Abstract
Protein kinase C-ε (PKC-ε) translocates to phagosomes and promotes uptake of IgG-opsonized targets. To identify the regions responsible for this concentration, green fluorescent protein (GFP)-protein kinase C-ε mutants were tracked during phagocytosis and in response to exogenous lipids. Deletion of the diacylglycerol (DAG)-binding εC1 and εC1B domains, or the εC1B point mutant εC259G, decreased accumulation at phagosomes and membrane translocation in response to exogenous DAG. Quantitation of GFP revealed that εC259G, εC1, and εC1B accumulation at phagosomes was significantly less than that of intact PKC-ε. Also, the DAG antagonist 1-hexadecyl-2-acetyl glycerol (EI-150) blocked PKC-ε translocation. Thus, DAG binding to εC1B is necessary for PKC-ε translocation. The role of phospholipase D (PLD), phosphatidylinositol-specific phospholipase C (PI-PLC)-γ1, and PI-PLC-γ2 in PKC-ε accumulation was assessed. Although GFP-PLD2 localized to phagosomes and enhanced phagocytosis, PLD inhibition did not alter target ingestion or PKC-ε localization. In contrast, the PI-PLC inhibitor U73122 decreased both phagocytosis and PKC-ε accumulation. Although expression of PI-PLC-γ2 is higher than that of PI-PLC-γ1, PI-PLC-γ1 but not PI-PLC-γ2 consistently concentrated at phagosomes. Macrophages from PI-PLC-γ2-/- mice were similar to wild-type macrophages in their rate and extent of phagocytosis, their accumulation of PKC-ε at the phagosome, and their sensitivity to U73122. This implicates PI-PLC-γ1 as the enzyme that supports PKC-ε localization and phagocytosis. That PI-PLC-γ1 was transiently tyrosine phosphorylated in nascent phagosomes is consistent with this conclusion. Together, these results support a model in which PI-PLC-γ1 provides DAG that binds to εC1B, facilitating PKC-ε localization to phagosomes for efficient IgG-mediated phagocytosis.
INTRODUCTION
Engagement of macrophage Fcγ receptors (FcγRs) with IgG-opsonized particles initiates a cascade of responses, including particle uptake, stimulation of respiratory burst, and up-regulation of gene expression. Because phagocytosis is a localized process, enzymes involved in FcγR signaling are likely to accumulate at the forming phagosome. We recently reported that protein kinase C (PKC)-ε localizes to phagosomes and enhances the rate of IgG-dependent phagocytosis (Larsen et al., 2002). Although recruitment to phagosomes is necessary for PKC-ε function, the mechanism by which it localizes is not known.
PKC-ε belongs to a family of related serine/threonine kinases characterized by their activation requirements: 1) classical isoforms (α, β I, β II, and γ) require Ca2+, diacylglycerol (DAG), and phosphatidylserine; 2) novel isoforms (ε, δ, θ, and η) require DAG and phosphatidylserine but are Ca2+ independent; and 3) atypical isoforms (ζ and ι/λ) require phosphatidylserine. Each isoform consists of a homologous C-terminal catalytic domain and a unique amino regulatory domain (RD). The regulatory domain directs isoform-specific localization in response to agonist stimulation (Newton, 1997). The regulatory domain of PKC-ε, εRD, contains a variable region (V1) with homology to the Ca2+ binding C2 domain of the classic isoforms, a pseudosubstrate (PS), and a conserved C1 region (Figure 2A). The latter region is involved in targeting the enzyme to subcellular membranes (Cho, 2001; Ananthanarayanan et al., 2003). The C1 region is ∼50 amino acids and is divided into tandem cysteine-rich motifs C1A and C1B, comprising two distinct lipid binding sites (Cho, 2001). Lipid binding to the C1 domain is essential for PKC membrane translocation. Of the three lipids known to facilitate PKC membrane translocation (DAG, arachidonic acid, and ceramide), only DAG promotes plasma membrane association of PKC-ε; arachidonic acid and ceramide stimulate PKC-ε Golgi accumulation in CHO-K1 cells (Shirai et al., 2000; Akita, 2002; Kashiwagi et al., 2002). DAG but not arachidonic acid or ceramide stimulates IgG-dependent phagocytosis, consistent with a role in activation of PKC-ε (Karimi and Lennartz, 1995; Suchard et al., 1997).
Figure 2.
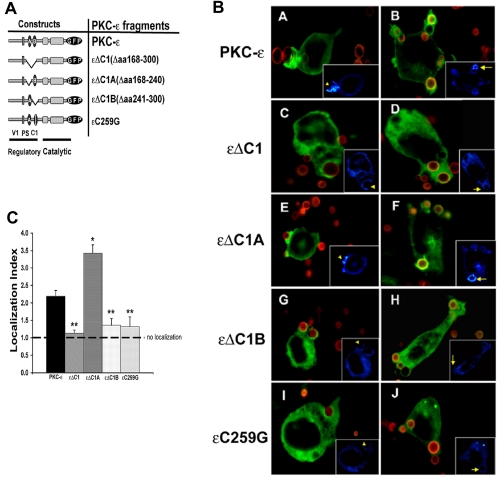
εC1B is necessary for localization to BIgG targets. (A) Constructs used in these studies: V1, variable region 1; PS, pseudosubstrate; C1, constant region 1 containing the lipid binding regions C1A and C1B. (B) Localization to BIgG. Indicated constructs were expressed in RAW 264.7 cells, and transfectants were incubated with Alexa 568-labeled BIgG. Phagocytosis was terminated at 5–7.5 min, the cells were fixed, and the fluorescence was analyzed with postacquisition deconvolution software. Protein kinase C-ε and εΔC1A concentrated at IgG-opsonized targets, whereas εΔC1, εΔC1B, and εC259G did not. Left, protein kinase C-ε distribution in cells containing forming phagosomes (concentration seen in protein kinase C-ε and εΔC1A expressing cells). Right, distribution of GFP conjugates in cells containing fully internalized particles (protein kinase C-ε and εΔC1A-expressing cells retain membrane localization). Insets, pseudocolor images of GFP pixel intensity (concentration visualized as white; blue represents minimal pixel intensity). (C) Quantitation of phagosomal localization. Localization index = GFP fluorescence of phagosomal membrane/nonphagosomal membrane in transfectants expressing the indicated protein kinase C-ε constructs. Data are presented as mean ± SEM (30–40 phagocytic events from four independent experiments). *p < 0.00003, **p < 0.00002 compared with full length protein kinase C-ε (Videos 1–3).
The activation of PKC is a multistep process (Newton, 1997). In the cytosol, the PS occupies the active site of the enzyme, maintaining it in an inactive conformation. On cell stimulation, lipid second messengers are generated that bind to C1, facilitating translocation to the membrane. In many cells, phosphatidylinositol-specific phospholipase C (PI-PLC) is responsible for the simultaneous production of DAG and inositol trisphosphate (IP3); IP3 then stimulates a rise in [Ca2+]. For conventional PKCs (cPKCs), coordinated binding of Ca2+ and lipid to the C2 and C1 domains, respectively, directs membrane association. The Ca2+/DAG binding exposes binding sites for phosphatidylserine on the PKC, leading to a conformational change in the enzyme that releases the PS from the active site. Exposure of the active site allows phosphorylation of downstream targets for signal propagation.
Although the activation steps have been well described for the cPKCs, less is known about the mechanism of activation/translocation of the novel family members. Recently, Jose Lopez-Andreo et al. (2003) reported that phosphatidic acid (PA) and DAG synergize to facilitate translocation of PKC-ε to the plasma membrane in RBL-2H3 mast cells. They present a model in which PA binds to the C2-like/V1 region and DAG binds the C1 domain, resulting in stable association of PKC-ε with the plasma membrane (Jose Lopez-Andreo et al., 2003). This provides a model, analogous to that of the cPKCs, in which synergistic binding to C1 and C2 leads to translocation. Given that PKC-ε concentrates at forming phagosomes and DAG is produced upon FcγR ligation that enhances phagocytosis, we tested the hypothesis that DAG, PA, or both mediate the phagosomal membrane association of PKC-ε. We report herein that DAG but not PA is necessary for PKC-ε translocation during phagocytosis.
DAG is generated via two major pathways: 1) PC→ PA→ DAG, catalyzed by the sequential action of phospholipase D (PLD) and phosphatidic acid phosphohydrolase-1 (PAP-1), respectively; and 2) PLC-dependent cleavage of phosphatidylinositol 4,5-bisphosphate to DAG and IP3 (Nishizuka, 1995; Banno, 2002; Fukami, 2002). Both PLC and PLD concentrate at forming phagosomes (Botelho et al., 2000; Iyer et al., 2004), and their products (DAG, IP3, and PA) are produced, demonstrating that the localized enzymes are active (Fallman et al., 1989; Della Bianca et al., 1990; Lennartz, 1999). We reasoned that if 1) DAG is required for PKC-ε translocation and 2) PKC-ε is necessary for efficient phagocytosis, then PLC, PLD, or both should coordinately modulate both PKC-ε localization and phagocytosis.
Support for a role for PLD in IgG-mediated phagocytosis is based on the following information. Inhibition of PLD with 2,3-diphosphoglyceric acid (2,3-DPG) blocks phagocytosis in human monocytes (Kusner et al., 1999), and ceramide blocks PLD activity and depresses phagocytosis in neutrophils (Suchard et al., 1997). Additionally, that macrophages from PLC-γ2 null mice do not flux Ca2+ in response to IgG-opsonized particles and that phagocytosis is unaffected (Wen et al., 2002) support a PLD-derived DAG model. However, pharmacological inhibition of PI-PLC blocks phagocytosis despite the fact that PI-PLC is an intracellular calcium concentration ([Ca2+]i)-dependent enzyme (Rhee and Bae, 1997) and phagocytosis occurs normally in the absence of cytoplasmic Ca2+ (McNeil et al., 1986; Di Virgilio et al., 1988; Della Bianca et al., 1990; Larsen et al., 2000; Wen et al., 2002). Thus, PLD, PLC, or both may produce the DAG necessary for PKC-ε localization. Our results are consistent with the hypothesis that PLC-derived DAG mediates PKC-ε membrane translocation for efficient phagocytosis.
