Abstract
When higher eukaryotic cells transition into mitosis, the nuclear envelope, nuclear pore complexes, and nuclear lamina are coordinately disassembled. The COPI coatomer complex, which plays a major role in membrane remodeling at the Golgi, has been implicated in the process of nuclear envelope breakdown and requires interactions at the nuclear pore complex for recruitment to this new site of action at mitosis. Nup153, a resident of the nuclear pore basket, was found to be involved in COPI recruitment, but the molecular nature of the interface between COPI and the nuclear pore has not been fully elucidated. To better understand what occurs at the nuclear pore at this juncture, we have probed the role of the nucleoporin Nup358/RanBP2. Nup358 contains a repetitive zinc finger domain with overall organization similar to a region within Nup153 that is critical to COPI association, yet inspection of these two zinc finger domains reveals features that also clearly distinguish them. Here, we found that the Nup358 zinc finger domain, but not a zinc finger domain from an unrelated protein, binds to COPI and dominantly inhibits progression of nuclear envelope breakdown in an assay that robustly recapitulates this process in vitro. Moreover, the Nup358 zinc finger domain interferes with COPI recruitment to the nuclear rim. Consistent with a role for this pore protein in coordinating nuclear envelope breakdown, Nup358-specific antibodies impair nuclear disassembly. Significantly, targeting either Nup153 or Nup358 for inhibition perturbs nuclear envelope breakdown, supporting a model in which these nucleoporins play nonredundant roles, perhaps contributing to COPI recruitment platforms on both the nuclear and cytoplasmic faces of the pore. We found that an individual zinc finger is the minimal interface for COPI association, although tandem zinc fingers are optimal. These results provide new information about the critical components of nuclear membrane remodeling and lay the foundation for a better understanding of how this process is regulated.
INTRODUCTION
The nuclear envelope creates a barrier that is critical to maintenance of the environment in the nucleus, specialized to support transcription and DNA replication. This double lipid membrane bilayer consists of an outer nuclear membrane, which is continuous with the endoplasmic reticulum (ER), and an inner nuclear membrane, which contains at least 78 different proteins, anchored via protein-protein interactions to the underlying nuclear lamina and chromatin (Gruenbaum et al., 2005). The nuclear envelope is perforated by nuclear pores, through which communication between the cytoplasm and nucleus occurs (Suntharalingam and Wente, 2003; Weis, 2003; Fahrenkrog et al., 2004; Pante, 2004). Despite the large size of the macromolecular nuclear pore complex (125 MDa in vertebrates), it is comprised of only ∼30 different proteins, or nucleoporins (Nups), each present in multiple copies (Rout et al., 2000; Cronshaw et al., 2002). In metazoan cells, all these components—the lamina, nuclear pores, as well as the membranes—are disassembled at mitosis. These remodeling events allow microtubules of the mitotic spindle access to the condensed chromosomes and ultimately ensure accurate division of both genomic DNA and nuclear envelope components between daughter cells.
Some of the events that occur during breakdown of the nuclear envelope have been characterized, but many steps remain poorly understood. During the transition into mitosis, cdk1-cyclinB (MPF) is activated and accumulates in the nucleus. The cdk1-cyclinB complex phosphorylates nucleoporins, lamins, and inner nuclear membrane proteins (Margalit et al., 2005). Such phosphorylation events are thought to weaken protein-protein interactions, compromising the integrity of higher order structures associated with the nuclear envelope.
The mechanism by which the nuclear membrane itself disperses is controversial (Prunuske and Ullman, 2006). Tension and subsequent tearing of the nuclear envelope by a microtubule-dependent mechanism has been observed (Beaudouin et al., 2002; Salina et al., 2002). At later stages of mitosis, inner nuclear membrane proteins colocalize with ER proteins, demonstrating that ER and nuclear membranes have merged (Ellenberg et al., 1997; Yang et al., 1997). Whether this is due to lateral diffusion within the membrane or to vesicle formation and fusion is an open question. In certain cases, extensively exemplified by Xenopus eggs, integral membrane proteins of the nuclear membrane are found in distinct vesicles (Vigers and Lohka, 1991; Buendia and Courvalin, 1997; Drummond et al., 1999). Indeed, recent reports suggest that there are multiple populations of specialized vesicles (Antonin et al., 2005).
A mechanism for mitotic vesicle formation, whether as an intermediate or a storage pool, was suggested with the identification of the coatomer complex COPI as an important player in nuclear envelope breakdown (Liu et al., 2003). COPI has been characterized in the context of Golgi trafficking to promote vesicle formation and may play a similar role at the nuclear envelope at a restricted window of the cell cycle. Understanding how COPI is recruited to nuclear membranes is of central importance to elucidating this aspect of mitotic nuclear remodeling. The pore component Nup153 was found to be important in the recruitment of COPI to nuclear envelope for this mitotic role, but given the large network of interactions that coordinate the role of COPI at the Golgi, there is likely to be more machinery involved in mobilizing COPI for action at the nuclear envelope.
The interaction between COPI and Nup153 was mapped to a distinctive zinc finger domain within Nup153. A related region is present in one other vertebrate nucleoporin, Nup358/RanBP2. The Nup153 and Nup358 zinc finger domains share tandem repeats of zinc fingers with a X4WXCX2CX3NX6CX2CX5 consensus, characteristic of a broader class of zinc fingers (Wang et al., 2003). Yet, many residues in the nucleoporin zinc fingers are not conserved (see Figure 1A), which is significant because a small number of amino acids within a zinc finger scaffold can dictate specific partnerships (Alam et al., 2004). Moreover, the linker regions between these zinc fingers, which comprise ∼45% of what is referred to as the zinc finger domain, are even less well conserved than the zinc finger motifs themselves. Notably, Nup358 is situated on the cytoplasmic-facing filaments of the pore, whereas Nup153 is a component of the nuclear-facing basket structure. The contrasting localizations of these two pore proteins could be consistent with a need to carry out specialized functions or with a role in coordinating a process that occurs on both sides of the nuclear pore complex. Thus, we wanted to address whether Nup358 plays a role similar to Nup153 in nuclear envelope breakdown or whether the distinctions between these Nup domains reflect a divergence in their specific functions. Here, we have determined that Nup358 participates in nuclear envelope breakdown, and we have gained further insight into the interaction between COPI and the nucleoporin zinc finger module.
Figure 1.
