Abstract
Toll-like receptors (TLRS) recognize pathogen-associated molecular patterns to enable innate immune responses. A number of genetic defects influencing the function of these receptors have been identified and are associated with recurrent and/or severe infection. Our goal was to develop a reproducible assay of TLR function in order to evaluate patients with recurrent infection who would be suspected of having a genetic defect affecting TLR signaling. We chose to study peripheral blood mononuclear cells (PBMCS) to avoid potential influences of soluble factors contained in whole blood, and we utilized ligands for TLRS 1/2, 2/6, 3, 4, 5, 6, 7, and 9. Tumor necrosis factor (TNF) production in PBMC supernatants was measured by an enzyme-linked immunosorbent assay after TLR ligand stimulation and was dependent on gene transcription and NF-κB activation. Some variables affecting the assay were assessed, including the effects of: blood anticoagulant, serum-containing media, incubation time, ligand storage, blood storage time, and cell cryopreservation. By using optimized assay conditions, effective concentrations of individual ligands and mean responses to those ligands were established for healthy control donors. Finally, three patients with a mutation in the IKBKG gene, encoding the NF-κB essential modulator (NEMO) protein, were evaluated as disease controls and were almost uniformly below the standard deviation of healthy donors for all ligands tested. Although a number of variables influence TLR ligand-induced TNF responses, this assay can be optimized for potential clinical use to screen patients with primary immunodeficiencies affecting TLR function.
Toll-like receptors (TLRs) are a critical component of the innate immune system and help to protect hosts from infectious disease through the recognition of pathogen-associated molecular patterns (PAMPs). The PAMPs recognized by TLRs are diverse but include molecules expressed by bacteria, fungi, viruses, and protozoa. Thus far, 11 TLRs have been identified in mice and humans and are expressed in both myeloid and lymphocytic lineages (1). The signaling that results after ligation of a TLR includes a complex series of intermediates that lead to activation of the inhibitor of NF-κB (IκB) kinase (IKK) complex and function of the transcription factor NF-κB. All TLRs have the capacity to initiate inflammation through their ability to induce the production of an important NF-κB target, tumor necrosis factor (TNF) (1). Although TLRs have the capacity to induce divergent signaling pathways resulting in the production of additional NF-κB-dependent as well as NF-κB-independent cytokines, the production of TNF provides a measure of the integrity of TLR itself and of the NF-κB-dependent pathway.
The significance of individual TLRs in protection against infectious disease has been clearly demonstrated in murine models. Specific TLRs have been genetically removed or altered in mice, resulting in increased susceptibility to pathogens expressing PAMPs recognized by the given TLR (10). For example, animals with a defective TLR 4 fail to produce TNF in response to the TLR 4 ligand lipopolysaccharide (LPS) and are more susceptible to infection with gram-negative bacteria (12, 21). The importance of the NF-κB-dependent pathway of TLR signaling has been specifically demonstrated by mice lacking interleukin-1 receptor-associated kinase 4 (IRAK-4), which is required in TLR signaling to activate IKK and induce NF-κB function; these animals demonstrate almost complete abrogation of TLR-induced functions (23). Similarly, mice deficient in NF-κB components also are unable to produce TNF in response to specific TLR ligands (25); such deficiencies have allowed dissection of the pathway from TLRs to effector cytokine production.
More recently, certain human infectious susceptibility syndromes have also been directly correlated with inappropriate TLR function. Most notably, a homozygous mutation in the gene encoding IRAK-4 results in recurrent pyogenic infection and completely abrogated TNF responses to TLR ligands (20). Since other measurable aspects of the innate and adaptive immunity are normal in this disorder, it presents a diagnostic challenge without the ability to evaluate TLR function. Similarly, several other human genetic abnormalities associated with increased susceptibility to infection that also impair, or would be expected to impair, TLR function have been described. These include mutations of the genes encoding TLR 2, TLR 4, the NF-κB essential modulator (NEMO), and an NF-κB inhibitor, IκBα (reviewed in reference 19). The latter two mutations can result in ectodermal dysplasia in addition to a combined immunodeficiency, but in some cases the phenotype is limited to the immunodeficiency of which TLR functional impairment can be a feature (18). Furthermore, mutations of individual TLRs might be recognized only by testing the response to the specific cognate ligand, since they may not otherwise impact immune function. For these reasons we sought to develop a reliable assay of TLR function to be used in consideration of patients who might have immunodeficiency resulting from genetic lesions that impair TLR signaling. Thus, we have examined several relevant clinical variables and established conditions that consistently provide reproducible quantification of TLR ligand-induced TNF production in human blood specimens.
MATERIALS AND METHODS
Human subjects.
Whole blood was collected from healthy adult donors after informed consent was obtained. The three patients with a mutation of the IKBKG gene encoding NEMO have been described previously (16-18). Patients 1, 2, and 3 had 458T→G, 1250G→A, and 1056-1118del alterations in their IKBKG genes, respectively. All three of these patients had been maintained on intravenous immunoglobulin therapy at the time when samples were collected. Blood collections and evaluations were approved by the Children's Hospital of Philadelphia internal review board for the protection of human subjects.
Cell isolation.