MATERIALS AND METHODS
Reagents and Antibodies
U73122, U73343, propranolol, and 1-hexadecyl-2-acetyl glycerol (EI-150) were purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA); 2,3-DPG and zymosan A were from Sigma-Aldrich (St. Louis, MO); and 1,2-dioctanoyl-sn-glycerol (DiC8) was from Avanti Polar Lipids (Alabaster, AL). Construction of green fluorescent protein (GFP)-protein kinase C-ε plasmids has been described previously (Larsen et al., 2000, 2002; Kashiwagi et al., 2002). GFP-PLD2 and dnPLD2 plasmids were a generous gift from Dr. Michael Frohman (State University of New York at Stony Brook, Stony Brook, NY) (Choi et al., 2002). The GFP-conjugated PLC-γ1 and PLC-γ2 constructs were described previously (Matsuda et al., 2001; Wang et al., 2001) and were kindly provided by Dr. M. Katan (The Institute of Cancer Research, London, United Kingdom). Antibodies to PLC-γ1 (sc-7290) and PLC-γ2 (sc-5283) and active PLC-γ1 (sc-12943) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture and Transfections
RAW 264.7 cells were maintained in DMEM (Invitrogen, Carlsbad, CA), 10% heat-inactivated bovine calf serum (BCS) (Hyclone Laboratories, Logan UT), 1% penicillin and streptomycin (Larsen et al., 2002). Bone marrow-derived macrophages (BMDMs) were generated from wild-type (C57BL/6) and PLC-γ2-/- bone marrows (Wen et al., 2002). The bones were flushed and cells recovered from the bone marrow were cultured in differentiation media (30% L-cell conditioned medium, 20% heat-inactivated BCS, and 50% DMEM) for 6 d. After differentiation, macrophages were seeded onto 35-mm plates (for phagocytosis), 35-mm glass-bottomed dishes (MatTek, Ashland, MA) for real-time confocal microscopy, or 12-mm coverslips (Erie Scientific, Portsmouth, NH) for immunofluorescence. RAW 264.7 cells (2 × 105) were transiently transfected with 0.5–1 μg of DNA in SuperFect (QIAGEN, Valcenia, CA). After 18–24 h, transfectants were incubated with IgG-opsonized glass bead (BIgG) targets and imaged.
Phagocytosis Assays
Phagocytosis of 51Cr-radiolabeled sheep erythrocytes opsonized with anti-sheep red blood cell IgG (EIgG) targets (PerkinElmer Life and Analytical Sciences, Boston, MA) was conducted as described previously (Larsen et al., 2000, 2002). All assays were done in Hank's balanced salt solution (HBSS) (Invitrogen) containing 4 mM sodium bicarbonate, 10 mM HEPES, and either 1.5 mM CaCl2 and MgCl2 (HBSS++) or 1 mM EGTA and 2 mM MgCl2 (Mg/EGTA). Cells were preincubated with inhibitors (15–45 min; 37°C) before addition of EIgG (10:1, targets:macrophages). Phagocytosis was carried out for 30 min at which time extracellular targets were lysed with 0.83% NH4Cl, and the cells were solubilized in 1 M NaOH. Cell-associated radioactivity was quantified, and the results are presented as percentage of uninhibited control. Background cell associated counts were determined using unopsonized sheep erythrocytes and were <5% of uninhibited controls. Synchronized time courses of BIgG were conducted as described previously (Larsen et al., 2002). Briefly, 4.0 × 105 cells were cooled to 4°C for 30 min, preincubated with BIgG targets on ice (15 min), and phagocytosis was conducted at 37°C. At varying times (0–15 min), cells were fixed and coverslips were imaged by confocal (LSM 510 microscope) or fluorescence microscopy (Olympus BX60 upright microscope with high-performance charge-coupled device camera). Ingestion of BIgG was quantified as phagocytic index, i.e., number of internalized targets/100 cells. Fluorescence images were subjected to postacquisition deconvolution analysis, using the SlideBook Digital microscope software (Intelligent Imaging Innovations, Denver, CO). Real-time imaging was done as detailed previously (Larsen et al., 2002). For exogenous DAG stimulation, 10 μM DiC8 (DAG) was added to live cells, and images were collected every 10 s for 10 min.
Reverse Transcription (RT)-PCR and Real-Time RT-PCR
PLD1 and PLD2 were analyzed by RT-PCR. Total RNA was isolated from RAW 264.7 cells using TRI Reagent (Molecular Research Center, Cincinnati, OH). RNA (5 μg) was transcribed into cDNA using oligo(dT) primers (Promega, Madison, WI) and 50 U of Invitrogen SuperScript III. PCR amplification was performed using primer sets specific for mouse PLD1 and PLD2; β-actin was used as the housekeeping gene. PCR (50 cycles for PLD1 and PLD2 and 28 cycles β-actin) products were separated by 1% agarose gel electrophoresis and stained with ethidium bromide. For real-time RT-PCR reactions, total RNA was prepared from RAW cells and bone marrow-derived macrophages by TriReagent and first-strand cDNA synthesized from total RNA using SuperScript III RNase H-reverse transcriptase per the manufacturer's instructions (Invitrogen). The primers were designed to generate products ≤200 bp with no dimer formation. Relative abundance of PLC-γ1 and PLC-γ2 message was normalized to β-actin in each sample and calculated as 2-(Ct PLC-γ1 or PLC-γ2 - Ct actin), where Ct represents the threshold cycle for each transcript. The specific primers used are as follows: mouse PLC-γ1 forward, 5′-GCCGAGGTGCTGCACCTC and reverse, 5′-GTAGCGGTCAAAGTCCCG; mouse PLC-γ2 forward, 5′-GTCCGGGAGCGGATGACC and reverse, 5′-CCAGTAGTGGGACAAGGG; PLD1 forward, 5′-AGCGAGACCAGGGTGAAC and reverse, 5′-GGTCTGGATGTTGGGCTC; PLD2 forward, 5′-GCCGCCGAGGATATACCCTCC and PLD2 reverse 5′-GGAGCCAAGGTCTGGGAT; and β-actin forward, 5′-TGCGTGGTACCACCATGTAC and β-actin reverse, 5′-AGGGGCCGGACTCATCGTACTC.
Immunoprecipitation and Western Blots
To assess endogenous levels of PLC-γ1 and PLC-γ2, RAW 264.7 cells (2 × 106) and BMDMs (2 × 106) were lysed with ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 12.7 mM deoxycholic acid, 10 mM sodium pyrophosphate, 25 mM sodium β-glycerophosphate, 1 mM sodium orthovanadate, 1% SDS, Triton X-100, and inhibitors 5 mM benzamidine, 50 μg/ml leupeptin, 50 μg/ml aprotinin, 50 μg/ml trypsin inhibitor, 5 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 1 mM Na3VO4, 1 mM para-nitrophenylphosphate, and 5 mM imidazole) and centrifuged (12, 000 × g at 4°C for 15 min) to remove debris. The resulting supernatant was immunoprecipitated with antibodies against PLC-γ1 or PLC-γ2 using protein G-Sepharose, subjected to Western blot analysis, and probed for PLC-γ1 and PLC-γ2 using enhanced chemiluminescence (Pierce Chemical, Rockford, IL). Phospho-PLC-γ1 was detected in phagocytic complexes using a modification of our published protocol (Larsen et al., 2002). Briefly, 12 × 106 RAW cells were seeded onto a 60-mm Petri dish. After adhesion, the cells were washed, 4 ml of HBSS++ was added, and the cells were chilled on ice. BIgG was added (33:1, targets:cells) and allowed to bind on ice. The plates were then put into a 37°C water bath to initiate synchronized phagocytosis. At varying times (0–7.5 min), the cells were scraped into sucrose lysis buffer (25 mM Tris-HCl, pH 7.4, 0.25 M sucrose, and 2.5 mM EDTA plus protease inhibitors), sonicated, and the beads were allowed to settle on ice. The supernatant was removed, and the soluble fraction was prepared by ultracentrifugation (100,000 × g; 60 min; 4°C). The nascent phagosome proteins were extracted from the beads by solubilization in 400 μl of ice-cold RIPA buffer. The supernatant and nascent phagosome solutions were precleared with protein A-agarose beads (2 h; 4°C) and immunoprecipiated with PLC-γ1 antibody (4 h; 4°C). The precipitated proteins were solubilized in sample preparation buffer, separated by SDS-PAGE, and probed for active PLC-γ1 using an antibody specific for the pY783 PLC-γ1 (Santa Cruz Biotechnology). The blots were then stripped and probed for total PLC-γ1.
