Abstract
To identify novel genes that play critical roles in mediating bone morphogenetic protein (BMP) signal pathways, we performed a yeast two-hybrid screen using Smad1 as bait. A novel mouse Krüppel-type zinc finger protein, mZnf8, was isolated. Interactions between mZnf8 and Smad proteins were further analyzed with various in vitro and in vivo approaches, including mammalian two-hybrid, in vitro glutathione S-transferase pulldown, and copurification assays. Results from functional analysis indicate that mZnf8 is a nuclear transcriptional repressor. Overexpression of mZnf8 represses activity of BMP and transforming growth factor beta (TGF-β) reporters. Silencing the expression of endogenous mZnf8 with an RNA interference approach caused a significant increase in the expression of one BMP reporter. These results suggest that mZnf8 negatively regulates the TGF-β/BMP signaling pathway in vivo. Transcription of mZnf8 is ubiquitous in mouse embryos, but high levels are specifically observed in adult mouse testes, with the same cell- and stage-specific transcription pattern as Smad1. Our data support the hypothesis that mZnf8 plays critical roles in mediating BMP signaling during spermatogenesis.
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor β (TGF-β) superfamily of growth factors and play critical roles in a number of developmental processes, including mesoderm formation and patterning, left-right asymmetry, neurogenesis, epithelial/mesenchymal interactions during organogenesis, and gametogenesis (reviewed in references 11 and 38). Binding of BMP/TGF-β cytokines to their receptors results in the phosphorylation of particular members of the Smad family of cytoplasmic proteins, which then translocate to the nucleus and function as transcriptional modulators (1, 38). Smad proteins are thus the direct downstream mediators of BMP/TGF-β signaling from the receptors to the nucleus.
Three classes of Smad proteins—receptor-activated Smads (R-Smads), common Smads (co-Smads), and inhibitory Smads (I-Smads)—have been identified in mammals. Smads 1, 5, and 8 are R-Smads that primarily mediate BMP signaling, while Smads 2, 3, are mainly involved in mediating TGF-β and activin signaling. Upon phosphorylation by type I receptors, R-Smads oligomerize with the co-Smad Smad4, which is essential for the BMP, TGF-β, and activin signaling pathways. The complex translocates to the nucleus and interacts with other cofactors to regulate the expression of downstream target genes. Smad6 and Smad7 are I-Smads that can interfere with R-Smad/receptor or R-Smad/co-Smad complex formation to inhibit BMP/TGF-β signaling pathways.
An intriguing question is how the relatively simple group of R-Smads mediates the many complex biological processes triggered in different tissues by BMP/TGF-β signaling. Accumulated evidence suggests that the different responses of cells to BMP/TGF-β ligands are mediated by interaction of R-Smads with cell-specific or stage-specific cofactors (1, 2, 38). Many nuclear proteins such as c-Jun/c-Fos (48), SnoN (31), Ski (23, 32), TGF-β induced factor (TGIF) (43), Sp1 (7), Lef1/Tcf1 (18), and GATA3 (5) have been identified as TGF-β Smad interaction partners. By contrast, relatively few BMP Smad interaction partners have been found.
It has been reported that Smad1 can interact with Hoxc-8 to dislodge it from its binding target, resulting in the activation of target genes (29). In another example, the zinc finger factor OAZ, originally identified as a nuclear partner of Olf-1/EBF, involved in olfactory epithelium and lymphocyte development, activates the homeobox gene Xvent-2 with a different group of zinc fingers than those for the Olf signaling pathway (10). Two negative transcriptional regulators of BMP Smads, Ski and Tob, have been identified recently (40, 46). We speculated that there are more transcription factors that can interact with BMP Smads, and identification of these Smad interaction partners is essential for elucidating the molecular mechanism for the various activities of BMPs during development.
Previous work from our laboratory has established that BMP signaling plays critical roles during male germ cell development. Bmp8b is required for the survival of germ cells in adult male mice, while Bmp8a is required for maintaining spermatogenesis and the integrity of the epididymis (50, 52). More recently, Zhao et al. (49) showed that mutation of Bmp7 enhances the defects in spermatogenesis caused by mutation of Bmp8a, further emphasizing the critical roles of BMPs during spermatogenesis. Smad1, a BMP-regulated R-Smad, is transcribed in the testes with a stage- and germ cell-specific pattern that is in contrast to its ubiquitous expression in mouse embryos (51). The molecular mechanism of BMP signaling during spermatogenesis is unknown.
In this paper, we describe the isolation of a mouse Krüppel-type zinc finger protein, mZnf8, as a novel interaction partner of Smad1 from a yeast two-hybrid screen. The interaction between mZnf8 and Smad1 was further confirmed with other experimental approaches. Functional analysis suggests that mZnf8 is a nuclear transcriptional repressor. Significantly, this gene is highly expressed in adult mouse testes, where it has an expression pattern similar to that of Smad1, as determined by section in situ hybridization. We propose that mZnf8 plays important roles in mediating BMP signaling during spermatogenesis.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The full-length Smad1 coding sequence was PCR amplified and cloned into the NdeI and BamHI sites of pGBKT7 (Clontech) to generate the Smad1 bait construct. Primer sequences used in this study will be provided upon request. All PCR constructs were sequenced to exclude potential mutations introduced during PCR. The NdeI/BamHI fragment containing the Smad1 coding region from the bait construct was cloned into pGADT7 (Clontech) to obtain the Smad1 prey construct. The Smad2 bait was obtained by PCR amplifying the coding region of Smad2 and cloning the PCR product into the NdeI and EcoRI sites of pGBKT7. The Smad2 coding region from the Smad2 bait plasmid was cloned into pGADT7 to generate the Smad2 prey construct.
The Smad1 bait plasmid was used to screen a mouse embryo (day of embryonic development 9.5 [E9.5] and E10.5) two-hybrid library (kindly provided by Stanley Hollenberg, Fred Hutchinson Cancer Research Center) with the Matchmaker Two-Hybrid System 3 (Clontech) following the manufacturer's instructions. Plasmids from positive clones were purified and cotransformed with the Smad2 bait construct into AH109 yeast cells to identify candidates that specifically interact with Smad1 but not with Smad2. Growth on selective medium (lacking Trp, Leu, His, and Ade) indicates positive interaction between bait and prey (Clontech). Two clones, S1 and S2, were identified from the screening. To perform the reciprocal two-hybrid assay, the insert from S1 was cloned into the bait vector pGBKT7, which was then cotransformed with different prey plasmids (as indicated in Fig. 1) into AH109 cells.
FIG. 1.
Clone S1 encodes a protein that specifically interacts with Smad1 in yeast two-hybrid assays. (A) The clone S1 was repeatedly isolated from a two-hybrid screen for gene products that interact with Smad1 but not with Smad2. In the forward two-hybrid assay, S1 was cotransformed with different bait constructs (GAL4DB domain fused with Smad1, Smad2, Lamintin, or the empty bait vector) into AH109 yeast cells. The growth of transformants on selective medium (lacking Trp, Leu, Ade, and His) indicates interaction between prey and bait. (B) In a reciprocal two-hybrid assay, the insert of S1 was fused with the GAL4DB domain in the bait vector, which was then cotransformed with the prey vector or prey vector containing Smad1, Smad2, or an unrelated gene (large T antigen) into AH109 yeast cells. Transformants were plated on selective medium (lacking Trp, Leu, Ade, and His). In both A and B, two independent colonies are shown for each interaction examined. DB, GAL4 DNA binding domain. AD, GAL4 activation domain.
The constructs used in this study were derived from mouse Smad1 and Smad2 and human SMAD3,4,5 genes.
Cloning full-length mZnf8.
The insert of clone S1, which encodes a partial cDNA sequence highly homologous to human ZNF8 (20), was used as a probe to screen an E8.5 to E9.0 mouse embryonic cDNA library (Stratagene) at high stringency as described before (51). Three positive clones were obtained out of 5 × 106 plaques. Sequencing analysis indicated that they were all identical clones containing the 5′ untranslated region and the coding sequence of the N-terminal part of mZnf8 (see below). The 5′-most ATG gives the longest reading frame and is surrounded by a Kozak consensus sequence. An in-frame stop codon is present upstream of the first ATG. Thus, the first ATG is probably the authentic start codon of mZnf8. No in-frame stop codon was identified in the 3′ end of three clones.
