Abstract
Dense-core secretory granule (DCG) biogenesis is a prerequisite step for the sorting, processing, and secretion of neuropeptides and hormones in (neuro)endocrine cells. Previously, chromogranin A (CgA) has been shown to play a key role in the regulation of DCG biogenesis in vitro and in vivo. However, the underlying mechanism of CgA-mediated DCG biogenesis has not been explored. In this study, we have uncovered a novel mechanism for the regulation of CgA-mediated DCG biogenesis. Transfection of CgA into endocrine 6T3 cells lacking CgA and DCGs not only recovered DCG formation and regulated secretion but also prevented granule protein degradation. Genetic profiling of CgA-expressing 6T3 versus CgA- and DCG-deficient 6T3 cells, followed by real-time reverse transcription-polymerase chain reaction and Western blotting analyses, revealed that a serine protease inhibitor, protease nexin-1 (PN-1), was significantly up-regulated in CgA-expressing 6T3 cells. Overexpression of PN-1 in CgA-deficient 6T3 cells prevented degradation of DCG proteins at the Golgi apparatus, enhanced DCG biogenesis, and recovered regulated secretion. Moreover, depletion of PN-1 by antisense RNAs in CgA-expressing 6T3 cells resulted in the specific degradation of DCG proteins. We conclude that CgA increases DCG biogenesis in endocrine cells by up-regulating PN-1 expression to stabilize granule proteins against degradation.
INTRODUCTION
Peptide hormones are stored and released from dense-core secretory granules (DCGs) in endocrine cells (Arvan and Castle, 1998; Dannies, 1999; Arvan and Halban, 2004). Although the mechanism involved in up-regulating DCG biogenesis to replenish the DCG store after stimulation of hormone secretion in endocrine cells remains largely unknown, DCG biogenesis seems to be regulated by a network of molecules functioning at multiple levels (Kim et al., 2001, 2003; Day and Gorr, 2003; Knoch et al., 2004). Molecules that specifically regulate biosynthesis and stability of cargo proteins at the transcriptional, posttranscriptional, or posttranslational level seem to participate in DCG biogenesis (Eiden et al., 1984; Rausch et al., 1988; Wan et al., 1991; Kilbourne et al., 1992; Hiremagalur et al., 1993; Rozansky et al., 1994; Tang et al., 1997; Kim et al., 2001). Such regulatory molecules would eventually increase the quantity of DCG proteins required for de novo synthesis of DCGs.
We and others have shown that chromogranin A (CgA) plays a critical role in DCG biogenesis in vitro and in vivo. Down-regulation of CgA in neuroendocrine PC12 cells severely impaired DCG formation (Kim et al., 2001). In CgA- and DCG-deficient endocrine 6T3 cells, of which regulated secretory pathway could be revived by cAMP treatment (Matsuuchi and Kelly, 1991; Day et al., 1995), exogenously expressed CgA restored DCG formation and the regulated secretory phenotype (Kim et al., 2001). These data indicate that CgA is a key molecule controlling DCG biogenesis in these cells (Kim et al., 2001, 2003). However, CgA does not seem to be able to rescue DCG formation in mutant PC12 (PC12-27) cells, where the expression of the regulated secretion program has been permanently lost at the genetic level (Grundschober et al., 2002; Malosio et al., 2004). Recently, the importance of CgA in DCG biogenesis in vivo has been demonstrated using CgA knockout (KO) mice (Mahapatra et al., 2005) and transgenic mice expressing antisense RNAs against CgA (Kim et al., 2005). Both in vivo mouse models showed significant reduction in DCG formation in adrenal medulla chromaffin cells, providing evidence that CgA is essential for normal DCG biogenesis. However, the detailed mechanism of how CgA regulates DCG biogenesis in (neuro)endocrine cells is not known.
In addition to the severe loss of DCG formation, we also observed that reduction of CgA led to the significant degradation of DCG proteins (chromogranin B [CgB], carboxypeptidase E [CPE], and synaptotagmin) in CgA-deficient PC12 cells (Kim et al., 2001). Reexpression of CgA in these cells prevented degradation of DCG proteins (Kim et al., 2001). Similarly, when CgB was expressed in a CgA- and DCG-deficient 6T3 cells, CgB was severely degraded in these cells (Kim et al., 2001). Moreover, the level of DCG proteins such as CgB, CPE, and dopamine β-hydroxylase in adrenal chromaffin cells were significantly decreased in CgA-antisense transgenic and CgA KO mice (Kim et al., 2005; Mahapatra et al., 2005). Thus, it seems that the level of CgA in the cells determines the fate of DCG proteins to either participate in DCG biogenesis or be degraded intracellularly. Therefore, we proposed that CgA regulates the stability of DCG proteins necessary for DCG biogenesis by controlling the degradation process in neuroendocrine and endocrine cells at the posttranslational level (Kim et al., 2003). In this study, we tested this hypothesis in endocrine 6T3 cells.
MATERIALS AND METHODS
Cell Culture and Chemicals
6T3, AtT-20, and human embryonic kidney (HEK)293 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM supplemented with 10% fetal bovine serum/penicillin/streptomycin. Brefeldin A (BFA), monensin, NH4Cl (Sigma-Aldrich, St. Louis, MO), and doxycycline (BD Biosciences, San Jose, CA) were purchased.
Plasmid Constructs
Rat protease nexin-1 (PN-1) cDNA was amplified by PCR using the following primers: 5′-ACGGGCTGTCCTTGTTG-3′ (PN1-5-UTR) and 5′-CAGCGATAGCAACAGCAACAGA-3′ (PN1-3-UTR) and AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). A full-length rat PN-1 cDNA or rat CgB cDNA (a gift from Dr. S. Mahata, University of California, San Diego, La Jolla, CA) was subcloned into pcDNA3.1(+). Transfection of these constructs was performed using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA). For adenoviral constructs, the rat PN-1 coding region was amplified from pcDNA3.1(+)-PN-1 using primers PN1-SacII (5′-GATTACCGCGGACGGGCTGTCCTTGTTGGAAGGAA-3′) and PN1-SalI (5′-ATTAAGTCGACTCAGGGCTTGTTCACCT-3′) and inserted into pTRE-Shuttle2 vector (BD Biosciences). Rat CgB coding sequence was amplified using primers CgB-SacII (5′-TATTACCGCGGAGCTCCTCCTACATGCAGCT-3′) and CgB-XbaI (5′-ATTAATCTAGATCAGCCCCGCTGGCTGAACTT-3′), and subcloned in pTRE-shuttle2 vector. A 226 base pairs-long coding sequence for PN-1 antisense was amplified from rat PN-1 cDNA using PN1-XbaI (5′-CACACGTCTAGATTTATTAGAGTGTGAACTTCCAGGACCC-3′) and PN1-SacII (5′-TAATTCCGCGGATTTTCCATCACCTGCCAC-3′) and subcloned into pTRE-Shuttle2 vector for Tet-On adenovirus for antisense PN-1 synthesis.
Adenoviral-inducible Expression System
Adeno-X Tet-On constructs expressing PN-1 (pAdenoX-PN-1) or CgB (pAde-noX-CgB) were generated according to the manufacturer (BD Biosciences). Adeno-X viral constructs were transfected into HEK293 cells for the generation of recombinant adenovirus expressing either PN-1 or CgB. Viruses were purified using Adeno-X purification system, and the viral titers were determined by Rapid titer kit (BD Biosciences). Doxycycline was added to the culture during the infection at various concentrations (2-10 μg/ml).