Confocal Analysis and Quantification
Quantitative analysis of DiC8 (DAG)-stimulated PKC-ε membrane localization was done with live cells imaged by confocal scanning microscopy (Larsen et al., 2002). RAW cells expressing GFP conjugated PKC-ε mutants were visualized and imaged (time 0). DiC8 was added to a final concentration of 10 μM, and images were collected every 10 s for 10 min. The “fold increase” of GFP fluorescence at the plasma membrane versus the cytosol was determined as follows: two 5-μm boxes were drawn on each cell in the first frame of the movie, one at the plasma membrane and the other in the cytosol. The INTERVISION analysis program propagated the boxes through the 120 frames of the movie and calculated the average GFP pixel intensity of the boxes. A localization index (ratio of the average pixel intensity of the plasma membrane/cytosol) was calculated for 1 and 5 min and normalized to the ratio before the addition of DAG (0 min) for each construct. A replot of the fold increase graph compared the localization of each mutant to that of GFP-PKC-ε. Quantitation of localization index: ratio of GFP-fluorescence of phagosomal membrane/nonphagosomal membrane. Images were prepared in Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Immunofluorescence
Endogenous PLC-γ1 and PLC-γ2 were visualized by immunofluorescence as described by Botelho et al. (2000) with minor modifications. Briefly, following phagocytosis, cells were fixed (3.7% formaldehyde; 5 min); permeabilized/blocked in 0.1% Triton X-100, 100 mM glycine, 5% horse serum in phosphate-buffered saline (30 min); stained with mouse anti-PLC-γ1 (overnight) or PLC-γ2 (1 h) (1:50) and visualized with Alexa 488-conjugated goat anti-mouse IgG (1:1000; 1 h).
H2O2 Production
H2O2 production was quantified by the production of oxidized homovanillic acid as described previously (Loegering and Lennartz, 2004). Macrophages were treated overnight with interferon-γ (100 U/ml) (Sigma-Aldrich). For each assay, 1.0 × 106 cells were pretreated with inhibitors (15–45 min; 37°C) or Ca2+ depleted by incubation in Mg/EGTA (Larsen et al., 2002) and stimulated with zymosan A (1 h; 37°C) followed by flourometric determination of oxidized homovanillic acid.
Statistical Analysis
All data are expressed as mean values ± SEM, and significant differences were calculated by analysis of variance. Results with p ≤ 0.05 were considered significant.
Online Supplemental Material
Phagocytosis of BIgG targets was followed with real-time laser scanning fluorescence microscope (LSM 410; Carl Zeiss MicroImaging, Thornwood, NY) with a heated stage and 40× oil objective. GFP was visualized using 488-nm argon excitation and a 505- to 555-barrier filter; Alexa 568 was detected using 543-nm HeNe excitation and a 560-long pass barrier filter. For each experiment, the media were removed and replaced with 800 μl of HBSS2+, followed by 200 μl of BIgG solution (4:1, targets:macrophages). Cells expressing GFP-conjugated PKC-ε, PLD2, PLC-γ1, and PLC-γ2 were located and BIgG was added. The GFP and Alexa images were collected simultaneously at 10-s intervals for 10 min. GFP-protein kinase C-ε, GFP-ε Δ C1A, and GFP-PLD2 localized to targets during IgG-mediated phagocytosis (Videos 1, 2, and 4, respectively); GFP-εΔC1B did not (Video 3).
RESULTS
We have previously shown that PKC-ε modulates the rate of IgG-mediated phagocytosis (Larsen et al., 2002). Because localization of PKC-ε to forming phagosomes is critical for its function, elucidating its mechanism of localization is central to understanding PKC-ε regulation during phagocytosis. Given that exogenous DAG enhances phagocytosis and that the PKC-ε εRD contains a C1 DAG-binding domain (Newton, 2003), we tested the hypothesis that binding of DAG to C1 is necessary for PKC-ε accumulation at IgG-opsonized targets.
εC1B Is Required for PKC-ε Localization to BIgG Phagosomal Membranes
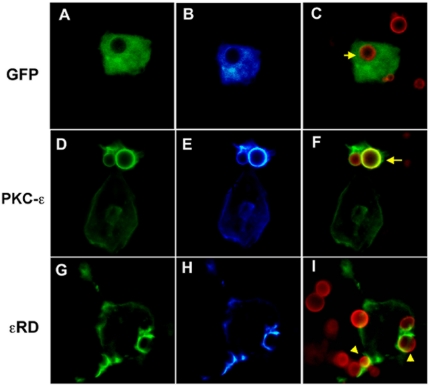
PKC-ε concentrates at forming phagosomes and dissipates soon after vesicle closure (Larsen et al., 2002). To verify that the phagosome localization region(s) reside in the PKC-ε εRD, we transiently expressed εRD-GFP (aa 1–900) and full-length PKC-ε-GFP in RAW 264.7 cells. Their localization was followed during synchronized phagocytosis of Alexa 568-labeled BIgG; control cells expressed unconjugated GFP. When analyzed by postacquisition deconvolution, εRD and PKC-ε exhibited similar patterns of localization (Figure 1). That εRD accumulates at the phagosome but inhibits phagocytosis (Larsen et al., 2002) is consistent with a model in which εRD contains the targeting information necessary to localize the catalytic domain for signal propagation. Given that DAG enhances phagocytosis (Larsen et al., 2002) and that εRD contains the DAG-binding C1 domain, we reasoned that εC1 contributes to PKC-ε membrane localization during phagocytosis. We used GFP-protein kinase C-ε deletion mutants to test this prediction (Figure 2A).
Figure 1.
Protein kinase C-ε and εRD localize to BIgG-containing phagosomes. RAW cells were transiently transfected with GFP (A–C), GFP-conjugated protein kinase C-ε (D–F), or GFP-εRD (G–I). Cells were incubated with BIgG and fixed after 5 min of ingestion. GFP-protein kinase C-ε and εRD accumulated at the phagosomal membrane. Column 1, GFP; column 2, pseudocolor of GFP pixel intensity: blue (low) and white (high); and column 3, merged image of Alexa 568-labeled BIgG and GFP. Note that although the timing of the images is similar, phagocytosis in εRD-expressing cells lags behind that in cells positive for GFP protein kinase C-ε (cups, arrowheads vs. closed phagosomes, arrows).
Deletion of the entire C1 region (εΔC1) abrogated localization in response to BIgG (Figure 2B, C and D). Because both εC1A and εC1B are lipid-binding motifs (Irie et al., 2002), we determined the effect of their individual deletion (εΔC1A and εΔC1B, respectively) on PKC-ε localization. Deletion of εC1A did not decrease PKC-ε localization (Figure 2B, E and F, and Videos 1 and 2). In contrast, εΔC1B-GFP failed to accumulate during phagocytosis, exhibiting a fluorescence pattern indistinguishable from that of the unconjugated GFP control (Figure 2B, G and H; compare with Figure 1 and Video 3). Quantitation of the GFP accumulation at phagosomes confirmed that deletion of εC1 or εC1B but not εC1A significantly decreased their concentration (Figure 2C). Indeed, deletion of C1A increased localization (see Discussion). These results support a role for εC1B in PKC-ε targeting to phagosomal membranes.
It could be argued that deletion of all or part of εC1 changes the conformation of the enzyme and thus interferes with translocation. To address this possibility, a point mutant was generated with a cysteine to glycine substitution at amino acid 259 in the εC1B region (εC259G). This mutation abrogates phorbol-12,13-dibutyrate binding (Kashiwagi et al., 2002). The εC259G mutant produced an expression pattern similar to εΔC1 and εΔC1B, i.e., εC259G did not concentrate at BIgG (Figure 2B, I and J; quantitation in 2C). Together, these results support the conclusion that the DAG-binding function of εC1B is necessary for PKC-ε localization to phagosomal membranes.
DAG Promotes PKC-ε Plasma Membrane Association via εC1B
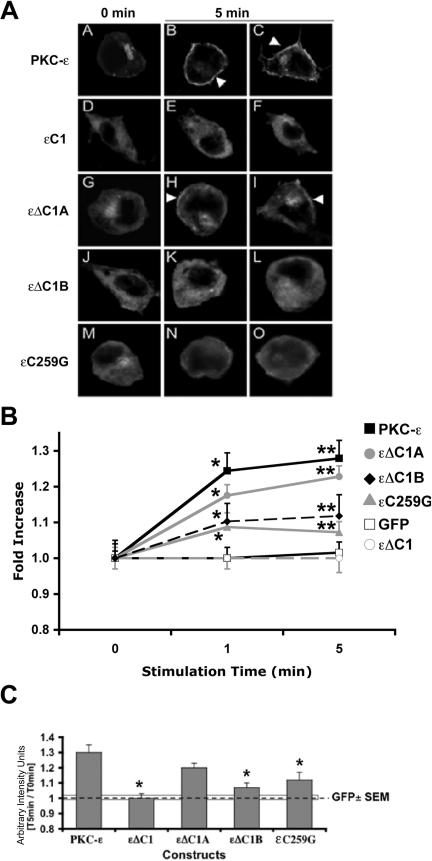
Of the three membrane lipids known to facilitate PKC-ε translocation to membranes (DAG, arachidonic acid, and ceramide), only DAG results in translocation of full-length PKC-ε to the plasma membrane; exogenous arachidonic acid and ceramide triggered PKC-ε movement to the Golgi (Shirai et al., 1998; Irie et al., 2002; Kashiwagi et al., 2002). Given the necessity for PKC-ε translocation to the plasma membrane for phagocytosis and the enhancement of phagocytosis by exogenous DAG, we predicted that DAG would mediate membrane targeting of PKC-ε through εC1B. To determine the effect of DAG on PKC-ε localization, DiC8, a membrane-permeant DAG, was added to cells expressing GFP-εΔC1, εΔC1B, εΔC1A, or the εC259G point mutant. The cells were fixed at 5 min, imaged, and analyzed with postacquisition deconvolution software. Before DAG addition, the constructs were predominantly cytosolic (Figure 3A, 0 min). By 5 min, PKC-ε had concentrated at the plasma membrane; the pattern of translocation was similar for εΔC1A (Figure 3A, B, C, H, and I). In contrast, little or no concentration was apparent in cells expressing εΔC1, εΔC1B, or εC259G (Figure 3A, E, F, K, L, N, and O).
Figure 3.
