Abstract
The molecular mechanisms by which Notch receptors induce diverse biological responses are not fully understood. We recently cloned a mammalian homologue of the Mastermind gene of Drosophila melanogaster, MAML1 (Mastermind-like-1 molecule) and determined that it functions as a transcriptional coactivator for Notch receptors. In this report, we characterize two additional genes in this Mastermind-like gene family: MAML2 and MAML3. The three MAML genes are widely expressed in adult tissues but exhibit distinct expression patterns in mouse early spinal cord development. All MAML proteins localize to nuclear bodies, share a conserved basic domain in their N termini that binds to the ankyrin repeat domain of Notch, and contain a transcriptional activation domain in their C termini. Moreover, as determined by using coimmunoprecipitation assays, each MAML protein was found to be capable of forming a multiprotein complex with the intracellular domain of each Notch receptor (ICN1 to -4) and CSL in vivo. However, MAML3 bound less efficiently to the ankyrin repeat domain of Notch1. Also, in U20S cells, whereas MAML1 and MAML2 functioned efficiently as coactivators with each of the Notch receptors to transactivate a Notch target HES1 promoter construct, MAML3 functioned more efficiently with ICN4 than with other forms of ICN. Similarly, MAML1 and MAML2 amplified Notch ligand (both Jagged2 and Delta1)-induced transcription of the HES-1 gene, whereas MAML3 displayed little effect. Thus, MAML proteins may modify Notch signaling in different cell types based on their own expression levels and differential activities and thereby contribute to the diversity of the biological effects resulting from Notch activation.
Notch receptors initiate a highly conserved signaling pathway that influences cell fate decisions within multiple tissues and regulates the ability of precursor cells to respond to other developmental signals (1). In mammals, Notch signaling has been shown to regulate neurogenesis (3, 51), myogenesis (29), vasculogenesis (28), hematopoiesis (27), skin development (32), and other aspects of organogenesis. In addition, Notch signaling is involved in other critical cellular processes such as proliferation and apoptosis (34, 35, 42, 45). Consistent with the ability to influence cellular differentiation in multiple tissues, mutations of Notch receptors and components of its signaling pathway have been associated with a number of diseases, including human T-cell leukemia (Notch1) (2, 9, 39), CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, Notch3) (22, 23), and Alagille syndrome (Jagged1) (31). The Notch pathway is also directly targeted by three proteins essential for Epstein-Barr virus transformation of B cells—EBNA2, EBNA3a, and EBNA3c—each of which binds to CSL and modifies Notch activity (17, 48). Also, the murine Notch4 gene has been identified as an integration site of mammary tumor virus (Int3), resulting in constitutive activation of Notch4 and breast carcinoma (12).
The components of the Notch signaling pathway appear to be highly conserved among species (36, 52). Activation of Notch receptors (Notch1, Notch2, Notch3, and Notch4 in mammals and Notch in Drosophila) by ligands (Jagged1, Jagged2, Delta1, Delta-like 1 [Dll1], Dll3, and Dll4 in mammals or Delta and Serrate in Drosophila) initiates the proteolytic processing events that result in the release of the intracellular domain of Notch (ICN). ICN subsequently translocates to the nucleus, and activates the major downstream nuclear target for Notch, the CSL family of DNA-binding transcription factors [CBF1/RBP-Jκ in mammals, Su(H) in Drosophila, and Lag-1 in Caenorhabditis elegans] by the displacement of the corepressors, including CIR (18), N-CoR/SMRT (25), and KyoT2 (49) and recruitment of coactivators, including PCAF and GCN5 (30), Mastermind-like-1 (MAML1) (26, 41, 55), and p300 (37). The transcription of CSL-dependent Notch target genes is then activated, including the well-studied basic-helix-loop-helix HES gene family (mammalian homologues of Drosophila Hairy and Enhancer of Split genes) such as HES-1 and HES-5 (8, 20, 24). These in turn regulate expression of tissue-specific transcription factors that influence lineage commitment and other events. Other potential Notch targets have been reported, including p21WAF1/Cip1 (42), cyclin D1 (44), HERP (19), and mitogen-activated protein kinase phosphatase LIP-1 (4). CSL-independent Notch signaling has also been demonstrated (46, 53), suggesting that some Notch effects can be mediated by other unidentified DNA-binding transcription factor(s).
One of the most intriguing questions in the Notch field is how a single pathway can be utilized effectively in so many diverse processes. Part of the diversity comes from the multiplicity of receptors and ligands, at least in mammals. However, genetic screens in Drosophila have also identified a number of genes capable of modifying Notch signaling (38). For example, Numb, a protein that becomes asymmetrically distributed between daughter cells, associates with ICN, and inhibits Notch signaling (15). Fringe limits Notch activity during boundary formation by glycosylating Notch and thereby modifying ligand binding (6). Another Notch modifier is the Drosophila mastermind gene and its mammalian homologue, Mastermind-like 1 (MAML1). LAG-3 was identified in C. elegans as a protein with functional similarity as Mastermind, although the sequence similarity is very low (40). The mastermind gene encodes a nuclear protein and was identified in multiple genetic screens for modifiers of Notch mutations in Drosophila (5, 14, 47, 57). Like Notch, loss-of-function mastermind mutations in flies result in “neurogenic” phenotypes, as well as dramatic interactions with different components of the Notch signaling pathway, including the ligand Delta and the effectors of Notch signals Su(H) and Deltex (10, 56). Consistent with a critical role of Mastermind in Notch signaling, the expression of truncated forms of Mastermind interferes with Notch functions in many tissues in Drosophila (16). Our previous studies demonstrated that MAML1 is a transcriptional coactivator for all four Notch receptors in mammals (55). MAML1 is a nuclear protein containing an N-terminal basic domain that binds to the ankyrin repeats of ICN1 and forms a DNA-binding transcriptional complex with ICN and CSL. Importantly, a transcriptional activation domain (TAD) was discovered in the more C-terminal portion of MAML1 that potentiates the activation of a well-characterized Notch target gene, HES-1. Endogenous MAML1 was shown to form a stable large protein complex with ICN and CSL in the nuclei of the ICN1-transformed RKE cells and a human T-cell leukemia cell line by size exclusion chromatography (21), demonstrating a physiological role of MAML1 in Notch signaling. It has also recently been demonstrated that MAML1 is required for chromatin-dependent transactivation by a recombinant ICN1-CSL enhancer complex in vitro, recruits p300/CBP to the Notch transcriptional complex, and may control the stability of ICN (11). Overall, these data suggest that MAML1 plays a critical regulatory role in Notch signaling.
