Abstract
Multiple binding sites for an initiator protein are a common feature of replicator sequences from various organisms. By binding to the replicator, initiators mark the site and contribute to melting or distortion of the DNA by largely unknown mechanisms. Here we analyze origin of DNA replication (ori) binding by the E1 initiator and show sequential binding to a set of overlapping binding sites. The assembly of these initiator complexes is controlled by a gradual reduction in the dependence of interactions between the initiator and DNA and a gradual increase in the reliance on interactions between initiator molecules, providing a mechanism for sequential and orderly assembly. Importantly, the binding of the initiator causes progressive structural alterations both in the sites and in the sequences flanking the sites, eventually generating severe structural alterations. These results indicate that the process of template melting may be incremental, where binding of each initiator molecule serves as a wedge that upon binding gradually alters the template structure. This mechanism may explain the requirement for multiple initiator binding sites that is observed in many ori's.
Initiator proteins, which have been identified for a variety of replicons, have critical functions in DNA replication and act by binding to specific sites at the origin of DNA replication (ori) (21). The role that the initiator plays, apart from the recognition of the replicator, varies considerably in different systems. Viral initiators such as E1 from papillomavirus and T-antigen (T-Ag) from simian virus 40 (SV40), in addition to ori binding, provide a little-understood activity that melts DNA, and these proteins also serve as the replicative DNA helicases (for reviews, see references 11 and 42). At the other end of the spectrum, for the origin recognition complex (ORC) proteins, no activity apart from DNA binding has been detected (for a review, see reference 20). Interestingly, the prokaryotic DnaA protein is intermediate in this regard. In addition to recognition of the replicator, DnaA is clearly involved in the melting of oriC (for a review, see reference 29). Thus, there are significant functional similarities between initiators from eukaryotic viruses and prokaryotic initiators, although the degree of relatedness between these initiator proteins is not known. Until recently, little connection to initiators from higher organisms has been found. Recent studies, however, demonstrate that RepA, the initiator from a Pseudomonas plasmid is related to CDC6/ORC proteins, thus forging a link between the prokaryotic and eukaryotic families of initiator proteins (16). This provides a strong justification for understanding how the relatively low-complexity viral initiators perform their function since this would likely provide information about how DnaA, and possibly even initiators from higher organisms, function. Furthermore, the viral initiators currently provide the most tractable systems to dissect the biochemical mechanisms involved in the melting of double-stranded DNA, a fundamental process for both transcription and replication that in spite of its importance is uncharacterized.
In both the viral ori's and in oriC, multiple binding sites (BS) for the initiator are present. Likely, the requirement for multiple BS is unrelated to recognition of the ori since a single site or a small number of sites could serve that purpose, as is apparent for Saccharomyces cerevisiae ORC, which utilizes a single BS (20). More likely, the function of the multiple sites is related to a function common to both the viral initiators and DnaA, such as ori melting, which requires the assembly of large DNA-protein complexes. Overall however, very little information about the role of the BS in the formation and function of these larger protein complexes is available. For most of the important model initiators such as DnaA, SV40 T-Ag, and ORC, only limited information about how they recognize and bind their target sequences is available. In recent years we have come to understand the DNA binding of the papillomavirus E1 initiator in great detail. This system now provides an excellent opportunity for the analysis of the function and importance of multiple initiator BS and the initiator complexes that are generated through the use of these sites.
The papillomavirus family constitutes a large group of viruses, many of which cause disease in humans, including cervical cancer (48). Viral proteins E1 and E2 are required for initiation of viral DNA replication (7, 45). E1 is a multifunctional protein that recognizes the origin of replication and that also melts the DNA duplex (see reference 42 for a review). In a hexameric form E1 also functions as the replicative DNA helicase (13, 36, 39, 47). E1 binds to an 18-bp palindromic sequence in the bovine papillomavirus (BPV) origin of DNA replication (ori) (17, 18, 28, 44, 46) (see Fig. 1). This sequence contains multiple BS for E1 arranged in an overlapping array (5, 6). This arrangement of BS is conserved in other papillomavirus ori's and shows distinctive similarities to the arrangement of large T-Ag BS in the SV40 ori (6, 43). E2 is a virus-encoded transcription factor (see reference 27 for a review) which binds to a site flanking the E1 palindrome and which interacts cooperatively with E1.
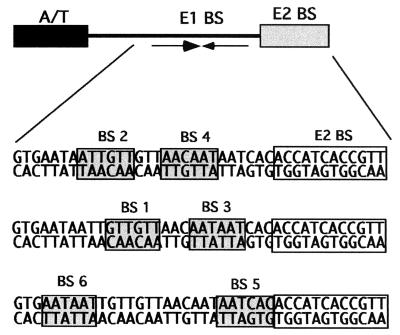
FIG. 1.
Schematic drawing of the 60-bp minimal origin of replication from BPV. The boxes highlight the sequences and positions of the six overlapping E1 BS (BS 1 to 6) as well as the position of the E2 BS.
The initial steps in the initiation process are relatively well understood. Cooperative binding of E1 and E2 results in the formation of a specific E12-E22-ori complex, where the two proteins interact with each other (3, 23, 24, 38, 46). The DNA binding domain (DBD) of E1 interacts with the DBD of E2 (3, 5). The E2 activation domain interacts with the E1 helicase domain (1, 2, 3, 4, 12, 26, 30, 33). As a result of these interactions the intervening DNA is sharply bent (15). This E12-E22-ori complex has the properties of an ori recognition complex, and E2 likely functions as a specificity factor for E1, allowing highly specific DNA binding by E1. Subsequently, in a second step E2 is displaced in an ATP-dependent manner and binding of additional E1 molecules generates a larger E1-ori complex (24, 31). In the E12-E22-ori complex E1 is bound to one pair of BS (E1 BS 2 and 4) (5). In addition, a second pair of E1 BS is present in the ori (E1 BS 1 and 3) (6). These sites overlap E1 BS 2 and 4 and have significantly lower affinity for E1.