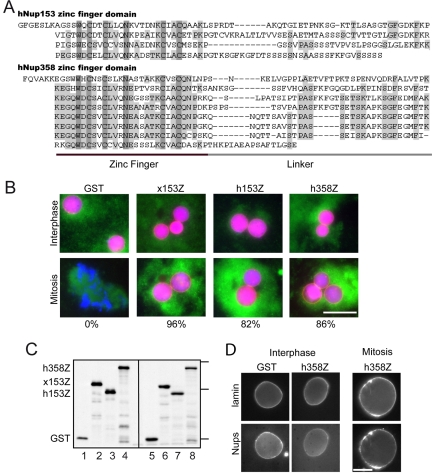
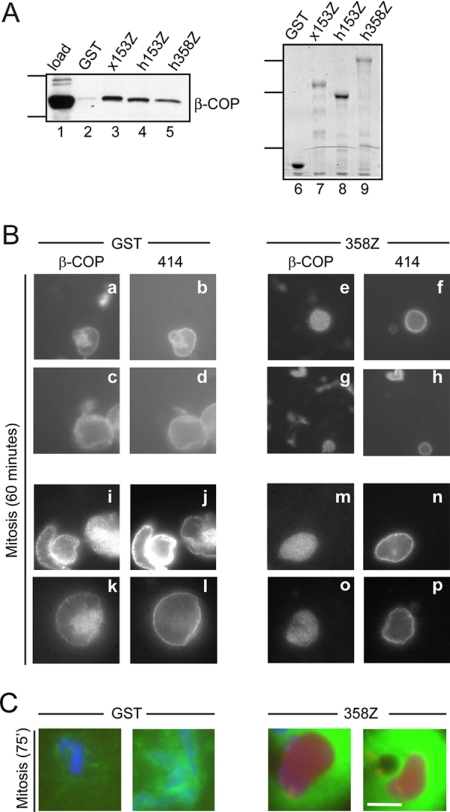
The zinc finger domain of Nup358 interferes with breakdown of the nuclear envelope and lamina. (A) Sequence alignment of the zinc finger domains from human Nup153 (h153Z) and Nup358 (h358Z). Darkly shaded residues are identical in all of the aligned sequences and lightly shaded residues are identical in more than half of the sequences. (B) Nuclei were assembled in the presence of the recombinant GST fusion proteins (1.7 μM final). The interphase samples were taken after 90 min of assembly, immediately before cyclin addition. The mitotic samples were taken 75 min after cyclin addition. Each picture is a merged image of DNA (Hoechst 33258 stain) shown in blue, NLS import (NLS-HSA-RITC) shown in red, and membrane (DHCC stain) shown in green. Numbers below the panels indicate the percentage of intact nuclei at the mitotic time point. Bar, 50 μm. (C) Immunoblot of recombinant proteins, GST, x153Z, h153Z, and h358Z before (lanes 1-4) and at the end (lanes 5-8) of the nuclei assembly/disassembly assay. Proteins were detected with antibodies directed against GST. Molecular-weight markers indicated correspond to 92, 52, and 28 kDa. (D) Recombinant protein fragments, GST (5 μg) and h358Z (5 μg, 1.7 μM), were added to egg extract 15 min before the assembly/disassembly assay. The interphase samples were taken 90 min after assembly, and the mitotic sample was taken 75 min after cyclin addition, when the nuclei containing the GST fragment were disassembled. Nuclei were processed for immunofluorescence to detect lamins and the 414-reactive nucleoporins: Nup358, Nup214, Nup153, and Nup62. Bar, 20 μm.
MATERIALS AND METHODS
Sequence Alignment
Nup358 and Nup153 sequences were aligned using the Vector NTI Align program.
Recombinant Protein Production and Purification
The hNup153 zinc finger domain (amino acids [aa] 651-912) was amplified from a pET28 plasmid containing full-length human Nup153 (Dimaano et al., 2001) and subcloned into pGEX-4T (Amersham, Piscataway, NJ) using BamHI/SmaI. The hNup358 zinc finger domain (aa 1346-1826) was amplified from hNup358 and subcloned into pGEX using BamHI/SmaI. The single human Nup153 zinc finger (aa 723-750) was a gift from Wes Sundquist. The glutathione-S-transferase (GST)-x153Z construct (aa 655-926) was a gift from Sundeep Shah and Douglass Forbes. The zyxin LIM domain (encompassing aa 349-542 from chicken zyxin) was a gift from Mary Beckerle. All constructs in pGEX vectors were induced with 1 mM isopropylthio-β-d-galactoside (IPTG) for 3 h at 37°C. Bacteria were lysed in the presence of deoxycholate. The fusion proteins were purified using glutathione-Sepharose 4B resin (Amersham). Protein concentrations were determined with a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA).
In vitro nuclear disassembly assay and microscopy. Interphase Xenopus egg extracts were prepared as previously described (Liu et al., 2003). Crude extract containing 5% glycerol was stored in liquid nitrogen, and fractionated extract was frozen in liquid nitrogen and stored at -80°C. Condensed sperm chromatin was isolated from Xenopus testes and demembranated with Triton-X (Powers et al., 2001) or lysolecithin (Lohka, 1998). Egg extracts were shifted into mitosis by the addition of recombinant Δ90 cyclinB. The nuclei assembly/disassembly assay was performed as previously described (Liu et al., 2003) except in some cases sperm chromatin was preincubated with nucleoplasmin. Samples were monitored by fluorescence microscopy using import substrate (NLS-HSA-RITC), DHCC (Calbiochem, San Diego, CA) to monitor membranes and Hoechst 33258 (Calbiochem) to monitor DNA. Images were acquired on a Zeiss Axioskop 2 (Carl Zeiss, Thornwood, NY) using FV12, a 12-bit monochrome digital camera, and Olympus MicroSuite software (Melville, NY). For the zyxin experiment, images were captured with the AxioCam HRm (Zeiss) using Axiovision software (Zeiss).
To examine the effects of protein fragments and antibodies on nuclear disassembly, the reagents were incubated with crude extract for 15 min at room temperature before the addition of sperm chromatin. To quantitate disassembly, samples were taken at the interphase time point (immediately before cyclin addition) and mitotic time point (75 min after addition of cyclin). Intact nuclei in two aliquots of each sample were counted and averaged. The relative number of nuclei remaining in each sample was calculated by dividing the number of nuclei at mitosis by the number of nuclei at interphase and multiplying by 100.
To monitor import after cyclin addition, 0.5 μg purified Nup358-2 antibody, 1 μg Xenopus Nup153 antibody, or 2.5 μg preimmune antibody were preincubated with crude extract for 15 min and assembled for 90 min after the addition of DNA and energy mix. Then both cyclin and NLS-HSA-RITC were added. After 25 min, 12 μl of each sample was fixed in 3.7% paraformaldehyde containing Hoechst. After 75 min, disassembly of samples was monitored by fluorescence microscopy. These samples were imaged using the Zeiss Axioskop2 and F view soft imaging system (Olympus).
Immunoprecipitation and Immunoblotting
The GST pulldown was performed as previously described (Liu et al., 2003). Immunoprecipitation and immunoblotting were performed following standard procedures. Antibodies used were obtained as follows: anti-GST (a kind gift from Dr. Sarah Guadagno); anti-Xenopus Nup358 (Saitoh et al., 1996); monoclonal antibody 414, which recognizes Nup62, Nup153, Nup214, and Nup358 (Covance, Richmond, CA); HRP secondary antibodies (Zymed Laboratories, South San Francisco, CA); and a second antibody (Nup358-2) against human Nup358 amino acids 2560-2971 (Joseph et al., 2004). Antibodies against β-COP and the zinc finger region of Nup153 were generated as previously described (Liu et al., 2003).