The expansion of the number of Notch receptor genes and other components, including Notch ligands, during evolution from the fly to the human led us to ask whether Mastermind-like coactivators of the Notch pathway had undergone a similar increase in number. We report here the cloning and characterization of two new genes, MAML2 and MAML3, that also function as transcriptional coactivators for Notch receptors. Both genes are widely expressed in different adult tissues, but we demonstrate that in one part of the developing central nervous system, the spinal cord, there is considerable variation in expression of the MAML genes. Also, the MAML proteins vary somewhat in their ability to cooperate with different Notch receptors. Overall, given that there is differential expression and signaling of receptors, multiple Notch ligands, and the differential expression and activity of multiple coactivators, Notch effects in mammals have an extraordinary potential for diversity.
MATERIALS AND METHODS
Plasmids.
KIAA1819 and KIAA1816 cDNA clones in pBluescript II SK(+) vector were obtained from KAZUSA DNA Research Institute in Japan. The GenBank accession numbers for KIAA1819 and KIAA1816 are XM_045716 and AB058719, respectively. Full-length MAML2 and MAML3 cDNAs were initially cloned from KIAA1819 and KIAA1816 into intermediate cloning vectors and eventually cloned as SalI-NotI fragments into pFLAG-CMV-2 (Sigma), pEGFP-C3 (Clontech), and pBIND (Promega) vectors to obtain FLAG-tagged, green fluorescent protein (GFP)-tagged and DNA-binding domain (DB)-tagged MAML2 or MAML3. Two chimeric cDNAs were generated—MM3/1, which fuses the N-terminal region of MAML3 (amino acids [aa] 1 to 155) and the C-terminal region of MAML1 (aa 288 to 1016), and MM1/3, which fuses the N-terminal region of MAML1 (aa 1 to 228) and the C-terminal region of MAML3 (aa 155 to 1133)—and cloned into pFLAG-CMV-2 and pBIND vectors. Further information regarding cloning is available upon request.
Expression constructs that encode FLAG-tagged MAML1, GFP-tagged MAML1, DB-tagged MAML1, hemagglutinin (HA)-tagged human ICN1, untagged human ICN2, HA-tagged murine ICN3, untagged human ICN4, and Myc-tagged CSL have been described (55). HES-1-luc contains the “−194 to +160” promoter fragment of the HES-1 gene cloned upstream of the firefly luciferase gene in the pGL2-basic vector (20). pRL-TK (Promega) encodes Renilla luciferase under the control of thymidine kinase (TK) promoter and was used to normalize firefly luciferase activities for transfection efficiency. pSG5-luc (Promega) is a firefly luciferase reporter plasmid that contains five copies of the GAL4-binding site upstream of a minimal TATA box.
Northern blot analysis.
Filter-immobilized polyadenylated RNAs from multiple human tissue blots (7780-1; Clontech) were hybridized with 32P-labeled MAML2 or MAML3 probes (nucleotides from 1 to 906 of the MAML2 open reading frame or nucleotides from 1 to 1568 of the MAML3 open reading frame) according to the manufacturer's instructions.
Antibodies.
The following antibodies were purchased from commercial sources: mouse anti-Flag antibody (clone M2; Sigma), mouse anti-HA antibody (clone HA.11; Babco), mouse anti-Myc antibody (clone 9E10; Clontech), horseradish peroxidase-coupled goat antimouse antibody (Amersham), and Rhodamine Red-X-conjugated F(ab′)2 fragment goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc.).
Cell culture and transient transfection.
Human U20S osteosarcoma cells were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% Fetalclone I serum (HyClone Laboratories, Inc.), COS7 cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), and HeLa cells and 293 cells were cultured in DMEM with 10% FCS. NIH 3T3 cells transduced by pBABE retrovirus encoding Jagged2, or empty pBABE retrovirus, were maintained in DMEM medium containing 10% FCS and 1 μg of puromycin/ml. Transfections were carried out by using Superfect transfection reagent (Qiagen) according to the manufacturer's instructions.
Immunofluorescence staining.
Cells grown on coverslips were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. After permeabilization in a solution containing 10 mM HEPES, 3 mM MgCl, 50 mM NaCl, 10% Triton, and 300 mM sucrose for 10 min, nonspecific binding sites were blocked with 2% nonimmune goat serum in PBS for 30 min. Cell were then incubated for 60 min with primary antibody in PBS, washed extensively with PBS, and then incubated for 60 min with secondary antibody in PBS. After an extensive washing in PBS and final wash with H2O, coverslips were then mounted in GEL/MOUNT medium (Biomeda Corp.) and photographed with an Olympus microscope and a SPOT camera (Diagnostic Instrument, Inc.).
Western blotting and immunoprecipitation.
Western blotting and immunoprecipitation were performed as described previously (55).
Luciferase assays.
U20S cells were seeded on the six-well plates at 105 cells per well 1 day before transfection and then transiently transfected with various combinations of expression plasmid DNA. The total amounts of plasmids were maintained constant by adding appropriate amounts of empty vectors without inserts. The transfected cells were harvested at 44 h posttransfection and luciferase activities were measured in a Berthold luminometer (Lumat LB9507) by using the dual luciferase reporter assay system (Promega). Relative luciferase activities were normalized to Renilla luciferase activity.
Mouse tissue preparation and in situ hybridization.