Here we demonstrate that, although the E1 BS 1 to 4 overlap, the E1 DBD can bind simultaneously to all four sites, generating a complex where two dimers of E1 are bound to two separate faces of the DNA helix. We show that the interference pattern for this complex is virtually identical to the interference pattern for the E1-ori complex, demonstrating that after formation of the E12-E22-ori complex the next step in the assembly is the binding of E1 to E1 BS 1 to 4. We propose that the multiple initiator BS function at several different levels. The number and the positions of the sites determine the architecture and composition of the initiator complexes. Successive binding to sites with different affinities generates a particular order of assembly, and, through a gradual reduction in the dependence on interactions between E1 and DNA and a gradual increase in the reliance on protein-protein interactions between E1 molecules, larger complexes can be assembled. This provides a means for the sequential and ordered addition of initiator molecules to the ori. We also conclude that the successive binding of E1 molecules results in progressive structural changes in the ori DNA, indicating that the E1 molecules may serve as “wedges” that progressively open the DNA duplex and contribute to melting.
MATERIALS AND METHODS
All ori constructs and mutants were derived from a plasmid containing the BPV minimal ori (nucleotides [nt] 7914 to 7927 of the BPV genome) cloned between the XbaI and HindIII restriction sites in pUC19 (37). In this context, the naturally occurring E2 BS 12 was replaced by the high-affinity E2 BS 9. Expression and purification of the E1 and E2 proteins have been described (5, 37). Probes were generated by PCR amplification of ori constructs cloned in pUC19 with the universal primers USP and RSP. Procedures for electrophoretic mobility shift assay (EMSA) and diethyl pyrocarbonate (DEPC) interference analysis have been described previously (5, 41). Briefly, a probe (5,000 cpm/sample) was mixed with the E1 or E2 protein or both in 10 μl of binding buffer (20 mM potassium phosphate [pH 7.4], 0.1 M NaCl, 1 mM EDTA, 0.1% NP-40, 3 mM dithiothreitol, 0.7 mg of bovine serum albumin/ml, 5% glycerol). After incubation at room temperature for 30 min, the samples were immediately loaded on 6% (40:1 acrylamide-bisacrylamide) polyacrylamide gels and subjected to polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA. Hydroxyl radical footprinting was performed essentially as described by Dixon et al. (8). Probes (25,000 cpm/reaction) were incubated with the respective proteins in 50 μl of binding buffer without glycerol, and cleavage was initiated by the addition of 1 mM sodium ascorbate, 0.01 mM ferrous ammonium sulfate, 0.02 mM EDTA, and 0.03% hydrogen peroxide. After 90 s, cleavage was quenched with 20 mM thiourea. KMnO4 reactivity assays were essentially performed as described previously (32, 34).
RESULTS
Simultaneous binding of E1 to overlapping sites.
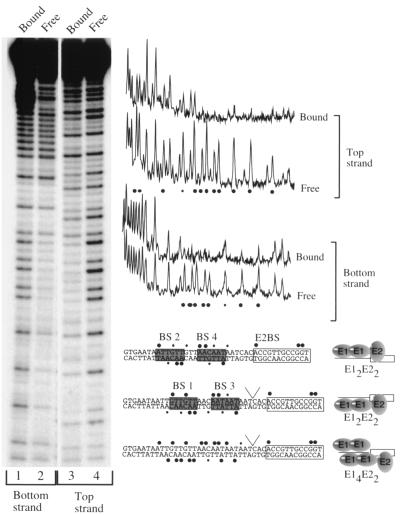
An interesting question regarding the role of initiator proteins in DNA replication is how multiple initiator binding sites orchestrate initiator complex formation. The BPV ori contains six hexanucleotide sequence elements that are putative E1 BS (E1 BS 1 to 6) (6) (Fig. 1). Four of these sites (E1 BS 2 and 4 and BS 1 and 3) form pairs. By directing binding of the E1 DBD through its interaction with the E2 DBD, we have demonstrated that the paired E1 BS 2 and 4 and E1 BS 1 and 3 can both bind dimers of E1 (6) (Fig. 1). However, because these pairs of sites are overlapping, 3 of the 6 bp in the recognition sequences are shared between E1 BS 1 and 2 and between E1 BS 3 and 4. Therefore simultaneous binding to these two pairs of sites cannot be taken for granted. To determine whether all four sites could be occupied simultaneously, we performed high-resolution footprinting with an OH radical to compare binding of the E2 DBD alone, the E1 DBD alone, and the E1 and E2 DBDs together (Fig. 2).
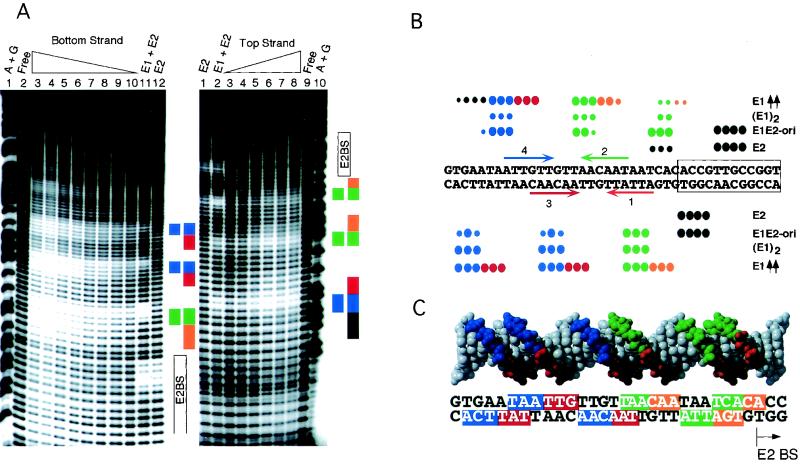
FIG. 2.