Peripheral blood mononuclear cells (PBMCs) were prepared from heparinized peripheral blood using Ficoll-Paque Plus density gradient separation (Amersham Biosciences, Uppsala, Sweden). As an alternative, cell preparation tubes (CPT) containing sodium citrate gel and density gradient media (Becton Dickinson, Franklin Lakes, NJ) were used where noted. Isolated PBMCs were resuspended in RPMI (GIBCO/BRL, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, UT), 0.1 mM nonessential amino acids, 10 mM HEPES, 1 mM sodium pyruvate, 50 U/ml penicillin G, 50 μg/ml streptomycin sulfate, and 2 mM l-glutamine (GIBCO/BRL), referred to as R10-FBS. Where specified, AB− human serum (Atlanta Biologicals, Atlanta, GA) was utilized instead of FBS (referred to as R10-HS). Also where specified, RPMI containing all the additives listed above but no serum was utilized (referred to below as RPMI). All media were passed through disposable 0.2-μm filter units (Millipore, Billerica, MA) after preparation. PBMCs from control donors or patients possessing a mutant NEMO were cryopreserved by resuspending cells in 0.2-μm filter-sterilized FBS containing 10% dimethyl sulfoxide (DMSO; Fisher Scientific, Fair Lawn, NJ) in a 1-ml polypropylene vial containing a neoprene gasket (Corning, Corning, NY). Vials were gradually reduced in temperature by approximately 1°C/min using a methanol-containing nonmechanical cryopreservation unit (Nalgene, Rochester, NY) in a −80°C freezer. Prior to rapid thawing for use in an experiment, cells were stored in liquid nitrogen. Where specified, cells were specially cryopreserved in FBS containing endotoxin-free DMSO (Sigma-Aldrich, St. Louis, MO). Upon thawing, cell viability was determined by trypan blue exclusion (GIBCO/BRL), and any sample with <95% viability was discarded. All cell and reagent manipulations were performed using sterile technique in a laminar flow hood.
TLR ligand stimulation.
TLR ligands used for cell stimulation were obtained from InvivoGen (San Diego, CA) and included the following: Pam3CSK4, a synthetic tripalmitoylated lipopeptide mimicking the acylated amino terminus of bacterial lipoproteins, to activate TLRs 1 and 2; zymosan, derived from Saccharomyces cerevisiae, for TLRs 2 and 6; poly(I:C), a synthetic analog of double-stranded RNA, for TLR 3; ultrapure LPS, the major component of gram-negative bacterial cell walls, derived from Salmonella enterica serovar Minnesota, for TLR 4; flagellin, a constituent of the flagellar filament, derived from Salmonella enterica serovar Typhimurium, for TLR 5; loxoribine, a guanosine analog derivative, for TLR7; and oligodinucleotide 2216 (ODN 2216 [5′-ggG GGA CGA TCG TCg ggg gg-3′ {lowercase letters indicate nuclease-resistant phosphothioates; capital letters indicate phosphodiesters}]), a synthetic oligonucleotide, for TLR 9. The use of these molecules in inducing the functions of the particular TLRs has been established previously for each of TLR 1/2 (3), TLR 2/6 (24), TLR 3 (2), TLR 4 (4), TLR 5 (7), TLR 7 (8), and TLR 9 (9). For TLR 9, the specificity of the ODN 2216 stimulant was ensured through the use of a non-CpG variant of ODN 2216 (5′-ggG GGA GCA TGC TGg ggg gc-3′). Individual ligands were prepared according to the manufacturer's recommendations in ultrapure pyrogen-free diluent provided along with the individual ligand. For use in cell stimulation, ligands were serially diluted in volumes of 100 μl R10-FBS in a sterile 96-well polystyrene microtiter plate. Medium without any added ligand was used to determine any baseline production of TNF. Each serial dilution was performed in duplicate. As a positive control, cells were treated with the NF-κB activator phytohemagglutinin (Murex Pharmaceuticals) at 2 μg/ml in R10-FBS. As controls for NF-κB-induced and transcription-dependent TNF production, cells were pretreated with 0.2-μm filter-sterilized solutions of 10 μM helenalin, to inhibit NF-κB function (14), or of 10 μM actinomycin D, to inhibit transcription, respectively, in R10-FBS for 30 min at 37°C prior to exposure to TLR ligands.
To determine TLR responses, 2 × 105 PBMCs in 100 μl R10-FBS (or R10-HS, or R10 without serum, where specified) were added to each of the duplicate ligand- or medium-containing wells and incubated at 37°C for 24 h with 5% CO2. This cell density was established by generating a TNF response curve for PBMCs at densities ranging from 3.1 × 104 to 1.0 × 106 cells/well; cells were stimulated with LPS for 24 h. The concentration of 2 × 105 cells/well corresponded to the linear phase of the TNF production curve and was ultimately chosen for subsequent experiments. All assay preparations were performed using sterile technique in a laminar flow hood.
To choose the TLR ligand concentrations used to evaluate experimental variables and obtain normative data, dose titration curves were generated for each TLR ligand. These ranges were based on manufacturers' recommendations, previously published results (5, 19), and our own pilot experiments. Using these sources, we chose the following concentrations of TLR ligands: 0.4 to 40 μg/ml Pam3CSK4, 0.02 to 2 μg/ml zymosan, 0.2 to 20 μg/ml poly(I:C), 0.02 to 2 μg/ml LPS, 0.02 to 2 μg/ml flagellin, 0.2 to 20 μg/ml loxoribine, and 0.4 to 40 μg/ml ODN 2216. Although these ligand ranges were utilized in the vast majority of the experiments presented in Results, the top of the dose-response curve was chosen to statistically assess the variables under study. In general, effects at high TLR ligand concentrations imparted by an experimental manipulation accurately reflected effects found at lower concentrations. Phytohemagglutinin (Murex Biotech, Dartford, United Kingdom) was used at 2 μg/ml as a positive control for TNF production and yielded 1,029 ± 206 pg/ml.
Enzyme-linked immunosorbent assay (ELISA).