Western Blotting Analysis and Antibodies
Cells lysate was prepared as described previously (Kim et al., 2001). Ten micrograms of total protein lysate of each sample was loaded onto 4-20% Tris/glycine polyacrylamide precast gels (Invitrogen) and transferred on nitrocellulose membrane. Blots were blocked in 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 and incubated with primary antibodies. Detection of protein signals was performed using chemiluminescent reagents (SuperSignal West Pico; Pierce Chemical, Rockford, IL). The radiographs were scanned, and the images were analyzed with ImageQuant 1.2 (Molecular Probes, Eugene, OR) or using secondary antibodies conjugated with infrared dyes (IRDye-700 or -800; LI-COR Biosciences, Lincoln, NE) followed by scanning signals using Odyssey infrared imaging system (LI-COR Biosciences). Monoclonal anti-CgB antibody (BD Biosciences) was used at dilution of 1:2000. Guinea pig polyclonal anti-CgA antibody (Kim et al., 2001) (1:3000), rabbit polyclonal anti-CgA antibody (TK-1; 1:5000) (Arnaoutova et al., 2003), rabbit polyclonal anti-proopiomelanocortin (adrenocorticotropin/β-lipotropin) (POMC) antibody (DP-4; 1:5000), and rabbit polyclonal anti-CPE antibody (Cool et al., 1997) (1:3000) were used. Mouse monoclonal anti-PN-1 antibody (4B3) (Mansuy et al., 1993) was a gift from Dr. D. Monard (Friedrich Miescher-Institut, Basel, Switzerland) (1:500). Rabbit anti-PN-1 antibody was kindly donated by Dr. K. X. Chai (University of Central Florida, Orlando, FL) and used at 1:3000. Mixture of monoclonal anti-synaptophysin antibodies against N and C termini of synaptophysin (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit polyclonal anti-VAMP2 antibody (Chemicon International, Temecula, CA) were used at 1:3000. Mouse monoclonal anti-Vti1a antibody (BD Biosciences) was used at 1:2000 as a trans-Golgi network (TGN) marker. Mouse monoclonal anti-α-tubulin antibody (Sigma-Aldrich) was used at 1:10,000 for normalization. Secondary antibodies conjugated with horseradish peroxidase were used at 1:3000.
Electron Microscopy and Morphometry
Cells were prepared for electron microscopy (EM) as described previously (Kim et al., 2001). The area of the cytoplasm was calculated by measuring the cytoplasmic area of cells less the area of nucleus in each EM micrograph. Each EM micrograph was scanned, and the digitalized image was analyzed using ImageJ program.
Affymetrix GeneChip Analysis
Gene expression profile was compared between 6T3 wild-type cells and 6T3-bCgA cells. cRNA probes from 6T3-WT, 6T3-bCgA, and AtT-20 cells were prepared. Total RNAs were purified from each cell line, and double-stranded cDNA was synthesized using SuperScript II reverse transcriptase and DNA polymerase I (SuperScript Choice system; Invitrogen) with T7-(dT)24 primer. Then, in vitro transcription was performed to generate biotin-labeled cRNA using RNA transcript labeling kit (Enzo Life Science, Farmingdale, NY), and the cRNA was fragmented by incubating at 94°C for 35 min in RNA fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc). Affymetrix murine genome U74Av2 chips (Affymetrix, Santa Clara, CA) were used for analysis. The cRNA probe was hybridized onto the array for 16 h at 45°C, and then the hybridized array was stained with streptavidin phycoerythrin conjugate using the GeneChip Fluidics Station 400 and scanned by the Hewlett-Packard GeneArray scanner at the excitation wavelength of 488 nm. Data analysis was carried out using Affymetrix Microarray Suite software.
Real-Time RT-PCR
Real-time RT-PCR was carried out for amplification and quantitative comparison of PN-1 mRNAs. Total RNAs were purified using RNeasy mini kit (Qiagen, Germantown, MD) and amplified with a primer set for PN-1 (PN1-B5; 5′-GAAGGAACCATGAATTGGCAT-3′ and PN1-C3; 5′-TGCAGCATGCCCAAGATGGAC-3′) using LightCycler and RNA master SYBR Green I (Roche Diagnostics, Indianapolis, IN). Glyceraldehyde-3-phosphate dehydrogenase was amplified using a primer set (Clontech, Mountain View, CA) and used for normalization of PN-1 mRNA levels in the samples. Analyses of PCR amplification were performed using the LightCycler3 software version 5.3 (Roche Diagnostics).
Subcellular Fractionation
Cells were harvested and homogenized in a buffer (150 mM NaCl, 20 mM HEPES, 1 mM EGTA, 0.1 mM MgCl2, and 0.32 M sucrose, pH 7.5), and a postnuclear supernatant was prepared. The resulting postnuclear supernatant was loaded on top of the linear sucrose gradient (0.6-1.6 M) prepared using a gradient maker (Gradient Master; Biocomp, Fredericton, New Brunswick, Canada), and centrifuged for 16 h at 4°C at 27,000 rpm in SW28 (Beckman Coulter, Fullerton, CA). In total, 17 fractions (1 ml/each) were collected from the top of the gradient using Auto Densi-Flow (Labconco, Kansas City, MO), and each fraction was precipitated in 10% trichloroacetic acid. Precipitates were then treated with SDS sample buffer, and an equal volume was loaded on Tris/glycine gels (Invitrogen) for Western blotting.
Immunofluorescence Microscopy
Cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Guinea pig polyclonal anti-CgA antibody (1:100) and monoclonal anti-CgB antibody (1:1000) in 3% normal goat serum/PBS were used at 1:100 and 1:1000, respectively. Monoclonal anti-PN-1 antibody was used at 1:100, and secondary antibody conjugated with either Alexa 488 or Alexa 568 (Molecular Probes) was used at 1:1000. A construct for GalT-CFP (a gift from Dr. J. Lippincott-Schwartz, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD) was expressed in 6T3 cells for double-labeling with CgB or PN-1. Lysosomal membrane glycoprotein (lgp120) conjugated with enhanced green fluorescent protein (EGFP-lgp120; a gift from Dr. J. Lippincott-Schwartz, National Institute of Child Health and Human Development) was expressed and detected directly. Rabbit polyclonal anti-GRASP65 antibody (Proteus Bioscience, Ramona, CA) was used at 1:1000 to detect endogenous GRASP65 in the Golgi apparatus. 4,6-Diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) was used for nuclear staining. Fluorescent images were taken using confocal microscopes (MRC 1024, Bio-Rad, Hercules, CA or LSM 510, Carl Zeiss, Thornwood, NY) at the microscopy and imaging core facility in National Institute of Child Health and Human Development and processed using Confocal Assistant and Adobe Photoshop 6.0 software.
Secretion Assay
For secretion studies, CgB was expressed in 6T3-WT or 6T3-bCgA cells by infecting with CgB/Tet-On viruses or CgB/PN-1/Tet-On viruses in the presence of doxycycline (10 μg/ml). At 48 h postinfection, cells were treated with DMEM supplemented with 0.01% BSA for 30 min three times in sequence (B1-B3) followed by stimulation in DMEM supplemented with 0.01% BSA, 50 mM KCl, 2 mM BaCl2 for 30 min at 37°C (S). Each secretion medium was collected and precipitated in 10% trichloroacetic acid (TCA) at 4°C. TCA precipitants were dissolved in 100 μl of sample buffer. An equal volume of each medium sample was analyzed by SDS-PAGE and Western blotting.