Hydroxyl radical footprints of the E1 and E2 DBDs. (A) End-labeled BPV minimal ori fragments were incubated together with (i) the E2 DBD alone (70 ng) (bottom strand, lane 12, and top strand, lane 1), (ii) the E1 DBD and the E2 DBD (0.5 μg and 0.7 ng, respectively; bottom strand, lane 11, and top strand, lane 2), and (iii) a titration of E1 DBD alone (16, 8, 4, 2, 1, and 0.5 μg of E1 DBD; bottom strand, lanes 3 to 10, and top strand, lanes 3 to 8). Lanes 3 and 4 on the bottom strand contain the same amount of E1 DBD (16 μg) but different amounts of hydroxyl radical cleavage reagent. Similarly, lanes 5 and 6 both contain 8 μg of E1 DBD but different amounts of cleavage reagent. Lanes A+G, marker generated by cleavage at A and G in the probe. Free indicates lanes where no protein was added. Blue and green boxes, protections generated by the binding of two monomers of the E1 DBD; orange and red boxes, protections generated by the binding of additional molecules of E1 DBD. (B) Summary of hydroxyl radical footprints. Black circles over the boxed E2 binding site, protections produced by the E2 DBD; blue and green circles, binding by two monomers at either low concentrations of the E1 DBD alone or together with E2 DBD. At higher concentrations of the E1 DBD, the protections extend as shown (red and orange circles). (C) Protected sequences at both low and high concentrations of the E1 DBD projected onto a double-helix model. The protections shown in blue and green correspond to the protections from one dimer of E1. The binding of an additional E1 dimer protects sequences on the side face of the helix (red and orange).
E2 DBD produced a strong protection over the E2 BS and a second weaker protection adjacent to the E2 BS on both strands (Fig. 2A, bottom strand, lane 12, and top strand, lane 1). In the presence of both E1 and E2, where E1 DBD is bound as a dimer to E1 BS 2 and 4, two additional protections on each strand were observed (blue and green). In addition, the weak protection between the E1 and E2 BS was intensified (bottom strand, lane 11, and top strand, lane 2,).
E1 DBD alone, at a low concentration, where E1 binds as a dimer to E1 BS 2 and 4, generates three protections on each strand (bottom strand, lane 10, and top strand, lane 3; blue and green bars). One of these protections coincides with the weak E2 protection, indicating that it is shared between E1 and E2. The binding of each monomer gave rise to two protections on one strand and one protection on the other strand, as expected from binding of a head-to-head dimer (protections resulting from binding of one monomer are shown in blue, and those resulting from the binding of the other are shown in green). At higher concentrations of E1, we observed an extension of these footprints, which repeats the pattern of protection observed for binding to E1 BS 2 and 4. This extension is in the 3′ direction (toward the top of the gel) on the top strand and in the 5′ direction (toward the bottom of the gel) on the bottom strand (red and orange, respectively). The repeating pattern strongly indicates that, at higher concentrations, two additional E1 molecules bind to the second pair of E1 BS (1 and 3). This is most clearly demonstrated by projecting the protections on a helix model (Fig. 2C). Protections produced at low concentrations of the E1 DBD representing two bound E1 molecules are located on one face of the DNA (blue and green). The nearly identical second set of protections (red and orange) appear at higher concentrations of E1 and are shifted by 3 bp, protecting a different face of the helix.
Comparison with the E1 DBD/DNA cocrystal structure.
The overall pattern of protection agrees very well with how E1 DBD binds DNA in the E1 DBD/DNA cocrystal structure in both the dimer and tetramer forms (9). However, some interesting differences, which likely are due to the very short DNA fragments used in the cocrystal structures, are apparent. Two sets of protections for the E1 DBD dimer, observed by hydroxyl radical footprinting, are not accounted for in the structure. These flanking protections (left-most set of blue dots on the bottom strand and right-most set of green dots on the top strand in Fig. 2B) do not correspond to sequences that are part of the E1 binding sites, and mutations in these sequences do not affect E1 binding (data not shown). These sequences are also outside the DNA sequence utilized in the cocrystal structures. Most likely these protections are the result of the significant bend in the template (40 to 50°), which can be observed by biochemical means upon the binding of an E1 DBD dimer (15) but which is notably absent in the cocrystal structures. The curving of the DNA around the bound E1 DBD dimer would account for the flanking protections. Such a bend would also explain the phenotype of some mutations in the E1 DBD that affect DNA binding but that are difficult to explain based on the crystal structure. Mutations in charged residues between α4 and β5, for example, K267A, affect DNA binding in a significant manner and would be in the correct position for interactions with the flanking DNA (E. Gillitzer and A. Stenlund, unpublished data.).
Simultaneous binding of four E1 DBD molecules generates structural changes in the template.