Immulon microtiter plates (ThermoLabSystems, Franklin, MA) were incubated overnight at 4°C with 100 μl monoclonal anti-human TNF capture antibody (BD Biosciences) in coating buffer (0.1 M sodium carbonate, pH 9.5). Individual wells were washed three times with phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBST), pH 7.4, and blocked with PBS with 10% bovine serum albumin, pH 7.0, for 1 h at room temperature. Individual wells were again washed three times with PBST, and the supernatants from individual wells of TLR ligand-stimulated PBMCs were transferred to individual antibody-coated wells. Supernatants were diluted twofold directly in the antibody-coated wells so that the amount of TNF measured would fall within the TNF standard curve range. To create a standard curve, a serially diluted standard preparation of TNF (R&D Systems, Minneapolis, MN) was added to a separate set of individual antibody-coated wells in duplicate. Plates containing samples were then incubated at room temperature for 2 h, followed by five washes of the individual wells with PBST. To detect captured TNF, 100 μl of a biotinylated anti-human TNF sandwich monoclonal antibody (BD Biosciences) along with an avidin-horseradish peroxidase conjugate (BD Biosciences) was added to each well and the plate was incubated for 1 h at room temperature. Individual wells were then washed five times with PBST and incubated with 100 μl tetramethylbenzidine plus hydrogen peroxide substrate solution for 30 min at room temperature. The colorimetric reaction was stopped with 50 μl of 1 M sulfuric acid, and absorbance was read at 450 nm using a microtiter plate spectrophotometer (Biotek, Winooski, VT). To generate values for TNF production, the serially diluted TNF standard was used to create a curve within the spectrophotometer software environment to which experimental readings were mathematically applied (KC junior; Biotek). TNF values obtained from duplicate samples were averaged and the mean value utilized in all subsequent considerations. Using this method the limit of detection for TNF in TLR ligand-stimulated cell supernatants was 15.6 pg/ml.
Statistical analysis.
All cell supernatants were analyzed in duplicate, and these biological replicates were averaged. A minimum of three separate donors were evaluated for each experimental condition. Resulting mean data were compared with a Student two-tailed t test using the statistical analysis functions of Microsoft Excel. The coefficient of variance (COV), or percent relative standard deviation, was calculated as 100 multiplied by the standard deviation divided by the mean.
RESULTS
Validation of TLR ligand-induced TNF response.
Evaluation of HEK293 cells coexpressing the individual TLR and an alkaline phosphatase NF-κB reporter plasmid was provided by the manufacturer as validation of the specificity of each TLR ligand. The induction of NF-κB activity, as demonstrated by alkaline phosphatase activity in the supernatants of these TLR-expressing HEK293 cells exposed to their cognate ligands, was certified for each lot of TLR ligand as being >0.6 OD unit, whereas unstimulated cells produced levels of <0.1 OD unit. As certification of specificity, noncognate TLR ligands or HEK293 cells expressing other TLRs also resulted in alkaline phosphatase levels of <0.1 OD unit. To further validate the chosen ligands for their ability to induce TNF in an NF-κB-dependent manner, the response of PBMCs to each ligand was determined for cells that had been pretreated with the transcriptional inhibitor actinomycin D or the NF-κB inhibitor helenalin. Although incubation of PBMCs in R10-FBS containing 10 μM of either inhibitor did not affect cell viability, both inhibitors completely abrogated production of TNF by all the ligands tested (Fig. 1). This demonstrates that production of TNF in the supernatant in response to this selected series of TLR ligands depends on NF-κB activation and gene transcription, thus inducing function through the expected pathway.
FIG. 1.
Effects of actinomycin D and helenalin on TLR ligand-induced TNF production. PBMCs were pretreated with media, 10 μM actinomycin D, or 10 μM helenalin for 30 min prior to incubation with TLR ligands. TNF levels in the supernatants were determined by ELISA. Values are means ± standard deviations for three control donors. Asterisks indicate undetectable levels of TNF.
Clinically relevant variables affecting TLR ligand-induced TNF response.
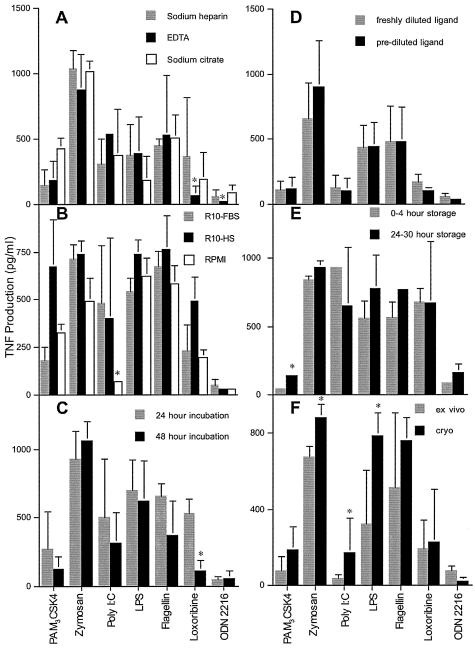
In an attempt to minimize interassay variability while maximizing sensitivity and specificity, experimental conditions that could potentially influence the assay performance in a clinical setting were critically evaluated. At each step, optimum assay conditions were chosen for the evaluation of additional variables. The overall goal was to establish reliable assay conditions with which to obtain some normative data. As a first variable, the specific anticoagulant in which whole-blood samples were collected was evaluated. For these studies, sterile Vacutainer tubes (Becton Dickinson) preloaded by the manufacturer with either sodium heparin, EDTA, or sodium citrate were utilized. Samples were otherwise collected and handled identically using R10-FBS as the assay medium. There were a number of variations of TLR ligand-induced TNF production among the anticoagulants, and significant differences (P < 0.05) were found in the responses to TLR 7 and 9 ligands, where the use of EDTA resulted in lower TNF production (Fig. 2A). As an alternative cell preparation technique, PBMCs isolated using CPT were compared directly to those isolated using a Ficoll gradient and responded similarly to TLR ligands (data not shown). However, there was a significantly higher poly(I:C) response in PBMCs isolated using CPT. In light of these results, and considering the utility of sodium heparin anticoagulant in other immunological function assays, these were ultimately chosen for blood collection to further standardize the assay.
FIG. 2.