RESULTS
CgA Expression Prevented Degradation of DCG Proteins
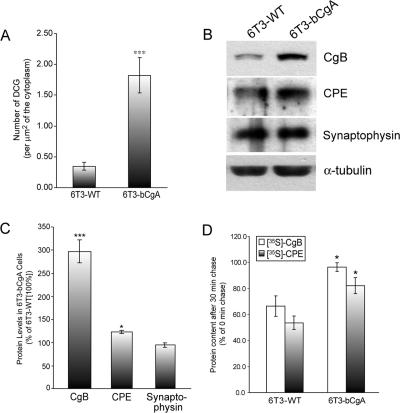
Endocrine 6T3 cells (6T3-WT) have been shown to lack DCG formation and expression of CgA and CgB (Matsuuchi and Kelly, 1991; Day et al., 1995; Kim et al., 2001). Exogenous expression of CgA in these cells recovered regulated secretion (Kim et al., 2001; Loh et al., 2004). Quantification of the number of DCGs formed in 6T3 cells stably expressing bovine CgA (6T3-bCgA) revealed a more than fivefold increase in DCGs (1.82 ± 0.29; scanning electron microscopy [SEM]; 10 EM micrographs covering ∼48 μm2 of the cytoplasmic area), compared with 6T3-WT cells (0.35 ± 0.06; SEM; 20 EM micrographs covering ∼112 μm2 of the cytoplasmic area) (Figure 1A). When rat CgB cDNA was transfected into 6T3-WT cells, CgB was significantly degraded (Kim et al., 2001). Because CgA expression recovered DCG formation in 6T3-WT cells, we predicted that it may also prevent degradation of DCG proteins in these cells. Indeed, the steady state level of transfected CgB was significantly increased in 6T3-bCgA cells (296.38 ± 24.64%; ±SEM; n = 5; p < 0.001) compared with 6T3-WT cells (100%) (Figure 1, B and C). In addition, the level of an endogenous DCG protein, CPE, was also increased in 6T3-bCgA cells (122.57 ± 3.78%; ±SEM; n = 3; p < 0.05) relative to 6T3-WT cells (100%) (Figure 1, B and C). Pulse-chase experiments showed that the levels of [35S]-CgB and [35S]-CPE after 10 min pulse followed by 30-min chase in 6T3-WT cells were ∼66 and ∼54%, respectively, of control (0-min chase). However, this reduction was significantly recovered in 6T3 cells expressing CgA (6T3-bCgA cells). In these cells, the levels of [35S]-CgB and [35S]-CPE were at ∼96% and ∼82% of the control levels (0 min chase), respectively, after 30 min chase (Figure 1D). These data showed that in the absence of CgA expression, DCG proteins such as CgB and CPE were actively degraded during the first 30 min postsynthesis, suggesting that the degradation occurs relatively early in their trafficking pathway (e.g., the endoplasmic reticulum [ER] and/or the Golgi apparatus). In the presence of CgA expression, this degradation was significantly inhibited in 6T3 cells. However, no significant difference was found in the steady-state levels of synaptophysin, a nongranule protein in 6T3-WT and 6T3-bCgA cells (Figure 1, B and C). These data indicate that CgA expression prevents DCG proteins from degradation.
Figure 1.
Recovery of granule proteins from degradation in CgA-expressing 6T3 cells. (A) EM morphometric analysis on the number of DCGs in 6T3-WT and 6T3-bCgA cells is shown as a bar graph. DCGs were counted from 20 (6T3-WT) or 10 (6T3-bCgA) individual EM micrographs, and the number of DCGs per square micrometer was calculated by dividing the number of DCGs with the cytoplasmic area measured from each micrograph. 6T3-WT cells had 0.35 ± 0.06 DCG/μm2 (total of ∼112 μm2 of the cytoplasmic area measured). 6T3-bCgA cells had 1.82 ± 0.29 DCG/μm2 (total of ∼48 μm2 of the cytoplasmic area measured) (***p < 0.0001). (B) Western blot analysis shows the intracellular level of CgB, CPE, synaptophysin, or α-tubulin from 6T3-WT and 6T3-bCgA cells. (C) Bar graph represents the percentage of CgB, CPE, or synaptophysin in 6T3-bCgA cells relative to 6T3-WT cells. CgB, CPE, and synaptophysin levels were 296.38 ± 24.64% (±SEM; n = 5; ***p < 0.001), 122.57 ± 3.78% (±SEM; n = 3; *p < 0.05), and 98.80 ± 4.75% (±SEM; n = 5), respectively, compared with 6T3-WT cells (100%). α-Tubulin was used for normalization. (D) 6T3-WT or 6T3-bCgA cells were metabolically labeled with [35S]Met/Cys for 10 min followed by 0- or 30-min chase. The levels of CgB (open bars) and CPE (closed bars) after 30-min chase were compared with those after 0-min chase as 100%. The levels of [35S]-CgB and [35S]-CPE after 30-min chase were ∼66 and 54%, respectively, in 6T3-WT cells. The levels of [35S]-CgB and [35S]-CPE were significantly (*p < 0.05) increased to ∼96 and ∼82%, respectively, in 6T3-bCgA cells.
Degradation of DCG Proteins Is Initiated in the Golgi Apparatus
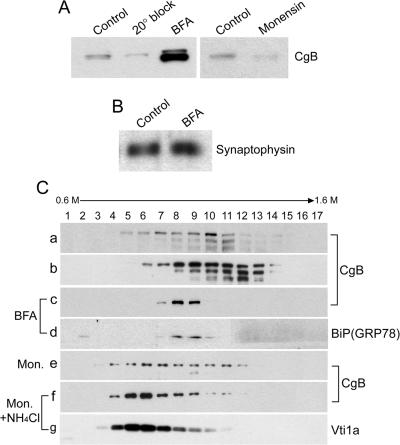
To identify the compartment responsible for degradation of DCG proteins in 6T3-WT cells, we tracked the degradation of exogenously expressed rat CgB as a representative DCG protein. When vesicular trafficking from the ER to the Golgi apparatus was blocked in these cells by BFA (5 μg/ml; 2 h), which redistributes the Golgi apparatus to the ER (Doms et al., 1989), the cellular level of CgB was significantly increased (Figure 2A). This result indicates that the degradation of CgB is a post-ER event. A nongranule protein, synaptophysin, was not affected by BFA treatment (Figure 2B), indicating no degradation of synaptophysin in these cells. Temperature block at 20°C prevents post-TGN trafficking and accumulates proteins in the TGN (Matlin and Simons, 1983). When 6T3-WT cells expressing CgB were incubated at 20°C for 2 h, the level of CgB was not recovered (Figure 2A), indicating that degradation of CgB was initiated in the Golgi complex. To substantiate this observation, another Golgi-perturbing agent, monensin, was used. Monensin treatment (10 μg/ml; 2 h), which blocked trafficking of viral glycoproteins from the medial- to the trans-Golgi cisternae (Griffiths et al., 1983) and also prevented sulfation of POMC occurring at the trans-Golgi cisternae in AtT-20 cells (Fernandez et al., 1997), did not recover the intracellular CgB level in 6T3-WT cells (Figure 2A), further indicating that degradation is initiated at the Golgi apparatus. To further analyze the localization of CgB after treatment of 6T3-WT cells with Golgi-perturbing reagents BFA and monensin, we carried out sucrose density gradient ultracentrifugation. CgB expressed in 6T3-WT cells was spread throughout the fractions 5-11, with slightly more accumulation in the heavier fractions 9-11 (Figure 2C, a). In contrast, CgB, when expressed in 6T3-bCgA cells, showed greater immunoreactivity than in 6T3-WT cells and was distributed toward the heavier density fractions (Figure 2C, 8-13), including fractions representing DCGs (Figure 2C, b, 12-14) (see also Figure 4C, d). Of note is the presence of two to three CgB-immunoreactive bands that are smaller than the intact CgB band (∼110 kDa; top band) in the heavier density fractions (8-14) of sucrose gradient (Figure 2C, a and b). These smaller fragments are likely not degradation products of CgB, because they were more evident under conditions where degradation was prevented, as in CgA-expressing 6T3 cells (Figure 2C, b), than in 6T3-WT cells. BFA treatment, which rescued degradation of CgB, concentrated CgB in fractions 7-9 in 6T3-WT cells (Figure 2C, c). These fractions were positive for an ER marker, BiP (GRP78), confirming that BFA treatment indeed accumulated CgB in the ER (Figure 2C, d). In the presence of monensin, CgB was distributed in fractions 4-12 with slightly more accumulation of immunoreactivity at the lighter density fractions (6-7) (Figure 2C, e) compared with nontreated control cells (Figure 2C, a). If the degradation occurred at the Golgi apparatus, blocking degradation with NH4Cl (Kim et al., 2001) in the presence of monensin would accumulate CgB in the Golgi fractions on sucrose density gradient. When degradation was blocked by 10 mM NH4Cl in the presence of monensin, there was significant accumulation of CgB in fractions 5-6 of 6T3-WT cells (Figure 2C, f), overlapping with the fractions for a TGN marker, Vti1a (Kreykenbohm et al., 2002; Mallard et al., 2002) (Figure 2C, g). There is no difference in the Vti1a localization in sucrose fractions with or without monensin or NH4Cl treatment (our unpublished data).