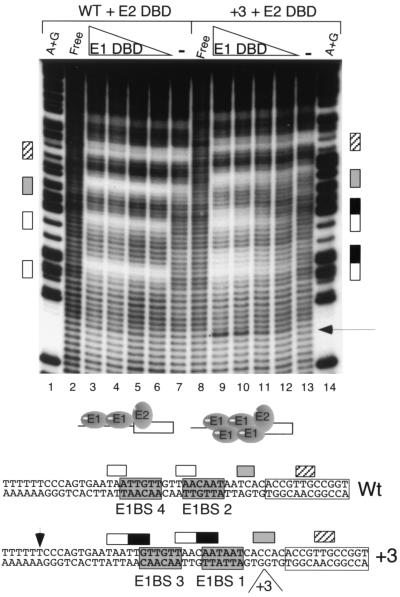
Specific DNA binding by the E1 DBD can be directed by the E2 DBD bound to an adjacent site through the interaction between the two DBDs. The interaction between E1 and E2 DBDs is highly position specific, and the E2 BS is located 6 bp from the E1 BS 2 and 4. By inserting an additional 3 bp between the E1 and E2 BS we were able to direct binding from the preferred pair of E1 BS (2 and 4) to the lower-affinity pair E1 BS 1 and 3 in EMSA (6). We reasoned that on this +3 template E2 DBD would be able to stimulate binding to E1 BS 1 and 3 specifically. We performed OH radical footprinting on the wild-type (wt) and +3 probes in the presence of E1 and E2 DBDs. In this case we analyzed the top strand only (Fig. 3). With both the wt and +3 probes, E2 gave rise to characteristic protections (lanes 7 and 13). In the presence of E1 DBD, two additional protections on the wt probe, corresponding to the binding of the E1 DBD dimer, were observed (lanes 3 to 6). The protections of the +3 probe showed an extension of these protections by 3 bp in the 3′ direction, resulting in 6-bp protections indicative of the binding of E1 to E1 BS 1 and 3 as well as 2 and 4 simultaneously. The failure to observe binding to E1 BS 1 and 3 alone is likely due to the higher affinity of E1 BS 2 and 4, which results in simultaneous occupancy of all four sites when E2 is present to stimulate binding to E1 BS 1 and 3. These footprints are completely consistent with the model for binding presented in Fig. 2.
FIG. 3.
Hydroxyl radical footprinting on wt and +3 templates. Footprinting was performed on the top strand of the wt (lanes 1 to 8) and the +3 (lanes 9 to 14) ori probes in the presence of E1 and E2 DBDs as indicated at the top. E1 DBD (0.8, 0.4, 0.2, and 0.1 μg) and E2 DBD (0.7 ng) were added in lanes 3 to 6 and 9 to 12. Lanes 7 and 13 contained 70 ng of E2 DBD alone. Below, the protections are shown projected onto the DNA sequence. Hatched boxes, protections generated by E2; open boxes, protections generated by the binding of two molecules of E1 DBD to the wt probe; black boxes, extensions of these protections observed on the +3 probe; gray boxes, protections shared between E1 and E2 DBDs. Arrow, hypersensitive site that appears upon the binding of four molecules of E1 DBD. Lanes A+G and Free are as defined for Fig. 2A.
Interestingly, the simultaneous binding of four E1 DBD molecules results in the appearance of an OH radical-hypersensitive site in the template 5′ to the binding sites (lanes 9 to 11). This position corresponds to the last base pair in the A/T-rich region, which is the site of strongest potassium permanganate reactivity when ATP-dependent ori melting is measured upon binding of full-length E1 (14, 32). This indicates that the binding of four E1 DBD molecules causes structural alterations that may be related to ori melting.
Stable E1 DBD tetramer complexes form only on short DNA probes.
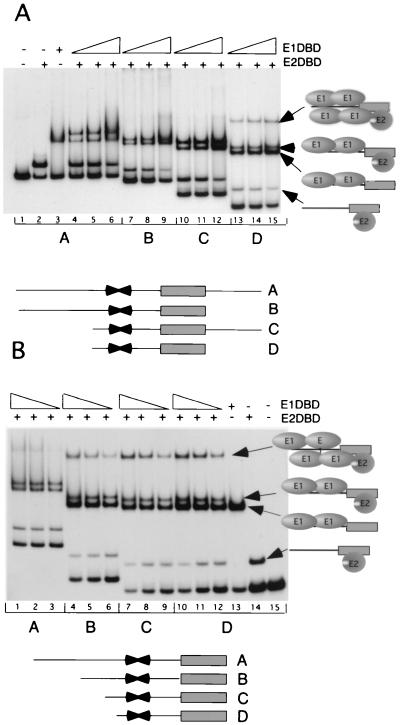
Our earlier attempts to isolate tetrameric E1 DBD complexes by gel shift analysis had not been successful although the footprinting results clearly demonstrated that simultaneous occupancy of the four sites could occur. The appearance of an OH radical-hypersensitive site upon the binding of four E1 molecules indicated to us that structural changes in the DNA could be a prerequisite for binding. We reasoned that a shorter probe may be easier to distort or may be easier to maintain in a distorted form and therefore might form a more stable E14 complex. We therefore tested complex formation by EMSA using a variety of shorter ori probes (Fig. 4A). We generated probes that deleted flanking vector sequences as well as sequences in the A/T-rich region. Probe A corresponds to our standard probe and contains the minimal ori sequence (approximately 60 bp) (nt 7914 to 7927) in addition to 48 bp of flanking polylinker sequences upstream and 25 bp at the downstream end. In probe B the polylinker sequences to the left of the ori were removed. In probe C the polylinker sequence to the right and the A/T-rich region were removed, and in probe D the polylinker sequences on both sides of the ori and the A/T-rich region were removed. For each probe we performed three fivefold titrations of E1 DBD in the presence of a constant level of the E2 DBD.
FIG. 4.