Impacts of selected experimental variables on TLR ligand-induced TNF. Values are means ± standard deviations for TLR responses to 0.1 mg/ml Pam3CSK4, 0.1 mg/ml zymosan, 2.5 mg/ml poly(I:C), 0.1 mg/ml LPS, 10 μg/ml flagellin, 10 mM loxoribine, and 50 μM ODN 2216. At least three repeat experiments were performed for each variable evaluated. (A) The effects of anticoagulants used for blood sample collection on TLR responses were evaluated for sodium heparin (gray bars), EDTA (black bars), and sodium citrate (white bars). Asterisks indicate values significantly different from those obtained using sodium heparin (P < 0.05). (B) The effect of serum used for cell incubation on TNF production was evaluated by incubating PBMCs with R10-FBS (gray bars), R10-HS (black bars), or RPMI containing no serum (white bars). Asterisks indicate values significantly different from those obtained using R10-FBS (P < 0.05). (C) Comparison of TNF production after 24-h (gray bars) and 48-h (black bars) incubations with TLR ligands. Asterisks indicate values significantly different from those obtained after a 24-h incubation (P < 0.05). (D) Effects of dilution and storage of ligands at −80°C (black bars) on TNF production compared to dilution of ligands at the time of the assay (gray bars). Asterisks indicate values significantly different from those obtained using ligands diluted at the time of the assay (P < 0.05). (E) Effects of PBMC isolation from blood stored at ambient temperature from 0 to 4 h (gray bars) and after 24 h (black bars) on TLR-induced TNF production. Asterisks indicate values significantly different from those obtained using blood stored 0 to 4 h (P < 0.05). (F) Differences in TNF production between PBMCs that were freshly isolated (gray bars) and those that were cryopreserved and then thawed (black bars). Asterisks indicate values significantly different from those obtained for freshly isolated PBMCs (P < 0.05).
We next sought to determine the effect of serum in the media in which cells were incubated during exposure to TLR ligands. This point was of potential concern because, although the serum used in these experiments was endotoxin tested by the manufacturer and filtered prior to use, it served as a potential source of TLR ligands. Also, we were concerned that exposure to the foreign proteins present in FBS may have had an indirect effect on TLR function through interactions with the ligands or the TLRs themselves. For this reason, a medium containing FBS was compared to a medium containing AB− HS, as well as to a serum-free medium. Although there were some significant differences in TNF production between serum-containing and serum-free media, these differences were not found for all ligands (Fig. 2B). The only significant difference found between cells stimulated in FBS-containing versus serum-free medium was for TLRs induced by poly(I:C). This difference, however, was not found between media containing FBS or HS, since the poly(I:C)-induced TNF in these media was indistinguishable. A single lot of poly(I:C) was used across all experiments, and thus the difference was not due to interlot variations. Although this pattern suggests that serum contained factors that may amplify TLR 3 ligation, this effect was not found with other ligands. To maximize sensitivity, however, serum-containing medium was preferred over serum-free medium. The comparison between FBS and HS, on the other hand, demonstrated increased TNF levels produced in response to Pam3CSK4 in HS-containing medium. Since the responses to Pam3CSK4 in FBS and serum-free media were nearly identical, this increased response was attributed to HS specifically and not to the ligand. Although this could have been due to a useful cofactor for Pam3CSK4 found in HS, the response in FBS-containing or serum-free medium was well within the measurable range. For these reasons, we opted to perform subsequent assays using an FBS-containing medium. This decision was based on the intent to increase sensitivity while maintaining specificity for the ligand. FBS was also chosen over HS due to differences in cost between the two.
Although 24 h of cell stimulation was considered a reasonable time frame for cytokine production dependent on new gene transcription, we wanted to determine if an advantage would be provided by allowing the cells either longer or shorter periods of TLR ligand stimulation. Thus, we performed a time course experiment comparing 10 min, 1 h, 2 h, 4 h, 6 h, 24 h, and 48 h of incubation with TLR ligands. TLR ligand-induced TNF production reached its peak across all ligands tested at 24 h of incubation. Pam3CSK4, LPS, flagellin, and ODN 2216 stimulation, however, achieved these levels after 6 h of incubation. To more specifically evaluate the TLR ligand incubation time that may be appropriate for a clinical immunology laboratory, 24 h and 48 h of incubation were specifically compared. Although TNF levels measured in supernatants in response to most ligands were not significantly different after 24 or 48 h of incubation (Fig. 2C), the amount of TNF measured after TLR 7 ligation was significantly reduced with the longer incubation. Due to the peak response for all ligands at 24 h, the increased sensitivity for TLR 7 ligation, and the convenient assay length, 24 h of incubation was considered adequate for evaluating TNF production. Therefore, the 24-h incubation time was utilized in all subsequent experiments (shorter incubations of >6 h, however, should also be acceptable).
In an effort to simplify the serial dilution of the TLR ligands for use in the assay, we next wanted to determine if freezing and storing prediluted ligands in R10-FBS at −80°C had an effect on TNF levels produced compared to ligands that were diluted just prior to their use in the assay. The ability to predilute ligands would allow numerous microtiter plates to be prepared simultaneously, conserving effort and potentially reducing variability. Dilution series of ligands were prepared in the same microtiter plates to which PBMCs would ultimately be added for cell stimulation. Plates were frozen at −80 °C and thawed for 1 h at room temperature prior to the addition of PBMCs on the day of the assay. The thawed prediluted ligands were then compared directly, using the same PBMC sample, to ligands that had just been diluted. We found no significant difference in the TNF levels produced in response to any of the ligands (Fig. 2D) and thus concluded that prediluted and frozen ligand was adequate for testing. All subsequent experiments utilized ligands that were either prediluted and frozen or diluted at the time of the assay.
