Abstract
In the Paramecium tetraurelia genome, 17 genes encoding the 100-kDa-subunit (a-subunit) of the vacuolar-proton-ATPase were identified, representing by far the largest number of a-subunit genes encountered in any organism investigated so far. They group into nine clusters, eight pairs with >82% amino acid identity and one single gene. Green fluorescent protein-tagging of representatives of the nine clusters revealed highly specific targeting to at least seven different compartments, among them dense core secretory vesicles (trichocysts), the contractile vacuole complex, and phagosomes. RNA interference for two pairs confirmed their functional specialization in their target compartments: silencing of the trichocyst-specific form affected this secretory pathway, whereas silencing of the contractile vacuole complex-specific form altered organelle structure and functioning. The construction of chimeras between selected a-subunits surprisingly revealed the targeting signal to be located in the C terminus of the protein, in contrast with the N-terminal targeting signal of the a-subunit in yeast. Interestingly, some chimeras provoked deleterious effects, locally in their target compartment, or remotely, in the compartment whose specific a-subunit N terminus was used in the chimera.
INTRODUCTION
The vacuolar-proton-ATPase (V-ATPase) is a multisubunit enzyme that translocates protons across membranes against their electrochemical potential through ATP hydrolysis. It is of crucial importance for many cellular processes through acidification of compartments and energization of membranes (Graham et al., 2003; Inoue et al., 2003). The V-ATPase consists of two subcomplexes, respectively, the cytoplasmic and the transmembrane V1 and V0 sectors. The V1 sector is built by eight different subunits (A-H) ranging from 13 to 70 kDa in a stoichiometry of A3B3CDEFG2H1-2, whereas the V0 sector consists of six different subunits (a, c, c′, c″, d, and e) of 10-100 kDa in a stoichiometry of ac4c′c″dex (Inoue et al., 2003). The V-ATPase was shown to act by a rotation mechanism (Hirata et al., 2003; Imamura et al., 2003). ATP is hydrolyzed by the V1 sector, and the energy liberated drives rotation of the rotor part of V0 (the c-subunits) relative to the stator (the a-subunit) in the membrane. During the rotation, protons are channeled across the membrane (Inoue et al., 2003).
In most organisms, the V-ATPase subunits are each encoded by a few genes, one to four in general. This also holds true for Paramecium whose genome is being currently sequenced and annotated, except for the V0 c-subunits, encoded by six genes, and the V0 a-subunits, the largest subunit of the V-ATPase (∼100 kDa), encoded by not <17 genes (Wassmer et al., 2005). This puzzling observation led us to investigate the consequence of such a diversity of sequences and to address the question of possibly different roles of a-subunits in Paramecium, compared with the generic role of the V-ATPase stator normally described.
In yeast, a-subunits are encoded by two genes, VPH1 and STV1 (Arata et al., 2002). These two different a-subunits were shown to be localized differentially, Stv1p in the Golgi and Vph1p in the membranes of vacuoles (Kawasaki-Nishi et al., 2001a). The N-terminal half of these proteins was shown to contain the localization signal for this differentiated targeting (Kawasaki-Nishi et al., 2001a). Knockout of either VPH1 or STV1 does not result in the vma-phenotype, typical of vacuolar dysfunction, like knockout of any of the other V-ATPase genes does. Only the double disruption of VPH1 and STV1 leads to such a phenotype (Manolson et al., 1994). Therefore, the a-subunit genes in yeast seem to be able to substitute for each other to some extent. V-ATPase complexes containing different a-subunits were shown to have different catalytic properties and different kinetics of association/dissociation of V1 to V0, especially in response to glucose depletion in the medium (Kawasaki-Nishi et al., 2001b; Kane and Smardon, 2003).
In mammals, the a-subunits are encoded by four genes that, in mice, were shown to have different expression levels in different tissues. For example, expression of the isoform 4 of the a-subunit is restricted to the kidney (Oka et al., 2001; Smith et al., 2001). The a4-containing protein is localized in the plasma membrane of renal intercalated cells. Loss-of-function mutations in that gene cause autosomal recessive distal renal tubular acidosis (Smith et al., 2000). The isoform 3 of the a-subunits was shown to be important for bone resorption by osteoclasts (Toyomura et al., 2000). Mutations in this isoform can lead to osteopetrosis (Li et al., 1999).
In Paramecium, the V-ATPase was shown to be most important for the processes of osmoregulation, phagocytosis, and the biogenesis of dense core secretory granules, called trichocysts (Fok and Allen, 1988; Allen and Naitoh, 2002; Wassmer et al., 2005). The major organelle for osmoregulation is the contractile vacuole complex that is known to contain a huge number of V-ATPase molecules, where the enzyme is used to create an electrochemical potential that is exploited to expel excess water from the cytosol (Grønlien et al., 2002; Stock et al., 2002). The radial arms of the organelle are built by two complex membrane systems, the smooth and the decorated spongiome (Allen and Naitoh, 2002). By using an antibody against the V1 B-subunit, the V-ATPase holoenzyme was demonstrated to be restricted to the decorated spongiome (Fok et al., 1995). In phagocytosis, the V-ATPase is required for the acidification that leads to the inactivation of ingested microorganisms and is necessary for enabling the fusion of phagosomes with lysosomes (Fok and Allen, 1988). Concerning the biogenesis of secretory granules, we have recently shown that it is affected, together with the osmoregulatory and phagocytotic pathways, by the inactivation of V-ATPase subunits c, A, or F, although the precise role the V-ATPase in this process is not yet known (Wassmer et al., 2005).
In this article, we show that the 17 isoforms of a-subunits define at least seven localizations within the Paramecium cell and suggest as many different functions. The necessity to maintain so many different a-subunits in a complex unicell such as Paramecium compared with multicellular organisms is discussed.
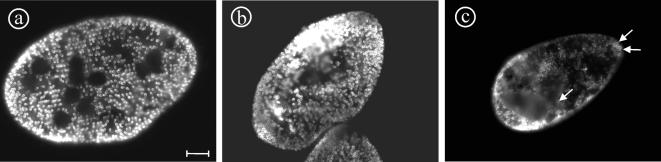
MATERIALS AND METHODS
Cell Culture and Phenotypical Tests
The wild-type strain of Paramecium tetraurelia used was stock d4-2, derived from stock 51. Cells were grown in wheat grass infusion (BHB; L'arbre de vie, Luçay Le Male, France), bacterized with Klebsiella pneumoniae the day before use, and supplemented with 0.4 μg·ml-1 β-sitosterol (Sonneborn, 1970). Trichocyst exocytosis capacity was visualized by a saturated solution of picric acid under low-magnification dark-field microscopy (Vayssié et al., 2000). Monitoring phagocytosis with India ink as well as following its inhibition by cytochalasin B were performed as described previously (Cohen et al., 1984). Contractile vacuole shape and pulsation rhythm were observed under phase microscopy.
Amplification, Cloning, and Sequencing from cDNA
A cDNA-library from P. tetraurelia was kindly provided by the laboratory of J. Schultz (University of Tübingen, Tübingen, Germany). The oligonucleotides (oligos) used for the amplification of the VATA1_1 cDNA from this library were 5′-aactgcagagtctttattagttgttgattccc-3′ and 5′-ggggtaccttacaaaggatattcaaacattac-3′, using the Advantage2-polymerase mixture (Clontech, Mountain View, CA). The PCR product was cloned into pBlueScript II SK- (Stratagene, La Jolla, CA) using the restriction enzymes PstI and Acc65I from New England Biolabs (Frankfurt, Germany) according to standard molecular biological protocols. Sequencing was performed by MWG-biotech (Ebersberg, Germany).
Construction of Green Fluorescent Protein (GFP)-Fusion Genes
Oligonucleotides used for amplification of a-subunits were a1f, 5′-cgaccggtcgccaccatgattagatctgaaggtatgag-3′ and a1r, 5′-gcaccggtggaagtttcattttatcttattatttcttcatc-3′; a2f, 5′-gcgactagtatgagtttctttagatcggagac-3′ and a2r, 5′-gcgctcgagatccttaatcctttggctgatttg-3′; a3f, 5′-gcgactagtatgagcttgtttagatcagagtag-3′ and a3r, 5′-gcgctcgagatccatcatcttattcattactttt-3′; a4f, 5′-gcgactagtatgttaagatcgtaggaaatgtc-3′ and a4r: 5′-gcgctcgagatcctaacgagtacacatgaaattcc-3′; a5f, 5′-gcgactagtatgagtttctttaggtctaaaaag-3′ and a5r, 5′-gcgctcgagatccttttgaatttgcctatttcag-3′; a6f, 5′-gcgactagtatgagtttctttaggtccaaac-3′ and a6r, 5′-gcgctcgagatcctttaagatgctcttataacattt-3′; a7f, 5′-gcgactagtatgttcagagcaaccgaaatacatc-3′ and a7r, 5′-gcgctcgagatcccattcttgtactaaattaaaac-3′; a8f, 5′-gcgactagtatgtttagatcacaagaaatgag-3′ and a8r, 5′-gcgctcgagatccctaagattctaaattgatgctttc-3′; and a9f, 5′-gcgactagtatgaacttctttaggtcccaaac-3′ and a9r, 5′-gcgctcgagatccttcctcctttggaactgcattg-3′. Underlined codons correspond to the start codon ATG and the reverse complement of the former stop codon TGA mutated to tcc (a1) or gga (a2-9). VATA1_1 was cloned from cDNA using AgeI into pPXV-GFP described in Hauser et al. (2000a). All other a-subunits were amplified using macronuclear DNA as template and cloned into pPXV-GFP described in Wassmer et al. (2005) using SpeI and XhoI according to standard protocols.