(A) Gel shift analysis using four different-size ori probes. Probe A contains the complete minimal ori sequence (∼60 bp) from nt 7914 to 7927 and in addition contains 48 bp of flanking sequence upstream (left) and 25 bp downstream (right) of the ori fragment derived from the pUC19 polylinker. Probe B lacks the flanking polylinker sequences on the downstream side of the ori fragment. Probe C lacks the flanking sequences on the upstream side of the ori fragment as well as the A/T-rich region from the ori. Probe D contains only the sequences of the E1 BS and the E2 BS and lacks any flanking sequences as well as the A/T-rich region. Lane 2, 40 pg of E2 DBD added; lane 3, 5 ng of E1 DBD added. For each probe three fivefold titrations of E1 DBD were added (0.2, 1, and 5 ng) in the presence of 40 pg of E2 DBD. The compositions of the different complexes are indicated on the right. (B) Gel shift analysis using probes lacking flanking polylinker sequences on the downstream side of the ori fragment but with sequences of different lengths on the upstream side. Probe A is identical to probe B in panel A, and probe D corresponds to probe D in panel A. Probe B lacks flanking sequences on both sides but maintains the minimal ori sequence. Probe C, in addition, lacks part of the A/T-rich sequence. The quantities of E1 and E2 DBD were identical to those used in panel A.
Based on our previous analysis of the complexes formed by E1 and E2 DBDs on the ori we can readily identify the complexes that were formed on these probes. For example, an E2 DBD dimer (Fig. 4A, lane 2), an E1 DBD dimer (lane 3), and the combined E12-E22 complex (lane 4) were formed on probe A. These same complexes were formed also on the shorter probes, B, C, and D, as expected. Strikingly, however, only on the shortest probe (probe D) could we detect significant levels of a complex larger than the E12-E22 complex (lanes 13 to 15).
To further analyze the sequence requirement for formation of the larger complex, we generated probes that each had a fixed end point at the end of the E2 BS and varied the lengths of the left (upstream) ends (Fig. 4B). Probe A (which corresponds to probe B in Fig. 4A) failed to give rise to a discrete large complex (lanes 1 to 3). However, removal of the flanking polylinker sequence resulted in the appearance of the larger complex (probe B, lanes 4 to 6), and further removal of sequences only slightly increased the efficiency of formation of this complex (probes C and D, lanes 7 to 15). These results indicate that a long probe (or possibly the presence of polylinker sequences) is deleterious for the formation of the larger E1-E2 complex.
Interference analysis of E1 tetramer complexes.
To verify that the large “new” complex corresponded to the expected E14-E22 complex, we performed DEPC interference analysis. We have previously used DEPC interference analysis to analyze the binding of E1 dimers to sites 2 and 4 and sites 1 and 3 as well as for analysis of the binding of full-length E1 (5, 6, 37). We modified probe D with DEPC, which modifies A and G bases at the N7 position, and used the modified probe for gel shift analysis. After excision of the putative E14-E22 band, the DNA was eluted, cleaved with piperidine, and analyzed on a sequencing gel (Fig. 5). The ladders obtained were quantitated with a Fuji BAS 1000 imager, and the tracings are shown at the right in Fig. 5. Below is shown a comparison with the interference patterns obtained for an E12-E22 complex bound to E1 BS 2 and 4 and to E1 BS 1 and 3 (6). The pattern of interference corresponded very strikingly to the sum of interferences obtained by the binding of E1 to both pairs of E1 BS, consistent with the notion that the large complex indeed corresponded to the E14-E22 complex, where E1 DBD is bound to E1 BS 1 to 4. Interestingly, the pattern of interference is almost identical to the pattern observed by using full-length E1 in a cross-linked E1-ori complex (37). This indicates that the binding to E1 BS 1 to 4 is similar to the binding of full-length E1 to the ori and that the transition from the E1-E2-ori complex to an E1-ori complex involves the binding of an additional pair of E1 molecules to E1 BS 1 and 3.
FIG. 5.
DEPC interference analysis of the putative E14-E22 complex. Probe B from Fig. 4B was modified with DEPC, annealed, and subsequently used for EMSA with E1 and E2 DBDs. The binding reactions were scaled up 20-fold under the conditions used in lane 10 in Fig. 4B. The putative E14-E22 complex and the free probe were excised, and the DNA was treated with piperidine and compared to the free probe on a sequencing gel. The patterns of interference for the top and bottom strands for shown. The gel was analyzed by a Fuji imager and quantitated, and a scan of the corresponding lanes is shown in to the left. Below is a summary of the positions where interference can be detected. As a comparison, the interference patterns for E12-E22 bound to sites 2 and 4 and 1 and 3 are shown (6).
E1 binding, in the absence of ATP, generates structural changes in the DNA that can be detected as permanganate reactivity.