All of the experiments presented thus far were performed with PBMCs prepared from blood obtained ≤4 h prior to use in the assay. Since blood samples obtained in a clinical setting cannot always be processed on the same day they are collected, we wanted to determine if storage time at room temperature would affect TLR ligand-induced TNF production. This point would also be relevant clinically, because samples occasionally need to be shipped from one clinical site to another via overnight courier. Thus, we compared TLR ligand-induced TNF production from PBMCs isolated from blood within 4 h of its being drawn from the donor to TNF production using blood stored at room temperature for 24 to 30 h prior to PBMC isolation. Although there was a significant difference only for Pam3CSK4 stimulation, there was some greater variability in TNF production from samples that had been stored for 24 to 30 h (Fig. 2E). Since the means of ligands other than Pam3CSK4, however, were not significantly different, blood storage was considered an acceptable alternative to immediate PBMC preparation. Thus, PBMCs were isolated from blood samples within 30 h in subsequent experiments.
Finally, we sought to validate this assay for cryopreserved cells, since this would potentially extend its application to appropriately shipped or archived specimens. All cryopreserved cells were rapidly thawed prior to use and washed twice in R10-FBS before being promptly added to the TLR ligands. For this evaluation, PBMCs from a given donor were prepared from peripheral blood and cryopreserved within 1 h of preparation. A portion of the whole blood was maintained at room temperature for later processing. Once the PBMCs had equilibrated at −80°C, they were transferred to liquid nitrogen for at least 4 h, after which they were rapidly thawed. At this time, PBMCs were prepared from the remaining whole blood and directly compared to the thawed and washed PBMCs in the assay. In general, cryopreserved cells produced a greater TLR ligand-induced TNF response than those isolated directly from blood at the time of assay performance (Fig. 2F). However, this was statistically significant only for zymosan, poly(I:C), and LPS. Conversely, the response to ODN 2216 was slightly but significantly reduced after cryopreservation. We hypothesize that the stress of freezing and thawing cells may have contributed to this altered and most frequently exaggerated stimulation. Regardless, we consider cryopreserved cells a reasonable source of PBMCs for evaluation of TLR ligand-induced TNF response, with the caveat that individual responses may be artificially elevated.
TLR ligand-induced TNF response in healthy donors.
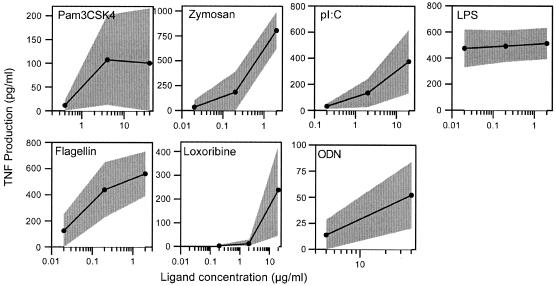
The preceding investigations allowed us to establish optimized and reproducible experimental conditions. Further studies were performed using PBMCs from whole blood collected in sodium heparin tubes that were processed within 24 h of collection and then incubated 18 to 24 h in RPMI containing 10% FBS with or without the TLR ligands. Ligand dilutions were either prepared at the time of the assay or prepared previously and stored at −80°C until the time of the assay. Using this method we wanted to establish a normative response range in healthy adult donors to several concentrations of TLR ligands. The ligand concentrations were established as described in Materials and Methods and were validated in the experiments described above. To simplify the analyses, however, only the highest ligand concentrations were presented in the preceding experiments. Normative data for TLR ligand-induced TNF responses were obtained for each of the ligands at three different ligand concentrations (two for ODN 2216) for 20 healthy adult donors. Although responses to Pam3CSK4, loxoribine, and ODN 2216 demonstrated a relatively high COV, the COV for each of the ligands tested was less than 100%. The best COVs were obtained for zymosan and LPS and were both 23%.
Individual variability in TLR ligand-induced TNF responses.
Biological assays of immune function typically reflect some degree of interassay variability within a given subject. This variation can be a feature of exogenous factors that impact immune function, such as viral infection or psychological or environmental stress. To obtain a measure of how much variability exists in TLR ligand-induced TNF production under the optimized conditions developed above, PBMCs isolated from successive blood collections from the same individual over a 9-month period were tested. Although there was little observed variation between mean responses for this individual and those for the healthy donors for whom results are shown in Fig. 3, there were differences in the amounts of TNF induced by given TLR ligands between PBMCs obtained from this individual at different times (Table 1). Although the COV was <100% at the highest concentrations of all ligands tested, it was greatest in the responses to loxoribine and ODN 2216 and lowest in the response to zymosan. Since the mean responses for this individual, however, were ultimately similar to those found in the control population, these findings suggest a utility in repeating tests when results fall outside of the control range. Regardless, the variability found in this one donor longitudinally was similar to the variability found among our 20 control donors sampled once each (Table 1).
FIG. 3.
TLR responses for control donors. A normal range for TLR ligand-induced TNF production was established by a 24-h incubation of PBMCs in varying concentrations of seven different TLR ligands. Individual points on black lines are means obtained for 20 healthy adult males and females. The gray regions surrounding the black lines represent standard deviations.
TABLE 1.