Exogenous DAG (DiC8) stimulates plasma membrane translocation of GFP-protein kinase C-ε and εΔC1A. (A) Localization of protein kinase C-ε and mutants in response to DAG. Cells were transiently transfected with GFP-conjugated deletion mutants and εC259G, the C1B point mutant. Cells were stimulated with 10 μM DiC8 and analyzed with postacquisition deconvolution software (A) or real-time confocal microscopy (B and C). 0 min, cells before DAG; 5 min, cells 5 min after DAG addition. Arrowheads show plasma membrane accumulation of construct. GFP-protein kinase C-ε and GFP-εΔC1A concentrated at the membrane in response to DAG; no change in localization is seen in cells expressing GFP-εΔC1. (B) Quantitation of DAG translocation. Confocal microscopy was used to quantify the membrane localization of GFP-conjugated constructs. Real-time imaging of cells was performed as detailed in Materials and Methods. GFP concentration was calculated as the ratio of plasma membrane/cytosol, and the results were normalized to the plasma membrane/cystol ratio calculated before DAG addition (0 min). Data are presented as average ± SEM; *p < 0.01, **p < 0.001 relative to 0 min. (C) Deletion/mutation of εC1B inhibits protein kinase C-ε concentration. Comparison of localization of protein kinase C-ε mutants to intact protein kinase C-ε. Five-minute time point in B was replotted as arbitrary intensity units for each construct (T5 min/T0 min). GFP bar is mean ± SEM of unconjugated GFP control cells. *p < 0.05 relative to protein kinase C-ε. (n > 50 events from at least four experiments). Deletion of C1A does not significantly affect localization to DAG. Deletion of εC1 or εC1B or the εC259G point mutant significantly reduced localization.
Real-time imaging by confocal microscopy was used to compare the kinetics of translocation of each mutant with respect to wild-type PKC-ε. The extent of accumulation was quantified as pixel intensity 1 and 5 min after DiC8 addition and normalized to the level of GFP expression in the cytosol (see details in Materials and Methods). With the exception of εΔC1 and the GFP control, all the constructs translocated to the plasma membrane with significant accumulation by 1 min. Depending on the construct, accumulation either plateaued or increased during the following 4 min (Figure 3, B and C). The extent of the accumulation varied among the mutants that did translocate. Although sometimes lower than full-length PKC-ε, εΔC1A concentration was not statistically different from the full-length construct (Figure 3C). However, loss of εC1, εC1B, or mutation of εC259G resulted in a dramatic decrease in translocation (Figure 3, B and C). These studies confirm the involvement of εC1B in the translocation of PKC-ε in response to DAG stimulation.
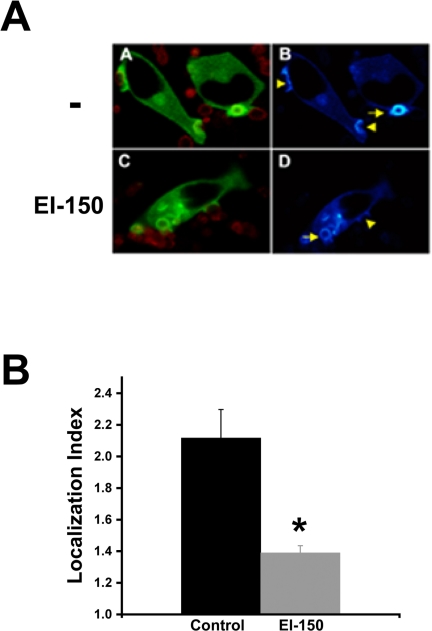
These results suggest that DAG, binding to εC1B, directs PKC-ε translocation during phagocytosis. This is consistent with the results in Figure 2 in which εC259G was significantly less responsive during phagocytosis and in response to exogenous DiC8. If it is true that DAG mediates PKC-ε localization, then blocking DAG should decrease membrane association. This was tested using EI-150 (Daniel and Fen, 1988), a DAG antagonist that binds to the C1 region of PKC, effectively blocking endogenous DAG association with those sites (Slater et al., 2001). Of relevance to these studies, we have previously shown that EI-150 blocks localization of PKC-ε in response to exogenous DiC8 in CHO-K1 cells (Shirai et al., 2000). GFP-protein kinase C-ε-expressing macrophages were pretreated with EI-150 (100 μM; 45 min) followed by real-time confocal imaging of BIgG-mediated phagocytosis. Quantitation of GFP accumulation in carrier control versus EI-150-treated cells revealed that EI-150 significantly decreased PKC-ε localization (Figure 4); phagocytosis was also substantially reduced. These data provide additional support for a model in which DAG binding to εC1B is necessary for PKC-ε accumulation during phagocytosis.
Figure 4.
DAG antagonist decreases protein kinase C-ε localization. GFP-protein kinase C-ε-expressing cells were preincubated with EI-150 (100 μM; 45 min) followed by BIgG phagocytosis. (A) GFP localization was followed by real-time confocal microscopy in control (-) or EI-150-treated cells (A and C, merge image; B and D, pseudocolor for GFP expression). Accumulation can be seen in forming phagosomes (arrowhead) and after target internalization (arrow). (B) Quantitation of GFP protein kinase C-ε localization in EI-150-treated cells. The Localization Index was calculated for phagosomes lying just beneath the plasma membrane (e.g., targets indicated by arrow in left panels). Data are presented as mean ± SEM (n > 35 events from four independent experiments). *p < 0.003 compared with untreated protein kinase C-ε-expressing cells.
PLD Is Not Necessary for IgG-mediated Phagocytosis
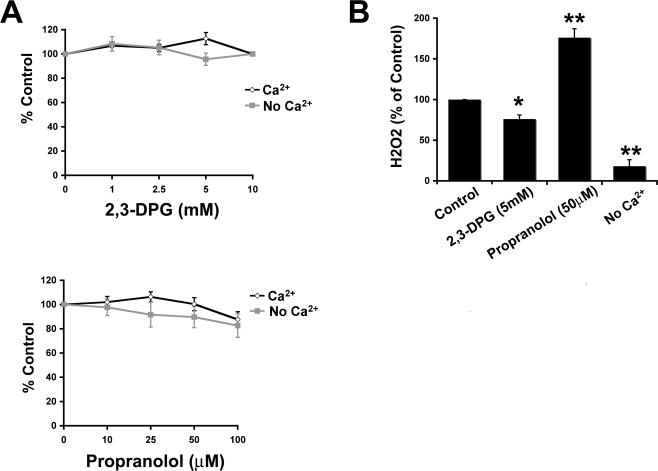
The above-mentioned results strongly support a role for DAG in the uptake of IgG-opsonized targets. Given that 1) phagocytosis occurs independently of Ca2+ (Lennartz et al., 1993); 2) Ca2+ depletion abrogates IP3 production in neutrophils (indicating a block in PI-PLC) (Fallman et al., 1989); and 3) FcγR-mediated phagocytosis is inhibited by 2,3-DPG in human monocytes (blocking PLD-derived DAG production (Kusner et al., 1999), we tested the hypothesis that PLD is necessary for PKC-ε localization during phagocytosis. RAW cells express mRNA for PLD1 and PLD2 (our unpublished data). Because phagocytosis is a localized response to FcγR cross-linking, we reasoned that enzymes that contribute to uptake should concentrate at the phagosome. In resting cells, GFP-PLD2 was distributed at the plasma membrane, whereas PLD1 was cytosolic. During phagocytosis, the distribution of PLD1 was not altered (our unpublished data), but PLD2 concentrated with a pattern resembling that of PKC-ε (compare Videos 1 and 4 and Figure 5A). Additionally, phagocytosis was enhanced in GFP-PLD2-expressing cells (Figure 5B), similar in extent to that produced upon expression of GFP-protein kinase C-ε (Larsen et al., 2002). Thus, PLD2 could provide the DAG necessary for PKC-ε localization. If true, blocking either production of PA or conversion of PA to DAG should decrease both localization of PKC-ε and phagocytosis. Neither 2,3-DPG, a direct inhibitor of PLD activity (Kanaho et al., 1993), nor propranolol, which blocks phosphatidic acid phosphohydrolase (Billah et al., 1989), altered target uptake (Figure 6A). To determine the effect of Ca2+ on the system, we tested 2,3-DPG in the presence and absence of Ca2+. Ca2+ depletion did not reveal a PLD requirement. Additionally, 2,3-DPG and propranolol (10 mM and 100 μM, respectively) did not significantly reduce GFP PKC-ε accumulation during phagocytosis. Because little or no inhibition was observed with these drugs, we tested their efficacy in RAW 264.7 cells by determining their effect on zymosan-stimulated respiratory burst. In this system, 2,3-DPG significantly inhibited the burst, whereas propranolol enhanced it (Figure 6B). These results are consistent with the literature demonstrating that the respiratory burst is PA dependent (Palicz et al., 2000). Together, these results argue against a primary role for PLD in phagocytosis/protein kinase C-ε localization in RAW 264.7 mouse macrophages.
Figure 5.