Figure 2.
Degradation of CgB in the Golgi apparatus. (A) Western blots of CgB expressing 6T3-WT cells comparing the level of CgB before (control) and after 20°C block, BFA, or monensin treatment. (B) Western blot of synaptophysin before and after BFA treatment in 6T3-WT cells. (C) Western blotting analyses on CgB (a, c, e, and f), BiP (GRP78) (d), or Vti1a (g) after subcellular fractionation of CgB-transfected 6T3-WT cells. CgB was also shown after subcellular fractionation of CgB-transfected 6T3-bCgA cells (b). CgB-transfected 6T3-WT cells were analyzed in the presence of BFA (5 μg/ml; 2 h; c and d), monensin (10 μg/ml; 2 h) (e), or monensin + NH4Cl (f and g).
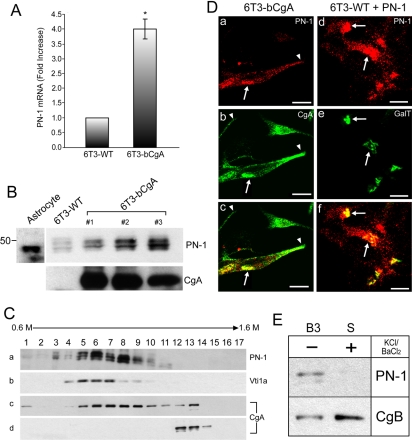
Figure 4.
Up-regulation of PN-1 expression by CgA. (A) Bar graph obtained by real-time PCR analyses shows that PN-1 mRNA levels in 6T3-bCgA cells were 4.01 ± 0.33-fold (±SEM; n = 3; *p < 0.05) higher than 6T3-WT cells. (B) Western blot analysis showing the expression of PN-1 in mouse astrocytes (a positive control), 6T3-WT, and three different clones of 6T3-bCgA cells. Astrocyte cell lysate was used for a positive control for PN-1 expression. CgA expression for 6T3-WT and 6T3-bCgA clones is shown. (C) Western blotting analyses of PN-1, Vti1a (a Golgi marker), and CgA (a DCG marker) after subcellular fractionation of 6T3-bCgA cells by sucrose density gradient ultracentrifugation. The distributions of PN-1 (a), Vti1a (b), and CgA (c) from 6T3-bCgA cells were in fractions 5-8, in fractions 5-7, and in fractions 5-9 and 12-13, respectively. Endogenous CgA from AtT-20 cells was in fractions 12-14 (d). (D) Immunofluorescence microscopy of PN-1 (red) in 6T3-bCgA (a) and 6T3-WT cells (d). For double labeling, CgA (b) or GalT-CFP (e) was visualized. Colocalization between PN-1 and CgA (b and c) or between PN-1 and GalT-CFP (e and f) is indicated by arrows. Bar, 10 μm. (E) Western blot analysis representing secretion assay on PN-1 and CgB from 6T3-bCgA cells. Three consecutive basal secretions (B1-B3; only B3 is shown) of 30 min each were followed by 30-min stimulation (S) with 50 mM KCl/2 mM BaCl2. Although no stimulated secretion of PN-1 was detected, secretion of CgB was stimulated to 2.72 ± 0.93-fold (±SEM; n = 5) in the presence of stimulus.
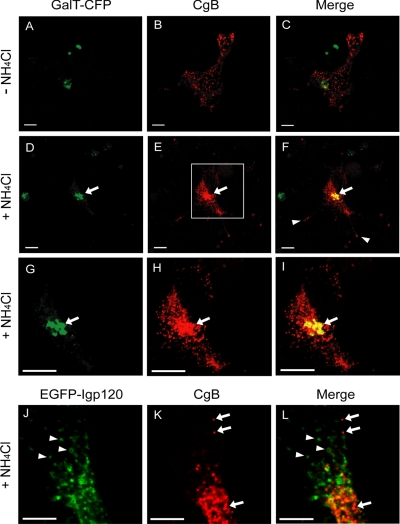
To further substantiate that the recovery of CgB degradation is at the Golgi apparatus, we carried out immunofluorescence microscopy on 6T3-WT cells expressing CgB with or without NH4Cl treatment. Immunoreactive CgB in 6T3-WT cells without NH4Cl treatment showed a very weak and sparse punctate distribution throughout the cell body (Figure 3, B and C). Furthermore, there was little or no colocalization of CgB with β-1,4-galactosyltransferase (GalT), a Golgi marker (Figure 3, A-C). However, when cells were treated with 10 mM NH4Cl for 2 h, the intensity and number of puncta of immunoreactive CgB were greatly increased, showing a strong perinuclear staining pattern that overlapped with GalT-CFP (Figures 3, D-I, arrows), and also staining at the tip of processes was detected (Figure 3F, arrowheads). This result indicates that the recovery of CgB in the presence of NH4Cl occurred at the Golgi apparatus. To determine whether CgB is also recovered in the lysosomes by NH4Cl, in addition to the Golgi apparatus, we coexpressed CgB along with a lysosomal membrane glycoprotein conjugated with EGFP (EGFP-lgp120) in 6T3-WT cells and carried out immunofluorescence microscopy after NH4Cl treatment. Surprisingly, there was no major colocalization of CgB with lgp120 (Figures 3, J-L, arrows and arrowheads), indicating that the recovery of CgB in the presence of NH4Cl occurred mostly in the Golgi apparatus rather than in lysosomes. However, we cannot rule out the possibility that once degradation is initiated at the Golgi apparatus, partially cleaved DCG proteins would then be shunted to lysosomes for complete degradation. Thus, the subcellular fractionation and immunocytochemical data indicate that degradation of CgB was initiated in the Golgi apparatus in 6T3-WT cells, in the absence of CgA expression and DCG biogenesis.
Figure 3.
Recovery of CgB degradation by NH4Cl at the Golgi apparatus. Immunofluorescence microscopy showing CgB immunoreactivity in 6T3-WT cells in the absence (A-C) or presence (D-L) of 10 mM NH4Cl treatment for 2 h. GalT-CFP (A, C, D, F, G, and I) or EGFP-lgp120 (J and L) was used as a Golgi or a lysosomal marker, respectively. Images at a higher magnification of the square box in E are shown in G, H, and I. Bar, 10 μm.
Protease Nexin-1 Expression Is Up-Regulated in 6T3 Cells Expressing CgA
Protection of DCG proteins from degradation in the Golgi apparatus in 6T3-bCgA cells, but not 6T3-WT cells, suggested that CgA might activate the expression of a protease inhibitor(s) to prevent proteolytic degradation in these cells. To identify potential protease inhibitor genes that may be up-regulated in the presence of CgA, we compared gene expression profiles from 6T3-bCgA and 6T3-WT cells using Affymetrix GeneChip microarray (murine U74A). A few genes were up- or down-regulated in 6T3-bCgA cells compared with 6T3-WT cells (Table 1). Among those up-regulated in 6T3-bCgA cells was an mRNA encoding a serine protease inhibitor, PN-1, showing a fold increase of 2.35 ± 0.07 (±SD; n = 2) (Table 1). Subsequent real-time PCR analyses confirmed that PN-1 mRNA was increased to 4.01 ± 0.33-fold (±SEM; n = 3; p < 0.05) in 6T3-bCgA cells compared with 6T3-WT cells (Figure 4A). Western blotting analysis consistently revealed an increase of PN-1 protein levels in three different clones (1-3) of 6T3-bCgA cells (Figure 4B). Thus, CgA expression in 6T3 cells up-regulated expression of a protease inhibitor, PN-1.