Microinjection Experiments
Microinjections were made under an inverted Nikon phase-contrast microscope, using a Narishige micromanipulation device, and an Eppendorf air-pressure microinjector. DNA for microinjection was prepared as described in Wassmer et al. (2005). When DNA was microinjected into wild-type cells, they were pretreated with a solution of the vital secretagogue aminoethyldextran at 0.2% (Plattner et al., 1985) to trigger trichocyst discharge and avoid disturbance during the microinjection procedure. To assess the degree of expression, a serial dilution (1/10, 1/100, and 1/1000) of the a1-GFP construct was injected with plasmid DNA diluted in herring sperm DNA (Sigma-Aldrich, St. Louis, MO) containing water to reach the viscosity of the solution necessary for successful injection. Mock-injected cells were only transformed with herring sperm DNA.
Expression of Enhanced (e)GFP in Escherichia coli and Production of Polyclonal Antibody
For heterologous expression of eGFP, we selected the amino acid sequence of eGFP (Hauser et al. 2000b). The coding region (M1-K238) was cloned into the XhoI/BamHI restriction sites of the pET16b expression vector from Novagen (Madison, WI). Purification of recombinant GFP was done by affinity chromatography on Ni2+-nitrilotriacetate agarose under native conditions, as recommended by the manufacturer (Merck, Bad Soden, Germany). Fractions containing recombinant GFP were used for immunization of rabbits. After several boosts, positive sera were taken and purified by two subsequent chromatography steps: the first step on a histidine (His)-tag peptide column (to remove His-tag-specific antibodies), followed by an affinity step on the recombinant GFP protein.
Expression of a1-1 Peptides in E. coli and Production of Polyclonal Antibody
The sequence coding for the amino acids P178-S328 of a-subunit 1-1 was amplified from macronuclear DNA with all the in-frame stop-codons (TAA and TAG) mutated to CAA or CAG by fusion PCR (Dillon and Rosen, 1990) and cloned into the pET16 vector system (Novagen) using XhoI restriction enzyme. For the expression of a smaller, isoform-specific peptide of the a-subunits 1-1, the DNA-coding sequence corresponding to a1-1(P228-E259) was PCR amplified from macronuclear DNA and cloned into the pET32 vector system (Novagen) using NcoI, XhoI. Recombinant expression and purification by histidine affinity chromatography on Ni2+-nitrilotriacetate agarose was done under denaturing conditions according to the manufacturer's protocol. The resulting peptide a1-1(P178-S328) was injected into a rabbit for immunization. The serum obtained was affinity purified first using a column with an immobilized His-tagged protein to remove His-tag-specific antibodies and in the second step against the recombinant protein. To ensure the isoform specificity of anti a1-1-antibody, serum obtained with a1-1(P178-S328) was affinity purified in the second step using a a1-1(P228-E259)-column.
Cell Fractionation and Western Blots
Whole cell homogenates were prepared as described in Kissmehl et al. (2004). Soluble and particulate fractions were separated by centrifugation of homogenates at 100,000 or 190,000 × g for 60 min at 4°C. Cell surface complexes (“cortices”) were prepared according to Lumpert et al. (1990). SDS-gel electrophoresis was carried out essentially as described by Laemmli et al. (1970), Western blotting was performed using a semidry blotter from Bio-Rad (Hercules, CA) according to the manufacturers protocol. Various sample buffers and boiling times for SDS-PAGE were tested, because the a1-1 protein signal was found to be very sensitive in sample preparation before gel electrophoreses, as described in yeast (Manolson et al., 1992). The protocol provided by Manolson et al. (1992) proved to be most suitable for the detection of endogenous a1-1 protein in Paramecium.
Protein/DNA Extraction and Slot Blots
Paramecium cells were grown in 300-ml culture until a density of ∼1500 cells/ml, checked for the absence of autogamy, concentrated by centrifugation, and lysed in 3 ml 6 M guanidine, 100 mM NaPi, and 10 mM Tris-Cl, pH 8,0. Five microliters of the lysates was loaded per slot when decorated with anti-a1-1 (P178-S328) or anti-actin1-1 serum (described in Kissmehl et al., 2004) as loading control, whereas 20 μl was loaded when decorated with the mouse monoclonal anti-GFP antibody (Roche Diagnostics, Mannheim, Germany). Slot blots were performed using the Bio-Dot apparatus according to the manufacturer's instructions (Bio-Rad). To prepare total DNA, 100 μl of the cell lysates was precipitated according to Wessel and Flügge (1984), and the aqueous, DNA-containing fraction was ethanol precipitated and redissolved in 100 μl of Tris-EDTA buffer.
PCR for the Detection of GFP-Plasmids in Paramecia
The successful transformation of injected paramecia was verified using PCR. The oligonucleotide located in the open reading frame of the GFP was 3′GFPseq 5′-aaagttaacttcaaaattagacac-3′ and the one located in the 3′-untranslated calmodulin region was TW-3 5′-catatgatgtctatgtattgtttg-3′, leading, in the case of successful transformation with pPXV-GFP, to the amplification of an ∼380-base pair fragment.
Immunolabeling of Paramecia and Fluorescence Microscopy
Individual paramecia were transferred as small pools in depressions slides and permeabilized with 1% Triton X-100 in PHEM buffer [60 mM piperazine-N, N′-bis(2-ethanesulfonic acid, 25 mM HEPES, 10 mM EGTA, and 2 mM MgCl2, pH 6.9] for 90 s, followed by rapid transfer to PHEM containing 2.5% formaldehyde for 10-min fixation. Alternatively, cells were fixed in 2.5% formaldehyde in PHEM buffer containing 0.5% digitonin or 0.5% Triton X-100 at room temperature for 10 min. Cells were then washed twice in Tris-buffered saline (TBS) + bovine serum albumin (BSA) (10 mM Tris-Cl, 150 mM NaCl, and 3% BSA, pH 7.4), followed by incubation with the primary antibody in TBS + BSA. The primary anti-trichocyst antibody was kindly provided by K. Klotz and F. Ruiz (Centre de Génétique Moleculaire, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) and used at 1/300 dilution. The primary anti-tubulin antibody was the monoclonal ID5 described in Wehland and Weber (1987), used at 1/300 dilution according to the digitonin permeabilization protocol for microtubular structures staining given above. The primary anti a1-1(P228-E259) was used at a concentration of 5 μg/ml. Afterward, cells were washed once in TBS + BSA, followed by the incubation with the secondary antibody, either Alexa568-coupled goat anti-mouse F(ab′)2 at 1/300 dilution or goat anti-rabbit F(ab′)2 (Invitrogen, Carlsbad, CA) at 1/400 dilution, for 30-45 min. Finally, cells were washed twice in TBS + BSA and mounted in Citifluor AF2 (Citifluor, London, United Kingdom). Fluorescence was analyzed under an Axioskop 2 Plus microscope (Carl Zeiss, Jena, Germany), equipped with a CoolSNAP cf. camera (IPS, North Reading, MA), using the plan-Neofluar objective 63× 1/1.25 (Carl Zeiss) with immersion oil ZEISS 518N or using an Axiovert 100TV microscope (Carl Zeiss) equipped with a plan-Neofluar 40×/1.30 objective and a ProgRes C10plus camera system from Jenoptik (Jena, Germany). Images were acquired using the MetaMorph software (Molecular Devices, Sunnyvale, CA).
Immunoelectron Microscopy
Cells derived from clones transformed with a1-, a4-, a7-, and a8-GFP were fixed for 2.5 h at room temperature with 2.5% formaldehyde + 0.15% glutaraldehyde in 50 mM cacodylate buffer, pH 7.4, followed by two washes in 50 mM cacodylate buffer, pH 7.4. Cells were trapped in 1% agarose and dehydrated in an ethanol series followed by embedding in LR-White resin according to standard protocols for immunoelectron microscopy. Ultrathin sections were decorated with affinity-purified, polyclonal anti-GFP antibody described above, followed by protein A-gold (5-nm) conjugates obtained from the Department of Cell Biology (University of Utrecht, Utrecht, The Netherlands).
Gene Silencing by Feeding
The open reading frames of the VATA2_1 and VATA3_1 genes were excised from the plasmids pPXV-a2-GFP and pPXV-a3-GFP with SpeI and XhoI and cloned into the double T7-promotor plasmid pL4440 (described in Timmons et al., 2001). Plasmids were introduced in the E. coli Ht115 strain, and Paramecium cells were fed with these strains as described in Wassmer et al. (2005) following the protocol of Galvani and Sperling (2002). After 38-48 h of feeding paramecia were analyzed.
Construction of Chimeras
The N- and C-terminal halves of the a2-1 subunit were amplified from the corresponding gene of the macronuclear DNA using oligonucleotides a2f/a2r-Xma (5′-catcccggggttcacttccttatacc-3′) and a2r/a2f-Xma (5′-gaaccccgggatgtttgccgttatg-3′) and cloned into pL4440 using SpeI/XmaI (yielding pL4440/a2v) and XmaI/XhoI (yielding pL4440/a2h), respectively. Afterward the 5′ and the 3′ halves of the gene encoding a3 were amplified using oligonucleotides a3f/a3r-Xma (5′-caacccgggattgatttccttgtatc-3′), cloned into pL4440/a2h using SpeI/XmaI (yielding pL4440/a3v/a2h), and a3r/a3f-Xma (5′-aatcccgggttgtctactatcataac-3′) cloned into pL4440/a2v using XmaI/XhoI (yielding pL4440/a2v/a3h). Both inserts were excised using SpeI/XhoI and cloned into pPXV-GFP, yielding pPXV-a2a3-GFP and pPXV-a3a2-GFP (compare Figure 11) with an artificially introduced XmaI site that does not change the amino acid sequence (ccc.ggg/Pro.Gly) between the N- and C-terminal halves of the protein. These two plasmids were used to construct other plasmids by cloning with SpeI, XmaI, and XhoI to test the localization of different N and C termini of the corresponding proteins. Amplification of the 5′ end corresponding to the N terminus of a4 was carried out using the oligos a4f and a4r-Xma (5′-tagcccggggttgatttctcgatatctagc-3′); for the 3′-end corresponding to the C terminus of a5 a5f-Xma (5′-aaccccgggttatttaccataatcacattc-3′) and a5r were used.
Figure 11.