Indirect Immunofluorescence Microscopy
In vitro assembled nuclei were prepared for immunofluorescence by freezing samples onto a slide with liquid nitrogen and then fixing with methanol (Liu et al., 2003). In contrast, nuclei processed for β-COP immunofluorescence were fixed 60 min after cyclin addition as described by Macaulay and Forbes (1996). Primary antibody dilutions used were anti-lamin at 1:500 (a kind gift from Dale Shumaker and Bob Goldman), anti-Nup153 at 1:2500, anti-xNup358 1:2500, anti-β-COP antibody at 1:500 and mAb414 at 1:2000 (Covance). Alexa Fluor secondary antibodies (Molecular Probes, Eugene, OR) were used at 1:500. Images for antibody localization experiment were imaged with an Olympus FVX IX70 confocal microscope. All other images were acquired on a Zeiss Axioskop 2.
DNA Replication Assay
In vitro nuclei containing recombinant protein (1.7 μM) or antibodies (2.5 μg of PI, 1 μg of αNup153, and 0.5 μg of αNup358) were assayed for the incorporation of α-32P-dCTP into genomic DNA as was previously described (Powers et al., 1995). At 0, 50, 90, 135, and 180 min, 5 μl of each reaction was fixed, digested with proteinase K (Fisher Scientific, Pittsburgh, PA), and run on a 0.8% agarose gel. Samples were quantitated with a Storm 860 scanner (Molecular Dynamics/GE Healthcare, Piscataway, NJ). Each sample was divided by the amount of radioactivity in the control sample at 180 min and then multiplied by 100. The average and SD of multiple experiments were graphed.
RESULTS
The Zinc Finger Domain of Nup358 Interferes with Nuclear Envelope Disassembly
To investigate whether the zinc finger domain of Nup358 helps to orchestrate nuclear disassembly, we first tested whether a recombinant protein fragment encompassing this domain can interfere with nuclear envelope breakdown. We had the human gene of Nup358 in hand, so we compared the effects of the zinc finger regions of human Nup358 and human Nup153 (Figure 1A). Recombinant GST fusion proteins were incubated in interphase Xenopus egg extract, which contained cycloheximide to prevent cyclin B synthesis. Sperm chromatin and an energy regeneration system were added to the extract to initiate in vitro assembly of nuclei. Import substrate was added after 60 min to monitor the integrity of the nuclear envelope and the functional status of the newly formed nuclear pores. After 90 min, an interphase time point was taken and analyzed (Interphase, Figure 1B). In each reaction, nuclei formed with closed nuclear envelopes and with equivalent ability to accumulate the import substrate. Cyclin was added to the remaining reaction in order to trigger the signaling cascade that shifts the conditions to a mitotic state. Seventy-five minutes later, a mitotic time point was analyzed. As expected, in the control reaction containing the GST fragment, nuclear envelopes had dispersed (Mitosis, Figure 1B). Breakdown of the nuclear envelope is most clearly visualized by loss of import substrate accumulation. As was seen previously, addition of the Xenopus Nup153 zinc finger domain (x153Z) interfered with nuclear disassembly (96% of nuclei remained intact). The human Nup153 zinc finger region (h153Z) was almost as potent, protecting 82% of the nuclei. Interestingly, the presence of the human Nup358 zinc finger domain (h358Z) prevented nuclear envelope breakdown, with 86% of the nuclei remaining intact at this time point. The recombinant protein fragments were stable in the reaction over the course of the experiment as assessed by immunoblot analysis of input proteins and of the reaction at the final time point (Figure 1C). These results suggest that the zinc finger region of Nup358 can associate with machinery required for dispersal of the nuclear envelope.
A hallmark of the phenotype seen with Xenopus Nup153 zinc finger domain was that mitotic signaling became uncoupled from breakdown of the lamina, perhaps reflecting a feedback mechanism that normally coordinates recruitment of membrane remodeling machinery with lamina dispersal (Liu et al., 2003). We were interested in determining whether the Nup358 zinc finger domain causes a similar phenotype or whether we could distinguish inhibitory mechanisms based on this. We therefore assessed lamina breakdown by indirect immunofluorescence in the presence of either the GST or h358Z fragment. At the interphase time point, both reactions showed a rim localization for lamins and nucleoporins (Figure 1D). Seventy-five minutes after cyclin addition, when nuclei were disassembled in the control reaction, samples containing h358Z continued to exhibit a nuclear rim localization for both lamins and nucleoporins. This result underscores the similarity between the inhibitory effects of the Nup358 zinc finger domain and the Nup153 zinc finger domain.
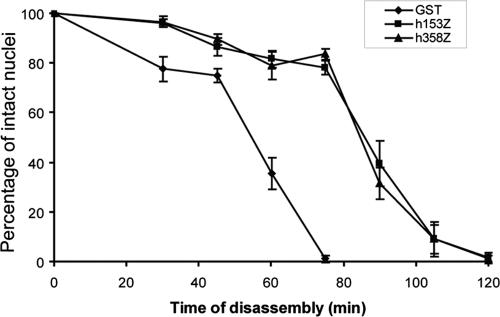
We next looked at a time course of nuclear envelope breakdown to address whether the h153Z and h358Z domains are equally potent as dominant negative inhibitors and whether nuclear envelope breakdown is absolutely dependent on the step targeted by these inhibitors. Nuclei were assembled in the presence of GST, h153Z, or h358Z. The zero time point was taken when cyclin was added, followed by subsequent time points as indicated. Each sample was quantitated to determine the number of nuclei remaining and the average of two independent experiments is plotted (Figure 2). Nuclei incubated with the GST fragment were completely disassembled after 75 min. Although the kinetics of breakdown (and protection) varies somewhat in different experiments, the presence of either h153Z or h358Z was seen here to delay completion of this process by ∼45 min. Significantly, these two nucleoporin zinc finger domains inhibit with similar potency, consistent with interference of a similar step. Because this is a delay, but not an absolute arrest, other mechanisms contributing to nuclear envelope breakdown are likely recapitulated in the egg extract and remain intact under these conditions.
Figure 2.
The zinc finger domains from Nup153 and Nup358 delay nuclear envelope breakdown with similar kinetics. Nuclei assembled in reactions containing GST, h153Z, or h358Z (0.9 μM final) for 90 min before cyclin addition. Samples were taken at 0, 30, 45, 60, 75, 90, 105, and 120 min after cyclin addition. Percent of intact nuclei at each time point was calculated. Data are from two independent experiments and presented as mean ± SD.