Swiss-Webster mouse embryos at embryonic day 9.5 (E9.5) and E11.5 were collected and fixed in 4% (wt/vol) paraformaldehyde in PBS overnight at 4°C. After fixation, embryos were cryoprotected in 20% sucrose in PBS for 24 h at 4°C and then embedded in OCT. The sections were then cut on a cryostat. Digoxigenin-labeled sense and antisense RNA probes for human MAML2 and MAML3, mouse Maml1, Hes1, and Notch1 were prepared by in vitro transcription. In situ hybridizations were performed overnight at 65°C, and hybrids were detected with alkaline phosphatase-conjugated anti-digoxigenin immunoglobulin G (IgG) with BM purple as a substrate (Roche).
RESULTS
Identification of two new members of the mammalian Mastermind-like family.
The most highly conserved regions between Drosophila Mastermind and its mammalian homologue MAML1 were located in their N-terminal basic domains (55). The basic domain of MAML1 was found to be necessary and sufficient for the binding of MAML1 with the ankyrin repeat motif of Notch1. Two truncated mutants of MAML1—one with the N-terminal basic domain only (capable of binding to ICN but defective in transcriptional activation) and the other without the N-terminal basic domain (defective in binding to ICN but containing intact TAD)—acted as dominant-negative inhibitors for Notch signaling. The role of this basic domain was further supported by the studies showing that a C-terminal truncation of Drosophila Mastermind functioned as a dominant-negative inhibitor of Notch functions in several tissues, including the peripheral nervous system (bristle), eye (imaginal disk), and wing (16), and that a naturally occurring alternative spliced form of the Drosophila mastermind gene lacking the basic domain functioned to repress Notch signaling (13). In light of the critical role of the basic domains, we performed database searches by using the basic domain sequence of MAML1 to identify additional MAML1 related genes. Two cDNAs were identified that encode N-terminal domains highly homologous to the MAML1 basic domain. These two previously uncharacterized cDNAs are represented by KIAA 1819 and KIAA 1816 (deposited in GenBank by the Kazuza DNA Research Institute in Japan). Based on the findings presented below, we designated these two genes MAML2 and MAML3.
The basic domains of MAML1, MAML2, and MAML3 are the most conserved regions among these three proteins (Fig. 1A), and the rest of the sequences are rather divergent. The amino acid sequence identity between MAML1 and MAML2, MAML1 and MAML3, and MAML2 and MAML3 are 60, 50, and 47% for the basic domains but only 21, 33, and 21% for the entire protein sequences, respectively. All three MAML proteins are proline and glutamine rich, which is commonly observed in transcriptional coactivators. MAML2 and MAML3 both contain several stretches of glutamine residues.
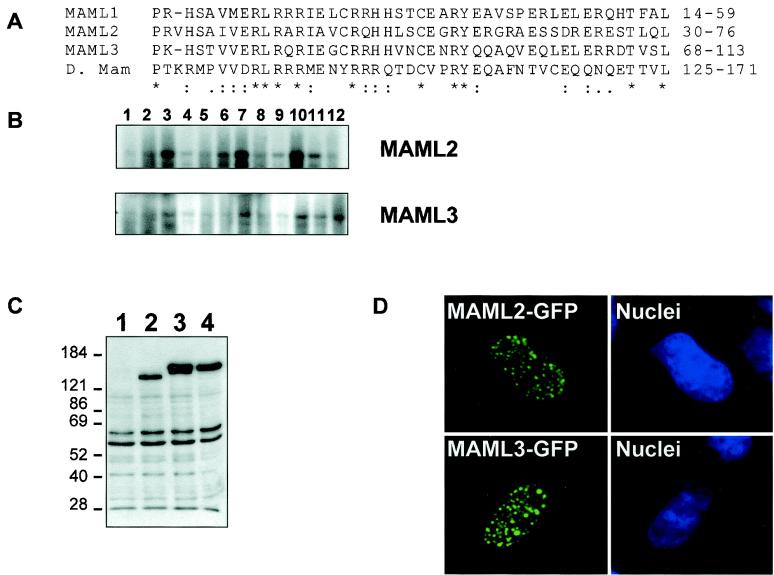
FIG. 1.
Sequence alignment and expression of the mammalian mastermind gene family. (A). Alignment of the conserved basic domains of three members of the mammalian Mastermind-like family—MAML1, MAML2, and MAML3—and Drosophila Mastermind by using CLUSTALW multiple sequence alignment software. The notations “✽”, “:”, and “.” indicate identical residues, conserved substitutions, and semiconserved substitutions in all sequences, respectively. The numbers after each sequence indicate the positions of the amino acids in each protein. (B) Northern blot analysis of MAML2 and MAML3 expression in human tissues. Lanes: 1, brain; 2, heart; 3, skeletal muscle; 4, colon; 5, thymus; 6, spleen; 7, kidney; 8, liver; 9, small intestine; 10, placenta; 11, lung; 12, peripheral blood leukocyte. A major 7.5-kb transcript was detected in these tissues for MAML2 and MAML3. (C) Expression of full-length MAML1, MAML2, and MAML3 proteins in COS7 cells. COS7 cells were transfected with vector alone (lane 1) or constructs encoding the full-length FLAG-tagged MAML1 (lane 2), MAML3 (lane 3), and MAML2 (lane 4) proteins for 44 h, and the cellular lysates were immunoblotted with anti-FLAG antibody. Numbers on the left are kilodaltons. (D). MAML2 and MAML3 are localized in nuclear dots. Transiently transfected COS7 cells expressing GFP-tagged MAML2 or MAML3 proteins (left) stained with DAPI (4′,6′-diamidino-2-phenylindole) (right) reveal the two proteins localize to the nucleus.
Like MAML1 (55), MAML2 and MAML3 are widely expressed in adult tissues as major transcripts of ca. 7.5 kb by Northern analysis (Fig. 1B). Based on the existing murine expressed sequence tag (EST) sequences, three human MAML genes are highly homologous to their murine counterparts with sequence identity ranging from approximately 85 to 90%. Database searches revealed that the MAML1, MAML2, and MAML3 genes are located on human chromosomes 5q35.3, 11q22.3, and 4q28.3, respectively.