Chimerical a-subunits molecules coupled to GFP. The chimeras are represented as box-schemes, drawn in red for the a2-1 subunit, in blue for the a3-1 subunit, in green for the a4-1 subunit and in yellow for the a5-1 subunit. The amino acid positions in the native proteins are indicated above/below the schemes. The alignment at the points of exchange is printed within the scheme. Fluorescence microscopy images (a-e) illustrate the different localizations observed. (a) The contractile vacuole complex (cvc), as observed using the a3a2 construct. (b) Trichocysts (tr) that are attached to the cell cortex, indicated by arrowheads, visualized with a2a3. (c) Membranes of phagosomes (ph), labeled by a3a5. (d) Vesicular and diffuse staining, produced by a3a2CH1. (e) Diffuse cytoplasmic staining, caused by a3a2Ch7. Bar, 10 μm.
Exchanges within the 3′ end corresponding to the C terminus of the a2/a3-proteins were done by fusion PCR. In a first round of PCR, the two fragments from a2 and a3 were amplified from plasmid or macronuclear DNA using oligos a2f-Xma, a3f-Xma, a2r, and a3r, and the respective internal primers. The two/three fragments were excised from the gel, and the gel-slices were frozen for 30 min, thawed, and centrifuged for 15 min at 4°C. From the supernatant, 2 μl of each fragment was used as template in a 50-μl PCR reaction using the oligos a2f-Xma/a3r or a3f-Xma/a2r, followed by cloning with XmaI/XhoI into the plasmid pPXV-a3a2-GFP. Oligo combinations used were as follows: pPXV-a3a2Ch1-GFP: first PCR: a2f-Xma/Ch1r (5′-cagtagctgcagagactcttgagactctaagtcaaac-3′)—first fragment, Ch1f (5′-gagtctctgcagctactgttattaaccatcataacc-3′)/a3r—second fragment. Fusion PCR: a2f-Xma/a3r. pPXV-a3a2Ch2-GFP: first PCR: a3f-Xma/Ch2r (5′-agattactgcaggaatctctattgctctccagattcg-3′)—first fragment, Ch2f (5′-agattcctgcagtaatctattctattttggtc-3′)/a2r—second fragment. Fusion PCR: a3f-Xma/a2r. pPXV-a3a2Ch3-GFP: first PCR: a2f-Xma/Ch3r (5′-aagaagtggttttgtaataagcatccaaggaatgc-3′)—first fragment, Ch3f (5′-tgcattccttggatgcttattacaaaaccacttct-3′)/a3r—second fragment. Fusion PCR: a2f-Xma/a3r. pPXV-a3a2Ch4-GFP: first PCR: a2f-Xma/Ch4r (5′-taaatagctagcagtattactaatagaaccaag-3′)—first fragment, Ch4f (5′-agtaatactgctagctatttaagattatgggcact-3′)/a3r—second fragment. Fusion PCR: a2f-Xma/a3r. pPXV-a3a2Ch5-GFP: first PCR: a2f-Xma/Ch5r (5′-acattccataagatccatgcacat-3′)—first fragment, Ch5f (5′-atgtgcatggatcttatggaatgt-3′)/a3r—second fragment. Fusion PCR: a2f-Xma/a3r. pPXV-a3a2Ch6-GFP: first PCR: a3f-Xma/Ch6r (5′-caaaacaacttccaaataagtttaag-3′)—first fragment, Ch6f (5′-aacttatttggaagttgttttgaaaatg-3′)/a2r—second fragment. Fusion PCR: a3f-Xma/a2r. pPXV-a3a2Ch7-GFP: first PCR: a3f-Xma/Ch7r (5′-cccaactctatccccatttcaaaaacaataagaaatc-3′)—first fragment, Ch7f (5′-ttttgaaatggggatagagttgggaaggtag-3′)/a2r—second fragment. Fusion PCR: a3f-Xma/a2r. pPXV-a3a2Ch8-GFP: first PCR: a2f-Xma/Ch8-1r (5′-ctaccaaattgtaacaacttccaaatatattccaag-3′)—first fragment, Ch8-1f (5′-tatttggaagttgttacaatttggtagatggtgaat-3′)/Ch8-2r (5′-ggcataaacttaattccatcggctttgtaaaatttg-3′)—second fragment, Ch8-2f (5′-caaagccgatggaattaagtttatgccattttcattc-3′)/a2r—third fragment. Fusion PCR: a2f-Xma/a2r.
RESULTS
Identification of 17 a-Subunit Genes
In a pilot sequencing project (Dessen et al., 2001), a clone of a genomic library containing a sequence homologue to a human a-subunit gene of the V-ATPase was identified. Sequencing of this clone, amplification from a cDNA library, cloning, and sequencing allowed us to identify the first a-subunit gene of Paramecium, called VATA1_1 (Table 1). During early steps of the Paramecium whole genome shotgun sequencing undertaken by Genoscope (http://www.genoscope.cns.fr), we were able to identify 15 contigs containing each a new a-subunit coding gene, and, by manual assembly of single reads excluded from the draft assembly we used, one more a-subunit (encoded by VATA5_2) could be found, giving a total of 17 a-subunit genes (Table 1). Note that the nomenclature does not correspond to the one used in mammals.
Table 1.
Molecular characteristics of the 17 a-subunit genes of the V-ATPase of P. tetraurelia.
| Gene | Protein | Length (bp) | Introns | Amino acids | Calculated Mr | Accession no. | GFP construct |
|---|---|---|---|---|---|---|---|
| VATA1_1 | a1-1 | 2545 | 2 | 832 | 97.3 | CR932830 | a1-GFP |
| VATA1_2 | a1-2 | 2557 | 2 | 836 | 97.6 | CR932831 | |
| VATA2_1 | a2-1 | 2849 | 5 | 906 | 104.3 | CR932832 | a2-GFP |
| VATA2_2 | a2-2 | 2852 | 5 | 908 | 104.7 | CR932833 | |
| VATA3_1 | a3-1 | 2530 | 5 | 800 | 92.8 | CR932834 | a3-GFP |
| VATA3_2 | a3-2 | 2529 | 5 | 800 | 93.0 | CR932835 | |
| VATA4_1 | a4-1 | 2440 | 5 | 772 | 89.6 | CR932836 | a4-GFP |
| VATA5_1 | a5-1 | 2651 | 4 | 850 | 99.6 | CR932837 | a5-GFP |
| VATA5_2 | a5-2 | 2655 | 4 | 850 | 99.2 | CR956351 | |
| VATA6_1 | a6-1 | 2624 | 5 | 831 | 96.5 | CR932838 | a6-GFP |
| VATA6_2 | a6-2 | 2613 | 5 | 828 | 96.4 | CR932839 | |
| VATA7_1 | a7-1 | 2462 | 4 | 788 | 92.4 | CR932840 | a7-GFP |
| VATA7_2 | a7-2 | 2464 | 4 | 788 | 92.4 | CR932841 | |
| VATA8_1 | a8-1 | 2534 | 6 | 793 | 93.1 | CR932842 | |
| VATA8_2 | a8-2 | 2539 | 6 | 795 | 93.3 | CR932843 | a8-GFP |
| VATA9_1 | a9-1 | 2768 | 7 | 860 | 99.0 | CR933341 | a9-1-GFP |
| VATA9_2 | a9-2 | 2760 | 7 | 859 | 99.0 | CR932845 | a9-2-GFP |
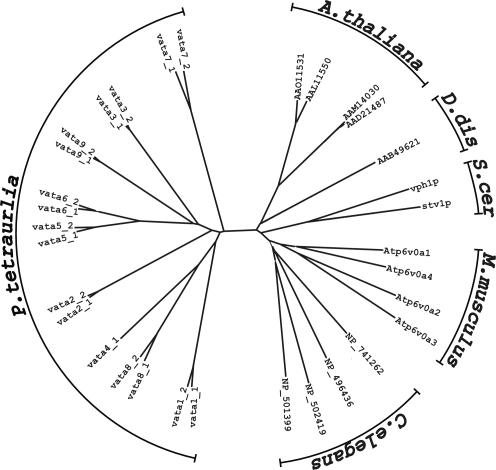
By alignment of the deduced protein sequences using the ClustalW algorithm, the a-subunits were found to cluster in pairs except the a-subunit 4 (encoded by VATA4_1) (Figure 1, compare with the alignments in Supplemental Materials). Within a pair, the two genes show a nucleotide identity of >80%, whereas the encoded proteins show sequence identities of 82.5-96.8%. Alignment and comparison of Paramecium a-subunit sequences with those of Mus musculus, Caenorhabditis elegans, Arabidopsis thaliana, Dictyostelium discoideum, and Saccharomyces cerevisiae resulted in amino acid identities between 18 and 28%.
Figure 1.
Unrooted tree constructed from V0 a-subunits of P. tetraurelia, C. elegans, M. musculus, S. cerevisiae, D. discoideum, and A. thaliana using the ClustalW algorithm. The a-subunits of Paramecium form a cluster that is well separated from any other organism. They form pairs generally with amino acid identity >82%, except VATA4_1.
Hydrophobicity plots of the a-subunits of Paramecium resemble those of Vph1p, one of the yeast a-subunits (Leng et al., 1999). Therefore, we propose a similar overall structure of the a-subunits in Paramecium, with a large N-terminal domain of ∼380-440 amino acids, followed by six to nine transmembrane segments, highly conserved among the a-subunits of Paramecium. Also, most residues identified in Vph1p being crucial for V-ATPase activity, e.g., D425 and R735 (Leng et al., 1996, 1998) are present in all Paramecium a-subunits.