Both Nup358 and Nup153 Play a Role in Nuclear Envelope Breakdown
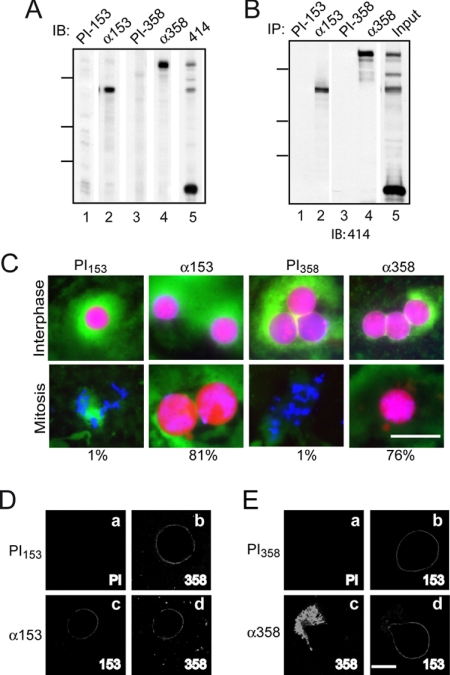
The ability of h358Z to inhibit nuclear disassembly similarly to h153Z lead us to test whether Nup358 plays a role in nuclear envelope breakdown. To target Nup358 directly, we used antibodies to interfere with its activity. Because of regions of similarity between Nup153 and Nup358, it was first important to establish that the antibodies clearly discriminate between these nucleoporins. Such specificity was observed in both immunoblot (Figure 3A) and immunoprecipitation (Figure 3B) analyses. We then tested these antibodies and corresponding preimmune antibodies in the nuclear assembly/disassembly assay. Nuclei assembled normally in the presence of the antibodies and accumulated the NLS import substrate (Interphase, Figure 3C). As we previously reported, at the mitotic time point, nuclear envelope breakdown was inhibited in the nuclei formed in the presence of the Nup153-specific antibodies, here with 81% of the nuclei intact (Mitosis, Figure 3C). Strikingly, 76% of the nuclei formed in the presence of the Nup358 antibodies were intact at the mitotic time point. This is in contrast to the samples containing the preimmune antibodies, where in each case only 1% of nuclei remained intact at the mitotic time point. The observation that interfering with either nucleoporin disrupts nuclear envelope breakdown supports the conclusion that the role of both nucleoporins is important.
Figure 3.
Both Nup358 and Nup153 play a role in nuclear envelope breakdown. (A) Immunoblot of fractionated egg extract probed with antibody against Nup153 (lane 2) and its matched preimmune (PI153; lane 1), Nup358 antiserum (lane 4) and its matched preimmune (PI358; lane 3), and 414-reactive Nups (lane 5). (B) Immunoprecipitation of egg extract with antibodies against Nup153 (lane 2) and its matched preimmune (lane 1), Nup358 antiserum (lane 4) and its matched preimmune (lane 3). Lane 5 was loaded with the equivalent of ∼20% input. The blot was probed with mAb414. For both A and B, molecular-weight markers indicated are 214, 118, and 92 kDa. (C) Antibodies against Nup153 or its matched preimmune (0.25 μg), or Nup358 antiserum or its matched preimmune (0.5 μl) was added 15 min before the beginning of the assembly/disassembly assay. Interphase samples were taken 90 min after assembly, and mitotic samples were taken 75 min after cyclin addition. Nuclear import cargo, DNA, and membrane are shown. The percentage of intact nuclei at mitosis is indicated below the panel. Bar, 50 μm. (D) In vitro nuclei were formed after a 15-min preincubation with preimmune antibody or antibody against Nup153. The added antibody was detected by indirect immunofluorescence with goat anti-rabbit secondary antibody (a and c). The localization of Nup358 was determined by incubating with primary and secondary antibodies postfixation (b and d). (E) In vitro nuclei were formed after a 15-min preincubation with preimmune or Nup358 antiserum. The added antibody was detected by indirect immunofluorescence with a goat anti-guinea pig secondary (a and c). The localization of Nup153 was determined by incubating with primary and secondary antibodies postfixation (b and d). Bar (D and E), 20 μm.
Nuclear pores form de novo in the in vitro assembly system and because Nup153 and Nup358 are both recruited relatively early in this process (Walther et al., 2003), we considered the possibility of interdependence between these nucleoporins for pore targeting. To test whether the Nup153 reactive antibodies affect Nup358 localization to the nuclear pore, these antibodies were again added at the beginning of the assembly assay and, in this case, the reactions were prepared for indirect immunofluorescence at the interphase time point. Antibodies to Nup153 were localized by probing with goat anti-rabbit secondary antibodies. The localization of Nup358 was simultaneously detected with Nup358-specific primary antibodies and a corresponding secondary antibody. Nup153 antibodies were detected at the nuclear rim, whereas the control preimmune antibodies showed no specific association with nuclei (Figure 3D, a and c). A rim localization of Nup358 was observed in the nuclei incubated with either preimmune or Nup153 antibody (Figure 3D, b and d). This suggests that the Nup153-specific antibodies do not interfere with Nup358 targeting to the pore.
Similarly, to test whether the Nup358 reactive antibodies affect Nup153 localization to the nuclear pore, we added Nup358-specific antibodies or matched preimmune antibodies at the beginning of the assembly assay and detected by immunofluorescence the localization of the added antibodies and Nup153. Targeting Nup358 with antibodies prevented its normal localization. Instead of a rim stain, the antibodies accumulated in a perinuclear pattern, which was not present in the reaction containing the preimmune antibodies (Figure 3Ec). This polyclonal antibody was generated using full-length Xenopus Nup358 so it may interact with the pore targeting domain of Nup358, preventing localization to the pore, or it may be more prone to forming a higher-order immune complex that precludes pore targeting. Regardless, the Nup358-specific antibody did not alter the localization of Nup153 to the nuclear envelope (Figure 3Ed). Thus, antibodies to Nup153 or to Nup358 do not cross-interfere with assembly of these nucleoporins into the pore. Rather, this data supports the hypothesis that Nup153 and Nup358 have nonredundant roles during nuclear envelope breakdown.