Table 1.
Genes regulated by CgA in 6T3 cells in comparison with CgA-deficient wild-type 6T3 cells
If PN-1 is responsible for the prevention of DCG protein degradation in 6T3-bCgA cells, PN-1 should localize to the Golgi apparatus. To determine the intracellular localization of PN-1 in 6T3-bCgA cells, we carried out subcellular fractionation using sucrose density gradient ultracentrifugation. Subcellular fractionation studies showed that endogenously expressed PN-1 in 6T3-bCgA cells was primarily localized in fractions 5-8 (Figure 4C, a), corresponding to the fractions (5-7) containing a TGN marker, Vti1a (Kreykenbohm et al., 2002) (Figure 4C, b). A DCG marker CgA in 6T3-bCgA cells showed two modes of distribution; fractions 5-9 and fractions 12-13 (Figure 4C, c), indicating the presence of CgA in the Golgi apparatus and DCGs, respectively. Endogenous CgA in AtT-20 endocrine cells was distributed in fractions 12-14, corresponding to DCGs (Figure 4C, d). Immunofluorescence microscopy for endogenous PN-1 in 6T3-bCgA cells consistently showed a perinuclear, Golgi-like distribution (Figure 4D, a and c, arrows), overlapping with CgA (Figure 4D, b and c, arrows). Similarly, when coexpressed with GalT, a Golgi marker, PN-1 (Figure 4D, d and f) colocalized with GalT in 6T3-WT cells (Figure 4D, e and f, arrows), indicating that exogenously expressed PN-1 is also localized to the Golgi apparatus. However, PN-1 did not colocalize with CgA along and at the tips of cell processes, where DCGs reside (Figure 4D, a-c, arrowheads). Furthermore, PN-1 secretion was not stimulated by 50 mM KCl/2 mM BaCl2 from 6T3-bCgA cells, but released constitutively, consistent with the lack of localization of PN-1 in DCGs, whereas transfected CgB was secreted in a regulated manner with a fold stimulation of 2.72 ± 0.93 (±SEM; n = 5) in 6T3-bCgA cells (Figure 4E). These data show that PN-1 is present in the Golgi apparatus and secreted through the constitutive pathway but not targeted to DCGs in the regulated secretory pathway.
Protease Nexin-1 Prevents Degradation of CgB at the Golgi Apparatus and Promotes DCG Biogenesis
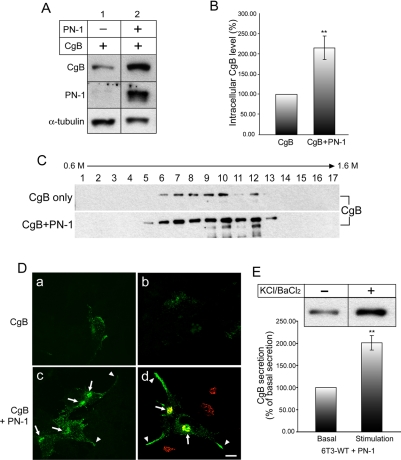
The localization of the degradation of DCG proteins to the Golgi apparatus where PN-1 resides supports a role of this protease inhibitor in regulating the stability of these proteins in this intracellular compartment. To test the role of PN-1 in protecting degradation of DCG proteins, we examined the stability of a DCG protein, CgB, before and after expression of PN-1 in 6T3-WT cells. When 6T3-WT cells were infected with CgB expressing Tet-On adenovirus, a small amount of CgB that survived degradation was detected in the cell lysates (Figure 5A, lane 1). When 6T3-WT cells were coinfected with PN-1 and CgB adenoviruses, the level of CgB in the cells was significantly increased to 216.61 ± 28.98% (±SEM, n = 7, p < 0.01), compared with the CgB level in control cells (Figure 5A, lane 2, and B). Subcellular fractionation study showed that in the presence of PN-1 (CgB+PN-1), overall CgB level was significantly increased throughout the gradients but particularly in the DCG fractions (12-13), compared with the nontransfected control (CgB) (Figure 5C). To determine whether expression of PN-1 could induce DCG biogenesis in 6T3-WT cells, we carried out immunocytochemical studies. When PN-1 was coexpressed with CgB in these cells, immunoreactive CgB was accumulated in the Golgi apparatus (Figure 5D, c and d, arrows), as evidenced by the staining pattern overlapping with a Golgi marker, GRASP65 (Figure 5D, d; red). More importantly, immunoreactive CgB was distributed in a punctate manner along the processes and tips (Figure 5D, c and d, arrowheads) of these cells, characteristic of localization in DCGs. Finally we determined whether the recovered CgB in the presence of PN-1 expression in 6T3-WT cells is in the secretion-competent DCGs. In the presence of 50 mM KCl/2 mM BaCl2, secretion of CgB was significantly stimulated to 201.36 ± 15.73% (±SEM, n = 4, p < 0.01) of basal secretion (100%) (Figure 5E), comparable with the fold stimulation of CgB (∼2.7-fold) shown in 6T3-bCgA cells (Figure 4E). Because CgB expressed in 6T3-WT cells was mostly degraded, CgB was barely detected in the medium. These data indicate that CgB recovered by PN-1 expression was packaged into regulated secretion-competent DCGs in 6T3-WT cells. Thus, PN-1 protected CgB against degradation at the Golgi apparatus, resulting in the promotion of DCG biogenesis in 6T3-WT cells.
Figure 5.
Recovery of CgB degradation by PN-1. (A) Western blotting analysis on CgB and PN-1 from 6T3-WT cells expressing CgB alone (1) or coexpressing CgB and PN-1 (2). (B) Bar graph represents CgB level in the cells expressing PN-1 (216.61 ± 28.98%, ±SEM; n = 7; **p < 0.01) compared with cells without PN-1 expression (100% as a control). α-Tubulin was used as a loading control in A and used for normalization of CgB level for the graph in B. (C) Western blotting analyses of CgB in 6T3-WT cells after subcellular fractionation by sucrose density gradient ultracentrifugation. Before subcellular fractionation, 6T3-WT cells were infected with CgB adenovirus only (top, CgB) or coinfected with CgB and PN-1 adenoviruses (bottom, CgB+PN-1). (D) Immunofluorescence microscopy on CgB in 6T3-WT cells was performed after infection with CgB adenovirus alone (a and b) or with both CgB and PN-1 adenoviruses (c and d). Arrows indicate the Golgi apparatus positive for GRASP65 (d; red), a Golgi marker. Arrowheads indicate CgB immunoreactivity in the processes. Bar, 10 μm. (E) Bar graph represents the normalized data from the Western blotting analysis of secretion assay on CgB from 6T3-WT cells coexpressing PN-1. CgB secretion was significantly increased to 201.36 ± 15.73% (±SEM; n = 4; **p < 0.01) with stimulation compared with basal secretion (100%).