Localization of the a-Subunits In Vivo by Tagging with GFP
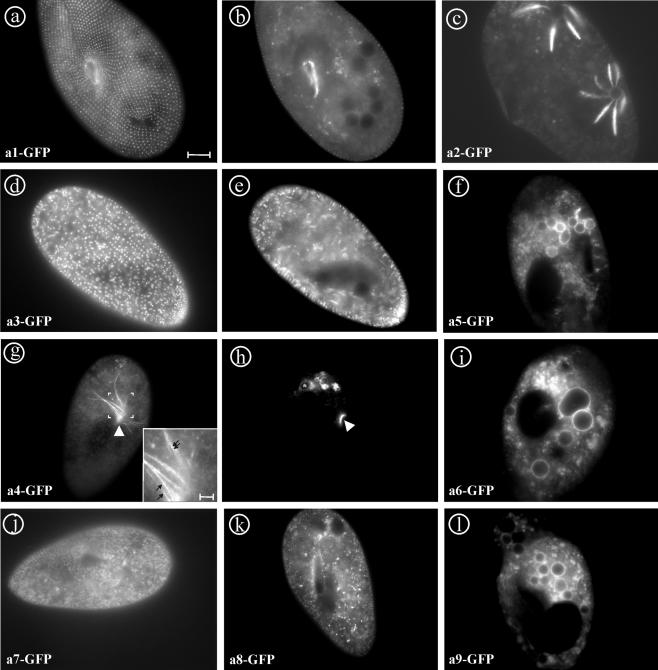
In yeast, the V-ATPase a-subunit Vph1p was found in the membrane of vacuoles, whereas Stv1p was found in the Golgi complex (Kawasaki-Nishi et al., 2001a). To test whether the 17 a-subunits in Paramecium also have differential localization, genes of each of the nine groups of isoforms were fused at their 3′ end to the 5′ of the GFP gene in the Paramecium expression vector pPXV-GFP. In general, always the first gene of a pair (VATA1_1, VATA 2_1, and so on) was selected, except the a-subunit 9 pair, from which both genes were GFP tagged (Table 1). After transformation with the different fusion constructs, intracellular labeling of resulting clones was observed by fluorescence microscopy. Interestingly, most of the fluorescent a-subunits labeled different organelles, giving a total of seven distinct cellular labeling (Figure 2).
Figure 2.
In vivo labeling of the a-subunits 1-9 by C-terminal GFP-tagging. (a and b) a1-GFP produces a punctate pattern at the cell cortex, and it labels the cytostome. (c) a2-GFP stains the radial arms and weakly the vacuole of the contractile vacuole complex. (d and e) a3-GFP labels trichocysts, the cytoplasmic background may correspond to the biogenic pathway of these organelles. (f) a5-GFP is targeted mainly to the membrane of phagosomes, but a “cloud” of vesicles in the posterior part of the cell is also stained. (g and h) a4-GFP localizes to a population of vesicles at the cytostome (arrowhead). Depending on the situation, “trains” of these vesicles (g, inset, arrows) from phagosomes back to the cytostome can be observed (g), whereas in other cells mainly the cytostome is strongly and the membrane of the youngest phagosomes is weakly stained (h; asterisk). (i) a6-GFP localized to the membrane of phagosomes as is a5-GFP. (j) a7-GFP shows mainly cytoplasmic localization that may correspond to the ER. (k) a8-GFP brightly labels “dot-like” structures throughout the cytoplasm that may represent the Golgi complex that in ciliates consists of small stacks dispersed throughout the cytoplasm. (l) both a9_1 and a9_2 fusion proteins show the same localization as a5- and a6-GFP. Bar, 10 μm; g inset, bar, 2 μm.
The a1-GFP fusion protein exclusively localized to a punctuate network at the cell cortex and the cytostome (Figure 2, a and b). Double labeling of paramecia with a monoclonal anti-tubulin antibody staining basal bodies and a1-GFP (Figure 3) showed that this protein is present in an organelle close to basal bodies, probably representing early endosomes or vesicles of the endocytotic route that are known to be located in proximity to basal bodies in Paramecium (Allen et al., 1992). To define the labeled structure more clearly, a1-GFP-transformed cells were prepared for immunoelectron microscopic analysis and decorated with anti-GFP antibody. Gold label was clearly found on vesicles in close relation to early endosomes, the so-called terminal cisternae in Paramecium (Figure 4, A-C).
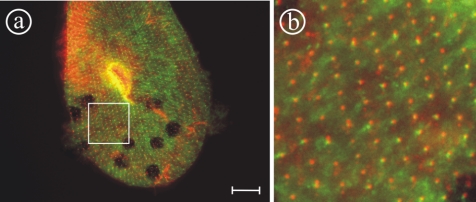
Figure 3.
Double labeling of paramecia using a1-GFP and the monoclonal anti-tubulin antibody ID5 to stain basal bodies (Wehland and Weber, 1987). The double staining shows that the organelles containing the a1-subunit (green) are positioned closely to basal bodies (red), although the position varies slightly. The proximity to basal bodies suggests that these organelles are associated with endocytotic elements, the so called “parasomal sacs” below basal bodies. (b) Enlargement of the square indicated in a. Bar, 10 μm.
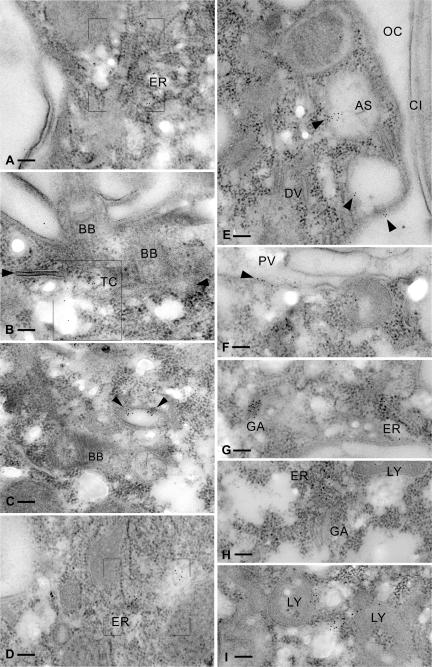
Figure 4.
Immunoelectron microscopy localization of a1-GFP (A-C), a7-GFP (D), a4-GFP (E and F), and a8-GFP (G-I). a1-GFP localizes to a cortical region (framed) enriched in ER (A), in a region (framed) located below ciliary basal bodies (BB) and containing terminal cisternae (TC) (B). (C) A cortical section displaying a basal body and a terminal cisternae with gold label (arrowheads) is shown. (D) Enrichment of a7-GFP, in an ER-rich region (framed). (E) a4-GFP label (arrowheads) is associated with the membranes of acidosomes (AS) closely attached to the oral cavity (OC) showing a cilium (CI), whereas no label occurs on discoidal vesicles (DV). (F) a4-GFP labeling along the membrane of a young phagocytic vacuole (PV), between arrowheads. (G) a8-GFP labels in the two framed areas, on one of the inconspicuous Golgi fields (GA; left) and in an ER-rich zone (right). (H) a8-GFP label in a domain located between a Golgi apparatus (GA) and an ER-rich zone. (I) a8-GFP label associated with polymorphous compact organelles, i.e., bona fide lysosomes (LY). Bars, 0.1 μm.
The a2-GFP fusion protein was exclusively targeted to the contractile vacuole complex where it strongly stained the radial arms (Figure 2c). Surprisingly, also the ampullae and the contractile vacuole are slightly stained by the fusion protein. Previously, by using an anti-B-subunit antibody of V1, the V-ATPase holoenzyme was shown to be restricted to the decorated spongiome of the radial arms of the contractile vacuole complex (Fok et al., 1995).
The a3-GFP fusion protein is localized in membranes of dense core secretory granules of Paramecium (Figure 2, d and e), known as trichocysts. Trichocysts' biogenesis occurs after massive discharge into the medium (Plattner et al., 1985; Garreau de Loubresse, 1993) or during cellular growth, in a process tightly regulated at the transcriptional level (Galvani and Sperling, 2000). The faint labeling of the cytoplasm therefore probably corresponds to the biogenetic pathway of these organelles. The V-ATPase was previously shown to be crucial for the formation of trichocysts (Wassmer et al., 2005).
The a4-GFP fusion protein was found in small vesicles at the cytostome and can be seen in the first one or two phagosomes just after pinching off of the cytostome, whereas older phagosomes are devoid of a4-GFP labeling (Figures 2h and 5a). In some cases, labeled “strings” emanating from phagosomes back to the cytostome can be observed (Figure 2g). These probably represent trains of small vesicles that are transported from phagosomes back to the oral cavity gliding along cytoskeletal elements. To investigate this phenomenon more closely, we inhibited phagosome formation by the addition of cytochalasin B to the a4-GFP-transformed cells for 60 min and analyzed them after fixation. The addition of cytochalasin B to Paramecium cells inhibits the process of phagocytosis almost instantaneously (Allen and Fok, 1983; Cohen et al., 1984). By light microscopy, it can be seen indeed that phagosome formation is inhibited by cytochalasin B and that the oral cavity is enormously enlarged (Figure 5d). In addition, the oral cavity is surrounded by a cloud of small vesicles that display a4-GFP fluorescence (Figure 5b). Because the membrane of the cavity is not continuously stained, this may indicate that fusion of the vesicles with the forming phagosome has not taken place. In contrast, control cells show staining of the membrane of the first food vacuole (asterisk) in a continuous manner, indicating that membrane fusion has taken place. Immunoelectron microscopy of the cytostome of a4-GFP cells showed gold label of vesicles docked at the oral cavity (Figure 4E). The physiology and the ultrastructure of these vesicles suggest that they represent acidosomes (Allen and Fok, 1983). Indeed, Paramecium acidosomes are known to have high V-ATPase activity, to be localized at the cytostome, and to fuse with phagosomes immediately after pinching off. This induces a rapid and strong acidification of the phagosomes' contents. In addition, excess membrane of these phagosomes is withdrawn and recycled after a few minutes, whereas lysosomes fuse in turn with the phagosomes (Fok and Allen, 1988).
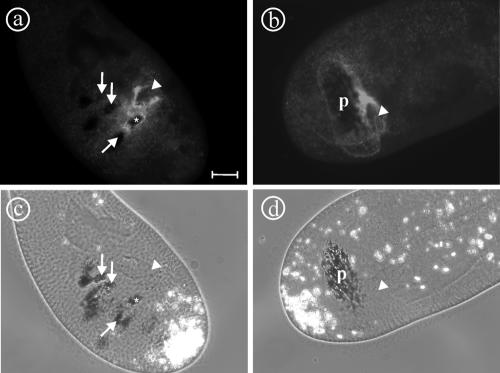
Figure 5.