To acquire a full-length mZnf8 transcript, 3′ rapid amplification of cDNA ends (RACE) was performed with Marathon-Ready cDNA (Clontech) following the manufacturer's instructions. A single DNA fragment was acquired and then sequenced. The putative full-length mZnf8 mRNA was assembled with the clones obtained from the cDNA library screening and the PCR product from 3′ RACE. The cDNA sequence of mZnf8 was submitted to GenBank under accession no. AF480861.
In vitro GST pulldown assay.
Full-length Smad1, Smad2, and Smad5 coding sequences or MH1 (amino acids 1 to 145), Linker (amino acids 146 to 266), and MH2 (amino acids 267 to 465) domains of Smad1 were PCR amplified and cloned into the BamHI site of pGEX-2T. The glutathione S-transferase (GST)-Smad3 and GST-Smad4 fusion constructs were gifts of X. Cao (University of Alabama) and R. Derynck (University of California at San Francisco), respectively. GST or GST fusion proteins were purified with glutathione-conjugated agarose beads (Pharmacia) from BL21 following the manufacturer's instruction. Full-length mZnf8 was labeled with [35S]methionine with the TNT coupled transcription system (Promega) according to the manufacturer's protocol. GST pulldown assays were performed as described before (26). Samples were separated via reducing sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-PAGE). Gels were first stained with Coomassie blue to visualize GST fusion proteins and then treated with Enhance (NEN) following the manufacturer's instructions. The treated gels were dried before autoradiography.
To quantify the probes pulled down with GST fusion proteins, bands corresponding to the mZnf8 probe were excised, and radioactivity was measured in a scintillation counter (Beckman LS3801).
Copurification of GST-mZnf8 and HA-Smad proteins from mammalian cells.
The full-length mZnf8 coding sequence was PCR amplified and cloned into the BamHI site of pCMV-GST (35), a eukaryotic GST expression vector. Smad3 or Smad4 coding sequence was PCR amplified and cloned into the pCMV-HA vector (Clontech). The MH1, Linker, and MH2 domains of Smad1 were removed from bacterial GST expression constructs (described above) and cloned into the BamHI site of the pGCN-HA vector (45). The GST-mZnf8 fusion construct was cotransfected with different hemagglutinin (HA)-tagged Smad constructs (as indicated in the figures) into COS-M6 cells (gift of L. Limbird, Vanderbilt University Medical Center) with the Fugene 6 transfection reagent (Roche). ALK6 (Q203D) or ALK5 (T204D) constructs encoding a constitutively active form of BMPRIB or TGF-βRI (gifts of L. Attisano, University of Toronto) were cotransfected as needed.
Purification of GST protein or GST-mZnf8 fusion protein was performed as described previously (35). Monoclonal GST (GST-2; Sigma) or HA (HA.11; Babco) antibodies were used to detect GST fusion proteins and HA-tagged Smad proteins, respectively. A horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Jackson) was used as the secondary antibody in Western analysis. A rabbit polyclonal anti-phospho-Smad1 serum (Upstate Biotechnology) and a horseradish peroxidase-conjugated anti-rabbit IgG (Jackson) were used to test whether the active form of Smad1 copurified with GST-mZnf8 from COS cells.
Cellular localization of mZnf8.
The full coding sequence of mZnf8 was PCR amplified and cloned into the BamHI site of pCGN-HA (45). This construct was transfected into COS-M6 cells as above. At 36 h after transfection, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 for 15 min. Then 0.5 μM propidium iodide (Molecular Probes) was used to stain the nuclei following the manufacturer's instructions. A rat monoclonal HA antibody (3F10; Roche) and Cy2-conjugated anti-rat immunoglobulin secondary antibody (Jackson) were used to stain the fusion protein. Cells were observed with a fluorescent microscope.
Transient-transfection and luciferase reporter assays.
In the mammalian two-hybrid assay, full-length Smad1 coding sequence was PCR amplified and cloned into the BamHI site of pFA-CMV (Stratagene) to acquire the GAL4 DNA binding domain (DB)-Smad1 fusion construct. To obtain the Znf8-vp16 construct, the potential Krüppel-associated box (KRAB; amino acids 25 to 96) coding region from pCGN-HA-mZnf8 (described above) was replaced with a sequence encoding the transcriptional activation domain of herpes simplex virus VP16. In the (Gal)5-E1b-lux reporter construct (gift of J. Massague, Memorial Sloan-Kettering Cancer Center), the transcription of the luciferase gene is driven by five copies of GAL4 DNA binding sites upstream of a basic E1b TATA box (43). Various constructs (as indicated in Fig. 4) were cotransfected into HEK293 cells; 0.2 μg of each plasmid was used for transfection unless otherwise indicated. The total amount of DNA used in all transfection assays was equalized by using pcDNA3 (Stratagene). Luciferase assays were performed with a dual luciferase reporter assay system (Promega) according to the manufacturer's protocol. pRL-TK (0.02 μg; Promega), which expresses Renilla luciferase (different from firefly luciferase) from a constant promoter, was cotransfected into HEK293 cells to normalize transfection efficiency in all assays.
FIG. 4.
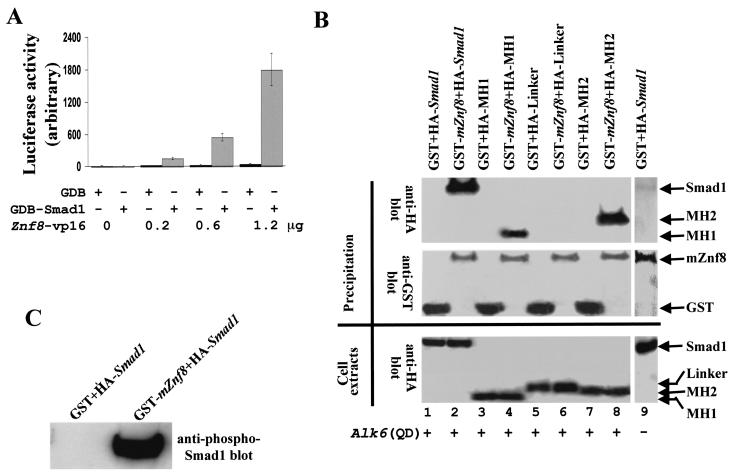
Association of Smad1 and mZnf8 in mammalian cells. (A) A total of 0.2 μg of pFA-CMV (GAL4DB [GDB] vector alone) or GAL4DB-Smad1 was cotransfected with various amounts of Znf8-vp16 (as indicated in the figure) as well as the reporter construct (Gal)5-E1b-lux into HEK293 cells. Luciferase activities were measured as described in Materials and Methods. The luciferase activity of cells with GAL4DB alone was defined as 10. Znf8-vp16 dramatically increased the reporter activity in cells transfected with GAL4DB-Smad1 in a dose-dependent fashion but had subtle effects on that of cells with the GAL4DB vector alone. Data were averaged from three independent cultures, with error bars indicating standard deviations. (B) mZnf8 fused with GST in a mammalian expression vector was cotransfected into COS-M6 cells with a plasmid expressing HA-tagged Smad1 or HA-tagged domains of Smad1. ALK6 (Q203D), encoding constitutively active BMPRI, was also cotransfected into COS cells in all experiments except for lane 9. The GST (negative control) and GST-mZnf8 fusion proteins were purified with GST beads (see Materials and Methods). The samples were divided into two equal parts. The first half was loaded onto an SDS-PAGE gel, and an HA antibody was applied to detect whether HA-tagged proteins copurified with GST-mZnf8 from cell lysates in Western analysis (top panel). The same filter was stripped and Western analysis with an anti-GST antibody was performed to confirm the precipitation of GST or GST-mZnf8 fusion proteins (middle panel). Expression of HA fusion proteins in COS cells was confirmed by Western analysis with an anti-HA antibody (bottom panel). (C) The left-half samples from lane 1 and lane 2 of panel A were loaded on another SDS-PAGE gel. Western analysis with anti-phospho-Smad1 antibody was performed to test whether the phosphorylated Smad1 (active form) copurified with GST-mZnf8.
To examine whether mZnf8 functions as a transcriptional repressor, full-length mZnf8 coding sequence or mZnf8 lacking the potential KRAB domain coding sequence was obtained by PCR and cloned into pFA-CMV to obtain GAL4DB fusion constructs. They were named pJK97 and pJK99, respectively. pFA-CMV, pJK97, or pJK99 was cotransfected with the (Gal)5-E1b-lux reporter construct into HEK293 cells. Luciferase assays were performed as described above. All proteins were expressed at a very similar level, as determined by Western analysis with an anti-GAL4DB antibody (data not shown).