The open reading frames for MAML1, MAML2, and MAML3 are 1016, 1153, and 1133 aa, with predicted molecular masses of 108, 125, and 115 kDa, respectively. Full-length cDNAs were obtained as described in Materials and Methods and are expressed as FLAG epitope-tagged fusion proteins in COS7 cells. By Western blot analysis with an antibody against the FLAG epitope (M2), MAML1 was detected as one band at ca. 130 kDa, MAML2 was detected as one band at ca. 160 kDa, and MAML3 was detected as two or more bands ranging from about 150 to 170 kDa (Fig. 1C), indicating some form of modifications for MAML3.
Nuclear body localization of MAML2 and MAML3.
MAML1 was previously found to localize in nuclear bodies (55). To determine whether MAML2 and MAML3 have a similar subcellular localization pattern, MAML2 and MAML3 cDNAs were fused to GFP and transiently expressed in COS7 and U20S cells. The subcellular localization of MAML2 and MAML3 was directly visualized by fluorescence microscopy. MAML2 and MAML3 displayed a speckled nuclear staining pattern, with considerable heterogeneity in the sizes of the dots (Fig. 1D). Cells expressing FLAG-tagged MAML2 and MAML3 proteins stained with anti-FLAG antibody also showed a similar nuclear staining pattern (data not shown). These results indicate that MAML1, MAML2, and MAML3 have a very similar subcellular localization, suggesting that the nuclear dot structure might be important for the function of this MAML family.
MAML1, MAML2, and MAML3 form complexes in vivo with ICN and CSL.
It has been shown that MAML1 binds directly to the ankyrin repeat region of Notch1 and forms a DNA-binding complex with ICN and CSL (55). Therefore, we sought to determine whether MAML2 and MAML3 are also able to form a multiprotein complex with ICN and CSL. As the first step, we looked for colocalization of MAML2 and MAML3 with ICN1 and CSL. Cells were cotransfected with different combinations of constructs encoding either GFP-tagged MAML2 or MAML3, HA-tagged ICN1, and Myc-tagged CSL and then analyzed them via staining with anti-HA or anti-Myc antibodies. Coexpression of MAML2 or MAML3 altered ICN1 staining from a diffuse pattern to the punctate pattern of MAML2 or MAML3 (data not shown). Moreover, both MAML2 and MAM3 colocalized with ICN1 in these dots. By itself, CSL had a diffuse nuclear staining pattern, but it also redistributed to nuclear dots with MAML2 or MAML3 in the presence, but not in the absence, of ICN1 (data not shown). These results were consistent with results previously reported for MAML1, suggesting that MAML2 and MAML3 were able to form complexes with CSL only in the presence of ICN1.
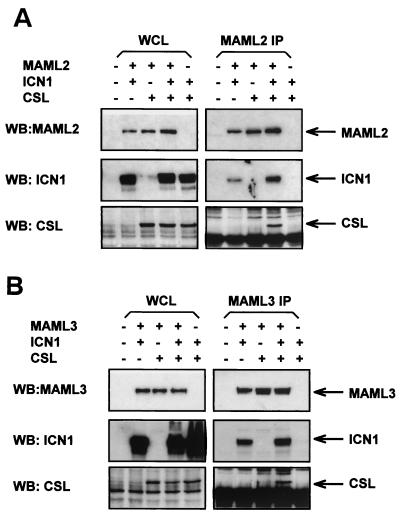
Next, we performed immunoprecipitation to determine whether MAML2 or MAML3 could form a complex with ICN and CSL. MAML2 immunoprecipitated with ICN1 (or ICN2, ICN3, or ICN4) and also immunoprecipitated with CSL, but only in the presence of ICN1 (or ICN2, ICN3, or ICN4) (Fig. 2A and not shown). MAML3 immunoprecipitated with ICN1 (or ICN2, or ICN3, or ICN4), and CSL in a manner similar to that with MAML2 (Fig. 2B and results not shown). Thus, all three members of the MAML family form nuclear complexes in vivo with the intracellular domain of all four Notch receptors (ICN1 through -4) and the DNA-binding protein CSL.
FIG. 2.
Formation of a ternary complex of MAML2 or MAML3, ICN1, and CSL in vivo. (A). MAML2, ICN1, and CSL form a ternary immunoprecipitate complex. COS7 cells were cotransfected with different combinations of three expression plasmids encoding FLAG-tagged MAML2, HA-tagged ICN1, and Myc-tagged CSL as indicated. Cellular lysates or anti-FLAG immunoprecipitates (IP) were immunoblotting with anti-FLAG, or anti-HA, or anti-Myc antibodies. (B) MAML3, ICN1, and CSL form a ternary immunoprecipitate complex. COS7 cells were cotransfected with different combinations of three expression plasmids encoding FLAG-tagged MAML3, HA-tagged ICN1, and Myc-tagged CSL. Analysis was performed as in panel A.
MAML2 and MAML3 bind to the ankyrin repeat domains of Notch1.
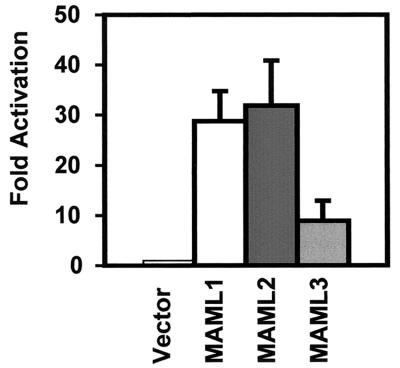
Since MAML1 is known to bind to the ankyrin repeats (ANK) of ICN1 through its N-terminal basic domain, we examined the ability of MAML2 and MAML3 to bind to the ANK domains of Notch1 by using a mammalian two-hybrid assay. As was the case for MAML1, the ANK domains of Notch1 were sufficient to bind to MAML2 and MAML3. Interestingly, MAML3 demonstrated less interaction with Notch1 ANK in this assay than did MAML1 and MAML2 (Fig. 3).