Effect of cytochalasin B on the distribution of a4-GFP. Cytochalasin B inhibits the fission of nascent phagosomes from the oral cavity, leading to the formation of a large pouch (p) originating from the buccal cavity (arrowheads), as visualized by the addition of India ink to the medium (d). This pouch is surrounded by vesicles that are labeled by a4-GFP and that apparently do not fuse with this structure (b). In contrast, in control cells the vesicles rapidly fuse with phagosomes that have been released from the cytostome (a). Note that only the newly formed phagosome (asterisk) shows GFP labeling, whereas the older phagosomes are devoid of any label (arrows; a). This finding suggests that the small vesicles are “acidosomes” that in Paramecium transport the V-ATPase to phagosomes via the cytostome (Allen and Fok, 1983). Bar, 10 μm.
The a5-GFP, a6-GFP, and both a9-1- and a9-2-GFP fusion proteins mainly localized to the membranes of phagosomes (Figure 2, f, i, and l) but also to clouds of small fluorescent vesicles, which may represent lysosomes. It is noteworthy that the cytostome is not labeled with any of these three a-subunits in contrast to a-subunit 4.
The a7-GFP fusion protein seemed to be in a continuous network that may represent at least a part of the endoplasmic reticulum (ER) (Figure 2j; Hauser et al., 2000a; Ramoino et al., 2000). Immunoelectron microscopy of a7-GFP cells showed labeling in the ER-rich cortical region, although any more strict structural assignment was not possible (Figure 4D).
The a8-GFP fusion protein gave strong staining of small dot-like structures in the cytoplasm (Figure 2k). These structures most likely represent the Golgi apparatus that, in Paramecium, consists of many stacks with very few cisternae that are dispersed throughout the cytoplasm (Estève, 1972; Garreau de Loubresse, 1993). In immunoelectron microscopy, we did find gold labeling on Golgi stacks but also in association with lysosomes (Figure 4, G-I).
Protein Level of a-Subunit-GFP Fusion Gene Products
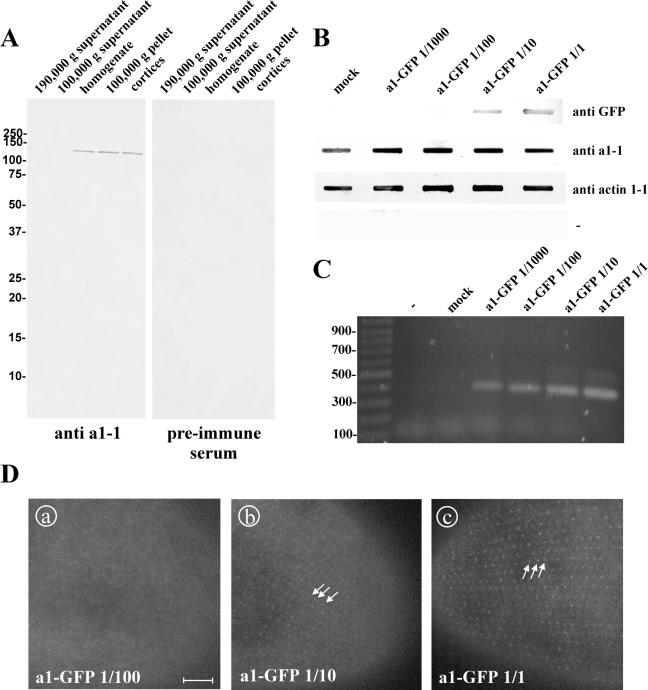
Because our fusion gene is expressed under the control of the calmodulin promotor and may reside in the macronucleus at a higher copy number than the endogenous gene, we tried to assess the degree of overexpression achieved by our system using a dilution series of plasmid DNA over a 1000-fold spectrum (Figure 6). Paramecia were transformed with ∼5 μg/μl and 1/10, 1/100, and 1/1000 serial dilutions thereof, grown to large culture volumes, harvested, and lysed. By PCR, the cell lysates were shown to contain the a1-GFP fusion gene (Figure 6C), whereas by slot-blots the content of the endogenous a1-1 was compared with that of recombinant a1-GFP (Figure 6B). The affinity-purified anti a1-1 antibody used to probe the slot blots was directed against the P178-S328 region of the a1-1 subunit (see Materials and Methods) and characterized by a Western blot analysis on Paramecium fractions (Figure 6A). Using PCR amplification, we were able to detect the a1-GFP fusion gene in all the clones of the dilution series, proving the actual transformation. In contrast, the slot-blot decoration with anti-GFP antibody was able to detect the presence of the a1-GFP protein only in clones transformed with 5 μg/μl and the 1/10 dilution thereof. Consequently, the GFP fluorescence signal is only visible in cells injected with the concentrations of 5 μg/μl and the 1/10 dilution (Figure 6D). Note that the localization in structures forming the regular, punctate, cortical pattern is the same at both concentrations, arguing for a localization that is independent of the concentration. Interestingly, decoration of the slot-blot with the a1-1 (P178-S328) antibody that is supposed to label the endogenous a1-1 as well as the recombinant a1-GFP, resulted in equally strong staining throughout the dilution series, meaning that the a1-1-plus the a1-GFP-content is constant in the range of the detection limit. A possible interpretation of this result is that the transformation system using the pPXV-GFP vector system produces only weak protein expression, which may not be detectable in the slot-blot system used. Another interpretation may be that Paramecium tightly controls the assembly of the V-ATPase and degrades excess a-subunits that are not incorporated in V0-complexes, as shown for Saccharomyces (Jackson and Stevens, 1997; Hill and Cooper, 2000). Therefore, using our transformation system for the expression of V-ATPase a-subunits apparently does not result in strong overexpression and was judged to be applicable for the localization of the a-GFP constructs.
Figure 6.
Assessment of the degree of expression of a1-GFP by the transformation of Paramecium cells in a dilution series. (A) characterization of the antibody directed against a1-1(P178-S328) in a Western blot of Paramecium cell fractions. Paramecia were homogenized and separated into a soluble and an isoluble fraction by centrifugation at 100,000 or 190,000 × g. “Cortices” were obtained by rupturing cells followed by washing, so that the cytoplasm is lost and only the insoluble structures of the cell cortex are retained. Fifty micrograms of protein was loaded per lane. A band of ∼120 kDa is visible in cell homogenate, 100,000 × g pellet, in cortices, and, very faintly, in 100,000 × g supernatant with the affinity-purified anti a1-1 antibody, whereas none is visible either in 190,000 × g pellet or when the preimmune serum was used. This is in accordance with the molecular characteristics of a1-1 as a transmembrane protein that is enriched in the cell cortex. (B) Slot-blot to estimate the degree of overexpression of a1-GFP. Cell lysates from clones injected with either a1-GFP at ∼5 μg/μl, 1/10, 1/100, 1/1000 dilutions thereof or the mock control were transferred on nitrocellulose membrane and probed with different antibodies. A monoclonal anti-GFP antibody was used to detect GFP, the anti a1-1(P178-S328) antibody was used to detect endogenous a1-1 and recombinant a1-GFP. Anti actin1-1 antiserum (described in Kissmehl et al. 2004) was used as loading control. To exclude cross reactivity of the secondary antibody with Paramecium proteins a control without a primary antibody was included (-). (C) PCR to confirm the successful transformation of the clones with a1-GFP. (D) GFP fluorescence of clones transformed with a1-GFP at a concentration of 5 μg/μl, 1/10 and 1/100 dilution. Using 5 μg/μl (D, c) or the 1/10 dilution (D, b), the pointed cortical pattern is visible (arrows), whereas no GFP fluorescence could be detected with the 1/100 (D, a) or 1/1000 dilution (our unpublished data). Note that the labeled structures are the same with 5 μg/μl and the 1/10 dilution, so the targeting of the GFP fusion protein is not dependent of the transformation degree. Bar, 10 μm.
Immunolocalization of Endogenous a1-1 Protein
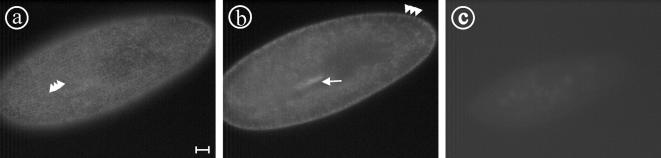
To confirm the localization of the a1-1 protein in the cortical organelles probably representing terminal cistaernae as suggested by GFP labeling of a1-1, immunolabeling with the anti a1-1(P228-E259) antibody was performed (Figure 7). The antibody stains the cell cortex and the cytostome (Figure 7, a and b), confirming the localization information obtained using a1-GFP, thus demonstrating that GFP-tagging is a reliable tool to acquire localization information on the V0-a-subunits.
Figure 7.
Localization of the endogenous a1-1 protein using the anti a1-1(P228-E259) antibody. a1-1 is localized in the cell cortex as visible in surface (a) or median sections (b) in a punctate pattern (arrowheads) with enrichment at the cytostome (arrow), paralleling the observations made by the transformation of paramecia with a1-GFP (compare Figure 2, a and b). Note that the network is in some places distorted and the punctate character less obvious in comparison to the GFP localization of a1-1 in living cells, probably because of the Triton X-100 permeabilization necessary for immunofluorescence. (c) Control in which the primary antibody was omitted. Bar, 10 μm.
Specificity of Function of the a-Subunits
Considering the specialized distribution of the different a-subunits to specific organelles, we wondered whether specialized functions could also be attributed to the different a-subunits and whether they can substitute for each other, as in yeast. To approach this question, we chose the two a-subunit genes VATA2_1 and VATA3_1, their gene products being specific to the contractile vacuole complex and trichocysts, respectively, to undertake RNAi experiments. RNAi was performed by delivery of double-stranded RNA to Paramecium cells by feeding with genetically engineered E. coli (Galvani and Sperling, 2002). A prerequisite for the inactivation of both genes (VATA2_1/VATA2_2 and VATA3_1/VATA3_2, respectively) using only one gene of the pair is a high degree of nucleotide identity (>85%) between the two genes (Ruiz et al., 1998). This is the case within each pair (VATA2_1/VATA2_2, 91% identity; VATA3_1/VATA3_2, 94%).