Constructs 12(GCCG)-lux (gift of K. Miyazono, Cancer Institute of the Japanese Foundation for Cancer Research), pTlx-lux (gift of J. Wrana, University of Toronto), and 3TP-lux (gift of J. Massague) were used to determine the effect of overexpression of mZnf8 on BMP and TGF-β reporters.
To stimulate P19 cells with the BMP4 ligand, cells were treated with 100 ng of hBMP4 (purchased from R&D Systems, Inc.) per ml or with bovine serum albumin alone in Dulbecco's modified Eagle's medium with 0.1% fetal bovine serum for 14 to 16 h. Luciferase assays were then performed as described above. The Flag-Smad1 and Flag-Smad2 constructs were gifts of L. Attisano and J. Massague, respectively. Western analysis confirms that expression of Flag-Smad proteins was not decreased by increasing the amount of mZnf8 in transfections (data not shown). The pM3VP16 (Clontech) construct encoding the GAL4DB-VP16 fusion protein was used in a control experiment to exclude that mZnf8 functions as a general transcriptional repressor.
RT-PCR and RNA interference (RNAi) assays.
Total RNA was isolated from cells with the RNeasy mini kit (Qiagen); 0.5 μg of total RNA was then subjected to reverse transcription (RT)-PCR analysis with the ThermoScript RT-PCR system (Life Technologies). Oligo(dT)20 instead of random hexamer was used for RT reactions. The expected PCR products for mZnf8 and β-actin (positive control for RT-PCR) are 380 and 490 bp, respectively.
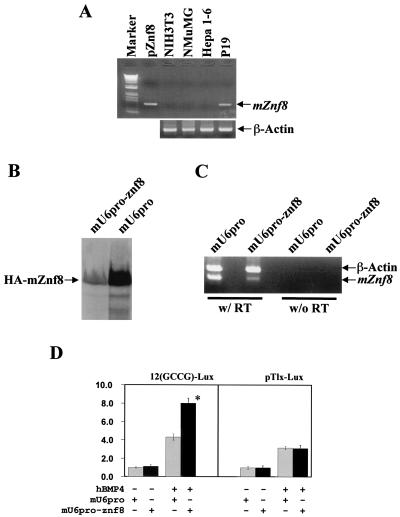
Synthetic oligonucleotides Primer2top (TTTGAGTCTTGAGCCCCGTCTACACGGTGTACGTGTAGACGGGGCTCAAGACTTTTTT) and Primer2bot (CTAGAAAAAAGTCTTGAGCCCCGTCTACACGTACACCGTGTAGACGGGGCTCAAGACT) were obtained from Integrated DNA Technologies, Inc. The two cDNA oligonucleotides were annealed and ligated into the vector mU6pro (gift of D. Turner, University of Michigan) to generate mU6pro-znf8, as described before (47). In mU6pro-znf8, a single short hairpin RNA corresponding to bp 1504 to 1529 of the mZnf8 coding region is transcribed from the U6 promoter. A total of 0.2 μg of reporter constructs with 1.0 μg of mU6proznf8 or 1.0 μg of mU6pro (vector alone control) were cotransfected into P19 cells, and luciferase assays were performed as described above.
Northern analysis.
Northern blots containing mouse mRNA from different embryonic stages or from different adult mouse tissues were purchased from Clontech. Northern analysis was performed with ExpressHyb (Clontech) according to the manufacturer's instructions and with the full-length mZnf8 coding sequence as the probe. The glyceraldehyde-3-phosphate dehydrogenase (G3PDH) probe (Clontech) was used for an mRNA loading control.
Section in situ hybridization.
The testes from an 8-week-old ICR male mouse and mouse embryos were sectioned at 5 μm as described before (51). The insert from clone S1 was cloned into pBluescript (Stratagene) for making sense and antisense probes. The Smad1 probe and in situ hybridization assay were described before (51).
RESULTS
Identification of Smad1 interaction proteins.
A yeast two-hybrid strategy was used to identify proteins that interact with Smad1. The full-length Smad1 cDNA sequence was fused with the GAL4 DNA binding domain (GAL4DB) and used as the bait to screen a mouse embryonic (E9.5 and E10.5) library (37). A total of 5 × 106 independent clones were screened, and about 400 clones were obtained from the first-round screening. Fifty clones were randomly picked and sequenced. Some of the 50 clones encoded known Smad1 interaction proteins, including the WW2 domain of Smurf1 (53) and its homologs, filamin (27) and ubiquitin (9).
To identify genes that are specifically involved in the BMP signaling pathway but not in the TGF-β or activin pathway, plasmids from positive clones obtained from the first-round screen were retested for their ability to interact with Smad2 (see Materials and Methods). Two clones, S1 (identified seven times) and S2 (identified twice), encoded proteins that interacted only with Smad1 but not with Smad2 in yeast cells. This paper focuses on the characterization of clone S1.
As shown in Fig. 1A, transformants containing both S1 (in the prey construct) and the Smad1 bait construct grew on selective medium deficient in Leu, Trp, Ade, and His, indicating positive interaction between prey and bait. Positive interaction was not observed between clone S1 and Smad2, empty bait vector, or an unrelated protein. To further confirm the specific interaction between clone S1 and Smad1 in yeast cells, a reciprocal two-hybrid assay was performed (Fig. 1B). As in the forward two-hybrid assay, the insert of clone S1 in a bait construct could still interact with the Smad1 prey but not with the empty prey vector, Smad2, or an unrelated protein.
Sequencing analysis indicated that clone S1 contained a partial cDNA sequence highly homologous to the zinc finger regions of human zinc finger protein 8 (hZNF8) (20). The complete coding sequence of the mouse gene was acquired by screening a mouse E9.0 embryo cDNA library with S1 as the probe and 3′ RACE (see Materials and Methods). The sequence of this cDNA (GenBank accession no. AF480861) contains an open reading frame of 1,719 nucleotides encoding a 573-amino-acid conceptual protein of 64 kDa.
Sequence alignment of the predicted mouse protein and human ZNF8 was performed with the program ClustalW (1.81) (Fig. 2). The extremely high conservation between the two proteins (74% identity and 91% similarity) indicates that the mouse gene is a true homolog of hZnf8. It encodes a Krüppel-type zinc finger protein with one potential KRAB domain at its N-terminal end and seven C2H2 zinc fingers in its C-terminal half. The KRAB domain has a transferable transcriptional repression activity that is required for the function of many Krüppel-type zinc finger proteins (6). The first six zinc fingers of mZnf8 are classical Krüppel-type fingers, with a conserved TGEKP(Y/F)X interfinger spacer (28), while the seventh finger is separated from the sixth by a longer spacer. The KRAB and zinc finger domains (including spacer regions) are the most highly conserved regions between the mouse and human proteins, consistent with the idea that they are critical for the function of Znf8 proteins. Regions outside of these two domains are less conserved, and no other potential functional domain was identified in the protein.
FIG. 2.
Sequence comparison between human ZNF8 and mouse Znf8. The sequences of mZnf8 and hZnf8 were aligned with the program ClustalW (1.81) (http://www.ebi.ac.uk/clustalw/). The potential KRAB domain is underlined. The predicted seven C2H2 zinc fingers are numbered at the bottom of the first conserved Cys of each finger. The sequence encoded by the original S1 clone is shown in italics. Amino acids identical between human and mouse Znf8 are marked with an asterisk, conserved residuesare marked with a colon, and semiconserved residues are marked with a period, as determined by the ClustalW program. The sequence of mZnf8 was submitted to GenBank under accession no. AF480861.
mZnf8 interacts with Smad1 in vitro and in mammalian cells.
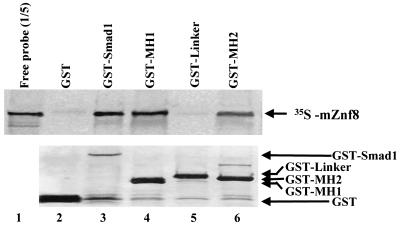
To further confirm the interaction between mZnf8 and Smad1 and to exclude that the two proteins interact with each other through a yeast protein(s) in two-hybrid assays, in vitro GST pulldown assays were performed (Fig. 3). The [35S]methionine-labeled full-length mZnf8 protein was pulled down by the GST-Smad1 fusion protein but not by the GST protein, indicating a direct interaction between the two proteins. The MH1, MH2, and Linker domains of Smad1 were fused with GST to determine which domain(s) of Smad1 can interact with mZnf8. Figure 3 shows that both the MH1 and MH2 domains but not the Linker domain can interact with in vitro-translated mZnf8.
