg, J. M., Harder, W., and Veenhuis, M. (1994). The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 127, 737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.