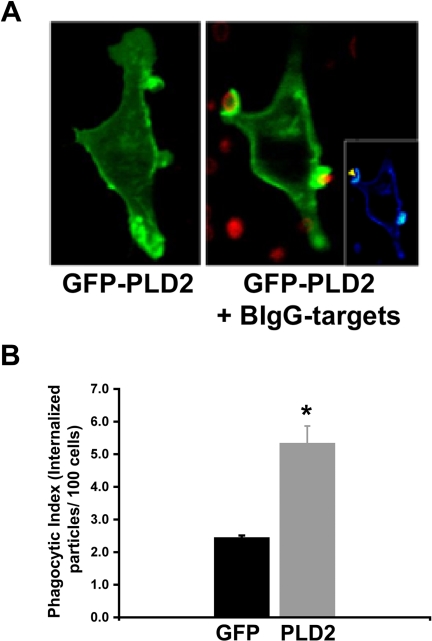
PLD2 localizes to forming phagosomes and enhances phagocytosis. (A) GFP-PLD2-expressing cells were incubated with BIgG, and ingestion was monitored by real-time confocal microscopy. PLD2 is associated with the plasma membrane before the addition of BIgG (left), concentrates at forming phagosomes (right), and dissipates upon phagosome closure (representative of >4 experiments). Images taken from online Video 4. (B) Expression of GFP PLD2 enhances BIgG phagocytosis. Cells were transfected with GFP-PLD2 or unconjugated GFP and incubated with BIgG, fixed at 7.5 min, and internalization was determined microscopically. Phagocytic index = number of internalized targets/100 cells. Data are presented as the mean ± SEM (>150 events/condition from 3 independent experiments). *p < 0.008.
Figure 6.
Inhibition of PLD does not alter phagocytosis. (A) Neither 2,3-DPG nor propranolol inhibits phagocytosis in the presence or absence of Ca2+. Cells were preincubated with Mg/EGTA (to deplete Ca2+) or HBSS++ (control) followed by 15- to 30-min incubation with inhibitors before the addition of EIgG. Phagocytosis was terminated by lysis of the extracellular erythrocytes at 30 min. Data are expressed as a percentage of the untreated control and presented as the mean ± SEM (n = 4). (B) 2,3-DPG and Ca2+ depletion depress H2O2 production, whereas propranolol enhances it. Cells were treated with the indicated inhibitors or Ca2+ depleted before incubation with zymosan (1 h; 37°C), which activates NADPH oxidase; H2O2 production was determined as in Materials and Methods. Data are presented as the mean ± SEM (n = 3). *p < 0.05, **p < 0.002 compared with control.
Involvement of PLC in Phagocytosis and PKC-ε Localization
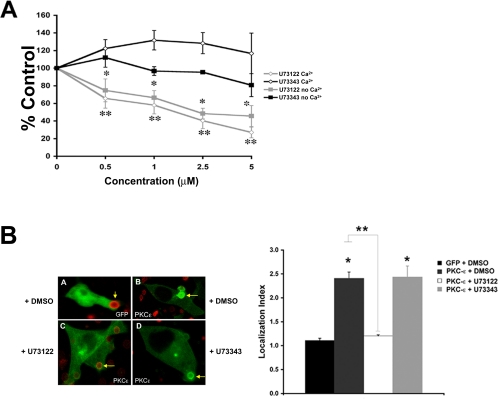
The simultaneous production of IP3 and DAG upon FcγR ligation implicates the activation of PI-PLC (Fallman et al., 1989). Additionally, the failure of PLC-γ2 null macrophages to flux Ca2+ during phagocytosis suggests that this is the predominant PI-PLC isoform linked to FcγR signaling (Wen et al., 2002). Thus, the role of PI-PLC in PKC-ε localization and phagocytosis was investigated. Cells were treated with U73122, a PI-PLC inhibitor, and U73343, its inactive analog. Phagocytosis of EIgG decreased in a dose-dependent manner upon U73122 treatment; U73443 did not inhibit uptake at any concentration tested (Figure 7A). Ca2+ depletion did not alter the dose dependency of U73122, suggesting that PLC, putatively a Ca2+-dependent enzyme (Rhee and Bae, 1997) is active at very low [Ca2+]i, below that necessary for translocation via the C2 domain (Larsen et al., 2000). Consistent with a role for PLC in PKC-ε localization, U73122 (1 μM) but not U73343 blocked PKC-ε translocation (Figure 7B).
Figure 7.
Inhibition of PLC blocks EIgG-mediated phagocytosis and localization of protein kinase C-ε. (A) Cells were Ca2+ depleted as described in Materials and Methods and preincubated with U73122 or U73343 (30 min; 37°C) before EIgG phagocytosis; control samples were calcium replete. U73122 but not U73343 inhibited phagocytosis to the same extent regardless of the presence of Ca2+. U73122 inhibition in Ca2+-depleted and Ca2+-replete cells is not significantly different for any concentration of drug used. Data are presented as the mean ± SEM (n = 3), *p < 0.04, **p < 0.009. (B) U73122 blocks localization of GFP protein kinase C-ε at phagosomes. Macrophages were transfected with GFP protein kinase C-ε or unconjugated GFP and treated with 1 μM U73122 (bottom left) or U73343 (bottom right) (30 min; 37°C) before the addition of BIgG. Real-time imaging was used to follow phagocytosis. Protein kinase C-ε concentrated at targets in dimethyl sulfoxide (DMSO) and U73343-treated cells, but localization was abolished by treatment with U73122. The Localization Index of GFP at fully formed phagosomes was quantified. Results are mean ± SEM for >40 events/condition from four independent experiments. *, **p < 0.001 relative to GFP and untreated GFP-protein kinase C-ε-expressing cells, respectively.
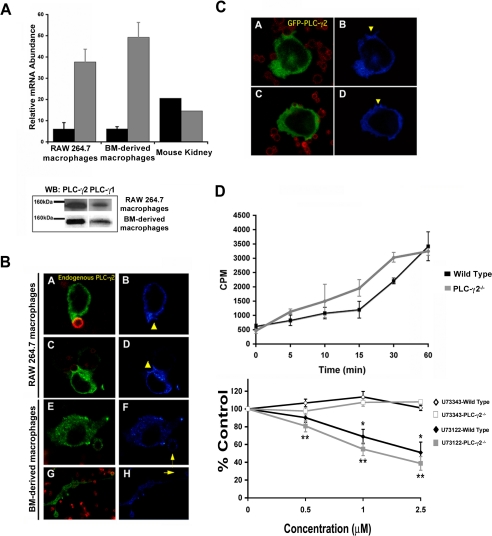
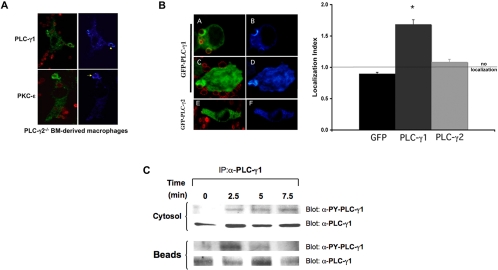
Macrophages express both PLC-γ1 and PLC-γ2 (Figure 8A). Because PLC-γ2 is a expressed selectively in hematopoietic cells, we tested its role in IgG-mediated phagocytosis. If PLC-γ2 was the major source of DAG for phagocytosis, we would predict that it would accumulate at the phagocytic cup during ingestion to generate the DAG necessary for PKC-ε localization. To test this, RAW 264.7 and bone marrow-derived macrophages were fixed at varying times during synchronized BIgG phagocytosis, and endogenous PLC-γ2 was localized by immunofluorescence. Surprisingly, little PLC-γ2 concentration was detected at regions of the membrane containing bound and/or internalized particles (Figure 8B). Similarly, GFP PLC-γ2 failed to concentrate during phagocytosis of BIgG in RAW cells (Figure 8C). These results argued against the involvement of PLC-γ2 in phagocytosis. We therefore examined phagocytosis in macrophages from PLC γ2-/- mice; loss of PLC-γ2 had no effect on either the rate or extent of phagocytosis (Figure 8D). Finally, PKC-ε concentration at the phagosome was normal in PLC-γ2-/- macrophages (Figure 9A). Together, these results argue against the involvement of PLC-γ2-derived DAG in the localization of PKC-ε to phagosomes. However, the observation that macrophages from PLC-γ2 null and wild-type mice were equally sensitive to U73122 inhibition (Figure 8D) supported the involvement of a (non-PLC-γ2) PI-PLC.
Figure 8.
PLC-γ2 is not required for phagocytosis. (A) Mouse macrophages contain significantly more PLC-γ2 than PLC-γ1 mRNA as determined by real-time RT-PCR. PLC-γ2 (gray bars) message levels were approximately threefold higher than PLC-γ1 (black bars) in RAW 264.7 macrophages and bone marrow (BM)-derived macrophages; expression of the two isoforms is similar in kidney. Data are presented as the mean ± SE for three independent experiments. Western blot confirming expression of PLC-γ2 and PLC-γ1 protein in BMDM and RAW cells. Blots are representative of three to five independent experiments. (B) Endogenous PLC-γ2 does not accumulate during IgG-mediated phagocytosis in RAW cells or BMDMs. Macrophages were incubated with BIgG targets, fixed at 5–7.5 min, and immunostained for PLC-γ2. (A, C, E, and G) Merge image Alexa 488 (PLC-γ2) and Alexa 568 (BIgG). (B, D, F, and H) Pseudocolor for Alexa 488. (C) GFP-PLC-γ2 does not accumulate at IgG-phagosomes. GFP-PLC-γ2-expressing cells were preincubated with BIgG targets, and phagocytosis was terminated at 5–7.5 min. (D) Phagocytosis is normal in PLC-γ2-/- BMDM but is sensitive to PLC inhibition. BMDMs from PLC-γ2-/- and littermate controls were incubated with EIgG, and phagocytosis was followed for 60 min. Phagocytosis in PLC-γ2-/- BMDMs was similar to that in wild-type cells. PLC-γ2-/- BMDMs were preincubated with the indicated concentrations of U73122 or U73343 (30 min; 37°C) followed by EIgG (60 min). U73122 but not U73343 inhibited phagocytosis to the same extent in the presence or absence of PLC-γ2. Data expressed as the mean ± SEM (n = 5 independent experiments; *p < 0.05 compared with control).
Figure 9.