FIG. 3.
Interaction of Smad1 with mZnf8 in vitro. Full-length or different regions of Smad were fused with GST in pGEX −2T, and fusion proteins were expressed and purified from 50 ml of bacterial culture. The mZnf8 protein was labeled with [35S]methionine by in vitro transcription and translation and incubated with the purified GST protein (negative control) or different GST fusion proteins for an in vitro pulldown assay (see Materials and Methods). Samples were separated via SDS-12.5% PAGE, and probes were visualized by autoradiography (top panel); 20% of input probes were loaded on the gel. The bottom panel shows the same gel stained with Coomassie blue. The positions of the GST protein and GST fusion proteins are indicated.
We then tested whether the interaction between mZnf8 and Smad1 occurs in mammalian cells. We first performed a mammalian two-hybrid assay, which detects interaction between bait and prey in mammalian cells instead of yeast cells. As shown in Fig. 4A, Znf8-vp16 dramatically increased the reporter activity in cells transfected with GAL4DB-Smad1 in a dosage-dependent fashion. More than 150-fold induction in reporter activity (1.2 μg versus 0 μg of Znf8-vp16) was observed in cells transfected with GAL4DB-Smad1, while under the same conditions less than fourfold induction was observed in control cells with GAL4DB alone. This result strongly suggests that mZnf8 and Smad1 interact in living mammalian cells.
In the next step, we applied a biochemical approach to test the interaction between the two proteins in mammalian cells. GST-mZnf8 and HA-Smad1 were coexpressed in COS-M6 cells, and the GST-mZnf8 fusion protein was purified from the cell lysate with GST binding beads. An anti-HA antibody was used to test whether HA-Smad1 copurified with GST-mZnf8 (Fig. 4B). In the absence of ALK6 (Q203D), which encodes a constitutively active form of BMPRI, only a trace amount of HA-Smad1 could be detected (lane 9 of Fig. 4B), but this was dramatically increased upon cotransfection of ALK6 (Q203D) (lane 2 of Fig. 4B). No HA-Smad1 was detected in the negative control lane (lane 1 of Fig. 4B), although equal amounts of HA-Smad1 were expressed (comparing lanes 1 and 2 in the bottom panel of Fig. 4B).
This result demonstrates that mZnf8 and Smad1 interact in mammalian cells and that interaction between the two proteins is greatly enhanced by BMP signaling. Since Smad1 is phosphorylated by BMPRI upon BMP stimulation (12, 48), we further confirmed that active (phosphorylated) Smad1 copurified with GST-mZnf8 by Western analysis with an anti-phospho-Smad1-specific antibody in Fig. 4C. Consistent with the result obtained from the in vitro pulldown assay (Fig. 3), the MH1 and MH2 domains but not the Linker domain of Smad1 were able to interact with mZnf8 in COS cells (lane 3 to lane 8 of Fig. 4B).
Interaction of mZnf8 with various Smad proteins.
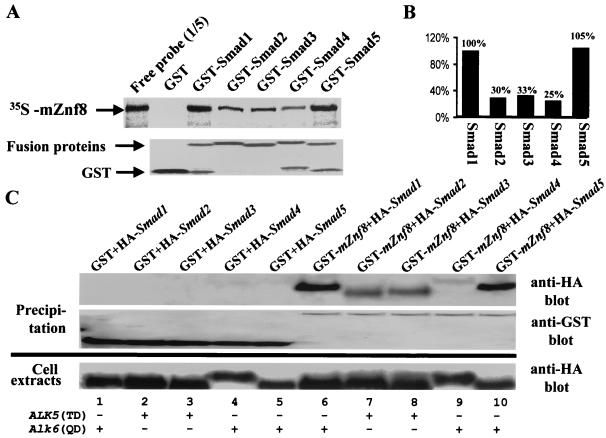
The results from yeast two-hybrid assays suggested that mZnf8 specifically interacts with BMP Smads but not with TGF-β Smads. Surprisingly, in the in vitro GST pulldown assay, all Smad proteins tested (Smads 1 to 5) interacted with the radiolabeled mZnf8 probe (Fig. 5A). However, quantitative analysis indicates that mZnf8 had a higher binding affinity for BMP Smads (Smad1 and Smad5) than for other Smads tested (Fig. 5B). Similar results were also obtained from the copurification experiment in mammalian cells (Fig. 5C); more Smad1 and Smad5 copurified with GST-mZnf8 from COS cells than Smad2, -3, and -4. The results from both in vitro and in vivo experiments were repeated three to five times.
FIG. 5.
Association of mZnf8 with different Smad proteins in vitro and in vivo. (A) The GST protein or GST-Smad fusion proteins were purified from bacteria, and 10 μg of GST or 5 μg of GST fusion proteins was then incubated with [35S]methionine-labeled mZnf8 for an in vitro pull-down assay (see Materials and Methods). Samples were separated via SDS-PAGE, and probes were visualized by autoradiography (top panel); 20% of input probes was loaded on the gel for comparison. The same gel was stained with Coomassie blue to visualize GST and GST fusion proteins (bottom panel). (B) The amount of probes that were pulled down by GST fusion proteins was quantitated via scintillation counting (see Materials and Methods). The value obtained for GST-Smad1 was defined as 100%. Data were averaged from three independent experiments. (C) mZnf8 was fused with GST in a eukaryotic expression vector, which was then cotransfected with various HA-tagged Smad constructs into COS-M6 cells. The empty GST vector was included as negative controls (lanes 1 to 5). For Smads 1 and 5 (lanes 1, 5, 6, and 10), a constitutively active form of BMPRI, ALK6 (Q203D), was included in cotransfection. For Smads 2 and 3 (lanes 2, 3, 7, and 8), a constitutively active form of TGF-βRI, ALK5 (T204D), was included. For Smad4 (lanes 4 and 9), both ALK6 (Q203D) and ALK5 (T204D) were examined, and the same results were obtained. In this figure, only the result from cotransfection of ALK6 (Q203D) is shown. An anti-HA antibody was used in Western analysis to test whether HA-Smad proteins copurified with GST-mZnf8 (top panel). Purification of GST and GST-mZnf8 proteins was confirmed by Western analysis with an anti-GST antibody (middle panel). The bottom panel shows that comparable amounts of HA-Smad proteins were expressed in each experiment.
The low binding affinity of mZnf8 to Smad2 may account for the failure to detect the interaction between the two proteins in yeast two-hybrid assays. We noticed that the interaction between mZnf8 and Smad4 was fairly weak in COS cells (lane 9 of Fig. 5C). Cotransfection of Smad1 and ALK6 (Q203D), encoding constitutively active BMPRI, slightly increased the amount of copurified Smad4 (data not shown).
mZnf8 is a nuclear transcriptional repressor.
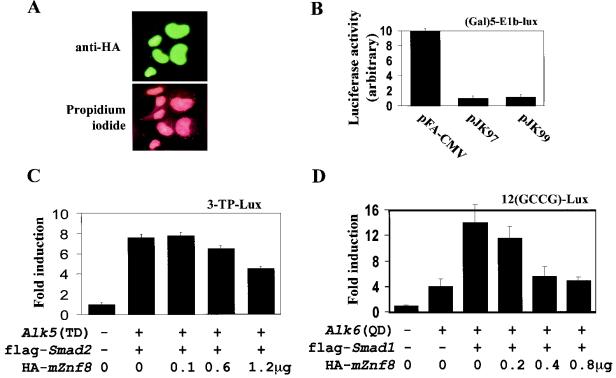
Since mZnf8 is a Krüppel-type zinc finger protein and interacts with phosphorylated, nucleus-localized Smad1, we speculated that mZnf8 is a nuclear protein, although no classical nuclear localization signal was identified from sequence analysis. To test this hypothesis, mZnf8 was tagged with HA and expressed in COS-M6 cells. As shown in Fig. 6A, HA-mZnf8 was localized exclusively in the nucleus, and its nuclear localization was not altered by stimulation with either 50 ng of hBMP4 or 5 ng of TGF-β1 per ml for 1 h (data not shown). The same results were also obtained in HEK293 cells (data not shown). The exclusive nuclear localization of mZnf8 may explain the low level of mZnf8/Smad1 complex formed in COS cells in the absence of BMP stimulation (see above).
FIG. 6.