Nup358 and Nup153 Inhibitors Do Not Prevent Import into the Nucleus
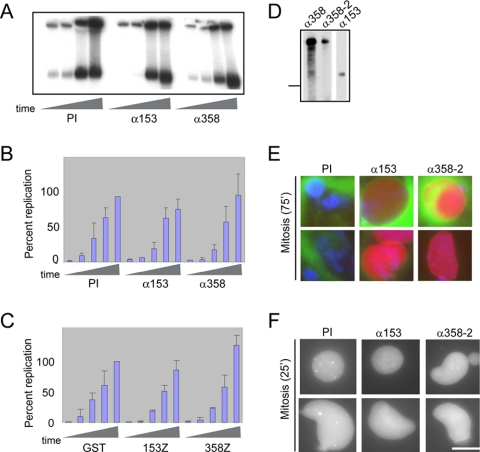
We next examined whether the Nup358- and Nup153-specific antibodies or the zinc finger fragments affect the transport function of the pore as such disruption could delay entry into mitosis. In experiments depicted in Figures 1 and 3, the RITC-HSA-NLS import substrate was added 60 min after initiating nuclear assembly. Thirty minutes later at the interphase time point, equivalent levels of import substrate had accumulated in all of the reactions, suggesting transport through the pore is not blocked. To determine whether a more subtle defect in the transport function of pores could be present and affect nuclear envelope breakdown, we looked at DNA replication, an interphase process that is dependent on nuclear import and a sensitive readout of pore function. Preimmune antibodies or antibodies reactive with either Nup153 or Nup358 were incubated with extracts. Assembly of nuclei was then initiated by adding sperm chromatin. DNA replication was monitored by the amount of radiolabeled nucleotide (α-32P-dCTP) incorporated into the DNA over time (Figure 4A). Quantitation of replication kinetics from several experiments demonstrated that neither Nup153- nor Nup358-specific antibodies alter DNA replication (Figure 4B). In addition, the kinetics of replication in the presence of the recombinant proteins, x153Z or h358Z, was similar to the kinetics of replication in control nuclei containing GST (Figure 4C). These results provide strong support that import is occurring with normal kinetics when either the antibodies or zinc finger fragments are present.
Figure 4.
Nup358 and Nup153 inhibitors do not prevent import into the nucleus. (A) DNA replication was monitored by following incorporation of α-32P-dCTP into genomic DNA during in vitro nuclear assembly. Reactions contained preimmune antibodies (lanes 1-5), antibodies to Nup153 (lanes 6-10), or Nup358 antiserum (lanes 11-15). Samples were analyzed at various times (0, 50, 90, 135, and 180 min). (B) Quantitation of nucleotide incorporated into genomic DNA in the presence of preimmune antibodies, antibodies to Nup153, or Nup358 antiserum. The graph shows the mean and SD of three experiments. (C) Graph of quantitation of replication in the presence of GST, x153Z, and h358Z (1.7 μM). The graph shows the mean and SD of two experiments. (D) Immunoblot of fractionated egg extract probed with antibody to Nup153 and two independent antibodies to Nup358. Molecular-weight marker 170 kDa. (E) Samples were formed in the presence of Nup153 preimmune antibodies, antibodies specific to Nup153, or antibodies specific to Nup358. Seventy-five minutes after addition, nuclear envelope breakdown was assessed by imaging DNA (blue), import substrate (red), and membranes (green). (F) Samples were formed in the presence of PI, αNup153, or αNup358-2. Ninety minutes after assembly, import substrate and cyclin were added. Twenty-five minutes after addition, nuclei were imaged for import substrate. For E and F, two nuclei representative of each sample are shown. Bar, 25 μm.
To look directly at import after cyclin addition, we monitored the accumulation of RITC-HSA-NLS added at the same time as cyclin. Nuclei were formed in the presence of the antibody panel. Cyclin and import substrate were added after 90 min. Twenty-five minutes later, we observed a slight reduction in the nuclear accumulation of the import substrate in nuclei incubated with antibodies to Nup358 compared with the preimmune (unpublished data). We therefore tested an independently generated antibody (358-2) against a discrete region of Nup358. We ensured that this antibody does not cross-react with Nup153 (Figure 4D) and determined that it also interferes with nuclear envelope breakdown (Figure 4E). Reactions containing preimmune, αNup358-2, or αNup153 accumulated similar levels of import substrate when assessed during the window after cyclin addition (Figure 4F). These collective results support the idea that Nup153 and Nup358 contribute to nuclear envelope breakdown in a manner independent of nucleocytoplasmic transport.
The Recruitment of β-COP to the Nuclear Envelope during Nuclear Disassembly Is Altered by the Zinc Finger Region of Nup358
To assess whether the zinc finger region of Nup358 participates in COPI recruitment to the nuclear envelope, we first examined whether the dominant negative fragment derived from this region has the ability to interact with COPI. We immobilized a panel of recombinant proteins, including GST, GST-x153Z, GST-h153Z, and GST-h358Z, on glutathione Sepharose beads. Following incubation in a fractionated Xenopus egg extract, proteins associated with each matrix were eluted and analyzed. An immunoblot of these samples revealed that β-COP associates with h358Z, but not with the GST control (Figure 5A, left panel). Coomassie stain was used to monitor the recovery of recombinant protein, which was fairly equivalent between samples (Figure 5A, right panel). We found that association of β-COP was somewhat lower with h153Z and h358Z than with x153Z; it is possible that differences across species result in a slightly lower affinity and loss of recovery in this biochemical assay. Overall, the association between β-COP and the ZnF region of Nup358 supports the notion that this nucleoporin aides in recruiting the COPI complex to the NPC.
Figure 5.
The zinc finger region of Nup358 associates with β-COP and alters β-COP recruitment to the nuclear envelope during nuclear disassembly. (A) The left panel is an immunoblot of GST pulldown samples probed with antibodies to β-COP. Five percent load of fractionated Xenopus egg extract is in lane 1. Lanes 2-5 show levels of β-COP recovery with purified recombinant proteins GST (lane 2), x153Z (lane 3), h153Z (lane 4), and h358Z (lane 5). Molecular-weight markers indicate 126 and 100 kDa. The right panel is a gel stained with Coomassie blue to detect recombinant protein recovered on the same glutathione-Sepharose pulldowns of GST (lane 6), x153Z (lane 7), h153Z (lane 8), and h358Z (lane 9). Molecular-weight markers indicate 72, 54, and 35 kDa. (B) Recombinant proteins, GST (5 μg), and 358Z (5 μg, 1.7 μM), were added 15 min before the addition of sperm chromatin. Cyclin was added after 90 min of assembly, and 60 min later β-COP (a, c, e, g, i, k, m, and o) and 414-reactive nucleoporins (b, d, f, h, j, l, n, and p) were detected by indirect immunofluorescence. Panels a-h and i-p are from independent experiments. (C) Seventy-five minutes after cyclin addition, nuclear envelope breakdown was assessed by imaging DNA (blue), import substrate (red), and membranes (green). Bar (A and B), 25 μm.
To further address this point, we examined whether COPI association with the nuclear rim at mitosis is disrupted by the Nup358 ZnF fragment. GST and h358Z fragments were added to the assembly/disassembly assay, and samples were processed for indirect immunofluorescence 60 min after cyclin addition and before breakdown of the nuclear envelope. In the reaction containing the control GST fragment, a rim localization of the COPI subunit β-COP similar to that of the 414-reactive nucleoporins was seen by indirect immunofluorescence (Figure 5B). When we looked at the same time point in reactions containing h358Z, the localization of β-COP was diffuse and no longer appeared enhanced at the nuclear rim, although 414-reactive nucleoporins were unaltered in localization. As expected, the nuclei in reactions containing the GST fragment were completely disassembled after 75 min, whereas most of the nuclei in reactions containing h358Z were still intact (Figure 5C). This change in β-COP localization indicates that, when present in excess, the Nup358 zinc finger domain inhibits COPI function at the nuclear envelope.