PLC-γ1 is involved in IgG-mediated phagocytosis. (A) Endogenous PLC-γ1 and protein kinase C-ε concentrate at phagosomes in PLC-γ2-/- BMDMs. PLC-γ2-/- BMDMs were incubated with BIgG targets (2.5–10 min), fixed and stained for PLC-γ2 (top) or protein kinase C-ε (bottom). Left column, merge image Alexa 488 (PLC-γ1protein kinase C-ε) and Alexa 568 (BIgG); right column, pseudocolor for Alexa 488. Representative of three experiments with similar results. (B) PLC-γ1 but not PLC-γ2 accumulates at forming phagosomes. RAW cells were transiently transfected with GFP-PLC-γ1 (A–D) or GFP-PLC-γ2 (E and F). Phagocytosis of BIgG was terminated at 5–7.5 min. Cells were fixed, and GFP distribution was detected by confocal microscopy. The localization index for GFP, GFP-PLC-γ1, or GFP-PLC-γ2 was calculated (>30 events from 3 independent experiments for each construct). Data are presented as mean ± SEM, *p < 0.0004 compared with GFP. (C) PLC-γ1 is phosphorylated during IgG phagocytosis. Bead-associated nascent phagosomes were separated from the nonbead cytosolic fraction at the indicated times during synchronized phagocytosis. Both fractions were solubilized and immunoprecipitated with antibody to PLC-γ1. The associated proteins were separated by SDS-PAGE and blotted for active PLC-γ1 using the activation-specific anti-PLC-γ1 antibody (recognizes phospho-tyrosine 783 on PLC-γ1) and PLC-γ1. Representative of three similar experiments.
PLC-γ1 is also expressed in RAW cells, albeit at much lower levels (Figure 8A). The most direct approach for determining the importance of PLC-γ1 in phagocytosis and PKC-ε localization would be to study PLC-γ1 down-regulated cells. However, because deletion of PLC-γ1 is an embryonic lethal and small interfering RNA is not effective in macrophages, we used alternative approaches to implicate PLC-γ1 in PKC-ε localization. Endogenous PLC-γ1 concentrated at phagosomes in wild-type and PLC-γ2-/- macrophages (Figure 9A). In contrast to GFP-PLC-γ2, GFP-PLC-γ1 transiently accumulated at targets (Figure 9B). Finally, the levels of tyrosine phosphorylated PLC-γ1 associated with targets increased at 2.5 min during synchronized phagocytosis (Figure 9D). Because tyrosine phosphorylation activates PLC (Wen et al., 2002), these findings suggest that PLC-γ1 is transiently activated at the phagosome during phagocytosis. This activation likely generates the DAG necessary for PKC-ε concentration. Surprisingly, neither expression of GFP PLC-γ1 nor GFP PLC-γ2 enhanced phagocytosis, suggesting that PLC activation is not a rate-determining step in particle ingestion.
Because U73122 did not abolish phagocytosis, we tested the hypothesis that PLD and PLC may each contribute to signaling. We thus combined varying concentrations of 2,3-DPG (0–10 mM) and 2.5 μM U73122 to determine whether there was a synergistic/additive effect of the drugs. No concentration of 2,3-DPG tested altered the 50% inhibition produced by 2.5 μM U73122, confirming that PLD has little or no impact on ingestion and that PI-PLC has a profound role in this process. That phagocytosis could not be eliminated with the combination of PLC and PLD inhibitors implicates an additional, as yet unknown, pathway that accounts for the balance of the ingestion signal.
DISCUSSION
Lipid-dependent Membrane Localization
We have previously shown that PKC-ε is required for efficient phagocytosis and that its regulatory domain, εRD, localizes to phagosomes. Although it concentrates at sites of target binding (Figure 1), εRD blocks internalization (Larsen et al., 2002), demonstrating that the function of εRD is to localize the PKC-ε catalytic domain for propagation of the internalization signal. Thus, we sought to determine how PKC-ε is retained at the phagosome.
Exogenous DAG promotes membrane translocation of PKC-ε in CHO-K1 cells (Shirai et al., 1998) and macrophages (Figure 3). Because DAG enhances phagocytosis (Karimi and Lennartz, 1995) and elicits translocation of GFP-protein kinase C-ε to the plasma membrane (Figure 3), we focused on the role of the lipid-binding C1 domain in localization. PKC-ε accumulation, in real time, is first apparent at the phagocytic cup as a “flash” upon phagosome closure (Video 1). This pattern is strikingly similar to that seen during phagocytosis in RAW-expressing PKC-δC1-YFP (a DAG reporter) (Botelho et al., 2000). In these studies, DAG concentration was maximal at phagosome closure, consistent with PKC-ε docking to DAG in the membrane. The failure of the εC259G point mutant to localize to targets and the ability of EI-150, the DAG antagonist, to block PKC-ε accumulation and phagocytosis is consistent with DAG, binding to εC1B, directing PKC-ε to the phagosome.
εC1B seems to be regulated by its counterpart εC1A. εΔC1A accumulation was approximately twice that of intact PKC-ε (Figure 2C), suggesting that εC1A may suppress translocation mediated by εC1B. Such control could impact the dwell-time of PKC-ε on the membrane. Because C1A has a lower affinity for lipids than C1B, one could imagine that at low concentrations of DAG (i.e., early in phagocytosis), C1B would be engaged, resulting in a tight association of PKC to the membrane. As DAG accumulates, it would bind C1A, decreasing enzyme affinity for the membrane and facilitating its eventual release (Szallasi et al., 1996; Bogi et al., 1998; Slater et al., 1998; Medkova and Cho, 1999; Cho, 2001).
It was reported that the PLD product PA synergizes with DAG for membrane translocation of PKC-ε in FcεR-activated RBL-2H3 cells (Jose Lopez-Andreo et al., 2003). Their data support a model in which PA (binding to the εV1 region) and DAG (binding to εC1) are both necessary for translocation. This model would predict that 1) PA should cause translocation, 2) propranolol should not alter this accumulation, and 3) deletion of εV1 should decrease/abrogate PKC-ε membrane association. In our cells, PA indeed causes plasma membrane accumulation of PKC-ε. However, the timing is much delayed (by >3 min) compared with DAG and is blocked by propranolol (Flax and Lennartz, unpublished data). Additionally, whereas point mutants in the PKC-ε V1 domain block ligand-mediated translocation of protein kinase C-ε in the RBL-2H3 mast cells, deletion of the entire εV1 region does not inhibit localization of protein kinase C-ε to IgG-containing phagosomes (Flax and Lennartz, unpublished data). Thus, it seems unlikely that either the V1 region or PA mediates protein kinase C-ε localization during IgG-mediated phagocytosis. These differences may reflect difference in mast cell versus macrophage signaling. However, it is also likely that these results reflect the ability of signaling molecules (i.e., protein kinase C-ε) to detect subtle differences in the second messengers (PA and DAG) produced upon ligation of different receptors.
PLD Versus PLC: Source of DAG
The data in Figures 1, 2, 3, 4 are consistent with the hypothesis that DAG, binding to εC1B, facilitates membrane translocation of protein kinase C-ε. DAG can be released the combined action of PLD and PAP-1 or directly via PI-PLC.
Results from several groups directed our attention to the PLD: 1) reports that PLD is necessary for IgG- and complement-mediated phagocytosis (Kusner et al., 1996, 1999; Suchard et al., 1997); 2) a [Ca2+]i rise mediates translocation of PI-PLC, and phagocytosis occurs in the absence of such a rise (McNeil et al., 1986; Di Virgilio et al., 1988); and 3) macrophages from PLC-γ2 null mice have no defect in IgG-mediated phagocytosis (Wen et al., 2002). RAW 264.7 cells express mRNA for PLD1 and PLD2 (Fenn and Lennartz, unpublished data). Expression of GFP-PLD2 revealed a concentration of the enzyme at phagosomes (Video 4) and increased phagocytosis in GFP-PLD2-expressing cells. However, expression of dominant negative PLD2 did not inhibit phagocytosis nor did 2,3-DPG or propranolol (Figure 6). Thus, our results argue against a role for PLD in protein kinase C-ε targeting in IgG-stimulated mouse macrophages. These results contrast with published reports (Kusner et al., 1999; Iyer et al., 2004) where these same PLD inhibitors at the same concentrations using the same targets block IgG-dependent phagocytosis in human monocytes. The explanation for this apparent discrepancy is unclear but may reflect differences in species or in the level of the PLD isoforms expressed in the two cell types.
PI-PLC is the other major cellular pathway for DAG release. Of the four PI-PLC families, only the PLC-γ isoforms have been linked to FcγR signaling (Bonilla et al., 2000; Botelho et al., 2000). Inhibition of phagocytosis by U73122 is consistent with the involvement of PI-PLC in target uptake. Because PLC-γ2 is selectively expressed in hematopoietic cells, it was reasonable to postulate that it plays a role in phagocytosis. We predicted that it would accumulate at phagosomes to generate the DAG for protein kinase C-ε localization. However, we were unable to detect concentration of endogenous PLC-γ2 in RAW cells. A slight accumulation was periodically detected in GFP-PLC-γ2-expressing RAW cells but did not reach statistical significance. Similarly, we could not detect concentration of endogenous PLC-γ2 in primary BMDMs. Extensive analysis of BMDMs from PLC-γ2-/- mice revealed that 1) neither the rate nor extent of phagocytosis was affected by the absence of PLC-γ2 (Figure 8D), 2) the U73122 sensitivity of both PLC-γ2-/- and controls was similar (Figure 8D), and 3) and protein kinase C-ε concentrated to phagosomes in macrophages lacking PLC-γ2 (Figure 9A). These results suggested that PLC-γ2 is not the major isoform contributing to protein kinase C-ε localization.