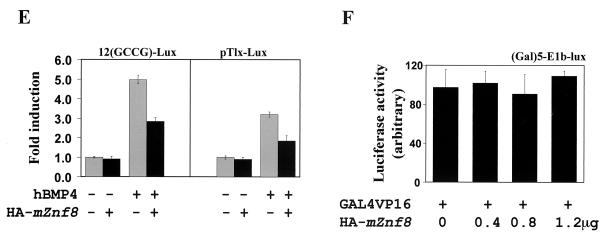
mZnf8 is a nuclear protein and negatively regulates BMP and TGF-β reporters. (A) mZnf8 was tagged with HA and expressed in COS-M6 cells. The localization of the fusion protein was detected with an anti-HA antibody (see Materials and Methods). Propidium iodide was used to stain the nuclei. The exclusive nuclear localization of mZnf8 was also observed in HEK293 cells (data not shown). (B) In the (Gal)5-E1b-lux reporter plasmid, the expression of luciferase is driven by five copies of the GAL4 DNA binding sites upstream of the basic E1b promoter. The reporter construct was cotransfected with pFA-CMV (expressing GAL4DB alone), pJK97 (expressing full-length mZnf8 fused with GAL4DB), or pJK99 (expressing mZnf8 lacking the KRAB domain fused with GAL4DB) into HEK293 cells. Luciferase activities were measured as described in Materials and Methods. The luciferase activity of pFA-CMV was defined as 10. (C) HEK293 cells were transfected with the 3-TP-lux reporter along with other constructs as indicated in the figure. The total amount of DNA used in each transfection was equalized with pcDNA3. Luciferase activities were determined, and induction relative to cells with 3-TP-lux alone is shown. For C and D, expression of Smad proteins was not altered by the increased amount of mZnf8 in the transfection, as determined by Western analysis (data not shown). (D) P19 cells were transfected with the 12(GCCG)-lux reporter together with other constructs as indicated in the figure. Luciferase activities were determined, and the induction relative to the reporter alone is shown. (E) A total of 0.2 μg of 12(GCCG)-lux or pTlx-lux reporter with or without 1.2 μg of the HA-mZnf8 construct (as indicated in the panel) was cotransfected into P19 cells. Cells were treated with 100 ng of hBMP4 per ml for 14 to 16 h, and luciferase assays were performed as described in Materials and Methods. (F) HEK293 cells were transfected with (Gal)5-E1b-lux, GAL4-VP16, and various amounts of HA-mZnf8 as shown in the figure. Luciferase activities were determined as described in Materials and Methods. Overexpression of mZnf8 has no effect on GAL4-VP16-mediated transcription. In panels B, C, D, E, and F, data were averaged from three independent cultures, and error bars indicate standard deviations.
To test whether mZnf8 is a transcriptional repressor, like other Krüppel-type zinc finger proteins (8, 22, 30, 34), we used a GAL4 heterologous system that has been used to study many other transcriptional repressors (4, 14). Consistent with a previous report that GAL4DB contains a cryptic activation domain when expressed in mammalian cells (21), the GAL4DB domain alone activated the (GAL4)5-E1b-lux reporter. When full-length mZnf8 was fused with the GAL4DB, transcription of the reporter was significantly reduced compared to the GAL4DB alone (Fig. 6B). Interestingly, the repression effect of mZnf8 did not absolutely depend on the presence of KRAB. Repression of the reporter was still observed even when KRAB was removed (Fig. 6B), suggesting that other sequences within mZnf8 also possess transcriptional repression activity.
To examine the potential function of mZnf8 in modulating transcriptional responses induced by BMP and TGF-β, BMP-responsive and TGF-β-responsive reporters were used in transient-transfection assays (Fig. 6C, D, and E). 3TP-lux contains TGF-β-responsive elements derived from the plasminogen activator inhibitor 1 and collagenase promoters (44). Kusanagi et al. (17) found that reporters with multiple copies of the GCCGnCGC motif (GCCG box) respond specifically to BMP stimulation but not to TGF-β or activin. The GCCG box was originally derived from Decapentaplegic (Dpp)-responsive promoters in Drosophila melanogaster (17). Later this motif was identified in the mouse Smad6 promoter and shown to be necessary for the transcriptional induction of mouse Smad6 by BMP stimulation (13).
Transcription of the luciferase reporter in pTlx-lux is driven by the BMP-specific responsive enhancer derived from Tlx-2 (33). As shown in Fig. 6C and D, overexpression of mZnf8 repressed both the TGF-β and BMP reporters in a dose-dependent fashion. This result is consistent with the idea that mZnf8 functions as a transcriptional repressor and interacts with Smad1 and Smad2 in mammalian cells. The fact that overexpression of mZnf8 had a more dramatic effect on a BMP reporter than on a TGF-β reporter is consistent with the observation that mZnf8 had a higher binding affinity for BMP Smads than for TGF-β Smads. The results in Fig. 6E indicate that overexpression of mZnf8 also repressed the expression of BMP reporters in BMP ligand-induced responses. In a control experiment, overexpression of mZnf8 had no effect on GAL4-VP16-mediated transcription (Fig. 6F), indicating that mZnf8 does not function as a general transcriptional repressor.
Involvement of endogenous mZnf8 in the BMP signaling pathway.
To directly test the role of mZnf8 in the BMP signaling pathway, we decided to silence (knock down) the expression of endogenous mZnf8 with an RNAi approach. It was reported recently that short hairpin RNA molecules can efficiently target corresponding mRNAs for degradation in mammalian cells (24, 47). For these experiments, we used P19 teratocarcinoma cells, which express endogenous mZnf8 (Fig. 7A). We made the construct mU6pro-znf8, which transcribes short hairpin RNA corresponding to bp 1504 to 1529 of the mZnf8 coding region (see Materials and Methods). Since no mZnf8 antibody is available, we could not examine the effect of this construct on the expression of the endogenous protein directly.
FIG. 7.
Involvement of endogenous mZnf8 in BMP signaling pathway. (A) To identify cell lines expressing endogenous mZnf8, RT-PCR was performed as described in Materials and Methods. The template in the PCR of pZnf8 is a plasmid containing the full-length mZnf8 cDNA. M refers to the 1kb+ DNA molecular size markers, purchased from Life Technologies. As a positive control, β-actin was detected in all lines. No signal was detected in no-RT controls (data not shown). (B) HA-mZnf8 was cotransfected with mU6pro-znf8 or with vector alone (mU6pro) into P19 cells. Total cell protein (20 μg) was analyzed by SDS-PAGE, and Western analysis was performed with an HA antibody. (C) P19 cells were transfected with either the vector alone (mU6pro) or mU6pro-znf8, and RT-PCR analysis was performed with mZnf8 and β-actin primers added to the same tube. Much less mZnf8 was amplified from cells transfected with mU6pro-znf8 compared to cells transfected with vector alone. (D) The 12(GCCG)-lux or pTlx-lux reporter (0.2 μg) was cotransfected with mU6pro or mU6pro-znf8 (1.0 μg) into P19 cells. Cells were treated with 100 ng of hBMP4 or bovine serum albumin per ml alone for 14 to 16 h, and luciferase assays were performed as described in Materials and Methods. Results are expressed as fold induction. The activity of cells with vector alone and without hBMP4 stimulation was defined as 1.0. Data were averaged from three independent cultures, with error bars indicating standard deviations. *, significant increase (P < 0.05).
To circumvent this, we first tested whether this construct efficiently decreases the expression of HA epitope-tagged mZnf8. As shown in Fig. 7B, expression of HA-mZnf8 in cells transfected with mU6pro-znf8 was dramatically reduced compared with that of cells transfected with vector alone. Since the transcript of HA-mZnf8 has the same coding sequence as the endogenous mZnf8 transcript, we speculated that the short hairpin RNA made by mU6pro-znf8 also efficiently reduced expression of the endogenous gene. This idea was confirmed by RT-PCR analysis, showing that the endogenous mRNA levels of mZnf8 were dramatically reduced but not eliminated in cells receiving the mU6pro-znf8 plasmid (Fig. 7C). Results in Fig. 7D indicated that the activity of the 12(GCCG)-lux reporter in response to hBMP4 stimulation was increased almost twofold by mU6pro-znf8. No significant alteration in the reporter activity of 12(GCCG)-lux was observed when hBMP4 stimulation was omitted. By contrast to the result with 12(GCCG)-lux, mU6pro-znf8 did not alter the activity of another BMP reporter, pTlx-lux (Fig. 7D). This result strongly suggests that endogenous mZnf8 is selectively involved in some but not all responses induced by BMP signaling (also see Discussion).
mZnf8 is ubiquitously transcribed in mouse embryos.