Down-Regulation of Protease Nexin-1 Expression Results in DCG Protein Degradation
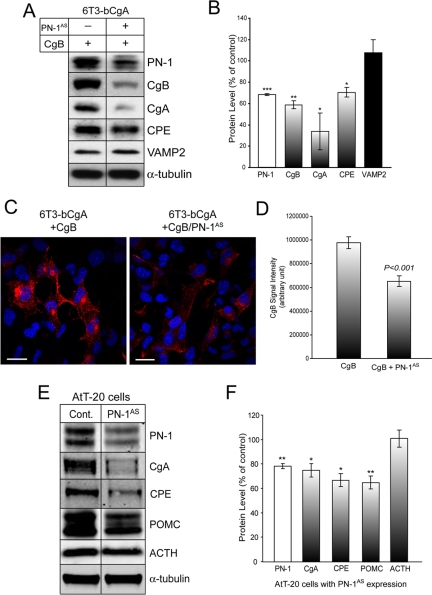
To verify that the inhibition of granule protein degradation by PN-1 is a specific event, we examined the effect of down-regulation of PN-1 in 6T3-bCgA cells that express PN-1 endogenously. PN-1 expression was down-regulated using antisense RNAs against PN-1 (PN-1AS). Expression of PN-1AS resulted in a significant reduction of PN-1 expression to 68.00 ± 1.08% (±SEM; n = 4; p < 0.0001), compared with control cells without PN-1AS viral infection. Concomitantly the level of exogenously expressed CgB was reduced to 58.5 ± 3.69% (±SEM; n = 4; p < 0.01) (Figure 6, A and B). These data further support the observations in Figure 5A, showing that the stability of CgB is dependent on the expression of PN-1 in 6T3-bCgA cells. Reduction of PN-1 also affected the stability of other DCG proteins, CgA and CPE (Figure 6, A and B). CgA and CPE levels in PN-1AS-expressing cells decreased to 33.75 ± 17.25% (±SEM; n = 4; p < 0.05) and 65.4 ± 9.99% (±SEM; n = 4; p < 0.05) of uninfected cells, respectively (Figure 6, A and B). The level of synaptobrevin-2 (VAMP2), a SNARE protein targeted to DCGs in endocrine cells (Eaton et al., 2000), was not affected by the reduction of PN-1 in these cells (107.47 ± 12.19%; ±SD; n = 2) (Figure 6, A and B). Immunofluorescence microscopy showed that the overall intensity of CgB staining was significantly decreased in PN-1 antisense virus-infected cells (Figure 6C), consistent with biochemical analyses shown in Figure 6, A and B. When the total CgB fluorescence signal intensity was measured, there was ∼33% reduction in CgB signal intensity in 6T3-bCgA cells infected with PN-1 antisense adenovirus compared with noninfected 6T3-bCgA cells (Figure 6D). Destabilization of secretory granule proteins caused by PN-1 depletion was also observed in AtT-20 cells. When the expression of endogenous PN-1 was reduced to 77.94 ± 2.07% (±SEM; n = 3) of total in AtT-20 cells by antisense RNAs against PN-1, total cellular levels of CgA, CPE, and POMC were significantly reduced to 74.75 ± 5.39% (±SEM; n = 4), 66.59 ± 5.33% (±SEM; n = 3), and 64.91 ± 5.2% (±SEM; n = 5), respectively (Figure 6, E and F). Because POMC processing to adrenocorticotropic hormone (ACTH) occurs in the DCGs, destabilization of secretory granule proteins by depleting PN-1 in the Golgi complex was expected not to affect the level of ACTH, which is already stored in the DCGs. Indeed, when the level of ACTH was measured in AtT-20 cells expressing PN-1 antisense RNAs, no reduction of ACTH (100.8 ± 6.92%; ±SEM; n = 5) was found in these cells (Figure 6, E and F). These results further show that PN-1 is necessary for the protection of DCG proteins at the Golgi complex.
Figure 6.
Depletion of PN-1 enhanced degradation of DCG proteins. (A) Western blotting analysis of PN-1, CgB, CgA, and CPE levels measured in CgB-expressing 6T3-bCgA cells with or without adenoviral PN-1 antisense (PN-1AS) expression. α-Tubulin was used for a loading control. (B) Bar graph represents quantitation of PN-1, CgB, CgA, CPE, and VAMP2 levels in 6T3-bCgA cells expressing PN-1AS in comparison with those in 6T3-bCgA cells as a control (100%). The reduction of PN-1 (68.00 ± 1.08%; ±SEM; n = 4; ***p < 0.0001) by antisense RNAs in these cells resulted in the decrease of CgB (58.5 ± 3.69%; ±SEM; n = 4; **p < 0.01), CgA (33.75 ± 17.25%; ±SEM; n = 4; *p < 0.05), and CPE (65.4 ± 9.99%; ±SEM; n = 4; *p < 0.05) but not VAMP2 (107.47 ± 12.19%; ±SD; n = 2). (C) Immunofluorescence microscopy on 6T3-bCgA cells with (bottom) or without (top) PN-1 antisense adenovirus infection. Immunoreactive CgB staining was labeled as red. Nuclei were stained with DAPI shown in blue. (D) Quantitation of CgB immunofluorescence intensity in 6T3-bCgA cells with or without PN-1 antisense expression. Intensity was measured using IPLab software of CgB-positive cells from 6T3-bCgA cells without PN-1 antisense expression (n = 291 cells) or 6T3-bCgA cells with PN-1 antisense expression (n = 266 cells). (E) Western blotting analysis of PN-1, CgA, CPE, POMC, and ACTH levels measured in AtT-20 cells with or without PN-1 antisense RNA expression. The blot represents at least three experiments. (F) Bar graph represents quantitation of secretory granule proteins from Western blotting analyses. The level of PN-1 (77.94 ± 2.03%; n = 3), CgA (74.75 ± 5.39%; n = 4), CPE (66.59 ± 5.33%; n = 3), POMC (64.91 ± 5.20%; n = 5), and ACTH (100.8 ± 6.92%; n = 5) measured from AtT-20 cells expressing PN-1 antisense RNAs was expressed as a percentage of control level (100%) in AtT-20 cells after normalization using α-tubulin (±SEM; *p < 0.05; **p < 0.01).
DISCUSSION
CgA plays a pivotal role in controlling the level of DCG biogenesis in neuroendocrine and endocrine cells (Kim et al., 2001). The importance of CgA as a regulator of DCG biogenesis was further emphasized by studies on CgA deficient mice. Transgenic mice down-regulated in CgA biosynthesis by antisense RNA (Kim et al., 2005) as well as CgA knockout mice (Mahapatra et al., 2005) showed a significant depletion of DCGs in the adrenals. We have now uncovered a mechanism of how CgA can regulate DCG biogenesis in endocrine cells. CgA stabilizes DCG proteins posttranslationally by up-regulating the expression of a serine protease inhibitor, PN-1 (Figure 4A). Furthermore, PN-1 was localized to the Golgi apparatus where degradation of DCG proteins was initiated (Figures 2, 3, 4). Depletion of PN-1 from 6T3-bCgA cells enhanced the degradation of not only CgB but also CgA and CPE and resulted in the loss of DCG formation (Figure 6, A-D). In addition, depletion of PN-1 from AtT-20 cells destabilized DCG proteins such as CgA, CPE and POMC but not ACTH that is stored in DCGs (Figure 6, E and F). In contrast, expression of PN-1 alone in the absence of CgA rescued CgB from degradation and induced DCG biogenesis in 6T3 cells (Figure 5). These data provide evidence that CgA up-regulates PN-1 expression, which then stabilizes DCG proteins necessary for DCG biogenesis. Such a regulatory mechanism has an advantage of being able to up- or down-regulate the levels of DCG proteins and therefore the quantity of DCGs formed by merely switching on or off the expression of one gene, PN-1.