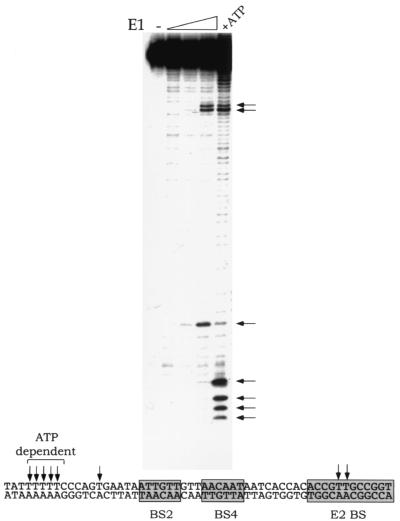
As discussed above, the E1 DBD fails to form complexes larger than the tetramer likely due to the lack of interaction domains involved in formation of larger complexes. To determine whether structural changes occur upon the binding of larger complexes, we used the full-length E1 protein. To determine the consequences of these structural changes, we utilized a permanganate reactivity assay. E1 can distort the sequences flanking the E1 binding site in an ATP-dependent manner, and this distortion or melting can be detected by increased reactivity of the DNA with permanganate (14, 32). Unpaired T residues can be specifically modified by the permanganate ion and subsequently detected by cleavage with piperidine. However, in the absence of ATP any distortion would have to result from DNA binding by E1. We performed permanganate reactivity assays in the absence and presence of ATP (Fig. 6). As expected, in the presence of ATP the characteristic pattern of permanganate reactivity was generated, with major reactivity in the A/T-rich region and within the E2 binding site. Surprisingly, even in the absence of ATP, permanganate reactivity was generated in two positions in response to the binding of E1. This reactivity is not due to contamination of the E1 preparation with ATP since the pattern of permanganate reactivity is distinctly different from the pattern obtained in the presence of ATP. Thus, these results indicate that the binding of E1 in itself generates structural changes in the ori, consistent with the structural changes we observe upon the binding of the E1 DBD. These changes are likely related to the ATP-dependent distortion (melting) but occur at partly different positions.
FIG. 6.
Potassium permanganate reactivity in the ori caused by binding of E1. A top-strand ori probe was incubated in the absence (−) or presence of increasing quantities of E1 protein (15, 30, and 60 ng). As a control, 60 ng of E1 was incubated in the presence of 5 mM ATP (+ATP). After 20 min at room temperature, potassium permanganate was added to 6 mM, and the reaction was terminated after 2 min by addition of 2-mercaptoethanol. After cleavage with piperidine, the resulting material was analyzed by 10% polyacrylamide gel electrophoresis. The positions of permanganate reactivity are indicated by arrows. Below, these positions are indicated in relation to the boxed E1 and E2 BS.
DISCUSSION
The binding of initiator proteins to origins of DNA replication functions to bring the initiator protein to the site of initiation and marks the ori. In addition, for viral initiators and DnaA from Escherichia coli the initial binding represents the first step in a process where the initiator ultimately melts the template DNA. The initial recognition of the ori by E1 requires cooperative binding with E2 and results in the binding of a dimer of E1 to the preferred E1 BS 2 and 4. Utilizing the E1 DBD, we demonstrate that simultaneous binding to two additional overlapping binding sites can occur. The interference pattern generated by the binding of four molecules of the E1 DBD is strikingly similar to the interference pattern obtained with complexes of full-length E1 bound to ori after cross-linking (37). This indicates that simultaneous binding to the overlapping sites is a function of E1 and that the binding sites do not represent alternative binding sites, as has been suggested for the four pentanucleotide T-Ag sites in the SV40 ori (19). Indeed, this apparent difference between E1 and T-Ag is very surprising since both the BS arrangements and the structures of the DBDs of these two proteins are strikingly similar (5, 10, 19, 22). We believe that the explanation for this difference may be technical. Solution footprinting at high resolution has not been performed with the T-Ag DBD, and gel shift experiments have only been performed after cross-linking (19). Further studies of the DNA binding properties of the T-Ag DBD will be required to resolve these differences.
The ability of E1 to bind to the four sites simultaneously provides a simple pathway for assembly of large complexes. Our data indicate that the specificity for ori recognition resides to a significant extent in the auxiliary factor E2 and that the binding of E1 to BS 2 and 4 may provide a particular positioning rather than ori recognition per se. Similarly, the binding of E1 to the second pair of sites provides little or no specificity. E1 BS 1 and 3 have significantly lower affinity for binding the E1 DBD (10- to 20-fold) than E1 BS 2 and 4 (G. Chen and A. Stenlund, unpublished data) (Fig. 2), and the binding of E1 DBD to these sites does not add significant specificity. Instead, the recruitment of the third and fourth E1 molecules to the ori relies to a significant extent on other interactions, most likely protein-protein interactions with E1 bound to sites 2 and 4, as indicated by the difference in behavior between the E1 DBD and the full-length E1. In contrast to the binding of the DBD to the four sites, which occurs sequentially, the binding of the full-length E1 protein to the four sites is cooperative; indeed, the binding of an E1 dimer cannot be observed in the absence of E2 (32). So, if the contribution from specific sequence recognition of the DNA is minimal, what functions do the sites have? Taken together, these results indicate that the primary functions of the sites are to determine positioning and order of assembly of the E1 molecules rather than to provide high-affinity binding. This implies that both positioning and order of assembly are of importance for formation of functional E1 complexes.
We have previously suggested that fifth and sixth E1 BS are present in the ori (6). Based on both the high-resolution footprints and on the cocrystal structure of four E1 DBD molecules bound to the ori (9) there clearly is room for another pair of E1 molecules on the third unoccupied face of the DNA helix. Binding to the putative sites 5 and 6 would complete a structure of two trimeric rings that encircle the DNA. Indeed, our previous data have demonstrated that full-length E1, after cross-linking with glutaraldehyde (which generates protein-protein cross-links but no protein-DNA cross-links under these conditions), is topologically linked to circular but not linear DNA, demonstrating that E1 forms a covalent circle around the template (35). Furthermore, under these same conditions E1 is cross-linked into a 200-kDa form consistent with a trimer of E1 (35). The failure to detect the binding of six molecules simultaneously is likely a consequence of the use of the E1 DBD in place of the full-length E1 protein. The formation of these larger complexes clearly depends on contributions from protein-protein interactions between domains that are not present in the E1 DBD. Thus, the assembly appears to represent a gradual reduction in the dependence on interactions between E1 and DNA and a gradual increase in the reliance on interactions between E1 molecules. As larger complexes are assembled, further oligomerization appears to rely exclusively on protein-protein interactions.