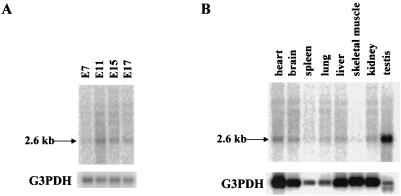
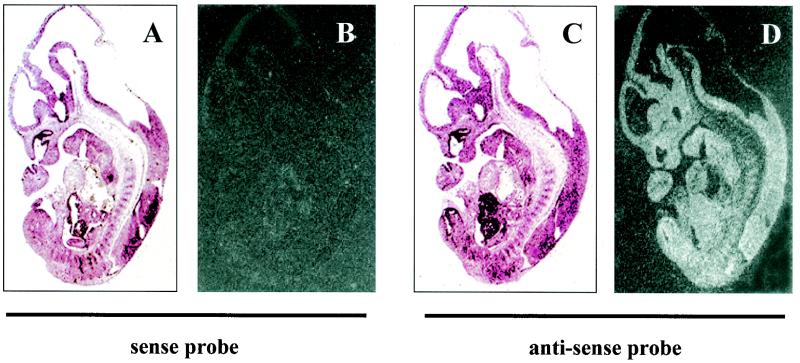
To begin to understand the in vivo function of mZnf8, we examined its expression pattern by Northern and section in situ hybridization assays. Results from Northern analysis indicated that transcription of mZnf8 was developmentally regulated during mouse embryogenesis (Fig. 8A). At E7, a very low level of mZnf8 transcripts could be detected. The mZnf8 transcript was most abundant at E11, and its level declined at later stages (E15 and E17). Section in situ hybridization analysis showed that mZnf8 was ubiquitously and evenly expressed in an E11.5 mouse embryo (Fig. 9). A similar expression pattern was also observed in E9.5 and E10.5 embryos (data not shown). The ubiquitous embryonic expression pattern of mZnf8 is identical to that of Smad1 (51) (data not shown) and is consistent with the possibility that mZnf8 interacts with Smad1 to modulate BMP signaling in many tissues during mouse embryogenesis.
FIG. 8.
Northern analysis of mZnf8 expression. (A) Northern blot containing mRNA from mouse embryos of different stages (Clontech) was hybridized with a [32P]dCTP-labeled mZnf8 cDNA probe. A 2.6-kb major band was detected (top panel). After exposure to X-ray film, the filter was stripped and probed with a G3PDH probe as an mRNA loading control (bottom panel). (B) Northern blot containing mRNAs from a variety of adult mouse tissues (Clontech) was probed as above.
FIG. 9.
mZnf8 is ubiquitously expressed in mouse embryos. In situ hybridization was performed on sections of an E11.5 mouse embryo with [35S]methionine-labeled sense (negative control, panels A and B) or antisense (panels C and D) probes of mZnf8. (A and C) Bright-field photomicrographs; (B and D) dark-field photomicrographs. No signal was detected with the sense control probe. Hybridization with the antisense probe revealed ubiquitous expression of mZnf8. The same results were obtained with E9.5 and E10.5 mouse embryo sections (data not shown).
mZnf8 has a stage- and cell-specific expression pattern similar to that of Smad1 in adult mouse testes.
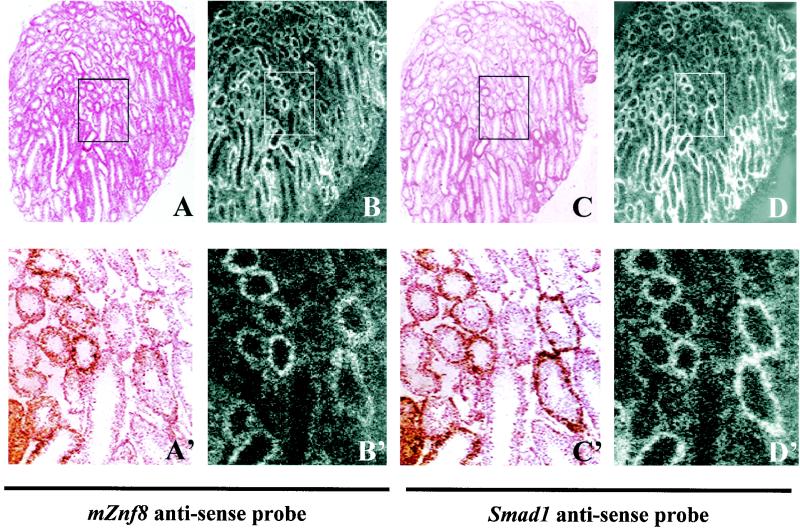
In contrast to its uniform pattern in all tissues of the mouse embryo (Fig. 9), Northern analysis showed that transcription of mZnf8 was high in adult mouse testes and low to moderate in other tissues examined (Fig. 8B). The very strong transcription in the testes implies potential functions of this gene during spermatogenesis. Previous work from our laboratory showed that Smad1 is expressed with a stage- and germ cell-specific pattern during the cycling of seminiferous epithelium (51). We reasoned that if interaction between mZnf8 and Smad1 is important for the function of mZnf8, these two genes should be coexpressed in the same population of cells in the testes.
To test this prediction, we performed in situ hybridization on adjacent sections of an 8-week-old ICR male mouse with an mZnf8 antisense probe and a Smad1 antisense probe (Fig. 10). Consistent with our previous report (51), Smad1 transcripts were detected in certain tubules with variable intensity in a sexually mature mouse testis when the steady-state cycling of the seminiferous epithelium was established (Fig. 10). Significantly, mZnf8 was transcribed in the same apparent population of cells and to the same apparent level as Smad1 (Fig. 10), indicating that mZnf8 has a similar stage- and cell-specific transcription pattern. This result supports the hypothesis that mZnf8 interacts with Smad1 at specific stages during spermatogenesis.
FIG. 10.
mZnf8 and Smad1 have a similar expression pattern in adult mouse testes. (A to D) Adjacent sections of an 8-week-old mouse testis were hybridized with a 35S-labeled mZnf8 antisense probe (A and B) or a Smad1 antisense probe (C and D). A′ to D′ are higher magnifications of the corresponding boxed regions in A to D, respectively. Panels A, A′, C, and C′ are bright-field photomicrographs. Panels B, B′, D, and D′ are dark-field photomicrographs. mZnf8 and Smad1 have a similar transcription pattern in mouse testis.
DISCUSSION
To identify new genes that play important roles in BMP signaling pathways, we performed a yeast two-hybrid screen with Smad1 as bait. mZnf8, encoding a Krüppel-type zinc finger protein, was isolated from the screen. Interaction between mZnf8 and Smad1 was further confirmed by a reciprocal yeast two-hybrid assay, a mammalian two-hybrid assay, in vitro GST pulldown assays, and copurification from mammalian cells. Transient-transfection and reporter assays indicate that overexpression of mZnf8 can repress BMP and TGF-β signaling. More significantly, knocking down the expression of endogenous mZnf8 significantly increased the activity of a BMP reporter. In situ hybridization analysis showed that mZnf8 and Smad1 have similar transcription patterns, both in mouse embryos and in adult male testes. Taken together, these results support the idea that mZnf8 is involved in modulating BMP signaling in vivo.
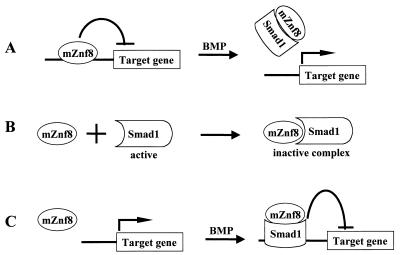
There are three possible models for mZnf8 function. First, it may bind a specific DNA sequence(s) to repress the transcription of target genes (Fig. 11A). This model is consistent with the observation that most of the known Krüppel zinc finger proteins interact with specific cis-regulatory DNA elements (reviewed in references 6 and 42). Upon BMP stimulation, Smad1 is phosphorylated and translocated into the nucleus. The binding of Smad1 to mZnf8 dislodges it from its binding sites and thus allows target genes to be transcribed. This derepression mechanism was proposed for the interaction between Hoxc-8 and Smad1 (29).
FIG. 11.
Three mechanisms by which mZnf8 may function. (A) In the first model, mZnf8 functions as a sequence-specific transcriptional repressor. Upon BMP stimulation, Smad1 translocates into the nucleus and interacts with mZnf8 to dislodge it from its binding sites, allowing the transcription of target genes. (B) In the second model, the interaction of mZnf8 with Smad1 blocks the activity of Smad1. Thus, the nucleus-localized mZnf8 may set a threshold for BMP signaling. (C) In the third model, mZnf8 functions as a corepressor of Smad1. Upon BMP stimulation, mZnf8 and Smad1 form a transcriptional repressor complex to actively shut down the transcription of specific target genes. To simplify this figure, other cofactors of Smad1 are not shown.