Because DCG formation emanates from the TGN, it would be appropriate for the cell to place a control locus at the level of the Golgi apparatus. Indeed, in this study, we have demonstrated the post-translational regulation of granule protein stability at the Golgi apparatus. Neither temperature block at 20°C nor monensin treatment of 6T3-WT cells rescued CgB from degradation in these cells, indicating that the degradation of DCG proteins in these cells is initiated in the Golgi apparatus. This proposal was further substantiated by sucrose density gradient centrifugation studies showing CgB accumulation in the Golgi fractions after monensin/NH4Cl treatment (Figure 2C), and the immunofluorescence microscopy showing that CgB recovered by NH4Cl was primarily accumulated in the Golgi apparatus, rather than lysosomes (Figure 3). Non-DCG protein such as synaptophysin (Figure 1B) was not degraded in 6T3-WT cells in the absence of CgA, similar to β-COP and ARF6, which were not degraded in the absence of CgA in PC12 cells (Kim et al., 2001). Thus, these findings indicate that the initial cleavage of granule proteins is in the Golgi apparatus. After cleavage the products may be further degraded either in the Golgi or sorted into lysosomes for complete degradation.
Selective degradation of DCG proteins may be achieved by proteases recognizing specific motifs on the granule cargo proteins. Several endoproteolytic proprotein processing enzymes (subtilisin-like proprotein convertases and furin) are known to be present and active in the Golgi apparatus (Nakayama, 1997; Steiner, 1998). The major active form of site-1 protease (S1P-C), also known as subtilisin kexin isozyme-1 (SKI-1) (Seidah et al., 1999; Elagoz et al., 2002), which cleaves sterol regulatory element binding proteins, has been shown to be in the Golgi apparatus, especially in the cis- or medial-Golgi cisternae (DeBose-Boyd et al., 1999; Espenshade et al., 1999; Brown et al., 2000). This raises SKI-1 as yet another candidate protease responsible for the initial degradation of DCG proteins in the Golgi apparatus in the absence of CgA. Finally, considering the role of PN-1 described in this report and the fact that it is a known protease inhibitor for plasminogen activator and its active product plasmin (Baker et al., 1980), these proteases could also be candidates, although they have generally been reported to function extracellularly (Murer et al., 2001). Identification of proteases involved in the degradation process of DCG proteins awaits future studies.
Our study shows that up-regulation of PN-1 expression in 6T3 cells is a consequence of CgA expression in these cells. However, it is not clear at present how CgA expression initiates expression of PN-1 in these cells. Unlike CgB (Yoo et al., 2002), there is no evidence of CgA being in the nucleus. Therefore, a role of CgA and/or its processed peptides acting directly as a transcriptional factor to activate gene expression, as in CgB (Yoo et al., 2002), seems unlikely. One plausible way is that secreted CgA or one of its processed peptide products, upon interaction with its cognate receptor(s) on the plasma membrane, may activate a signaling cascade that can then transmit information to the nucleus to up-regulate PN-1 transcription. Such a pathway for the up-regulation of PN-1 expression in CgA-expressing cells will be the focus of our future investigations.
The level of DCG biogenesis is dictated by the availability of DCG proteins in the cells. The amount of DCG proteins available for DCG biogenesis can be regulated at multiple levels: transcriptional, posttranscriptional, and posttranslational. Stimulation (e.g., nicotine, corticosterone, or depolarization) of chromaffin as well as PC12 cells has been shown not only to release DCG content but also to trigger transcriptional up-regulation of mRNAs encoding DCG proteins, including CgA, enkephalin, and enzymes involved in catecholamine synthesis in these cells (Eiden et al., 1984; Wan et al., 1991; Kilbourne et al., 1992; Tang et al., 1996). Although these studies did not analyze DCG biogenesis, it is expected that transcriptional up-regulation of CgA and other DCG protein expression would eventually lead to increased DCG biogenesis. However, because such transcriptional activation to increase the levels of mRNAs encoding multiple DCG proteins is a relatively slow process, this would not be a dynamic regulatory mechanism to rapidly replenish the DCG pool after exocytotic events. Recently, a posttranscriptional mechanism for up-regulating DCG protein expression was reported (Knoch et al., 2004). In that study, polypyrimidine tract-binding protein (PTB), a nuclear-resident protein, was shown to promote DCG biogenesis in pancreatic β cells by stabilizing mRNAs encoding DCG proteins (Knoch et al., 2004). On glucose stimulation in insulin-secreting cells, PTB was translocated to the cytoplasm where it bound to the specific sequence of mRNAs encoding DCG proteins, including CgA and insulin. Stabilization of these mRNAs to increase the level of DCG proteins led to the promotion of DCG biogenesis within hours of stimulation. In our study, we describe yet another unique mechanism, operating at the posttranslational level, which can regulate the amount of DCGs in the cells by maintaining a balance between degradation and stabilization of DCG proteins at the Golgi apparatus. We propose that when increased formation of DCGs is required subsequent to stimulation of endocrine cells to secrete, CgA up-regulates PN-1 expression, which then protects DCG proteins from degradation in the Golgi apparatus and promotes DCG biogenesis. When there are sufficient quantities of DCGs in the cells, excess DCG proteins would be degraded at the Golgi apparatus, and DCG biogenesis would be down-regulated to a steady state level. Because endocrine cells are poised to secrete large amounts of hormones upon demand, and to terminate secretion abruptly upon removal of the stimulus, such a mechanism involving proteolytic degradation and stabilization by a protease inhibitor, PN-1, to modulate the levels of DCG proteins, could constitute a highly efficient way to either replenish or down-regulate DCG formation under various physiological conditions.
Acknowledgments
We thank Dr. J. Lippincott-Schwartz and Dr. L. E. Eiden (National Institute of Mental Health, National Institutes of Health, Bethesda, MD) for the critical reading of this manuscript. We thank Dr. D. Monard for a gift of the anti-PN-1 monoclonal antibody, Dr. K. X. Chai for rabbit PN-1 antibody, Dr. J. Lippincott-Schwartz for GalT-CFP and EGFP-lgp120 constructs, Dr. Susan Cheng (National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD) for EM analyses, and Dr. S. Mahata and D. O'Connor (University of California, San Diego) for the rat CgB construct. We thank S. Imamura and A. Goel for assistance in carrying out some experiments. We also thank the members of SCN for support. This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0755) on November 30, 2005.