FIG. 3.
MAML1, MAML2, and MAML3 have differential binding to the ankyrin repeats of Notch1. U20S cells in a six-well plate were transfected with 25 ng of pRL-TK plasmid encoding Renilla luciferase, 0.5 μg of a firefly luciferase construct containing four GAL4-binding sites in the promoter (pG5luc), 0.5 μg of the plasmid encoding DB fused to the ankyrin repeats of Notch1, and 0.5 μg of pFLAG-CMV-2 empty vector or pFLAG-CMV-2 encoding MAML1, MAML2, or MAML3. Cells were harvested at 44 h posttransfection. Firefly luciferase activity, normalized to Renilla luciferase, was expressed as the fold activation (relative to the background level of firefly luciferase expression in the presence of an empty pFLAG-CMV-2 vector).
Like MAML1, MAML2 and MAML3 function as transcriptional coactivators for Notch signaling.
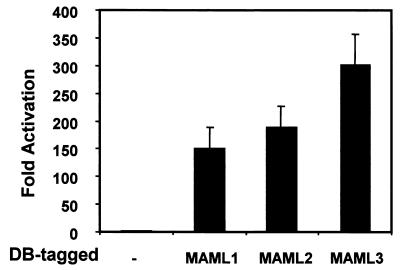
MAML1 serves as a transcriptional coactivator for four mammalian Notch receptors (55). Therefore, we sought to determine whether MAML2 and MAML3 can also function as transcriptional coactivators by using the following several approaches. First, full-length MAML2 and MAML3 were expressed as fusion proteins with the GAL4 DB and examined for their ability to activate a luciferase reporter that contains GAL4-binding sites in the promoter. Both MAML2 and MAML3 exhibited transcription activation activity similar to or higher than that of MAML1 (Fig. 4). The transcriptional activation domain was found on the C-terminal regions of MAML2 and MAML3 since the truncated mutants that lacked the N-terminal basic domain still retained transcriptional activation activity (not shown).
FIG. 4.
Like MAML1, MAML2, and MAML3 are transcriptional coactivators. U20S cells were transfected with 0.5 μg of a firefly luciferase construct containing four GAL4-binding sites in the promoter (pG5luc) and 0.5 μg of pBIND plasmid encoding either the GAL4 DB only or the DB fused to full-length MAML1, MAML2, or MAML3. Firefly luciferase activity, normalized to Renilla luciferase expressed from the pBIND plasmid, was expressed as the fold activation (relative to the background level of firefly luciferase expression in the presence of an empty pBIND vector).
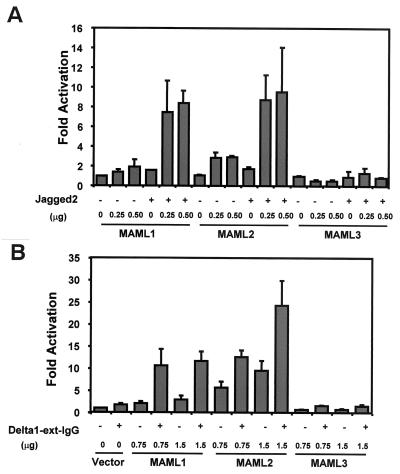
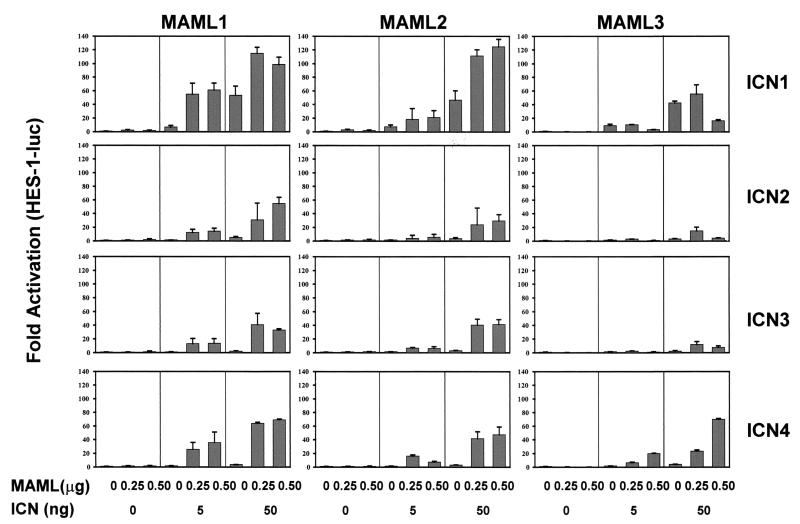
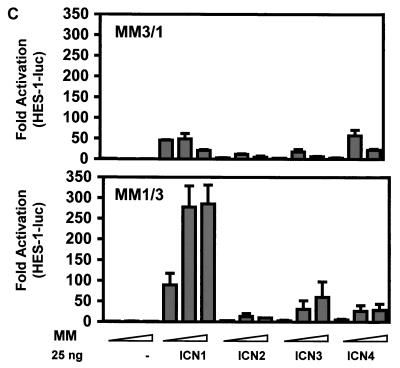
Second, the functional significance of the MAML-ICN-CSL interaction was examined by evaluating Notch-induced activation of a HES-1 promoter construct. The activation of Notch signaling was achieved here either by ligand-dependent stimulation or by ligand-independent stimulation through expression of constitutively active forms of Notch receptors, ICN. U20S cells were cotransfected with a HES-1 promoter reporter construct and increasing amounts of MAML1, MAML2, or MAML3 constructs. After 24 h, the transfected cells were then cocultured with NIH 3T3 cells expressing the Jagged2 ligand or control NIH 3T3 cells expressing an empty vector. In the absence of Notch activation, MAML1 only slightly increased HES-1 promoter activity. However, in the presence of Notch ligand stimulation, MAML1 dramatically enhanced HES-1 transcription (Fig. 5A). Therefore, the activation of the HES-1 promoter by MAML1 is dependent on Notch ligand Jagged2 stimulation. We found that MAML2 behaved similarly to MAML1. However, MAML3 displayed little effect when the Jagged2 ligand was used to stimulate Notch signaling in these cells (Fig. 5A). Similar results were obtained when Notch signaling was stimulated by using the soluble extracellular domain of Delta1 (50), another ligand for Notch (Fig. 5B). Moreover, we found that MAML1 and MAML2 cooperated with all four Notch receptors to activate HES-1 transcription. However, MAML3 acted as a robust transcriptional coactivator for ICN4 but was much less effective for ICN1, ICN2, and ICN3 (Fig. 6, note that high levels of ICN1 have significant activity alone that is not further augmented by MAML3). U20S cells were found to express Notch4 as determined by Western blot analysis (data not shown), although we did not formally compare the level of Notch4 expression to that of other Notch receptors. It is possible that Jagged2 and Delta1 might not be efficient activators of the Notch4 receptors in U20S cells.