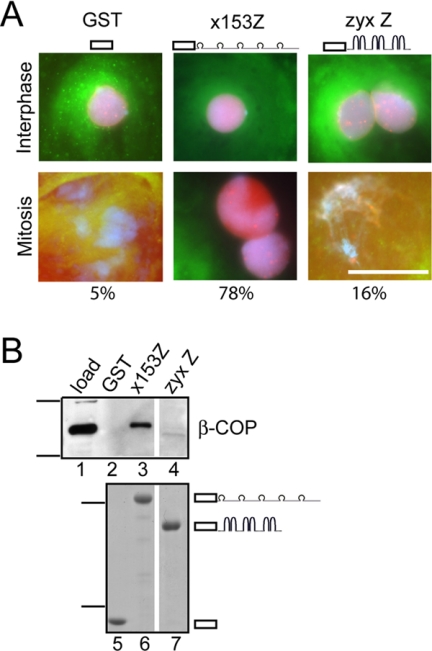
The similarity in results obtained with Nup153 and Nup358 ZnF regions in both protein partner analysis and nuclear disassembly phenotype brings up the question of whether these are general attributes of repetitive zinc fingers. To address this, we used a recombinant protein encompassing six zinc fingers from the LIM domain of zyxin. In assays similar to those described above, this ZnF fragment did not significantly perturb nuclear disassembly (Figure 6A) and did not associate with β-COP (Figure 6B). Thus, the zinc finger domains unique to these two nucleoporins have a specific dominant negative effect on nuclear disassembly, which correlates with their ability to associate with β-COP.
Figure 6.
The zyxin LIM domain (zyx Z), which contains six zinc fingers, does not interfere with nuclear envelope breakdown or bind β-COP. (A) Nuclei were assembled from extracts preincubated with GST, x153Z, or zyx Z (1.7 μM). An interphase time point was taken before cyclin addition and a mitosis time point was taken 75 min later. Nuclear import cargo, DNA, and membrane are shown. The percentage of intact nuclei at mitosis is indicated below the panel. Bar, 50 μm. (B) The top panel is an immunoblot of GST pulldown samples probed with antibodies to β-COP. Five percent load of fractionated Xenopus egg extract is in lane 1. Lanes 2-4 show levels of β-COP recovery with purified recombinant proteins GST (lane 2), x153Z (lane 3), and zyx Z (lane 4). Molecular-weight markers indicated are 126 and 100 kDa. The bottom panel is a gel stained with Coomassie blue to detect recombinant protein recovered on the same glutathione-Sepharose pulldowns: GST (lane 5), x153Z (lane 6), and zyx Z (lane 7). Molecular-weight markers indicated are 54 and 35 kDa.
A Single Zinc Finger Contains Important Determinants for Regulating Nuclear Envelope Breakdown
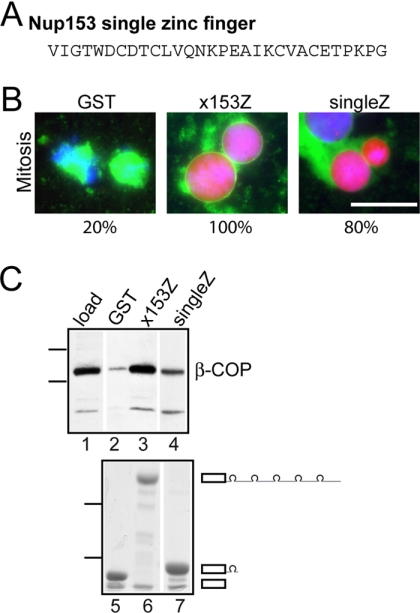
The zinc finger domains of both Nup153 and Nup358 contain multiple zinc fingers separated by substantial linker regions (Figure 1A). The zinc fingers are more highly conserved than the linker region; thus this discrete structural motif seemed a good candidate to be the primary functional determinant within this region. To test whether a single zinc finger contains the interface important to nuclear envelope breakdown, we used a recombinant fragment encompassing only one Nup153 zinc finger in the nuclear assembly/disassembly assay (Figure 7A). We found that the single Nup153 zinc finger fragment was inhibitory, with 80% of nuclei still intact at the mitotic time point (Figure 7B). Notably, this effect was seen only when the single zinc finger fragment was present at greater molar levels than the entire zinc finger domain, indicating that an individual zinc finger is not as potent as the entire domain. This result suggests that a Nup zinc finger contains essential functional determinants, but presentation in tandem is optimal, because of the contribution made by the linker region and/or the repetitive nature itself.
Figure 7.
An individual zinc finger motif contains the critical determinant for dominant negative activity. (A) Sequence of the second human Nup153 zinc finger used in experiment. (B) Recombinant proteins (32 μM of GST, 1.7 μM of x153Z, and 32 μM of single Nup153 Zinc finger) were added 15 min before the beginning of the assembly/disassembly assay. The mitotic samples were taken 75 min after cyclin addition. DNA (blue), NLS import substrate (red), and membrane (green) are shown. Numbers below the panels indicate the percentage of intact nuclei at the mitotic time point. Bar, 50 mm. (C) The top panel is an immunoblot of GST pulldown samples probed with antibodies to β-COP. Five percent load of fractionated Xenopus egg extract is in lane 1. Lanes 2-4 show levels of β-COP recovery with purified recombinant proteins GST (lane 2), x153Z (lane 3), and single Nup153 zinc finger (lane 4). Molecular-weight markers indicated are 118 and 92 kDa. The bottom panel is Coomassie blue staining of protein recovered in the same samples of glutathione-Sepharose: GST (lane 5), x153Z (lane 6), and single Nup153 zinc finger (lane 7). Molecular-weight markers indicate 52 and 36 kDa.
The ability of the single zinc finger to perturb nuclear envelope breakdown leads to the further prediction that the COPI complex can associate with this minimal zinc finger motif. To test this, the GST fusion proteins: GST, x153Z, and the single Nup153 zinc finger, were immobilized onto glutathione beads and a pull-down assay was performed as described above. We found that β-COP was indeed recovered with the single zinc finger albeit in lower levels than with an intact x153Z domain (Figure 7C). Recovery of the recombinant proteins was equivalent. The difference in potency between the native zinc finger domain and a single zinc finger is more striking in the functional assay than the pulldown assay. The pulldown assay may not fully reflect the potency of partnerships with the zinc fingers because of more dilute conditions or because of the loss of a regulatory factor or modification. Nonetheless, this qualitative assay demonstrates the single zinc finger is sufficient to interact with COPI and that the presence of tandem zinc fingers or the linker regions enhances association with COPI.
DISCUSSION
Here we report that Nup358 plays an important role in nuclear envelope breakdown. A recombinant protein encompassing the zinc finger domain of Nup358 has dominant interfering activity in a nuclear envelope breakdown assay. Presence of the Nup358 zinc finger domain protected nuclei from mitotic disassembly with similar potency as the Nup153 zinc finger domain. This result suggests that these regions have a shared capacity to interface with machinery involved in nuclear envelope breakdown and that, in its native context at the pore, the Nup358 zinc finger module serves to help recruit factors into proximity with the nuclear pore and the envelope itself. Consistent with this, two independent antibodies directed against Nup358 were found to interfere with normal progression of nuclear envelope breakdown in the nuclear disassembly assay.