Longitudinal variation in TLR-induced TNF production
| Ligand | Ligand concn (μg/ml) | Mean TNF production (pg/ml)a | SD | COV (%) for:
|
|
|---|---|---|---|---|---|
| Single donor | Control populationb | ||||
| Pam3CSK4 | 40 | 205.8 | 157.4 | 77 | 75 |
| 4.0 | 155.8 | 103.8 | 67 | 82 | |
| Zymosan | 2.0 | 955.9 | 198.7 | 21 | 22 |
| 0.2 | 168.7 | 169.2 | 100 | 84 | |
| 0.02 | 49.3 | 100.2 | 203 | 236 | |
| Poly(I:C) | 20 | 508.3 | 374.1 | 74 | 31 |
| 2.0 | 220.1 | 142.0 | 65 | 84 | |
| 0.2 | 43.3 | 19.8 | 46 | 60 | |
| LPS | 2.0 | 558.1 | 121.7 | 22 | 25 |
| 0.2 | 478.2 | 155.1 | 32 | 23 | |
| 0.02 | 459.5 | 142.3 | 31 | 21 | |
| Flagellins | 2.0 | 660.4 | 193.5 | 29 | 39 |
| 0.2 | 416.2 | 230.3 | 55 | 31 | |
| 0.02 | 249.4 | 312.6 | 125 | 38 | |
| Loxoribine | 20 | 156.5 | 139.7 | 89 | 87 |
| 2.0 | BLD | BLD | 207 | ||
| 0.2 | BLD | BLD | 238 | ||
| ODN 2216 | 40 | 46.3 | 36.4 | 79 | 75 |
| 4.0 | BLD | BLD | 80 | ||
Mean TLR ligand-induced TNF values were derived from 10 successive blood samples obtained over 38 weeks from a single donor. BLD signifies values that contained more than one repeat that fell below the limit of detection for TNF in the ELISA.
COV for the single evaluations performed on the 20 donors for whom results are shown in Fig. 3.
Evaluation of TLR ligand-induced TNF production in patients with NEMO deficiency.
TLR ligand-induced TNF production is likely to be most useful in identifying deficient TLR responses resulting from genetic lesions that impair the TLRs themselves or downstream signaling components. Although a number of human gene mutations that affect or would be expected to affect these responses have been defined, we chose to evaluate PBMCs from patients with immunodeficiency resulting from a rare mutation in the IKBKG gene (incidence, 1:250,000 [17]), which encodes the NEMO protein. Since NEMO plays a central role in transmitting the signal from all TLRs to affect TNF production, we considered patients with suspected NEMO deficiency to be ideal candidates for evaluation by TLR ligand-induced TNF production. To determine whether this assay could detect interrupted TLR function in this population, mononuclear cells from three patients known to have IKBKG mutations were evaluated (Table 2). Mean TNF responses were significantly impaired across almost all TLR ligands tested, the exception being one patient's TLR 9 response (patient 3), which produced means within the control donor normative range. This result may be a feature of the particular IKBKG mutation found in this individual, as proposed previously (18). Despite this, the patient responses across the different ligands can be easily distinguished from the control donor means presented in Fig. 3. Although these results cannot make a conclusive statement about either the sensitivity or the specificity of this assay, due to the very limited availability of IKBKG patient samples, depressed TLR responses within a population with a high pretest probability of abnormal TLR function suggest that the assay can be useful in identifying abnormal patterns in TLR function.
TABLE 2.
TLR ligand-induced TNF production in PBMCs from three patients with NEMO deficiency
| Ligand | Ligand concn (μg/ml) | TNF production (pg/ml)a
|
Meanb | SDc | ||
|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | ||||
| Pam3CSK4 | 40 | BLD | BLD | BLD | BLD | |
| 4.0 | BLD | BLD | BLD | BLD | ||
| Zymosan | 2.0 | 17.0 | 25.3 | 18.7 | 20.3 | 4.3 |
| 0.2 | BLD | BLD | BLD | BLD | ||
| 0.02 | BLD | BLD | BLD | BLD | ||
| Poly(I:C) | 20 | BLD | BLD | BLD | BLD | |
| 2.0 | BLD | BLD | BLD | BLD | ||
| 0.2 | BLD | BLD | 18.0 | |||
| LPS | 2.0 | BLD | 256.3 | 22.1 | <98.0 | |
| 0.2 | BLD | 51.1 | 22.6 | <29.8 | ||
| 0.02 | BLD | 28.7 | BLD | <19.7 | ||
| Flagellins | 2.0 | BLD | 161.4 | 44.4 | <73.8 | |
| 0.2 | BLD | 34.0 | 20.8 | <23.5 | ||
| 0.02 | BLD | 27.1 | 26.3 | <23.0 | ||
| Loxoribine | 20 | BLD | BLD | BLD | BLD | |
| 2.0 | BLD | BLD | BLD | BLD | ||
| 0.2 | BLD | BLD | BLD | BLD | ||
| ODN 2216 | 40 | BLD | BLD | 41.9 | <30.1 | |
| 4.0 | BLD | BLD | 16.1 | <15.7 | ||
BLD, below the limit of detection.
To calculate a mean, the limit of detection was used for values that fell below the limit of detection. In these cases the mean is marked as “less than” the calculated mean obtained using the limit of detection value.
Provided only for means that contained three values that fell within the detectable range of the ELISA.
DISCUSSION
An expanded appreciation of innate immunity in host defense has allowed for the definition of a number of human infectious susceptibility syndromes resulting from genetic lesions presumably affecting TLR function. Patients with these defects typically have recurrent or severe pyogenic infections which are associated with significant morbidity and mortality. Although these types of immunodeficiencies would have previously escaped specific diagnosis, they can now be defined genetically. One example is the homozygous deficiency of IRAK-4, which completely abrogates TLR function (15, 20). This loss of function results from the requirement for IRAK-4 in inducing NF-κB activation after TLR ligation. There are a number of other genetic lesions that would also presumably interrupt TLR function but have not yet been definitively demonstrated to do so in cells from patients who carry them. These include hypermorphic mutation of IκBα and diverse hypomorphic mutations of NEMO that result in susceptibility to infection and would be predicted to interrupt TLR signaling, since these molecules are required to activate NF-κB downstream of IRAK-4. Furthermore, a number of mutations of the TLRs themselves, including TLR 2 (13) and TLR 4 (22), have been associated with severe or recurrent infection and would be presumed to affect the function of the given TLR in response to its ligand. Finally, any number of signaling intermediates between TLRs and NF-κB have the potential to be mutated in syndromes of infectious susceptibility and interrupted TLR function. Given the potentially numerous molecular defects affecting TLR function that could result in recurrent infection, a reliable assay to screen selected patients for TLR function is needed. This is particularly important considering that some of these defects, such as that of IRAK-4, do not appreciably affect other measures of immunity typically taken in clinical laboratories (20).