Involvement of PLC-γ1 in phagocytosis is implicated by early studies linking FcγR IIA ligation to tyrosine phosphorylation of PLC-γ1 (Shen et al., 1994). More recently, Jones et al. (2005) demonstrated that PLC-γ1 is required for cell migration, a process that may be similar to pseudopod extension. In RAW cells, endogenous PLC-γ1 localized to BIgG phagosomes in both wild-type and PLC-γ2-/- cells. Its concentration was significantly higher than that of PLC-γ2 despite the fact that there is substantially less PLC-γ1 than PLC-γ2 protein in the RAW cells (Figure 8A). Phosphorylated PLC-γ1 associated transiently with the phagosome, peaking around 2.5 min, earlier than protein kinase C-ε (Larsen et al., 2002), suggesting that PLC-γ1 enters the phagosome before protein kinase C-ε. Real-time microscopy experiments are required to address this issue. Botelho et al. (2000) demonstrated that the tandem Src homology (SH)2 domains of PLC-γ1 inhibit IgG-mediated phagocytosis, consistent with our data implicating PLC-γ1 in protein kinase C-ε localization of and propagation of the uptake signal. Additionally, their studies suggest that the SH2 domains dock to the phosphorylated intracellular tyrosine-based activation motifs in the cytoplasmic tail of activated FcγR, providing a Ca2+-independent mechanism for localizing PLC to phagosomes.
An intriguing question is why PLC-γ1 but not PLC-γ2 is recruited to phagosomes. The low accumulation of GFP PLC-γ2 and occasional detection of endogenous PLC-γ2 at phagosomes suggest that, although PLC-γ2 may be recruited to phagosomes, it is not retained there. The SH2 and pleckstrin homology (PH) domains can bind to phosphorylated ITAMs and polyphosphoinositides, both of which are present in the phagosome. Although the PH and SH2 domains of PLC-γ2 and PLC-γ1 are 50 and 58% identical, respectively, it is possible that these differences decrease the stability of PLC-γ2 at the phagosome. Chimeric PLCs expressing different SH2 and PH domains may be necessary to address this question.
Model of Protein Kinase C Activation and Membrane Localization during IgG-mediated Phagocytosis
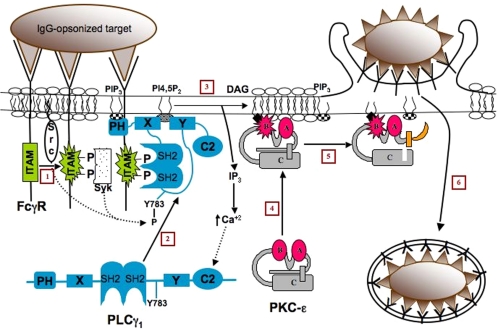
Collectively, the pharmacological and molecular data, in conjunction with published reports, suggest the following model (Figure 10): IgG-opsonized targets cross-link FcγR on the macrophage cell surface. The FcγR ITAM domains are phosphorylated by a Src family kinase (Figure 10, 1) (Garcia-Garcia and Rosales, 2002), providing a docking site for the tandem SH2 domains on PI-PLC-γ1 (Figure 10, 2) (Botelho et al., 2000). Docking of PLC-γ1 to the phosphorylated ITAMs brings it into proximity with the nonreceptor tyrosine kinases src and syk that phosphorylate tyrosine 783, an event critical for PI-PLC-γ activation (Rhee, 2001; Jones et al., 2005). Interaction of PLC-γ1 with membrane phosphatidylinositol-(3,4,5)-trisphosphate may allow it to be retained and further increase its activity (Sekiya et al., 2004). PI-PLC-γ1 hydrolyzes phosphatidylinositol to DAG and IP3. The liberated DAG binds to εC1B, facilitating localized accumulation of protein kinase C-ε. PIP3 can also enhance the catalytic activity of protein kinase C-ε (Toker et al., 1994). Active protein kinase C-ε phosphorylates as yet unidentified downstream targets, thereby amplifying a signaling cascade that culminates in internalization of IgG-opsonized particles. The specificity of the system lies in the ITAMs of the FcγR. The phosphorylated ITAMs are scaffolds for numerous SH2-containing molecules (e.g., PI3Kinase, syk, and PLC-γ) and serve to concentrate them, and their products, at the phagosome for target internalization. Other protein kinase C-ε domains may also be necessary to facilitate the conformational change that fully activates protein kinase C-ε (Figure 10, 5). This process concentrates protein kinase C-ε at the forming phagosome where it propagates the uptake signal (Figure 10, 6).
Figure 10.
Model of macrophage protein kinase C-ε activation during phagocytosis. Activation of protein kinase C-ε involves the following steps: FcγR cross-linking results in Src activation and phosphoryaltion of the FcγR ITAM (1). The phosphorylated ITAMs provide a docking site for the tandem SH2 motifs of PLC-γ1 (2). PI-PLC generates DAG in the phagosomal membrane (and IP3 in Ca2+-containing cells) (3). This DAG binds to the C1B domain of protein kinase C-ε, thus concentrating it near bound targets (4). The IP3-mediated rise in [Ca2+]i may facilitate recruitment or retention of PI-PLC at the phagosomal membrane. The binding of DAG (and possibly other docking molecules in the phagosome) results in a conformational change in protein kinase C-ε that exposes its active site (5) for phosphorylation of downstream targets and completion of ingestion (6).
Supplementary Material
Acknowledgments
We thank Shoua Yang for genotyping the PLC-γ2 null mice, Kathleen Fenn for the PCR analysis, David Chrostowski for the phosphotyrosine blots, Drs. N. P. Jones and M. Katan for the generous gift of GFP-PLC-γ1 and GFP-PLC-γ2, Dr. Joseph Mazurkiewicz for confocal assistance, Dr. James Drake for critical reading of the manuscript and discussions, and Rachel Hathaway and Ami Baxi for technical assistance. Confocal studies were done in the Albany Medical Center Imaging Core and the Biosignal Research Center. This study was supported by National Institutes of Health Grant GM-50821 (to M.R.L.) and T32HL-07194.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1100) on November 30, 2005.
Abbreviations used: 2,3-DPG, 2,3-diphospho-d-glyceric acid; BIgG, IgG-opsonized glass beads; BMDM, bone marrow-derived macrophage; DAG, diacylglycerol; EIgG, IgG-opsonized sheep red blood cell; GFP, green fluorescent protein; IP3, inositol trisphosphate; PA, phosphatidic acid; PAP-1, phosphatidic acid phosphohydrolase-1; PI-PLC, phosphatidylinositol-specific phospholipase C; PKC, protein kinase C; PLD, phospholipase D.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Akita, Y. (2002). Protein Kinase C-epsilon (protein kinase C-epsilon): its unique structure and function. J. Biochem. 132, 847-852. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan, B., Stahelin, R. V., Digman, M. A., and Cho, W. (2003). Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 278, 46886-46894. [DOI] [PubMed] [Google Scholar]
- Banno, Y. (2002). Regulation and possible role of mammalian phospholipase D in cellular functions. J. Biochem. 131, 301-306. [DOI] [PubMed] [Google Scholar]
- Billah, M. M., Eckel, S., Mullmann, T. J., Egan, R. W., and Siegel, M. I. (1989). Phosphatidylcholine hydrolysis by phospholipase D determines phosphatidate and diglyceride levels in chemotactic peptide-stimulated human neutrophils. Involvement of phosphatidate phosphohydrolase in signal transduction. J. Biol. Chem. 264, 17069-17077. [PubMed] [Google Scholar]
- Bogi, K., Lorenzo, P. S., Szallasi, Z., Acs, P., Wagner, G. S., and Blumberg, P. M. (1998). Differential selectivity of ligands for the C1a and C1b phorbol ester binding domains of protein kinase Cdelta: possible correlation with tumor-promoting activity. Cancer Res. 58, 1423-1428. [PubMed] [Google Scholar]
- Bonilla, F. A., Fujita, R. M., Pivniouk, V. I., Chan, A. C., and Geha, R. S. (2000). Adapter proteins SLP-76 and BLNK both are expressed by murine macrophages and are linked to signaling via Fcgamma receptors I and II/III. Proc. Natl. Acad. Sci. USA 97, 1725-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho, R. J., Teruel, M., Dierckman, R., Anderson, R., Wells, A., York, J. D., Meyer, T., and Grinstein, S. (2000). Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151, 1353-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, W. (2001). Membrane targeting by C1 and C2 domains. J. Biol. Chem. 276, 32407-32410. [DOI] [PubMed] [Google Scholar]
- Choi, W. S., Kim, Y. M., Combs, C., Frohman, M. A., and Beaven, M. A. (2002). Phospholipases D1 and D2 regulate different phases of exocytosis in mast cells. J. Immunol. 168, 5682-5689. [DOI] [PubMed] [Google Scholar]
- Daniel, T. O., and Fen, Z. (1988). Distinct pathways mediate transcriptional regulation of platelet-derived growth factor B/c-sis expression. J. Biol. Chem. 263, 19815-19820. [PubMed] [Google Scholar]
- Della Bianca, V., Grzeskowiak, M., and Rossi, F. (1990). Studies on molecular regulation of phagocytosis and activation of the NADPH oxidase in neutrophils. J. Immunol. 144, 1411-1417. [PubMed] [Google Scholar]