These results indicate that there are significant similarities not only between E1 and viral initiators such as T-Ag, as is expected, but also between E1 and cellular initiator proteins, such as E. coli DnaA. DnaA binds first to a subset of high-affinity DnaA binding sites (DnaA boxes R1 and R4). Subsequently, these DnaA molecules serve as anchors for the binding of additional DnaA molecules to R2 and R3 through a combination of the binding of DnaA to lower-affinity sites and interactions with the anchored DnaA molecules, eventually resulting in a large cooperative complex bound to oriC (25, 40). Although this process for DnaA is not understood in detail, the similarities suggest that a gradual transition from the sequence-specific binding of the initiator to binding through protein-protein interactions may be a conserved strategy to generate DNA-bound oligomeric initiator complexes.
Viral initiators and DnaA, in addition to marking the ori, are designed to alter DNA structure. How the distortion of the DNA structure comes about is understood only at the most primitive level. Our analysis demonstrates that the binding of the E1 DBD results in structural changes in the DNA. From the E1 DBD/DNA cocrystal structures we know that these changes are progressive, i.e., the DNA structure is more distorted in the tetramer/DNA complex than in the dimer/DNA complex and certain characteristic changes in the tetramer are augmented compared to those in the dimer (9). The changes that are observed in these structures are not of a kind that can be readily detected by biochemical means; however, using hydroxyl radical footprinting we can detect a hypersensitive site which is located on the flank of the binding sites for E1. This demonstrates that, in the context of larger DNA fragments, the binding of four molecules of E1 DBD has structural consequences outside the E1 binding site. The small size of this cleaving reagent makes OH radical hypersensitivity in double-stranded DNA highly unusual, and it is therefore not clear what type of structural alteration the hypersensitivity corresponds to. The OH radical reacts with the 3 and 5 positions in the sugar ring that is accessible mainly from the minor groove of the DNA, indicating that a widening of the minor groove may be a consequence of the formation of the complex containing four E1 DBD molecules. However, since the OH radical hypersensitivity affects only a single position, the structural change most likely is local rather than a general widening of the minor groove.
Similarly, the dependence on probe length of the formation of tetrameric E1 complexes is consistent with structural changes. Very short DNA segments may be more flexible than long fragments, and structural alterations in the DNA may therefore be more easily generated or maintained. The binding of a dimer of E1 DBD to E1 BS 2 and 4 results in the generation of a significant bend (40 to 50°) in the DNA (15). Assuming that the binding of the second dimer on the other face of the helix generates a similar bend, very significant strains on the DNA involving two perpendicular bends would be expected, and a final structure is difficult to predict. Unfortunately, in this regard the cocrystal structure provides little information since the DNA in the structure is not bent, likely due to the very short oligonucleotide used as discussed above (9).
To probe structural changes in the DNA generated by the binding of more than four E1 molecules, we resorted to using the full-length E1 protein, which forms the larger complexes that the E1 DBD is not capable of forming. Under these conditions we detect the appearance of permanganate reactivity on the flanks of the E1 BS, indicating that the DNA is either melted or distorted. This indicates that the binding of E1 results in a progression of structural changes in the DNA that are of increasing severity. They range from changes generated within the BS by an E1 dimer and an E1 tetramer which are detectable only by structural determination to changes that we can detect by biochemical means. These include the OH radical hypersensitivity for the tetramer and melting or distortion for a larger E1 complex. An intriguing possibility is that E1 molecules function as wedges that, as the molecules are bound successively, result in the unraveling of the DNA molecule. The binding of one pair of E1 molecules could change the DNA structure slightly, favoring the binding of a second dimer, which in turn alters the structure more, etc. In this way, significant changes in the DNA structure could be generated in an energy-independent manner. Our data thus indicate that melting may be a gradual process rather than an ATP-dependent one-step switch. This model also provides a possible explanation for the presence of multiple initiator binding sites in many ori sequences. If progressive alterations in the DNA structure as a consequence of initiator binding constitute a general feature of initiation, ordered binding to multiple binding sites for the initiator would be a simple way to accomplish these alterations.
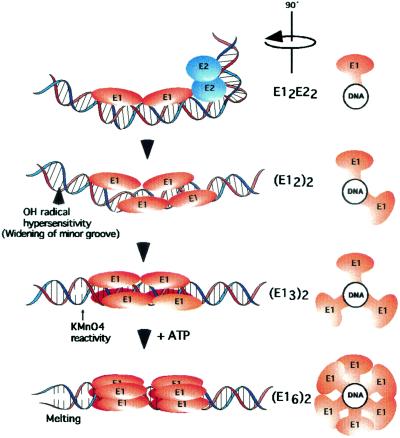
We can now provide fairly good descriptions of how E1 is assembled on the ori (Fig. 7). E1 binds initially together with E2 on one face of the helix to form the sequence-specific ori recognition complex E12-E22, where E1 is bound to the high-affinity E1 BS 2 and 4. The interaction between E1 and E2 generates a sharp bend in the DNA, and the E1 dimer adds an additional bend. Upon ATP hydrolysis, E2 is displaced and additional E1 molecules are added to the complex partly by sequence-specific binding to E1 BS 1 and 3 but mainly by interactions with the E1 molecules already bound to E1 BS 2 and 4. Under these conditions, due to the “wedging” action of the E1 DBDs, alterations in template structure, resulting in OH radical hypersensitivity flanking the binding sites, are observed. In the next step, which is largely based on indirect evidence (see above), the E1 molecules 5 and 6 are added. The addition of these molecules further augments the prior structural changes, resulting in alterations that are severe enough to generate permanganate reactivity in the sequences flanking the E1 binding sites, possibly signaling the first indications of melting. Finally, full-scale melting, likely involving the ATP-dependent formation of larger E1 complexes, such as E1 hexamers, is observed.