FIG. 5.
MAML family members exhibit differential effects on HES-1 activation upon stimulation with ligands of Notch receptors, Jagged2, or Delta1. (A) U20S cells were transfected with 25 ng of a pRL-TK control plasmid expressing Renilla luciferase, 0.5 μg of HES-1-luc, and increasing amounts of expression plasmids encoding either MAML1, MAML2, or MAML3. At 20 h posttransfection, 105 NIH 3T3 cells expressing Jagged2 or 105 NIH 3T3 cells infected with empty pBABE virus as control were added to each well. Cell extracts were prepared 44 h posttransfection. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, is expressed as the fold activation relative to cells not expressing MAML family members that were cocultured with control NIH 3T3 cells. (B). U20S cells in the 60-mm plates were transfected with 75 ng of pRL-TK control plasmid encoding Renilla luciferase, 1.5 μg of HES-1-luc, and different amounts of expression plasmids encoding either MAML1, MAML2, or MAML3. At 24 h posttransfection, cells were split into four wells in the 24-well plates, two wells coated with human IgG and two wells coated with Delta ligand, and cultured for another 20 h. Cellular extracts were then prepared, and the luciferase activity was measured. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, was expressed as the fold activation relative to cells transfected with control vector plasmid and cultured in the wells coated with human IgG.
FIG. 6.
MAML family members exhibit differential effects on HES-1 activation induced by ICN1 to -4. U20S cells were transfected with 25 ng of pRL-TK plasmid encoding Renilla luciferase, 0.5 μg of HES-1-luc, and increasing amounts of pFLAG-CMV-2 plasmid encoding MAML1, MAML2, or MAML3 (each at 0, 0.25, and 0.5 μg) in the presence of 0 to 50 ng of pcDNA3 plasmids encoding human ICN1, human ICN2, murine ICN3, or human ICN4. Cellular extracts were prepared 44 h posttransfection, and the luciferase activity was measured. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, is expressed as the fold activation relative to cells not expressing MAML1, MAML2, MAML3, and ICN.
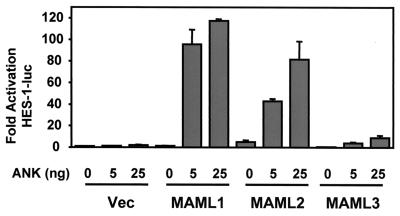
As above in Fig. 3, the ankyrin repeats in Notch1 contain important binding sites for all three MAML proteins, and we have previously shown that MAML1 can cooperate with a minimal ANK domain of Notch 1 (Notch1-ANK) to transactivate the HES-1 reporter gene (55). We therefore examined the ability of MAML2 and MAML3 to activate HES-1 activity with Notch1-ANK. MAML1 and MAML2, but not MAML3, were able to strongly cooperate with the Notch1-ANK to activate expression of the HES-1 reporter (Fig. 7). These results suggest that the reduced binding between Notch1-ICN and MAML3 observed in a mammalian two-hybrid assay (Fig. 3) is correlated with a decreased ability to function as a coactivator in this context, even though we were unable to see differential binding in vitro through coimmunoprecipitation experiments (Fig. 2). These results suggested that the activation of ANK-induced HES-1 promoter by three MAML proteins might be related to the differential ability of these MAML proteins to bind to the ankyrin repeats in vivo.
FIG. 7.
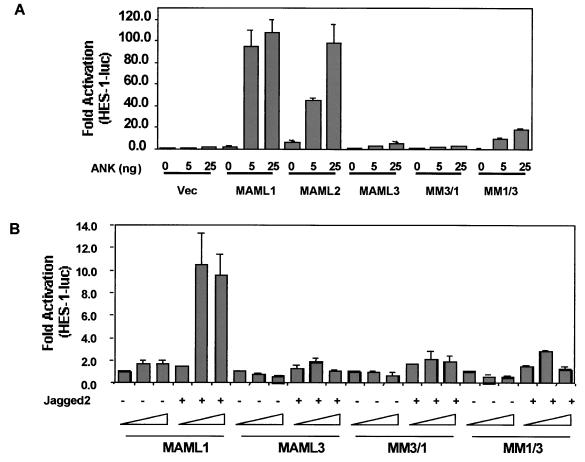
MAML1, MAML2, and MAML3 have differential effects on HES-1 activation induced by the ankyrin repeats of Notch1. U20S cells were transfected with 25 ng of pRL-TK plasmid encoding Renilla luciferase, 0.5 μg of HES-1-luc, and increasing amounts of pFLAG-CMV-2 plasmid encoding MAML1, MAML2, or MAML3 in the presence of 0, 5, or 25 ng of pcDNA3 plasmid encoding the ankyrin repeats (ANK) of human ICN1. Cellular extracts were prepared 44 h posttransfection, and the luciferase activity was measured. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, is expressed as the fold activation relative to cells not expressing MAML and ANK.