Given the importance of nucleocytoplasmic trafficking at the transition into mitosis, we closely examined whether reagents designed to block the role of Nup358 had an effect on transport that could account for the striking persistence of nuclei under mitotic conditions. We routinely monitor overall accumulation of an NLS cargo before addition of cyclin in our nuclear breakdown assay and had observed no apparent changes in this parameter. Assessing import at this end point, however, could mask a kinetic difference in transport that in turn impinges on nuclear envelope breakdown. We therefore monitored the kinetics of DNA replication, a process sensitive to transport levels, and again observed no indication of alterations in the transport function of the NPC. We also assessed whether a potential alteration in transport might occur only after the mitotic signaling cascade had been initiated, and, in this case, one of the antibodies directed against Nup358 seemed to result in some delay. Equivalent accumulation was seen, however, in the presence of an independent antibody specific for Nup358 that was active in perturbing progress of nuclear envelope breakdown. Our conclusion that Nup358 activity can be blocked without significantly altering import through the pore is consistent with reports in the literature in which this nucleoporin was depleted by various means without impacting nuclear import (Walther et al., 2002; Salina et al., 2003, Forler et al., 2004, Bernad et al., 2004). Nup358 has been suggested to play a supporting role in exportin-1 (CRM1)-dependent export (Bernad et al., 2004); however, this is unlikely to contribute to inhibition of nuclear disassembly as a potential decrease in nuclear export sequence (NES) export would be predicted to increase nuclear levels of cyclin B and speed, rather than delay, mitotic entry (Yang et al., 1998).
If the Nup358 zinc finger domain serves as a cytoplasmically facing scaffold for COPI recruitment to the pore at mitosis, then this domain module would be predicted to interfere with the nuclear rim localization of COPI observed in the in vitro nuclear assembly/disassembly assay. This is, in fact, what we observed. Nup358 has been previously identified as a mitotic regulator, although not at the stage of nuclear envelope breakdown but rather during the process of attachment between chromosomes and the mitotic spindle. Nup358 plays an important role at the kinetochore, a site where it is targeted at mitosis. RNAi-mediated depletion of Nup358 results in striking defects in spindle formation and chromosome segregation and, in some cases, the appearance of micronuclei (Salina et al., 2003; Joseph et al., 2004). The mislocalization of Mad1 and Mad2, as well as certain other kinetochore proteins in the absence of Nup358, suggests a role for this pore protein in kinetochore assembly and attachment to microtubules. The reciprocal localization of Nup358, either at the nuclear pore or the kinetochore, previously led to the suggestion that this nucleoporin may integrate the completion of nuclear envelope breakdown— and the concomitant disassembly of nuclear pore complexes—with the downstream mitotic events involved in chromosome segregation (Joseph et al., 2002; Salina et al., 2003). The results described here indicate that Nup358 is involved in coordinating the events of nuclear envelope breakdown very early in mitosis and, once completed, the arrival of Nup358 at the kinetochore provides a signal that aides in coordinating attachment to microtubules.
Is an early role in mitosis consistent with the reported phenotypes of Nup358 depletion in cultured cells? These cells are not noticeably blocked in late prophase, at the stage of nuclear envelope breakdown. The embryonic cell cycle, recapitulated in Xenopus egg extract, may differ from the somatic cell cycle in requirements for nuclear disassembly. Alternatively, the multilayered nature of nuclear envelope breakdown may rescue Nup358-depleted cells from arresting at late prophase. For instance, microtubule-dependent tearing mechanisms, which themselves are not absolutely required to accomplish nuclear envelope breakdown, may push the process sufficiently for progression into mitosis. Additional mechanisms, including more passive events, may contribute to nuclear envelope breakdown, albeit less efficiently. Indeed, the observation made here that dominant inhibition of zinc finger function does not lead to a persistent block in nuclear disassembly supports the idea that other means are available to achieve dispersal of the nuclear membrane. Similarly, there are mechanisms in place to compensate for alterations in the timing of nuclear envelope breakdown. For example, migration of microtubule asters to sites at polar sides of the nucleus can be mediated by events that are both nuclear envelope dependent and independent (Rosenblatt et al., 2004). Rather than blocking nuclear envelope breakdown, depletion of Nup358, and the consequent disruption of efficiently recruiting COPI to the nuclear envelope, would be predicted to result in defects in downstream events due to delayed membrane dispersal from the chromosomal surfaces. Given the additional roles that Nup358 plays specifically in the context of the kinetochore, it is therefore not surprising that miscoordination of chromosome division is the predominant phenotype detected in cells depleted of Nup358.
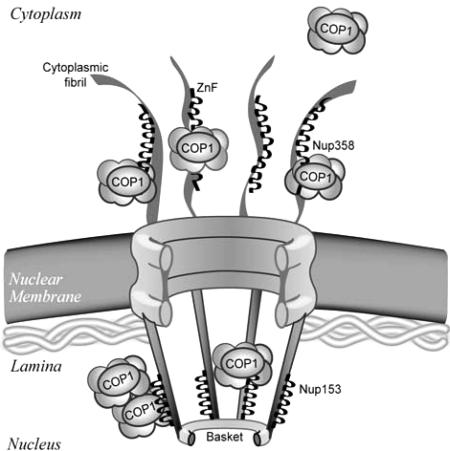
Integrating our data here of a newly identified role for Nup358 in nuclear disassembly and the finding that this nucleoporin contains a motif which can associate with COPI brings us to a working model of COPI recruitment to the nuclear pore (Figure 8). Specifically, we propose that the zinc finger modules found in both Nup153 and Nup358 serve as dual scaffolds on which COPI becomes locally concentrated in proximity to the nuclear membranes. A single zinc finger appears to contain the determinants critical for COPI association, but it is clear that the tandem arrangement of the zinc finger motifs is important. The requirement for both Nup153 and Nup358 in efficient nuclear envelope disassembly further suggests that having such a scaffold on both sides of the pore is important. Indeed, because of the eightfold symmetry of the nuclear pore, at least 32-64 individual zinc fingers are present on each face of the nuclear pore complex. It will be important to understand COPI recruitment to these zinc fingers and how it is regulated in molecular detail, as well as to identify what other proteins serve downstream roles in facilitating COPI loading on to the nuclear membrane.
Figure 8.
Model of COPI recruitment to the nuclear pore. Efficient nuclear envelope breakdown requires recruitment of COPI to both faces of the nuclear pore via zinc finger scaffolds present in Nup153 and Nup358. Note that, although the zinc finger domains are resident on both faces of the pore, their precise location and arrangement within the architecture of the pore is unknown and is represented schematically here. Antibodies specific for the zinc finger region of Nup153 decorate the distal ring of the nuclear pore basket (Fahrenkrog et al., 2002), although zinc finger exposure may change during early phases of mitotic pore remodeling (Ball and Ullman, 2005).