To design a reliable assay of NF-κB-dependent response to TLR ligands that could be performed by a standard clinical laboratory, we chose to measure TNF production, due to the well-established relationship of TLR ligation with TNF production. We also decided to measure TNF by ELISA, because this assay is cost-effective, widely available, and easily performed and provides quantitative data. We could have also measured TLR ligand-induced alpha/beta interferon (IFN-α/β), as this response utilizes only partially overlapping signaling mechanisms. In preliminary experiments we found that IFN-α production paralleled TNF production, but we did not pursue large-scale measurement due to cost concerns and our specific interest in NF-κB-dependent pathways. Ultimately TLR ligand-induced IFN-α production may allow for the expanded clinical application of this assay and result in the identification of unappreciated molecular defects associated with recurrent infection.
To ensure the specificity of TNF responses, we used only high-quality and validated TLR ligands due to the need for reagent purity and reproducibility. The intensive certification methodologies used by the manufacturer, in combination with our demonstration of the dependence of these ligands on NF-κB and transcriptional function (Fig. 1), gave us confidence in a range of stimuli for assessing TLR ligation. We also chose to perform our assays of TLR response using PBMCs. Some authors have effectively evaluated TLR responses in whole blood (6, 11, 18), but soluble inhibitors of TLR function (11) and preexisting plasma cytokine levels could negatively impact the ability to discern specific TLR function. To specifically isolate TLR signals in immune cells resulting after TLR ligation and to exclude the effects of exogenous serum factors, we opted against performing whole-blood assays. PBMC-based assays, therefore, will be most useful in identifying inherent defects of TLR function, such as those interrupting the intracellular TLR signaling pathway (Table 2).
In order to design an assay of TLR function that would be clinically useful, we wanted to minimize interassay variability while maximizing potential sensitivity and specificity. Thus, we evaluated a number of variables that may affect the response of human PBMCs to TLR ligands. These included (i) the anticoagulant used in blood collection, (ii) the presence or type of serum in the assay medium, (iii) incubation time with TLR ligands, (iv) TLR ligand storage, (v) ambient temperature during blood storage, and (vi) cell cryopreservation. Optimal conditions included the collection of blood in sodium heparin-containing tubes from which PBMCs were isolated within 30 h of phlebotomy. Once isolated, preferably PBMCs were incubated with TLR ligands for 24 h in a medium containing 10% FBS. TLR ligands could be diluted at the time of the assay or stored prediluted at −80°C, and PBMCs could be used immediately after isolation or after cryopreservation, with the caveat that cryopreservation can artificially increase some TLR responses. It should be noted that the PBMCs used to generate the normative ranges presented in Fig. 3 were not cryopreserved, and therefore we cannot say if it is appropriate to compare responses obtained using cryopreserved cells to these ranges. In general it would be best to compare similarly prepared PBMCs; however, grossly abnormal results with absent TLR responses (such as those in Table 2) are likely to be apparent regardless of the cell preparation technique. Even though there are a large number of variables that can be evaluated in this assay, we believe that these were particularly important and allowed for an assay that was both reproducible and potentially useful in the clinical laboratory. The issues of reproducibility and clinical utility were suggested by Fig. 3 and Table 2, in which normative TLR response data were established, and ligand-induced TNF production by patients with NEMO mutation and infectious susceptibility could be easily distinguished from that by controls.
Although we present a reproducible assessment of TLR function, there are a number of outstanding issues. First, it is possible that donor age will be an important factor in interpretation of the values produced by this assay. Our normative data measured TNF only in adults. It has been reported previously that TLR ligand-induced TNF production differs significantly in neonatal and adult patient populations (11). Although this was identified as a feature contributed by soluble plasma proteins, and it may be a feature of whole-blood-based evaluation of TLR responses, normative data for pediatric populations will need to be collected before this assay can be truly applied to children undergoing immunologic assessments due to recurrent infection. Second, we are unaware of the specific external factors that will affect TLR functional assessments. All of the control donors in our study were healthy at the time of participation. It is possible that acute infection may increase baseline TNF production from PBMCs while increasing or decreasing the specific response to ligand. Similarly, the effects of medications, environmental factors, and genetic factors, including common TLR polymorphisms, are also unknown. Since many of these are likely to affect assay results, the best present approach would be to repeat abnormal results with a new blood sample. Moreover, evaluation of patients with other diagnoses by using this assay may allow for the consideration of the effects of those conditions on TLR function.
In this study we have developed a reproducible assay of TLR function that could be used to screen patients with a history of recurrent or severe infection for the absence of single or multiple TLR responses. We believe that this assay will allow for the identification of patients who may have one of the previously described genetic defects in the TLR signaling pathway and could help select those who warrant additional molecular diagnosis. This approach will be particularly useful because many of these patients will not have otherwise measurable impairments of immune function. Further investigation of patients with abnormal TLR responses will also be likely to reveal previously unappreciated genetic defects that result in infectious susceptibility.
A control sample that has been collected, handled, and prepared identically to the unknown sample must be tested in each experiment for accurate comparison of values.
Acknowledgments
This study was supported by grants from the Ethel Brown Foerderer Fund and the Jeffery Model Foundation.