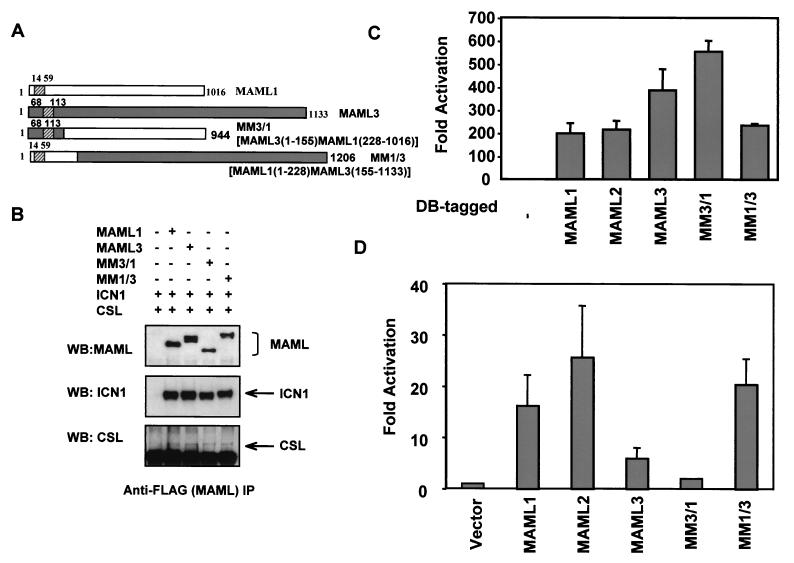
To determine whether the strength of binding between various MAML proteins and Notch might account for differential ability of MAML proteins to activate the HES-1 promoter, we performed domain-swapping experiments between MAML1 and MAML3. Since the basic domains of MAML proteins are responsible for binding to the ankyrin repeats of ICN, we generated two chimeric fusions: one is MM3/1, in which the MAML1 basic domain (aa 1 to 228) was replaced by the corresponding basic domain of MAML 3 (aa 1 to 155), and the other is MM1/3, in which the MAML3 basic domain (aa 1 to 155) was replaced by that of MAML1 (aa 1 to 228) (Fig. 8A). These two chimeric proteins were expressed at the expected sizes (Fig. 8B). Similar to MAML1 and MAML3, MM3/1 and MM1/3 were able to form complexes with ICN1 and CSL by as determined by immunoprecipitation (Fig. 8B) and also retained transcriptional activation domains (Fig. 8C). As described above, although differential ability of MAML proteins to interact with ICN were indistinguishable in immunocoprecipitation assays, differential binding between three MAML proteins and the ankyrin repeats of Notch1 was observed in mammalian two-hybrid assays, with weakest binding seen between MAML3 and Notch1 ANK. We therefore repeated the mammalian two-hybrid assays and found that MM3/1 binding to Notch1-ANK was reduced (compared to MAML1), whereas the binding of MM1/3 to Notch1-ANK was increased compared to MAML3. This result showed that the basic domain of MAML1 is likely to bind better to Notch1-ANK (Fig. 8D) than that the basic domain of MAML3. We then examined the ability of these two chimeric proteins to enhance the ANK-induced and Jagged2-induced HES-1 promoter activation in U20S cells (Fig. 9). MM3/1 became a weak activator similar to MAML3. However, MM1/3 only weakly activated either Jagged2- or ANK-induced HES-1 promoter activity and was not as active in this assay as was MAML1 (Fig. 9A and B). These data suggest that, although MAML1 and MAML3 differ significantly in their ability to bind to the ankyrin repeats of Notch 1, the reduced ability of MAML3 to function as an efficient coactivator for Notch 1 in vivo is not solely due to the differential binding activity and that sequences C-terminal of the basic domain are also important in specifying the function of each MAML.
FIG. 8.
MM3/1 and MM1/3 were able to form complexes with ICN1 and CSL, exhibited transcriptional activities, and showed differential binding determined by the type of the basic domain. (A) Diagram showing two chimeric proteins, MM3/1 and MM1/3, in which MAML1 and MAML3 have their basic domains swapped. (B) COS7 cells were cotransfected with different combinations of three expression plasmids encoding FLAG-tagged MAML1, MAML3, MM3/1, MM1/3, HA-tagged ICN1, and Myc-tagged CSL as indicated. Cellular lysates or anti-FLAG immunoprecipitates (IP) were immunoblotted with anti-FLAG, anti-HA, or anti-Myc antibodies. (C). U20S cells were transfected with 0.5 μg of a firefly luciferase construct containing four GAL4-binding sites in the promoter (pG5luc) and 0.5 μg of pBIND plasmid encoding either the GAL4 DNA- binding domain (DB) only or DB fused to full-length MAML1, MAML2, MAML3, MM3/1, or MM1/3. Firefly luciferase activity, normalized to Renilla luciferase expressed from the pBIND plasmid, was expressed as the fold activation (relative to the background level of firefly luciferase expression in the presence of an empty pBIND vector). (D) U20S cells were transfected with 25 ng of pRL-TK plasmid encoding Renilla luciferase, 0.5 μg of a firefly luciferase construct containing four GAL4-binding sites in the promoter (pG5luc), 0.5 μg of the plasmid encoding DB fused to the ankyrin repeats of Notch1, and 0.5 μg of pFLAG-CMV-2 empty vector or pFLAG-CMV-2 encoding MAML1, MAML2, MAML3, MM3/1, or MM1/3. Cells were harvested at 44 h posttransfection. Firefly luciferase activity, normalized to Renilla luciferase, was expressed as the fold activation (relative to the background level of firefly luciferase expression in the presence of an empty pFLAG-CMV-2 vector).
FIG. 9.