A second possible model is that mZnf8 serves as a general inhibitor that blocks the trans-activity of Smad proteins (Fig. 11B). Such a mechanism has been proposed to account for the activity of another Smad nuclear interaction protein, Snip1 (15). According to this hypothesis, mZnf8 is involved in setting a threshold for BMP signaling. Low levels of BMP signaling will be blocked by the interaction of mZnf8 and Smad1; the level of active Smad1 has to pass a threshold to overcome the presence of mZnf8 in the nucleus to activate BMP downstream genes. In this scenario, it is particularly interesting that mZnf8 has different binding affinities for different R-Smads. Since TGF-β Smads have lower binding affinity to mZnf8, the threshold set by mZnf8 for TGF-β signaling will be lower than that for BMP signaling. This provides a new mechanism for cells to have different sensitivities to different extracellular signals. The result from the transient-transfection assay, that overexpression of mZnf8 has a more dramatic effect on a BMP reporter than on a TGF-β reporter, is mostly consistent with this model.
In mammalian cells, Snip1 preferentially interacts with Smad4. Since this common Smad4 is required for the BMP, TGF-β, and activin pathways (15), Snip1 is unable to set distinct thresholds for different TGF-β superfamily cytokines. Thus, this potential appears to be unique to mZnf8. It is particularly interesting that mZnf8 can interact with both the MH1 and MH2 domains of Smad1. The MH1 domain is generally believed to be responsible for binding DNA, while the MH2 domain interacts with other partner proteins (1, 3, 25). Thus, the inhibition of Smad1 activity by mZnf8 can be twofold. First, mZnf8 interacts with the MH1 domain of Smad1 to block its DNA binding activity. Second, mZnf8 binds to the MH2 domain to block its trans-activation activity. Further studies are required to test this hypothesis.
Third, mZnf8 may function as a corepressor of Smad1 to actively repress the transcription of BMP downstream genes (Fig. 11C). A similar mechanism was proposed for a Smad2 interaction partner, TGIF (43). It was proposed that upon TGF-β stimulation, Smad2 can interact either with coactivators, forming a transcriptional active complex, or with TGIF, forming a transcriptional repressor complex (43). mZnf8 may function in a similar way to form a transcriptional repressor complex with Smad1 to repress target genes. In addition to TGIF, SnoN and Ski were also identified as corepressors of TGF-β Smads (23, 31, 32). Ski and Tob are two other known corepressors of BMP Smads (40, 46). mZnf8 is the third candidate as a corepressor of BMP Smads. Further experiments are required to distinguish among these three models.
Applying the RNAi knocking-down technique, we showed that decreasing expression of endogenous mZnf8 significantly increased the activity of one BMP reporter [12(GCCG)-lux] but not the other (pTlx-lux) in response to hBMP4 (Fig. 7). A simple explanation for this is that endogenous mZnf8 is selectively involved in certain responses of BMP signaling but not others. However, as we noticed in Fig. 7D, transcription of endogenous mZnf8 was not completely eliminated; likely some mZnf8 is still expressed, though we do not have the mZnf8-specific antibody to test this directly. Thus, we cannot exclude the possibility that a low level of endogenous mZnf8 is sufficient to suppress the activity of pTlx-lux. Another possible explanation is that an unknown corepressor of BMP Smad functions redundantly with mZnf8 in the pTlx-lux reporter but not in the 12(GCCG)-lux assay.
Krüppel-type zinc finger proteins belong to a very large family; it has been estimated that there are close to 300 Krüppel-type zinc finger proteins in humans (6, 16). About one third of these zinc finger proteins also contain a KRAB domain at their N-terminal ends. So far, mZnf8 is the only member of the KRAB transcriptional repressor family that may interact with Smad1 in vivo. It will be of interest in the future to determine what properties of mZnf8 make it a Smad1 interaction partner.
One interesting feature of mZnf8 is that its transcriptional repression activity was only slightly affected by deletion of the KRAB domain (Fig. 6B). This is different from most, if not all, other Krüppel zinc finger proteins studied (19, 30, 36, 41). The isolation of clone S1, which encodes the first five Krüppel-type zinc fingers, from the yeast two-hybrid screen suggests that these Krüppel-type zinc fingers do not possess transcriptional repression activity. Sequence comparison between the mouse and human proteins indicates that the seventh zinc finger sequence and the last 41 amino acids are highly conserved (Fig. 2). We speculate that one or both of the regions harbor the transcriptional repression activity. This will be tested in future studies.
BMP signaling plays critical roles in male germ cell development; deletion of Bmp8a or Bmp8b causes severe defects in spermatogenesis, and mutation in Bmp7 enhances the mutant phenotype of Bmp8a (49, 50, 52). However, the underlying molecular mechanism for BMP signaling remains unknown. In this study, we show that mZnf8 is highly transcribed in the testes of adult mice, in contrast to its ubiquitous transcription in mouse embryos. Furthermore, mZnf8 has a cell- and stage-specific transcription pattern similar to that of Smad1, the transcripts of which are mainly detected in the midstage pachytene spermatocytes (from stage V to stage XII) during cycling of the seminiferous epithelium, with the peak at stage X (51). This result is consistent with the hypothesis that the two proteins interact with each other at specific stages during spermatogenesis.
Smad2, a TGF-β Smad protein, is expressed in preleptotene to pachytene spermatocytes (39). The different expression patterns of Smad1 and Smad2 suggest that BMP and TGF-β are involved in different aspects of spermatogenesis (39). The similar transcription pattern of mZnf8 and Smad1 suggests that mZnf8 mainly mediates BMP signaling but not TGF-β signaling during spermatogenesis. This is consistent with the discovery that mZnf8 has a higher binding affinity for BMP Smads (Smads 1 and 5) than for TGF-β Smads (Smads 2 and 3). Thus, mZnf8 is so far the only candidate gene for mediating BMP signaling during mouse spermatogenesis. Future conditional gene inactivation experiments will help determine the exact function of mZnf8 during male germ cell development.
Acknowledgments
We thank L. Attisano, X. Cao, R. Derynck, J. Massague, D. Turner, and J. Wrana for providing various expression constructs, L. Limbird for providing the COS-M6 cell line, and S. Hollenberg for providing the mouse embryo two-hybrid library. We thank members of the Hogan laboratory, especially H. Kulessa, and J. Topczewksa, for suggestions and assistance on the project. We thank M. de Caestecker, H. Crawford, N. Bhowmick, and M. Weaver for critically reading and commenting on the manuscript. We also thank K. Tompkins for excellent research assistance during the early stages of this work.