References
- Arnaoutova, I., Smith, A. M., Coates, L. C., Sharpe, J. C., Dhanvantari, S., Snell, C. R., Birch, N. P., and Loh, Y. P. (2003). The prohormone processing enzyme PC3 is a lipid raft-associated transmembrane protein. Biochemistry 42, 10445-10455. [DOI] [PubMed] [Google Scholar]
- Arvan, P., and Castle, D. (1998). Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem. J. 332, 593-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan, P., and Halban, P. A. (2004). Sorting ourselves out: seeking consensus on trafficking in the beta-cell. Traffic 5, 53-61. [DOI] [PubMed] [Google Scholar]
- Baker, J. B., Low, D. A., Simmer, R. L., and Cunningham, D. D. (1980). Protease-nexin: a cellular component that links thrombin and plasminogen activator and mediates their binding to cells. Cell 21, 37-45. [DOI] [PubMed] [Google Scholar]
- Brown, M. S., Ye, J., Rawson, R. B., and Goldstein, J. L. (2000). Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391-398. [DOI] [PubMed] [Google Scholar]
- Cool, D. R., Normant, E., Shen, F., Chen, H. C., Pannell, L., Zhang, Y., and Loh, Y. P. (1997). Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpe(fat) mice. Cell 88, 73-83. [DOI] [PubMed] [Google Scholar]
- Dannies, P. S. (1999). Protein hormone storage in secretory granules: mechanisms for concentration and sorting. Endocr. Rev. 20, 3-21. [DOI] [PubMed] [Google Scholar]
- Day, R., Benjannet, S., Matsuuchi, L., Kelly, R. B., Marcinkiewicz, M., Chretien, M., and Seidah, N. G. (1995). Maintained PC1 and PC2 expression in the AtT-20 variant cell line 6T3 lacking regulated secretion and POMC: restored POMC expression and regulated secretion after cAMP treatment. DNA Cell Biol. 14, 175-188. [DOI] [PubMed] [Google Scholar]
- Day, R., and Gorr, S. U. (2003). Secretory granule biogenesis and chromogranin A: master gene, on/off switch or assembly factor? Trends Endocrinol. Metab. 14, 10-13. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd, R. A., Brown, M. S., Li, W. P., Nohturfft, A., Goldstein, J. L., and Espenshade, P. J. (1999). Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703-712. [DOI] [PubMed] [Google Scholar]
- Doms, R. W., Russ, G., and Yewdell, J. W. (1989). Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J. Cell Biol. 109, 61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, B. A., Haugwitz, M., Lau, D., and Moore, H. P. (2000). Biogenesis of regulated exocytotic carriers in neuroendocrine cells. J. Neurosci. 20, 7334-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiden, L. E., Giraud, P., Dave, J. R., Hotchkiss, A. J., and Affolter, H. U. (1984). Nicotinic receptor stimulation activates enkephalin release and biosynthesis in adrenal chromaffin cells. Nature 312, 661-663. [DOI] [PubMed] [Google Scholar]
- Elagoz, A., Benjannet, S., Mammarbassi, A., Wickham, L., and Seidah, N. G. (2002). Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277, 11265-11275. [DOI] [PubMed] [Google Scholar]
- Espenshade, P. J., Cheng, D., Goldstein, J. L., and Brown, M. S. (1999). Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 22795-22804. [DOI] [PubMed] [Google Scholar]
- Fernandez, C. J., Haugwitz, M., Eaton, B., and Moore, H. P. (1997). Distinct molecular events during secretory granule biogenesis revealed by sensitivities to brefeldin A. Mol. Biol. Cell 8, 2171-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, G., Quinn, P., and Warren, G. (1983). Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J. Cell Biol. 96, 835-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundschober, C., Malosio, M. L., Astolfi, L., Giordano, T., Nef, P., and Meldolesi, J. (2002). Neurosecretion competence. A comprehensive gene expression program identified in PC12 cells. J. Biol. Chem. 277, 36715-36724. [DOI] [PubMed] [Google Scholar]
- Hiremagalur, B., Nankova, B., Nitahara, J., Zeman, R., and Sabban, E. L. (1993). Nicotine increases expression of tyrosine hydroxylase gene. Involvement of protein kinase A-mediated pathway. J. Biol. Chem. 268, 23704-23711. [PubMed] [Google Scholar]
- Kilbourne, E. J., Nankova, B. B., Lewis, E. J., McMahon, A., Osaka, H., Sabban, D. B., and Sabban, E. L. (1992). Regulated expression of the tyrosine hydroxylase gene by membrane depolarization. Identification of the responsive element and possible second messengers. J. Biol. Chem. 267, 7563-7569. [PubMed] [Google Scholar]
- Kim, T., Tao-Cheng, J.-H., Eiden, L. E., and Loh, Y. P. (2001). Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell 106, 499-509. [DOI] [PubMed] [Google Scholar]
- Kim, T., Tao-Cheng, J. H., Eiden, L. E., and Loh, Y. P. (2003). The role of chromogranin A and the control of secretory granule genesis and maturation. Trends Endocrinol. Metab. 14, 56-57. [DOI] [PubMed] [Google Scholar]
- Kim, T., Zhang, C.-F., Sun, Z. Q., Wu, H. L., and Loh, Y. P. (2005). Chromogranin A deficiency in transgenic mice leads to aberrant chromaffin granule biogenesis. J. Neurosci. 25, 6958-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch, K. P., Bergert, H., Borgonovo, B., Saeger, H. D., Altkruger, A., Verkade, P., and Solimena, M. (2004). Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 6, 207-214. [DOI] [PubMed] [Google Scholar]
- Kreykenbohm, V., Wenzel, D., Antonin, W., Atlachkine, V., and von Mollard, G. F. (2002). The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81, 273-280. [DOI] [PubMed] [Google Scholar]
- Loh, Y. P., Kim, T., Rodriguez, Y. M., and Cawley, N. X. (2004). Secretory granule biogenesis and neuropeptide sorting to the regulated secretory pathway in neuroendocrine cells. J. Mol. Neurosci. 22, 63-72. [DOI] [PubMed] [Google Scholar]
- Mahapatra, N. R., et al. (2005). Targeted ablation of the chromogranin A gene: elevated blood pressure rescued by the human ortholog. J. Clin. Investig. 115, 1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard, F., Tang, B. L., Galli, T., Tenza, D., Saint-Pol, A., Yue, X., Antony, C., Hong, W., Goud, B., and Johannes, L. (2002). Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malosio, M. L., Giordano, T., Laslop, A., and Meldolesi, J. (2004). Dense-core granules: a specific hallmark of the neuronal/neurosecretory cell phenotype. J. Cell Sci. 117, 743-749. [DOI] [PubMed] [Google Scholar]
- Mansuy, I. M., van der Putten, H., Schmid, P., Meins, M., Botteri, F. M., and Monard, D. (1993). Variable and multiple expression of Protease Nexin-1 during mouse organogenesis and nervous system development. Development 119, 1119-1134. [DOI] [PubMed] [Google Scholar]
- Matlin, K. S., and Simons, K. (1983). Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell 34, 233-243. [DOI] [PubMed] [Google Scholar]
- Matsuuchi, L., and Kelly, R. B. (1991). Constitutive and basal secretion from the endocrine cell line, AtT-20. J. Cell Biol. 112, 843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murer, V., Spetz, J. F., Hengst, U., Altrogge, L. M., de Agostini, A., and Monard, D. (2001). Male fertility defects in mice lacking the serine protease inhibitor protease nexin-1. Proc. Natl. Acad. Sci. USA 98, 3029-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, K. (1997). Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327, 625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, D. M., Iacangelo, A. L., and Eiden, L. E. (1988). Glucocorticoid- and nerve growth factor-induced changes in chromogranin A expression define two different neuronal phenotypes in PC12 cells. Mol. Endocrinol. 2, 921-927. [DOI] [PubMed] [Google Scholar]
- Rozansky, D. J., Wu, H., Tang, K., Parmer, R. J., and O'Connor, D. T. (1994). Glucocorticoid activation of chromogranin A gene expression. Identification and characterization of a novel glucocorticoid response element. J. Clin. Investig. 94, 2357-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah, N. G., et al. (1999). Mammalian subtilisin/kexin isozyme SKI-1, a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA 96, 1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, D. F. (1998). The proprotein convertases. Curr. Opin. Chem. Biol. 2, 31-39. [DOI] [PubMed] [Google Scholar]
- Tang, K., Wu, H., Mahata, S. K., Mahata, M., Gill, B. M., Parmer, R. J., and O'Connor, D. T. (1997). Stimulus coupling to transcription versus secretion in pheochromocytoma cells. Convergent and divergent signal transduction pathways and the crucial roles for route of cytosolic calcium entry and protein kinase C. J. Clin. Investig. 100, 1180-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, K., Wu, H., Mahata, S. K., Taupenot, L., Rozansky, D. J., Parmer, R. J., and O'Connor, D. T. (1996). Stimulus-transcription coupling in pheochromocytoma cells. Promoter region-specific activation of chromogranin a biosynthesis. J. Biol. Chem. 271, 28382-28390. [DOI] [PubMed] [Google Scholar]
- Wan, D. C., Marley, P. D., and Livett, B. G. (1991). Coordinate and differential regulation of proenkephalin A and PNMT mRNA expression in cultured bovine adrenal chromaffin cells: responses to secretory stimuli. Brain Res. Mol. Brain Res. 9, 103-111. [DOI] [PubMed] [Google Scholar]
- Yoo, S. H., You, S. H., Kang, M. K., Huh, Y. H., Lee, C. S., and Shim, C. S. (2002). Localization of the secretory granule marker protein chromogranin B in the nucleus. Potential role in transcription control. J. Biol. Chem. 277, 16011-16021. [DOI] [PubMed] [Google Scholar]