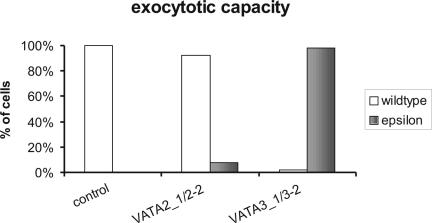
Silencing of VATA2_1,2_2 genes led to heavy perturbations of the contractile vacuole complex. The cells became swollen within 38 h and died within 48 h of feeding, whereas control cells and cells with silenced VATA3_1,3_2 genes showed no impairment of the contractile vacuole shape or activity and continued to divide. In contrast, silencing of the VATA3_1,3_2 genes completely blocked trichocyst exocytosis, as tested by picric acid, whereas control cells and cells silenced for VATA2_1,2_2, even shortly before death, showed a wild-type exocytosis phenotype (Figure 8). Immunolabeling in silenced cells showed an almost total absence of trichocysts in VATA3_1,3_2-silenced cells, whereas control- or VATA2_1,2_2-silenced cells showed a normal number of trichocysts docked at the cell cortex (Figure 9). This experiment shows that silencing of one of these a-subunit pairs cannot be complemented by any other a-subunit gene, suggesting that not only the localization of the a-subunits differs but also that they differentiated to exert specific biological functions.
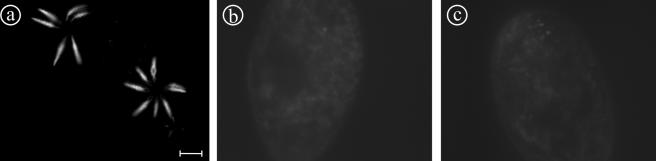
Figure 8.
Exocytotic capacity of cells silenced for VATA2_1/2_2 or VATA3_1/3_2, as analyzed by the picric acid test after 48 h of feeding. Samples of ∼35 cells were analyzed in each experiment. Cells silenced for the VATA3_1/3_2 showed a massive impairment of exocytotic capacity (“epsilon” phenotype corresponds to <50 expelled trichocysts per cell), whereas control cells and cells silenced for VATA2_1/2_displayed wild-type phenotype (500-1000 expelled trichocysts per cell).
Figure 9.
Immunolabeling of trichocysts in cells silenced for VATA2_1/2_2 or VATA3_1/3_2 shows that the silencing of VATA3_1/3_2 selectively blocks the formation of trichocysts. To characterize the exocytosis deficiency of VATA3_1/3_2-silenced cells in the picric acid test in detail, an immunofluorescence analysis was performed to test whether functional trichocysts are formed and attached to the cell cortex. Although control- (a) and VATA2_1/2_2-silenced paramecia (b) showed extremely high density of trichocysts docked at the cell cortex, their number was greatly reduced in VATA3_1/3_2-silenced cells (c); arrows indicate remaining trichocysts. Bar, 10 μm.
Cross-Regulation between V1 and V0
We then asked what would happen to V0 during the assembly of the holoenzyme if single subunits are present in substoichiometric amounts or absent. The answer to this question may provide valuable insight into regulatory processes both at the posttranscriptional and posttranslational level. Therefore, Paramecium cells were transformed with the a2-GFP fusion construct, and gene silencing of all c-subunits of V0 or the F-subunits of V1 (Wassmer et al., 2005) was carried out. Cosilencing of all c-subunits almost eliminated fluorescence of a2-GFP within 48 h (Figure 10), meaning that the expression of c-subunit interaction partners within V0 has to be down-regulated. This regulation probably occurs at the translational level, because a2-GFP is expressed under the control of the constitutive calmodulin promotor, not the endogenous promotor, so a transcriptional regulation is not possible.
Figure 10.
Effect of gene silencing of V0-c-subunits or V1-F-subunits on the a2-1 subunit expression. Paramecia transformed with a2-GFP were subjected to gene silencing by feeding of all six c-subunits of V0 (b) or both F-subunits of V1 (c) (Wassmer et al., 2005). After 48 h of feeding, cells were analyzed for fluorescence. Silencing of c- or F-subunits led to the total absence of GFP fluorescence of a2-GFP (b and c) in contrast to control cells (a). This suggests that the protein level of a-subunits is posttranslationally controlled and that excess a-subunits are degraded if other subunits are missing for the correct assembly of the holoenzyme. Bar, 10 μm.
Surprisingly, silencing of the F-subunits of V1 also led to near elimination of fluorescence within 48 h. This suggests that excess V0 subunits are degraded if not enough functional V1 complexes are available for building the holoenzyme. So, the assembly of the V-ATPase in Paramecium seems to be tightly controlled at the protein level. This contrasts with the situation in yeast in which V0 is assembled and targeted correctly despite the loss of any of the V1-subunits (Parra et al., 2000).
The Search for the Targeting Signal
In contrast to the c-subunit isoforms that all seem to be targeted to all organelles containing V0 sectors (Wassmer et al., 2005), a-subunit isoforms are specific to particular organelles with little or no overlap in localization between the different isoforms. We wondered whether the addressing signal was contained within the primary sequence of the a-subunits and tried to unravel the nature of this signal. The principle of the experiment was to construct chimerical molecules between subunits in tandem with GFP and to follow their subcellular localization. The junctions between peptides of different origin were always carefully chosen in regions that are well conserved between all Paramecium a-subunits, to avoid the fusion of nonhomologues regions.
The first experiment consisted in joining N-terminal halves to C-terminal halves of different a-subunits. The N-ter./C-ter. chimeras tested combined respectively a2-1 with a3-1 (a2a3), a3-1 with a2-1 (a3a2), a3-1 with a5-1 (a3a5), a4-1 with a5-1 (a4a5), and a4-1 with a2-1 (a4a2). Consistently, all chimerical peptides localized to the compartment corresponding to the target of the a-subunit composing their C terminus: a2a3 went to the trichocysts (Figure 11b), a3a2 and a4a2 went to the contractile vacuole complex (Figure 11a), whereas a3a5 and a4a5 are located in the membranes of phagosomes (Figure 11c). This means that the targeting information is within the C terminus of the proteins, a very surprising finding because it represents the opposite situation compared with yeast, in which the targeting signal is contained within the N terminus (Kawasaki-Nishi et al., 2001a).
To get a better idea of the nature of the localization signal within the C-terminal half of the a-subunits, we focused on the a2- and a3-subunits and constructed a series of chimeras, also fused to GFP, keeping the N terminus of a3-1 and exchanging several portions of the C termini of a2-1 and a3-1 (Figure 11). First, halves of the a2-1 C terminus were introduced in the a3-1 subunit. Then, the a3-1/a2-1 middle junction or the a2-1/a3-1 C-terminal junction was displaced toward the end of the molecule to include varying parts of a2-1 in the a3-1 C terminus. In all these cases (Figure 11), the localization of the chimeras was mainly found in the ER (Figure 11e) and in vesicle-like structures that are normally not visible in cells (Figure 11d), and cells were unable to divide and died within 48 h. This result may indicate that there is no particular region shorter than the C-terminal half of the molecule with targeting efficiency. The localization signal seems to be much more than a short signal sequence and seems to depend on the overall organization of the C-terminal region.
Local and Remote Effects Caused by Chimeras
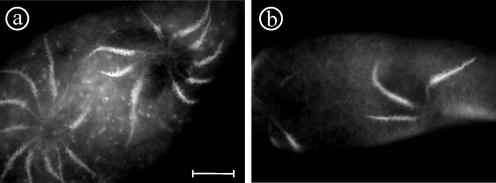
Cells transformed with the a3a2 chimera showed some slight alterations of the contractile vacuole complex. The frequency of contraction of the contractile vacuole was somewhat prolonged and cells looked inflated, compared with noninjected control cells or cells injected with a2-GFP or a3-GFP, indicating a general insufficiency of the contractile vacuole complex. The number and length of radial canals in these chimerical cells were also larger than in control cells (Figure 12a). The effects seemed to depend on the quantity of the material injected. From these observations, we conclude that the composite a3a2-subunit differs in its physiological properties from the a2-subunit, although the localization in the contractile vacuole complex is the same. It should be kept in mind that besides the expression of a3a2, the endogenous forms a2-1 and a2-2 are also expressed, possibly obscuring a clear-cut phenotype that would be provoked by the selective expression of the chimera. We call this phenomenon “local effect” because the phenotype touches the organelle in which the protein is present.
Figure 12.
Local effect obtained with the a3a2 chimera. Cells expressing a3a2 (a), which is targeted to the contractile vacuole complex, seemed inflated and the fluid expulsion cycle of the contractile vacuole was prolonged. The number and length of the radial arms also seemed to be increased compared with cells transformed with the a2-GFP fusion (b). Bar, 10 μm.
In contrast, the presence of the a2a3 chimera provoked a different effect. Although this chimera is targeted to the trichocysts, as a3-GFP is, it gave a surprisingly dramatic effect on the contractile vacuole complex to which it is not targeted but in which the N-terminal part of a2-1 should normally be present. Indeed, the contractile vacuole complex shows severe disturbances in the organization of its cycle. Some transformed clones did not show any contractile vacuole beating for >10 min, compared with the 8- to 12-s period in control cells, also correlated with exaggerated swelling (Figure 13, b and c). Cells injected with a large DNA quantity of the chimera generally made between zero and two divisions and died within 48 h. We propose to call this phenomenon “remote effect,” because the phenotype caused by the chimera does touch a remote organelle, not the organelle to which the protein is targeted. In cells transformed with this chimera, the trichocyst biogenesis seemed to be unaffected.
Figure 13.
Remote effect obtained with the a2a3 chimera (b-d) compared with cells expressing a3-GFP (a). Different clones expressing a2a3 (b-d), which is targeted to trichocysts, show nevertheless heavy defects in contractile vacuoles, such as enormous inflation (b and c) or an increased number of contractile vacuole complexes (d). Arrowheads indicate the contractile vacuole. Bar, 10 μm.