K.J. is an Associate and B.L.M.H. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Attisano, L., and S. T. Lee-Hoeflich. 2001. The Smads. Genome Biol. 2:3010.1-3010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attisano, L., and J. L. Wrana. 2002. Signal transduction by the TGF-β superfamily. Science 296:1646-1647. [DOI] [PubMed] [Google Scholar]
- 3.Attisano, L., and J. L. Wrana. 2000. Smads as transcriptional comodulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 4.Ayer, D. E., C. D. Laherty, Q. A. Lawrence, A. P. Armstrong, and R. N. Eisenman. 1996. Mad proteins contain a dominant transcription repression domain. Mol. Cell. Biol. 16:5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blokzijl, A., P. ten Dijke, and C. F. Ibanez. 2002. Physical and functional interaction between GATA-3 and Smad3 allows TGF-β regulation of GATA. Target Genes Curr. Biol. 12:35-45. [DOI] [PubMed] [Google Scholar]
- 6.Dang, D. T., J. Pevsner, and V. W. Yang. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 32:1103-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, X. H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-β. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebelein, B., and R. Urrutia. 2001. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol. Cell. Biol. 21:928-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruendler, C., Y. Lin, J. Farley, and T. Wang. 2001. Proteasomal degradation of Smad1 induced by bone morphogenetic proteins. J. Biol. Chem. 276:46533-46543. [DOI] [PubMed] [Google Scholar]
- 10.Hata, A., J. Seoane, G. Lagna, E. Montalvo, A. Hemmati-Brivanlou, and J. Massague. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229-240. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, B. L. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580-1594. [DOI] [PubMed] [Google Scholar]
- 12.Hoodless, P. A., T. Haerry, S. Abdollah, M. Stapleton, M. B. O'Connor, L. Attisano, and J. L. Wrana. 1996. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell 85:489-500. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, W., T. Hamamoto, K. Kusanagi, K. Yagi, M. Kawabata, K. Takehara, T. K. Sampath, M. Kato, and K. Miyazono. 2000. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 275:6075-6079. [DOI] [PubMed] [Google Scholar]
- 14.Kaczynski, J., J. S. Zhang, V. Ellenrieder, A. Conley, T. Duenes, H. Kester, B. van Der Burg, and R. Urrutia. 2001. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co repressors and competing with Sp1. J. Biol. Chem. 276:36749-36756. [DOI] [PubMed] [Google Scholar]
- 15.Kim, R. H., D. Wang, M. Tsang, J. Martin, C. Huff, M. P. de Caestecker, W. T. Parks, X. Meng, R. J. Lechleider, T. Wang, and A. B. Roberts. 2000. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-β signal transduction. Genes Dev. 14:1605-1616. [PMC free article] [PubMed] [Google Scholar]
- 16.Klug, A., and J. W. Schwabe. 1995. Protein motifs 5. Zinc fingers. FASEB J. 9:597-604. [PubMed] [Google Scholar]
- 17.Kusanagi, K., H. Inoue, Y. Ishidou, H. K. Mishima, M. Kawabata, and K. Miyazono. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 11:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe, E., A. Letamendia, and L. Attisano. 2000. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl. Acad. Sci. USA 97:8358-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange, R., A. Christoph, H. J. Thiesen, G. Vopper, K. R. Johnson, L. Lemaire, M. Plomann, H. Cremer, D. Barthels, and U. A. Heinlein. 1995. Developmentally regulated mouse gene NK10 encodes a zinc finger repressor protein with differential DNA-binding domains. DNA Cell Biol. 14:971-981. [DOI] [PubMed] [Google Scholar]
- 20.Lania, L., E. Donti, A. Pannuti, A. Pascucci, G. Pengue, I. Feliciello, G. La Mantia, L. Lanfrancone, and P. G. Pelicci. 1990. cDNA isolation, expression analysis, and chromosomal localization of two human zinc finger genes. Genomics 6:333-340. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Y. S., M. F. Carey, M. Ptashne, and M. R. Green. 1988. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell 54:659-664. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz, P., D. Koczan, and H. J. Thiesen. 2001. Transcriptional repression mediated by the KRAB domain of the human C2H2 zinc finger protein Kox1/ZNF10 does not require histone deacetylation. Biol. Chem. 382:637-644. [DOI] [PubMed] [Google Scholar]
- 23.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus, M. T., C. P. Petersen, B. B. Haines, J. Chen, and P. A. Sharp. 2002. Gene silencing with micro-RNA designed hairpins. RNA 8:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazono, K. 2000. TGF-β signaling by Smad proteins. Cytokine Growth Factor Rev. 11:15-22. [DOI] [PubMed] [Google Scholar]
- 26.Richman, J. G., A. E. Brady, Q. Wang, J. L. Hensel, R. J. Colbran, and L. E. Limbird. 2001. Agonist-regulated Interaction between alpha2-adrenergic receptors and spinophilin. J. Biol. Chem. 276:15003-15008. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki, A., Y. Masuda, Y. Ohta, K. Ikeda, and K. Watanabe. 2001. Filamin associates with Smads and regulates transforming growth factor-beta signaling. J. Biol. Chem. 276:17871-17877. [DOI] [PubMed] [Google Scholar]
- 28.Schuh, R., W. Aicher, U. Gaul, S. Cote, A. Preiss, D. Maier, E. Seifert, U. Nauber, C. Schroder, R. Kemler, et al. 1986. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell 47:1025-1032. [DOI] [PubMed] [Google Scholar]
- 29.Shi, X., X. Yang, D. Chen, Z. Chang, and X. Cao. 1999. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem. 274:13711-13717. [DOI] [PubMed] [Google Scholar]
- 30.Skapek, S. X., D. Jansen, T. F. Wei, T. McDermott, W. Huang, E. N. Olson, and E. Y. Lee. 2000. Cloning and characterization of a novel Kruppel-associated box family transcriptional repressor that interacts with the retinoblastoma gene product, Rb. J. Biol. Chem. 275:7212-7223. [DOI] [PubMed] [Google Scholar]
- 31.Stroschein, S. L., W. Wang, S. Zhou, Q. Zhou, and K. Luo. 1999. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science 286:771-774. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Y., X. Liu, E. N. Eaton, W. S. lane, H. F. Lodish, and R. A. Weinberg. 1999. Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell 4:499-509. [DOI] [PubMed] [Google Scholar]
- 33.Tang, S. J., P. A. Hoodless, Z. Lu, M. L. Breitman, R. R. McInnes, J. L. Wrana, and M. Buchwald. 1998. The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development 125:1877-1887. [DOI] [PubMed] [Google Scholar]
- 34.Thiel, G., M. Lietz, K. Bach, L. Guethlein, and G. Cibelli. 2001. Biological activity of mammalian transcriptional repressors. Biol. Chem. 382:891-902. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, R. Y., and R. R. Reed. 1997. With a eukaryotic GST fusion vector for proteins difficult to express in E. coli. BioTechniques 23:794-796., 798: 800. [DOI] [PubMed] [Google Scholar]
- 36.Vissing, H., W. K. Meyer, L. Aagaard, N. Tommerup, and H. J. Thiesen. 1995. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 369:153-157. [DOI] [PubMed] [Google Scholar]
- 37.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 38.von Bubnoff, A., and K. W. Cho. 2001. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev. Biol. 239:1-14. [DOI] [PubMed] [Google Scholar]
- 39.Wang, R. A., and G. Q. Zhao. 1999. Transforming growth factor beta signal transducer Smad2 is expressed in mouse meiotic germ cells, Sertoli cells, and Leydig cells during spermatogenesis. Biol. Reprod. 61:999-1004. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W., F. V. Mariani, R. M. Harland, and K. Luo. 2000. Ski represses bone morphogenic protein signaling in Xenopus and mammalian cells. Proc. Natl. Acad. Sci. USA 97:14394-14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witzgall, R., E. O'Leary, A. Leaf, D. Onaldi, and J. V. Bonventre. 1994. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl. Acad. Sci. USA 91:4514-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe, S. A., L. Nekludova, and C. O. Pabo. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29:183-212. [DOI] [PubMed] [Google Scholar]
- 43.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 44.Wrana, J. L., L. Attisano, J. Carcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang, and J. Massague. 1992. TGF- beta signals through a heteromeric protein kinase receptor complex. Cell 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 45.Yingling, J. M., P. Das, C. Savage, M. Zhang, R. W. Padgett, and X. F. Wang. 1996. Mammalian dwarfins are phosphorylated in response to transforming growth factor beta and are implicated in control of cell growth. Proc. Natl. Acad. Sci. USA 93:8940-8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, Y., S. Tanaka, H. Umemori, O. Minowa, M. Usui, N. Ikematsu, E. Hosoda, T. Imamura, J. Kuno, T. Yamashita, K. Miyazono, M. Noda, T. Noda, and T. Yamamoto. 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103:1085-1097. [DOI] [PubMed] [Google Scholar]
- 47.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., X. Feng, R. We, and R. Derynck. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383:168-172. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, G. Q., Y. X. Chen, X. M. Liu, Z. Xu, and X. Qi. 2001. Mutation in Bmp7 exacerbates the phenotype of Bmp8a mutants in spermatogenesis and epididymis. Dev. Biol. 240:212-222. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, G. Q., K. Deng, P. A. Labosky, L. Liaw, and B. L. Hogan. 1996. The gene encoding bone morphogenetic protein 8B is required for the initiation and maintenance of spermatogenesis in the mouse. Genes Dev. 10:1657-1669. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, G. Q., and B. L. Hogan. 1997. Evidence that Mothers-against-dpp-related 1 (Madr1) plays a role in the initiation and maintenance of spermatogenesis in the mouse. Mech. Dev. 61:63-73. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, G. Q., L. Liaw, and B. L. Hogan. 1998. Bone morphogenetic protein 8A plays a role in the maintenance of spermatogenesis and the integrity of the epididymis. Development 125:1103-1112. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, H., P. Kavsak, S. Abdollah, J. L. Wrana, and G. H. Thomsen. 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400:687-693. [DOI] [PubMed] [Google Scholar]