- Di Virgilio, F., Meyer, C. B., Greenberg, S., and Silverstein, S. C. (1988). Fc Receptor-mediated phagocytosis occurs in macrophages at exceedingly low cytosolic Ca+2 levels. J. Cell Biol. 106, 657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallman, M., Lew, D. P., Stendahl, O., and Andersson, T. (1989). Receptor-mediated phagocytosis in human neutrophils is associated with increased formation of inositol phosphates and diacylglycerol. Elevation in cytosolic free calcium and formation of inositol phosphates can be dissociated from accumulation of diacylglycerol. J. Clin. Investig. 84, 886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami, K. (2002). Structure, regulation, and function of phospholipase C isozymes. J. Biochem. (Tokyo) 131, 293-299. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia, E., and Rosales, C. (2002). Signal transduction during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 72, 1092-1108. [PubMed] [Google Scholar]
- Irie, K., Nakahara, A., Nakagawa, Y., Ohigashi, H., Shindo, M., Fukuda, H., Konishi, H., Kikkawa, U., Kashiwagi, K., and Saito, N. (2002). Establishment of a binding assay for protein kinase C isozymes using synthetic C1 peptides and development of new medicinal leads with protein kinase C isozyme and C1 domain selectivity. Pharmacol. Ther. 93, 271-281. [DOI] [PubMed] [Google Scholar]
- Iyer, S. S., Barton, J. A., Bourgoin, S., and Kusner, D. J. (2004). Phospholipases D1 and D2 coordinately regulate macrophage phagocytosis. J. Immunol. 173, 2615-2623. [DOI] [PubMed] [Google Scholar]
- Jones, N. P., Peak, J., Brader, S., Eccles, S. A., and Katan, M. (2005). PLC{gamma}1 is essential for early events in integrin signalling required for cell motility. J. Cell Sci. 118, 2695-2706. [DOI] [PubMed] [Google Scholar]
- Jose Lopez-Andreo, M., Gomez-Fernandez, J. C., and Corbalan-Garcia, S. (2003). The simultaneous production of phosphatidic acid and diacylglycerol is essential for the translocation of protein kinase Cepsilon to the plasma membrane in RBL-2H3 cells. Mol. Biol. Cell 14, 4885-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaho, Y., Nakai, Y., Katoh, M., and Nozawa, Y. (1993). The phosphatase inhibitor 2,3-diphosphoglycerate interferes with phospholipase D activation in rabbit peritoneal neutrophils. J. Biol. Chem. 268, 12492-12497. [PubMed] [Google Scholar]
- Karimi, K., and Lennartz, M. R. (1995). Protein kinase C activation precedes arachidonic acid release during IgG-mediated phagocytosis. J. Immunol. 155, 5786-5794. [PubMed] [Google Scholar]
- Kashiwagi, K., Shirai, Y., Kuriyama, M., Sakai, N., and Saito, N. (2002). Importance of C1B domain for lipid messenger-induced targeting of protein kinase C. J. Biol. Chem. 277, 18037-18045. [DOI] [PubMed] [Google Scholar]
- Kusner, D. J., Hall, C. F., and Jackson, S. (1999). Fc gamma receptor-mediated activation of phospholipase D regulates macrophage phagocytosis of IgG-opsonized particles. J. Immunol. 162, 2266-2274. [PubMed] [Google Scholar]
- Kusner, D. J., Hall, C. F., and Schlesinger, L. S. (1996). Activation of phospholipase D is tightly coupled to the phagocytosis of Mycobacterium tuberculosis or opsonized zymosan by human macrophages. J. Exp. Med. 184, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, E. C., DiGennaro, J. A., Saito, N., Matha, S., Loegering, D. J., Mazurkiewicz, J. M., and Lennartz, M. R. (2000). Differential requirement for classic and novel protein kinase C isoforms in respiratory burst and phagocytosis in RAW 264.7 cells. J. Immunol. 165, 2809-2817. [DOI] [PubMed] [Google Scholar]
- Larsen, E. C., Ueyama, T., Brannock, P. M., Shirai, Y., Saito, N., Larsson, C., Loegering, D., Weber, P. B., and Lennartz, M. R. (2002). A role for protein kinase C-varepsilon in FcgammaR-mediated phagocytosis by RAW 264.7 cells. J. Cell Biol. 159, 939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz, M. R. (1999). Phospholipases and phagocytosis: the role of phospholipid-derived second messengers in phagocytosis. Int. J. Biochem. Cell Biol. 31, 415-430. [DOI] [PubMed] [Google Scholar]
- Lennartz, M. R., Lefkowith, J. B., Bromley, F. A., and Brown, E. J. (1993). IgG-mediated phagocytosis activates a calcium-independent, phosphatidylethanolamine specific phospholipase. J. Leukoc. Biol. 54, 389-398. [DOI] [PubMed] [Google Scholar]
- Loegering, D. J., and Lennartz, M. R. (2004). Signaling pathways for Fcγ receptor-stimulated tumor necrosis factor-α secretion and respiratory burst in RAW 264.7 macrophages. Inflammation 28, 23-31. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Paterson, H. F., Rodriguez, R., Fensome, A. C., Ellis, M. V., Swann, K., and Katan, M. (2001). Real time fluorescence imaging of PLC gamma translocation and its interaction with the epidermal growth factor receptor. J. Cell Biol. 153, 599-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, P. L., Swanson, J. A., Wright, S. D., Silverstein, S. C., and Taylor, D. L. (1986). Fc-receptor-mediated phagocytosis occurs in macrophages without an increase in average [Ca++]i. J. Cell Biol. 102, 1586-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova, M., and Cho, W. (1999). Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem. 274, 19852-19861. [DOI] [PubMed] [Google Scholar]
- Newton, A. C. (1997). Regulation of protein kinase C. Curr. Opin. Cell Biol. 9, 161-167. [DOI] [PubMed] [Google Scholar]
- Newton, A. C. (2003). Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem. J. 370, 361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka, Y. (1995). Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 9, 484-496. [PubMed] [Google Scholar]
- Palicz, A., Foubert, T. R., Jesaitis, A. J., Marodi, L., and McPhail, L. C. (2000). Phosphatidic acid and diacylglycerol directly activate NADPH oxidase by interacting with enzyme components. J. Biol. Chem. 276, 3090-3097. [DOI] [PubMed] [Google Scholar]
- Rhee, S. G. (2001). Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70, 281-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S. G., and Bae, Y. S. (1997). Regulation of phosphoinositide-specific phospholipase C isozymes. J. Biol. Chem. 272, 15045-15048. [DOI] [PubMed] [Google Scholar]
- Sekiya, F., Poulin, B., Kim, Y. J., and Rhee, S. G. (2004). Mechanism of tyrosine phosphorylation and activation of phospholipase C-gamma 1. Tyrosine 783 phosphorylation is not sufficient for lipase activation. J. Biol. Chem. 279, 32181-32190. [DOI] [PubMed] [Google Scholar]
- Shen, Z., Lin, C.-T., and Unkeless, J. C. (1994). Correlations among tyrosine phosphorylation of Shc, p72syk, PLC-γ1, and [Ca2+]i flux in FcγRIIA signaling. J. Immunol. 152, 3017-3023. [PubMed] [Google Scholar]
- Shirai, Y., Kashiwagi, K., Yagi, K., Sakai, N., and Saito, N. (1998). Distinct effects of fatty acids on translocation of gamma- and epsilon-subspecies of protein kinase C. J. Cell Biol. 143, 511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai, Y., Segawa, S., Kuriyama, M., Goto, K., Sakai, N., and Saito, N. (2000). Subtype-specific translocation of diacylglycerol kinase alpha and gamma and its correlation with protein kinase C. J. Biol. Chem. 275, 24760-24766. [DOI] [PubMed] [Google Scholar]
- Slater, S. J., Seiz, J. L., Stagliano, B. A., Cook, A. C., Milano, S. K., Ho, C., and Stubbs, C. D. (2001). Low- and high-affinity phorbol ester and diglyceride interactions with protein kinase C: 1-O-alkyl-2-acyl-sn-glycerol enhances phorbol ester- and diacylglycerol-induced activity but alone does not induce activity. Biochemistry 40, 6085-6092. [DOI] [PubMed] [Google Scholar]
- Slater, S. J., Taddeo, F. J., Mazurek, A., Stagliano, B. A., Milano, S. K., Kelly, M. B., Ho, C., and Stubbs, C. D. (1998). Inhibition of membrane lipid-independent protein kinase Calpha activity by phorbol esters, diacylglycerols, and bryostatin-1. J. Biol. Chem. 273, 23160-23168. [DOI] [PubMed] [Google Scholar]
- Suchard, S. J., Hinkovska-Galcheva, V., Mansfield, P. J., Boxer, L. A., and Shayman, J. A. (1997). Ceramide inhibits IgG-dependent phagocytosis in human polymorphonuclear leukocytes. Blood 89, 2139-2147. [PubMed] [Google Scholar]
- Szallasi, Z., Bogi, K., Gohari, S., Biro, T., Acs, P., and Blumberg, P. M. (1996). Non-equivalent roles for the first and second zinc fingers of protein kinase Cdelta. Effect of their mutation on phorbol ester-induced translocation in NIH 3T3 cells. J. Biol. Chem. 271, 18299-18301. [DOI] [PubMed] [Google Scholar]
- Toker, A., Meyer, M., Reddy, K. K., Falck, J. R., Aneja, R., Aneja, S., Parra, A., Burns, D. J., Ballas, L. M., and Cantley, L. C. (1994). Activation of protein kinase C family members by the novel polyphosphoinositides PtdIns-3,4-P2 and PtdIns-3,4,5-P3. J. Biol. Chem. 269, 32358-32367. [PubMed] [Google Scholar]
- Wang, X. J., Liao, H. J., Chattopadhyay, A., and Carpenter, G. (2001). EGF-dependent translocation of green fluorescent protein-tagged PLC-gamma1 to the plasma membrane and endosomes. Exp. Cell Res. 267, 28-36. [DOI] [PubMed] [Google Scholar]
- Wen, R., Jou, S. T., Chen, Y., Hoffmeyer, A., and Wang, D. (2002). Phospholipase C gamma 2 is essential for specific functions of Fc epsilon R and Fc gamma R. J. Immunol. 169, 6743-6752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.