Acknowledgments
We thank members of the Ullman lab for helpful discussion and Brian Bennion in particular for reagents. We are grateful to Dale Shumaker and Robert Goldman for the gift of the anti-lamin antibodies. We also thank Wes Sundquist and Mary Beckerle for constructs. We acknowledge expert assistance with graphics from Diana Lim. This work was supported by National Institutes of Health (NIH) Grant GM61275 to K.S.U. as well as funds from the Leukemia and Lymphoma Society and the Huntsman Cancer Fund. A.J.P. was supported by a predoctoral fellowship from the National Science Foundation and University of Utah Graduate Research Fellowship. University of Utah core facilities used in this study are partially supported by NIH Grant P30CA42014.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0485) on November 28, 2005.
Abbreviations used: Nup, nucleoporin; IB, immunoblot; IP, immunoprecipitation; PI, preimmune; aa, amino acids; ER, endoplasmic reticulum; NLS, nuclear localization signal; COPI, coatomer complex I.
References
- Alam, S. L., Sun, J., Payne, M., Welch, B. D., Blake, B. K., Davis, D. R., Meyer, H. H., Emr, S. D., and Sundquist, W. I. (2004). Ubiquitin interactions of NZF zinc fingers. EMBO J. 23, 1411-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin, W., Franz, C., Haselmann, U., Antony, C., and Mattaj, I. W. (2005). The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell 17, 83-92. [DOI] [PubMed] [Google Scholar]
- Ball, J. R., and Ullman, K. S. (2005). Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 114, 319-330. [DOI] [PubMed] [Google Scholar]
- Beaudouin, J., Gerlich, D., Daigle, N., Eils, R., and Ellenberg, J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83-96. [DOI] [PubMed] [Google Scholar]
- Bernad, R., van der Velde, H., Fornerod, M., and Pickersgill, H. (2004). Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24, 2373-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia, B., and Courvalin, J. C. (1997). Domain-specific disassembly and reassembly of nuclear membranes during mitosis. Exp. Cell Res. 230, 133-144. [DOI] [PubMed] [Google Scholar]
- Cronshaw, J. M., Krutchinsky, A. N., Zhang, W., Chait, B. T., and Matunis, M. J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano, C., Ball, J. R., Prunuske, A. J., and Ullman, K. S. (2001). RNA association defines a functionally conserved domain in the nuclear pore protein Nup153. J. Biol. Chem. 276, 45349-45357. [DOI] [PubMed] [Google Scholar]
- Drummond, S., Ferrigno, P., Lyon, C., Murphy, J., Goldberg, M., Allen, T., Smythe, C., and Hutchison, C. J. (1999). Temporal differences in the appearance of NEP-B78 and an LBR-like protein during Xenopus nuclear envelope reassembly reflect the ordered recruitment of functionally discrete vesicle types. J. Cell Biol. 144, 225-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., Koser, J., and Aebi, U. (2004). The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 29, 175-182. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., Maco, B., Fager, A. M., Koser, J., Sauder, U., Ullman, K. S., and Aebi, U. (2002). Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140, 254-267. [DOI] [PubMed] [Google Scholar]
- Forler, D., Rabut, G., Ciccarelli, F. D., Herold, A., Kocher, T., Niggeweg, R., Bork, P., Ellenberg, J., and Izaurralde, E. (2004). RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol. 24, 1155-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K., and Wilson, K. L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell. Biol. 6, 21-31. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Liu, S. T., Jablonski, S. A., Yen, T. J., and Dasso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611-617. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Tan, S. H., Karpova, T. S., McNally, J. G., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Prunuske, A. J., Fager, A. M., and Ullman, K. S. (2003). The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev. Cell 5, 487-498. [DOI] [PubMed] [Google Scholar]
- Lohka, M. (1998). Analysis of nuclear envelope assembly using extracts of Xenopus eggs. In: Nuclear Structure and Function, ed. M. Berrios, San Diego: Academic Press, 371-374. [DOI] [PubMed]
- Macaulay, C., and Forbes, D. J. (1996). Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J. Cell Biol. 132, 5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit, A., Vlcek, S., Gruenbaum, Y., and Foisner, R. (2005). Breaking and making of the nuclear envelope. J. Cell. Biochem. 95, 454-465. [DOI] [PubMed] [Google Scholar]
- Pante, N. (2004). Nuclear pore complex structure: unplugged and dynamic pores. Dev. Cell 7, 780-781. [DOI] [PubMed] [Google Scholar]
- Powers, M. A., Evans, E. K., Yang, J., and Kornbluth, S. (2001). Xenopus egg extracts. In: Current Protocols in Cell Biology, vol. 2, New York: John Wiley & Sons, 11.10.11-11.11.24. [DOI] [PubMed] [Google Scholar]
- Powers, M.A., Macaulay, C., Masiarz, F., and Forbes, D. J. (1995). Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128, 721-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske, A. J., and Ullman, K. S. (2006). The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Rosenblatt, J., Cramer, L. P., Baum, B., and McGee, K. M. (2004). Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell 117, 361-372. [DOI] [PubMed] [Google Scholar]
- Rout, M. P., Aitchison, J. D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B. T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, H., Cooke, C. A., Burgess, W. H., Earnshaw, W. C., and Dasso, M. (1996). Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol. Biol. Cell 7, 1319-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., Bodoor, K., Eckley, D. M., Schroer, T. A., Rattner, J. B., and Burke, B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97-107. [DOI] [PubMed] [Google Scholar]
- Salina, D., Enarson, P., Rattner, J. B., and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S. R. (2003). Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775-789. [DOI] [PubMed] [Google Scholar]
- Vigers, G. P., and Lohka, M. J. (1991). A distinct vesicle population targets membranes and pore complexes to the nuclear envelope in Xenopus eggs. J. Cell Biol. 112, 545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T. C., Askjaer, P., Gentzel, M., Habermann, A., Griffiths, G., Wilm, M., Mattaj, I. W., and Hetzer, M. (2003). RanGTP mediates nuclear pore complex assembly. Nature 424, 689-694. [DOI] [PubMed] [Google Scholar]
- Walther, T. C., Pickersgill, H. S., Cordes, V. C., Goldberg, M. W., Allen, T. D., Mattaj, I. W., and Fornerod, M. (2002). The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158, 63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., Alam, S. L., Meyer, H. H., Payne, M., Stemmler, T. L., Davis, D. R., and Sundquist, W. I. (2003). Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 278, 20225-20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K. (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441-451. [DOI] [PubMed] [Google Scholar]
- Yang, L., Guan, T., and Gerace, L. (1997). Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137, 1199-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Bardes, E. S., Moore, J. D., Brennan, J., Powers, M. A., and Kornbluth, S. (1998). Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 12, 2131-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]