After the discovery of this distant effect, cells transformed with the a3a2-chimera were reexamined and analyzed for exocytosis by the picric acid test. We found a mild impairment of exocytotic capacity, estimated to be decreased down to 50%. So, the remote effect seems to exist for other chimeras as well, although it is less pronounced than with the a2a3-chimera.
DISCUSSION
Specialization of V-ATPase a-Subunits
In P. tetraurelia, we found 17 genes encoding a-subunits of the V-ATPase representing nine families that are targeted to at least seven different compartments. Seventeen a-subunit genes is by far the largest number of a-subunits found in any organism, followed by mammals with four a-subunit genes (Nelson, 2003). This raises the question why so many subunits have been maintained in Paramecium. One answer could be the high degree of cellular organization with many differentiated organelles in a single cytoplasm. Multicellular organisms perform different functions by distributing them on different cellular populations with varying gene expression levels, alternative splicing, and posttranscriptional and translational modifications. In Paramecium, these regulation elements would have global effects that would be unable to discriminate between subcellular compartments. It is thus understandable that it is beneficial for Paramecium to keep different isoforms of one gene that allow a high degree of specialization within one cell.
If we assume that all different localizations actually correspond to different functions, this means that a-subunits bear both localization signals specific for a compartment and functional domains specific for their activity in this compartment. They should thus act as a bridge between two interaction domains, the one interacting with compartment-specific partners, the other one with functional partners.
C-Terminal Localization Domain
We found the localization signal in the C-terminal half of the molecule, a region containing transmembrane domains. This contrasts with the observations made in yeast in which both V-ATPase a subunits are targeted to their compartment through their N-terminus (Kawasaki-Nishi et al., 2001a). Using various composite chimeras within the C-terminal half of a2 and a3, we found that the signal is unlikely to be restricted to short sequences in the molecule. Indeed, all the tested chimeras led to retention of the composite a-subunit in the ER, to growth arrest and cell death, so it was not possible to define more precisely the signal. The block of the chimerical proteins in the ER could either be because of defective targeting by “incompatible mixing” of signal domains or because of subunit trapping because of incorrect association with other V-ATPase subunits. Regardless, the localization signal seems to encompass several sites over the C terminus, most probably as loops between the highly conserved transmembrane segments. The nature of this localization signal is novel, because it differs from the classical signals, e.g., for secretion, mitochondrial presequences, or nuclear localization that are short stretches of amino acids that target any protein to the cognate compartment. Here, within V-ATPase a-subunits, the signal seems to be spread over a large region in which several sequences or structural motifs cooperate to define the efficient conformation. The interesting goal would be to identify the proteins that interact with this domain and direct each a-subunit to the correct compartment.
Different Functions of V-ATPases According to Cellular Compartments
It is clear from the chimera experiments that at least a part of the functional domain of the a-subunits is localized within their N-terminal halves, and it is likely that different a-subunits have different functions and properties in each compartment. So, V-ATPases containing a1-subunits are probably involved in endocytosis. Acidification of early endosomes is necessary for uncoupling of receptor/ligand complexes internalized from the plasma membrane and for vesicle trafficking via late endosomes (Stevens and Forgac, 1997). The V-ATPase containing the a2-subunit is localized in the osmoregulatory system and is crucial for the process of osmoregulation in Paramecium (Fok et al., 1995; Allen and Naitoh, 2002). The proton potential is used for secondary active transport mainly of K+ and Cl- but also of Ca2+ ions into the lumen of the contractile vacuole (Stock et al., 2002), although the exact mechanism and its correlation with the ultrastructural differentiation into the decorated and the smooth spongiome are unclear. The role of the V-ATPase containing a3-subunits in trichocysts is also unclear. The V-ATPase was shown previously to be essential for the biogenesis of these secretory organelles (Wassmer et al., 2005), although they were found not to be acidic (Lumpert et al., 1992; Garreau de Loubresse et al., 1994). So, it is likely that the electrochemical potential created by the V-ATPase is deployed for the accumulation/depletion of other ions, thus creating the environment required for the crystallization of trichocyst matrix proteins to the highly structured paracrystaline cores of trichocysts. In contrast, nascent phagosomes that contain the V-ATPase with the a4-subunit are rapidly acidified by the V-ATPase after its delivery by acidosomes immediately after pinching off the cytostome (Fok and Allen, 1988). These V-ATPase molecules are quickly withdrawn from the phagosomes and transported back to the cytostome. After the withdrawal, these “young” phagosomes were reported to fuse with lysosomes (Allen and Fok, 2000). Our results suggest that these lysosomes deliver V-ATPases containing a5, a6, and a9. Therefore, within the digestive cycle of Paramecium should exist two cycles of delivery and withdrawal of V-ATPase enzymes, both being distinguishable by the presence of different a-subunits within the V-ATPase. This exchange may explain the biphasic acidification profile of phagosomes, from a rapid and strong acidification burst to pH <5 within the first 3 to 5 min, followed by an increase to ∼pH 6 throughout the rest of the life span of a phagosome (Fok and Allen, 1988). Also in the trans-Golgi network, a mildly acidic milieu that is necessary for protein targeting (Sun-Wada et al., 2004) is established by the V-ATPase containing a8-2.
Chimerical a-Subunits Reveal Local and Remote Interactions
In addition to revealing the localization domain in the V-ATPase a-subunits, the use of chimeras allowed to distinguish two kinds of deleterious effects introduced by the combination of different N- and C-terminal halves. The first effect was observed by providing the contractile vacuole complex with a chimera containing the N terminus of the trichocyst-specific a3-1-subunit. As expected, this disturbed its functioning in an easily understandable way, because abnormal molecules are introduced in its structure, thus causing a local effect. The more intriguing effect was observed in the reciprocal chimera in which the N terminus of the contractile vacuole-specific a2-1-subunit was targeted to trichocysts. Indeed, in this case, the strongest effect was observed in the contractile vacuoles not in trichocysts. There are two explanations for this remote effect, which have now to be experimentally tested. The simplest explanation would be to assume that V1-complexes exist with different affinities for different V0-complexes, depending on the a-subunit it contains. Localization of an a-subunit N terminus to a wrong compartment would lead to the titration of the V1-sectors specific for this a-subunit, because they are redirected to this compartment. So, we have to assume that there are at least as many V1-sectors as there are V0 a-subunit localizations, i.e., seven. In previous work, we could identify for the V1-sector four A-, B-, E-, and H-, two F-, and one C- and D-subunit-encoding genes in the Paramecium draft genome (Wassmer et al., 2005). It may be possible that, by the combination of the different paralogues, several distinct V1-complexes are formed, thus conferring organelle specificity to the V1-subcomplexes. However, the different V1-subunit sequences are highly conserved: they often differ by a single or few amino acids. Thus, it remains speculative whether these variations are sufficient to yield functionally different V1-complexes. Another explanation of the remote effect would be to assume a competition for potential assembly factors necessary during V-ATPase biogenesis. In such a case, the mistargeting of an N terminus because of chimerical proteins would unroute bona fide assembly molecules and drive them to the wrong place, thus preventing the correct assembly of other V-ATPase molecules.
The system we describe here will facilitate addressing specific questions about the V-ATPase and its role in acidification and membrane energization and targeting in an absolutely compartment-specific way by investigating the different a-subunits.
Supplementary Material
Acknowledgments
We thank Denise Menay (Centre National de la Recherche Scientifique [CNRS], Gif-sur-Yvette, France) for oligonucleotide synthesis, Maud Silvain (CNRS) for sequencing, Oluscha Traub (Universität Konstanz, Konstanz, Germany) for technical assistance with cell fractionation and Western blotting, and Lauretta Nejedli (Universität Konstanz) for technical assistance with the electron microscopy. We also thank K. Klotz and F. Ruiz (CNRS) for providing the anti-trichocyst antiserum. Financial support by the Deutsche Forschungsgemeinschaft (Grant P178/20-1) and the TR-SFB11 (project C4) as well as support from the Ministèrede l'Education Nationale de la Recherche et de la Technologie, Program “Centre de Ressources Biologiques” are gratefully acknowledged. We also thank the French Government and the Deutscher Akademischer Austauschdienst for the fellowships of T. W.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0511) on November 28, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Allen, R. D., and Fok, A. K. (1983). Nonlysosomal vesicles (acidosomes) are involved in phagosome acidification in Paramecium. J. Cell Biol. 97, 566-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, R. D., and Fok, A. K. (2000). Membrane trafficking and processing in Paramecium. Int. Rev. Cytol. 198, 277-318. [DOI] [PubMed] [Google Scholar]
- Allen, R. D., and Naitoh, Y. (2002). Osmoregulation and contractile vacuoles of protozoa. Int. Rev. Cytol. 215, 351-394. [DOI] [PubMed] [Google Scholar]
- Allen, R. D., Schroeder, C. C., and Fok, A. K. (1992). Endosomal system of Paramecium: coated pits to early endosomes. J. Cell Sci. 101, 449-461. [DOI] [PubMed] [Google Scholar]
- Arata, Y., Nishi, T., Kawasaki-Nishi, S., Shao, E., Wilkens, S., and Forgac, M. (2002). Structure, subunit function and regulation of the coated vesicle and yeast vacuolar (H(+))-ATPases. Biochim. Biophys. Acta 1555, 71-74. [DOI] [PubMed] [Google Scholar]
- Cohen, J., Garreau de Loubresse, N., and Beisson, J. (1984). Actin microfilaments in Paramecium: localization and role in intracellular movements. Cell Motil. 4, 443-468. [DOI] [PubMed] [Google Scholar]
- Dessen, P., et al. (2001). Paramecium genome survey: a pilot project. Trends Genet. 17, 306-308. [DOI] [PubMed] [Google Scholar]
- Dillon, P. J., and Rosen, C. A. (1990). A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques 9, 298-300. [PubMed] [Google Scholar]
- Estève, J. C. (1972). Golgi apparatus in ciliates. Ultrastructure, with special reference to Paramecium. J. Protozool. 19, 609-618. [DOI] [PubMed] [Google Scholar]
- Fok, A. K., Aihara, M. S., Ishida, M., Nolta, K. V., Steck, T. L., and Allen, R. D. (1995). The pegs on the decorated tubules of the contractile vacuole complex of Paramecium are proton pumps. J. Cell Sci. 108, 3163-3170. [DOI] [PubMed] [Google Scholar]
- Fok, A. K., and Allen, R. D. (1988). The lysosomal system. In: Paramecium, ed. H. D. Görtz, Heidelberg: Springer-Verlag, 301-324.