We thank Steven Douglas, Michele Paessler, and Dmitri Monos for helpful discussions.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 4.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 5.Currie, A. J., D. J. Davidson, G. S. Reid, S. Bharya, K. L. MacDonald, R. S. Devon, and D. P. Speert. 2004. Primary immunodeficiency to pneumococcal infection due to a defect in Toll-like receptor signaling. J. Pediatr. 144:512-518. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg, J., C. Fieschi, R. Doffinger, M. Feinberg, T. Leclerc, S. Boisson-Dupuis, C. Picard, J. Bustamante, A. Chapgier, O. Filipe-Santos, C. L. Ku, L. de Beaucoudrey, J. Reichenbach, G. Antoni, R. Balde, A. Alcais, and J. L. Casanova. 2004. Bacillus Calmette Guerin triggers the IL-12/IFN-γ axis by an IRAK-4- and NEMO-dependent, non-cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol. 34:3276-3284. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 8.Heil, F., P. Ahmad-Nejad, H. Hemmi, H. Hochrein, F. Ampenberger, T. Gellert, H. Dietrich, G. Lipford, K. Takeda, S. Akira, H. Wagner, and S. Bauer. 2003. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur. J. Immunol. 33:2987-2997. [DOI] [PubMed] [Google Scholar]
- 9.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 10.Kopp, E., and R. Medzhitov. 2003. Recognition of microbial infection by Toll-like receptors. Curr. Opin. Immunol. 15:396-401. [DOI] [PubMed] [Google Scholar]
- 11.Levy, O., K. A. Zarember, R. M. Roy, C. Cywes, P. J. Godowski, and M. R. Wessels. 2004. Selective impairment of TLR-mediated innate immunity in human newborns: neonatal blood plasma reduces monocyte TNF-α induction by bacterial lipopeptides, lipopolysaccharide, and imiquimod, but preserves the response to R-848. J. Immunol. 173:4627-4634. [DOI] [PubMed] [Google Scholar]
- 12.Li, Q., and B. J. Cherayil. 2004. Toll-like receptor 4 mutation impairs the macrophage TNF-α response to peptidoglycan. Biochem. Biophys. Res. Commun. 325:91-96. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz, E., J. P. Mira, K. L. Cornish, N. C. Arbour, and D. A. Schwartz. 2000. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect. Immun. 68:6398-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyss, G., A. Knorre, T. J. Schmidt, H. L. Pahl, and I. Merfort. 1998. The anti-inflammatory sesquiterpene lactone helenalin inhibits the transcription factor NF-κB by directly targeting p65. J. Biol. Chem. 273:33508-33516. [DOI] [PubMed] [Google Scholar]
- 15.Medvedev, A. E., A. Lentschat, D. B. Kuhns, J. C. Blanco, C. Salkowski, S. Zhang, M. Arditi, J. I. Gallin, and S. N. Vogel. 2003. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J. Exp. Med. 198:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orange, J. S., S. R. Brodeur, A. Jain, F. A. Bonilla, L. C. Schneider, R. Kretschmer, S. Nurko, W. L. Rasmussen, J. R. Kohler, S. E. Gellis, B. M. Ferguson, J. L. Strominger, J. Zonana, N. Ramesh, Z. K. Ballas, and R. S. Geha. 2002. Deficient natural killer cell cytotoxicity in patients with IKK-γ/NEMO mutations. J. Clin. Investig. 109:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orange, J. S., A. Jain, Z. K. Ballas, L. C. Schneider, R. S. Geha, and F. A. Bonilla. 2004. The presentation and natural history of immunodeficiency caused by nuclear factor κB essential modulator mutation. J. Allergy Clin. Immunol. 113:725-733. [DOI] [PubMed] [Google Scholar]
- 18.Orange, J. S., O. Levy, S. R. Brodeur, K. Krzewski, R. M. Roy, J. E. Niemela, T. A. Fleisher, F. A. Bonilla, and R. S. Geha. 2004. Human nuclear factor κB essential modulator mutation can result in immunodeficiency without ectodermal dysplasia. J. Allergy Clin. Immunol. 114:650-656. [DOI] [PubMed] [Google Scholar]
- 19.Orange, J. S., O. Levy, and R. S. Geha. 2005. Human disease resulting from gene mutations that interfere with appropriate nuclear factor-κB activation. Immunol. Rev. 203:21-37. [DOI] [PubMed] [Google Scholar]
- 20.Picard, C., A. Puel, M. Bonnet, C. L. Ku, J. Bustamante, K. Yang, C. Soudais, S. Dupuis, J. Feinberg, C. Fieschi, C. Elbim, R. Hitchcock, D. Lammas, G. Davies, A. Al-Ghonaium, H. Al-Rayes, S. Al-Jumaah, S. Al-Hajjar, I. Z. Al-Mohsen, H. H. Frayha, R. Rucker, T. R. Hawn, A. Aderem, H. Tufenkeji, S. Haraguchi, N. K. Day, R. A. Good, M. A. Gougerot-Pocidalo, A. Ozinsky, and J. L. Casanova. 2003. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299:2076-2079. [DOI] [PubMed] [Google Scholar]
- 21.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 22.Smirnova, I., N. Mann, A. Dols, H. H. Derkx, M. L. Hibberd, M. Levin, and B. Beutler. 2003. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc. Natl. Acad. Sci. USA 100:6075-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki, N., S. Suzuki, G. S. Duncan, D. G. Millar, T. Wada, C. Mirtsos, H. Takada, A. Wakeham, A. Itie, S. Li, J. M. Penninger, H. Wesche, P. S. Ohashi, T. W. Mak, and W. C. Yeh. 2002. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416:750-756. [DOI] [PubMed] [Google Scholar]
- 24.Underhill, D. M., A. Ozinsky, A. M. Hajjar, A. Stevens, C. B. Wilson, M. Bassetti, and A. Aderem. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401:811-815. [DOI] [PubMed] [Google Scholar]
- 25.Weih, F., G. Warr, H. Yang, and R. Bravo. 1997. Multifocal defects in immune responses in RelB-deficient mice. J. Immunol. 158:5211-5218. [PubMed] [Google Scholar]