FIG. 7.
Model for the sequential assembly of E1 initiator complexes on the origin of DNA replication. See text for explanations.
Acknowledgments
This work was supported by Public Health Service grant CA 13106 from the National Cancer Institute
REFERENCES
- 1.Abroi, A., R. Kurt, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, J. D., and P. M. Howley. 1995. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 proteins participate in complex formation. J. Virol. 69:4364-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blitz. I. L., and L. Laiminis. 1991. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J. Virol. 65:649-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., and A. Stenlund. 1998. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 72:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, G., and A. Stenlund. 2001. The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication. J. Virol. 75:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, W. J., J. J. Hayes, J. R. Levin, M. F. Weidner, B. A. Dombroski, and T. D. Tullius. 1991. Hydroxyl radical footprinting. Methods Enzymol. 208:380-413. [DOI] [PubMed] [Google Scholar]
- 9.Enemark, E. J., A. Stenlund, and L. Joshua-Tor. 2002. Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J. 21:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enemark, E. J., G. Chen, D. E. Vaughn, A. Stenlund, and L. Joshua-Tor. 2000. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell 6:149-158. [PubMed] [Google Scholar]
- 11.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, M. K., and M. R. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 14.Gillette, T. G., M. Lusky, and J. A. Borowiec. 1994. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc. Natl. Acad. Sci. USA 91:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillitzer, E., G. Chen, and A. Stenlund. 2000. Separate domains in E1 and E2 proteins serve architectural and productive roles for cooperative DNA binding. EMBO J. 19:3069-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraldo, R., and R. Diaz-Orejas. 2001. Similarities between the DNA replication initiators of gram-negative bacteria plasmids (RepA) and eukaryotes (Orc4p)/archaea (Cdc6p). Proc. Natl. Acad. Sci. USA 98:4938-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt, S. E., G. Schuller, and V. G. Wilson. 1994. DNA binding specificity of the bovine papillomavirus E1 protein is determined by the sequences contained within an 18-base-pair inverted repeat element at the origin of replication. J. Virol. 68:1094-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt, S. E., and V. G. Wilson. 1995. Mutational analysis of the 18-base-pair inverted repeat element at the bovine papillomavirus origin of replication: identification of critical sequences for E1 binding and in vivo replication. J. Virol. 69:6525-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumarr, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg, A., and T. A Baker. 1992. DNA replication. W. H. Freeman, New York, N.Y.
- 22.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Structure of the origin-specific DNA binding domain from simian virus 40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 23.Lusky, M., J. Hurwitz, and Y. S. Seo. 1993. Cooperative assembly of the bovine papillomavirus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J. Biol. Chem. 268:15795-15803. [PubMed] [Google Scholar]
- 24.Lusky, M., J. Hurwitz, and Y. S. Seo. 1994. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc. Natl. Acad. Sci. USA 91:8895-8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margulies, C., and J. A. Kaguni. 1996. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 271:17035-17040. [DOI] [PubMed] [Google Scholar]
- 26.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase α-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride, A., and G. Myers. 1997. The E2 proteins, p. 37-53. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 28.Mendoza, R., L. Gandhi, and M. R. Botchan. 1995. E1 recognition sequences in the bovine papillomavirus type 1 origin of DNA replication: interaction between half-sites of the inverted repeats. J. Virol. 69:3789-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messer, W., and C. Weigel. 1996. Initiation of chromosome replication, p. 1579-1601. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington D.C.
- 30.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson., and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 31.Sanders, C. M., and A. Stenlund. 1998. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 17:7044-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders, C. M., and A. Stenlund. 2000. Transcription factor-dependent loading of the E1 initiator reveals modular assembly of the papillomavirus origin melting complex. J. Biol. Chem. 275:3522-3534. [DOI] [PubMed] [Google Scholar]
- 33.Sarafi, T. R., and A. A. McBride. 1995. Domains of the BPV-1 E1 replication protein required for origin-specific DNA-binding and interaction with the E2 transactivator. Virology 211:385-396. [DOI] [PubMed] [Google Scholar]
- 34.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 264:8074-8081. [PubMed] [Google Scholar]
- 35.Sedman, J., and A. Stenlund. 1996. The initiator protein E1 binds to the bovine papillomavirus origin of replication as a trimeric ring-like structure. EMBO J. 15:5085-5092. [PMC free article] [PubMed] [Google Scholar]
- 36.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedman, T., Sedman, J., and A. Stenlund. 1997. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 71:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo, Y. S., F. Muller, M. Lusky, E. Gibbs, H. Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papillomavirus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo, Y. S., F. Muller, M. Lusky, and J. Hurwitz. 1993. Bovine papillomavirus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturm, R., T. Baumruker, B. R. Franza, Jr., and W. Herr. 1987. A 100-kD HeLa cell octamer binding protein (OBP 100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes. Dev. 1:1147-1160. [DOI] [PubMed] [Google Scholar]
- 42.Sverdrup, F., and G. Myers. 1997. The E1 proteins, p. 37-53. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 43.Tjian, R. 1978. The binding site of SV40 DNA for a T-antigen-related protein. Cell 13:165-179. [DOI] [PubMed] [Google Scholar]
- 44.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 47.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papillomavirus 1 is an ATP dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.zur Hausen, H. H. 1991. Viruses in human cancers. Science 254:1167-1173. [DOI] [PubMed] [Google Scholar]