- Galvani, A., and Sperling, L. (2000). Regulation of secretory protein gene expression in Paramecium role of the cortical exocytotic sites. Eur. J. Biochem. 267, 3226-3234. [DOI] [PubMed] [Google Scholar]
- Galvani, A., and Sperling, L. (2002). RNA interference by feeding in Paramecium. Trends. Genet. 18, 11-12. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse, N. (1993). Early steps of the secretory pathway in Paramecium: ultrastructural, immunocytochemical and genetic analysis of trichocyst biogenesis. In: Membrane Traffic in Protozoa, ed. H. Plattner, Greenwich: JAI Press, 27-59.
- Garreau de Loubresse, N., Gautier, M. C., and Sperling, L. (1994). Immature secretory granules are not acidic in Paramecium: implications for sorting to the regulated pathway. Biol. Cell 82, 139-147. [Google Scholar]
- Grønlien, H. K., Stock, C., Aihara, M. S., Allen, R. D., and Naitoh, Y. (2002). Relationship between the membrane potential of the contractile vacuole complex and its osmoregulatory activity in Paramecium multimicronucleatum. J. Exp. Biol. 205, 3261-3270. [DOI] [PubMed] [Google Scholar]
- Graham, L. A., Flannery, A. R., and Stevens, T. H. (2003). Structure and assembly of the yeast V-ATPase. J. Bioenerg. Biomembr. 35, 301-312. [DOI] [PubMed] [Google Scholar]
- Hauser, K., Haynes, W. J., Kung, C., Plattner, H., and Kissmehl, R. (2000b). Expression of the green fluorescent protein in Paramecium tetraurelia. Eur. J. Cell Biol. 79, 144-149. [DOI] [PubMed] [Google Scholar]
- Hauser, K., Pavlovic, N., Klauke, N., Geissinger, D., and Plattner, H. (2000a). Green fluorescent protein-tagged sarco(endo)plasmic reticulum Ca2+-ATPase overexpression in Paramecium cells: isoforms, subcellular localization, biogenesis of cortical calcium stores and functional aspects. Mol. Microbiol. 37, 773-787. [DOI] [PubMed] [Google Scholar]
- Hill, K., and Cooper, A. A. (2000). Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19, 550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, T., Iwamoto-Kihara, A., Sun-Wada, G. H., Okajima, T., Wada, Y., and Futai, M. (2003). Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J. Biol. Chem. 278, 23714-23719. [DOI] [PubMed] [Google Scholar]
- Imamura, H., Nakano, M., Noji, H., Muneyuki, E., Ohkuma, S., Yoshida, M., and Yokoyama, K. (2003). Evidence for rotation of V1-ATPase. Proc. Natl. Acad. Sci. USA 100, 2312-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Wilkens, S., and Forgac, M. (2003). Subunit structure, function, and arrangement in the yeast and coated vesicle V-ATPases. J. Bioenerg. Biomembr. 35, 291-299. [DOI] [PubMed] [Google Scholar]
- Jackson, D. D., and Stevens, T. H. (1997). VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J. Biol. Chem. 272, 25928-25934. [DOI] [PubMed] [Google Scholar]
- Kane, P. M., and Smardon, A. M. (2003). Assembly and regulation of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 35, 313-321. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi, S., Bowers, K., Nishi, T., Forgac, M., and Stevens, T. H. (2001a). The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276, 47411-47420. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi, S., Nishi, T., and Forgac, M. (2001b). Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunits differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276, 17941-17948. [DOI] [PubMed] [Google Scholar]
- Kissmehl, R., Sehring, I. M., Wagner, E., and Plattner, H. (2004). Immunolocalization of actin in Paramecium cells. J. Histochem. Cytochem. 52, 1543-1559. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Leng, X. H., Manolson, M. F., and Forgac, M. (1998). Function of the COOH-terminal domain of Vph1p in activity and assembly of the yeast V-ATPase. J. Biol. Chem. 273, 6717-6723. [DOI] [PubMed] [Google Scholar]
- Leng, X. H., Manolson, M. F., Liu, Q., and Forgac, M. (1996). Site-directed mutagenesis of the 100-kDa subunit (Vph1p) of the yeast vacuolar (H+)-ATPase. J. Biol. Chem. 271, 22487-22493. [DOI] [PubMed] [Google Scholar]
- Leng, X. H., Nishi, T., and Forgac, M. (1999). Transmembrane topography of the 100-kDa a subunit (Vph1p) of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 274, 14655-14661. [DOI] [PubMed] [Google Scholar]
- Li, Y. P., Chen, W., Liang, Y., Li, E., and Stashenko, P. (1999). Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 23, 447-451. [DOI] [PubMed] [Google Scholar]
- Lumpert, C. J., Glas-Albrecht, R., Eisenmann, E., and Plattner, H. (1992). Secretory organelles of Paramecium cells (trichocysts) are not remarkably acidic compartments. J. Histochem. Cytochem. 40, 153-160. [DOI] [PubMed] [Google Scholar]
- Lumpert, C. J., Kersken, H., and Plattner, H. (1990). Cell surface complexes ('cortices') isolated from Paramecium tetraurelia cells as a model system for analysing exocytosis in vitro in conjunction with microinjection studies. Biochem. J. 269, 639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson, M. F., Proteau, D., Preston, R. A., Stenbit, A., Roberts, B. T., Hoyt, M. A., Preuss, D., Mulholland, J., Botstein, D., and Jones, E. W. (1992). The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267, 14294-14303. [PubMed] [Google Scholar]
- Manolson, M. F., Wu, B., Proteau, E., Taillon, B. E., Roberts, B. T., Hoyt, M. A., and Jones, E. W. (1994). Stv1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J. Biol. Chem. 269, 14064-14074. [PubMed] [Google Scholar]
- Nelson, N. (2003). A journey from mammals to yeast with vacuolar H+-ATPase (V-ATPase). J. Bioenerg. Biomembr. 35, 281-289. [DOI] [PubMed] [Google Scholar]
- Oka, T., Murata, Y., Namba, M., Yoshimizu, T., Toyomura, T., Yamamoto, A., Sun-Wada, G. H., Hamasaki, N., Wada, Y., and Futai, M. (2001). a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J. Biol. Chem. 276, 40050-40054. [DOI] [PubMed] [Google Scholar]
- Parra, K. J., Keenan, K. L., and Kane, P. M. (2000). The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275, 21761-22177. [DOI] [PubMed] [Google Scholar]
- Plattner, H., Stürzl, R., and Matt, H. (1985). Synchronous exocytosis in Paramecium cells. IV. Polyamino compounds as potent trigger agents for repeatable trigger-redocking cycles. Eur. J. Cell Biol. 36, 32-37. [PubMed] [Google Scholar]
- Ramoino, P., Diaspro, A., Fato, M., Beltrame, F., and Robello, M. (2000). Changes in the endoplasmic reticulum structure of Paramecium primaurelia in relation to different cellular physiological states. J. Photochem. Photobiol. 54, 35-42. [DOI] [PubMed] [Google Scholar]
- Ruiz, F., Vayssié, L., Klotz, C., Sperling, L., and Madeddu, L. (1998). Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell 9, 931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. N., et al.. (2000). Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 26, 71-75. [DOI] [PubMed] [Google Scholar]
- Smith, A. N., Finberg, K. E., Wagner, C. A., Lifton, R. P., Devonald, M. A., Su, Y., and Karet, F. E. (2001). Molecular cloning and characterization of Atp6n1b: a novel fourth murine vacuolar H+-ATPase a-subunit gene. J. Biol. Chem. 276, 42382-42388. [DOI] [PubMed] [Google Scholar]
- Sonneborn, T. M. (1970). Methods in Paramecium research. Methods Cell Physiol. 4, 242-335. [Google Scholar]
- Stock, C., Grønlien, H. K., Allen, R. D., and Naitoh, Y. (2002). Osmoregulation in Paramecium: in situ ion gradients permit water to cascade through the cytosol to the contractile vacuole. J. Cell Sci. 115, 2339-2348. [DOI] [PubMed] [Google Scholar]
- Stevens, T. H., and Forgac, M. (1997). Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell. Dev. Biol. 13, 779-808. [DOI] [PubMed] [Google Scholar]
- Sun-Wada, G. H., Wada, Y., and Futai, M. (2004). Diverse and essential roles of mammalian vacuolar-type proton pump ATPase: toward the physiological understanding of inside acidic compartments. Biochim. Biophys. Acta 1658, 106-114. [DOI] [PubMed] [Google Scholar]
- Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- Toyomura, T., Oka, T., Yamaguchi, C., Wada, Y., and Futai, M. (2000). Three subunit a isoforms of mouse vacuolar H(+)-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J. Biol. Chem. 275, 8760-8765. [DOI] [PubMed] [Google Scholar]
- Vayssié, L., Skouri, F., Sperling, L., and Cohen, J. (2000). Molecular genetics of regulated secretion in Paramecium. Biochimie 82, 269-288. [DOI] [PubMed] [Google Scholar]
- Wassmer, T., Froissard, M., Plattner, H., Kissmehl, R., and Cohen, J. (2005). The vacuolar-H+-ATPase plays a major role in several membrane bounded organelles in Paramecium. J. Cell Sci. 118, 2813-2825. [DOI] [PubMed] [Google Scholar]
- Wehland, J., and Weber, K. (1987). Turnover of the carboxy-terminal tyrosine of α-tubulin and means of reaching elevated levels of de-tyrosination in living cells. J. Cell Sci. 88, 185-203. [DOI] [PubMed] [Google Scholar]
- Wessel, D., and Flügge, U. I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141-143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.