Effect of MM3/1 and MM1/3 on ANK-induced (A) or Jagged2-induced (B) HES-1 promoter activation. (A). U20S cells were transfected with 25 ng of pRL-TK plasmid encoding Renilla luciferase, 0.5 μg of HES-1-luc, and increasing amounts of pFLAG-CMV-2 plasmid encoding MAML1, MAML2, MAML3, MM3/1, or MM1/3 in the presence of 0, 5, or 25 ng of pcDNA3 plasmid encoding the ankyrin repeats (ANK) of human ICN1. Cellular extracts were prepared 44 h posttransfection, and the luciferase activity was measured. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, is expressed as the fold activation relative to cells not expressing MAM and ANK. (B). U20S cells were transfected with 25 ng of a pRL-TK control plasmid expressing Renilla luciferase, 0.5 μg of HES-1-luc, and increasing amounts of expression plasmids encoding either MAML1, MAML3, MM3/1, or MM1/3. At 24 h posttransfection, 105 NIH 3T3 cells expressing Jagged2 or 105 NIH 3T3 cells infected with empty pBABE virus as control were added to each well. Cell extracts were prepared 44 h posttransfection. HES-1 reporter firefly luciferase activity, corrected for Renilla luciferase activity, is expressed as the fold activation relative to cells not expressing MAML family members that were cocultured with control NIH 3T3 cells.
Since different profiles were observed for the ability of MAML proteins to cooperate with all four Notch receptors to activate the HES-1 transcription (Fig. 6). The abilities of MM3/1 and MM1/3 to cooperate with different Notch genes to activate the HES-1 promoter were therefore compared. We found that MM3/1 was a more efficient coactivator for ICN4 than other three forms of ICN (Fig. 9C). No cooperation between MM3/1 and ICN1 was observed. The overall profile for MM3/1 is similar to MAML3, although the levels of activation for ICN4 may be less. MM1/3 showed a profile similar to that for MAML1, although MM1/3 may be a better coactivator for ICN1 than for ICN2, -3, and -4 (compared to MAML1). These results again suggest that the basic domains are important for determining the specificity of MAML to serve as coactivators for various Notch receptors but that other domains of MAML are likely to be important as well. It will be of interest to determine in the future if the MAML proteins differ with respect to the functions of one of the two transcriptional activation domains recently identified by Fryer et al. (11), particularly the N-terminal “TAD1” (aa 75 to 301) of MAML1.
Expression of MAML genes in the developing mouse spinal cord.
The above data suggested that the MAML family serves as transcriptional coactivators for Notch and exhibits differential ability to cooperate with different Notch receptors. Notch signaling induces a broad range of biological responses in mammals, and the effect is mainly manifested at the transcriptional level. Additional diversity of signaling could be obtained through variable expression of Mastermind-like genes in various tissues or at different times during development. In adult tissues, as shown in Fig. 1 and in previous studies (55), the MAML genes appear to be very widely expressed based on Northern blots of various organs. To look into detailed temporal and spatial expression of Maml genes in vivo, we examined Maml expression by in situ hybridization in the spinal cord of developing Swiss-Webster mouse embryos, with the highly homologous human MAML2 and MAML3 cDNAs and mouse Maml1 cDNA as probes.
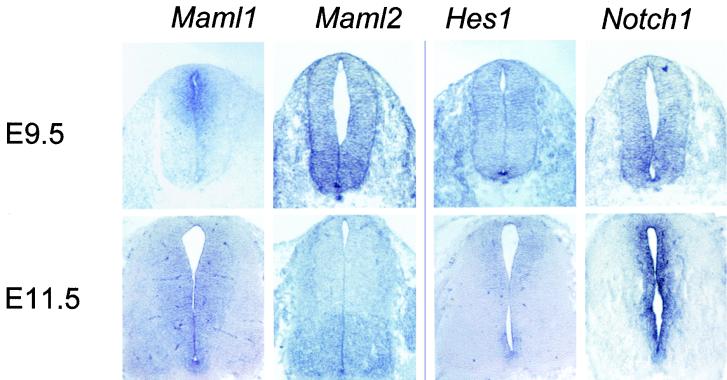
At E9.5 of mouse development, mouse Maml1 is strongly expressed in the dorsal spinal cord (Fig. 10), whereas Maml2 is mostly expressed in the ventral spinal cord. However, no detectable expression for Maml3 was found (not shown). At the same age, Hes1 expression was observed in the dorsal spinal cord and also in the floor plate. In contrast, Notch1 is mostly expressed in the ventral spinal cord. Maml expression was further determined in E11.5 spinal cord, the time when neurons have differentiated. Maml1 is evenly expressed in the ventricular zone (VZ) in the spinal cord at E11.5, whereas Maml2 still displayed a ventral high-expression pattern. Maml3 was again not detectable in the E11.5 spinal cord (not shown). Hes1 is strongly expressed in the dorsal and ventral VZ but weakly expressed in the medial region. Compared to the ventral expression at E9.5, Notch1 is expressed in both the dorsal and ventral VZ at E11.5 (Fig. 10). Taken together, Maml gene expression, like that of Hes1 and Notch1, is dynamic and spatially specific in the early developing spinal cord. The differential expression of Maml genes relative to Notch1 and Hes1 suggests that Maml gene expression provides an additional level of potential signaling diversity in the Notch pathway, at least in the central nervous system. We do not know whether the undetectable expression of Maml3 in the spinal cord is due to use of the human probe for in situ hybridization or to a low level of expression in the spinal cord that is not detectable under these conditions.
FIG. 10.
Maml genes, Hes1 and Notch1 expression in early developing mouse spinal cord. In situ hybridization of cross sections through the cervical spinal cord from E9.5 and E11.5 Swiss Webster mouse embryos was performed with mouse Maml1, human MAML2 and MAML3, and mouse Hes1 and Notch1 mRNA antisense probes and sense probes (not shown). Maml1 and Hes1 are expressed in the dorsal spinal cord at E9.5, whereas Maml2 and Notch1 are expressed in the ventral spinal cord. Hes1 expression also was observed in the floor plate at this age. At E11.5, ventral expression of Maml2 is maintained, whereas Maml1 and Notch1 expression is evenly distributed in the VZ in the spinal cord. At this stage, Hes1 is strongly expressed in both dorsal and ventral VZ but not in the medial region. Maml3 expression was not detected in either E9.5 or E11.5 spinal cords (not shown).
DISCUSSION
Mastermind was identified as one of the original group of “neurogenic” loci in Drosophila, along with Notch, and has since been demonstrated to be a positive regulator of the Notch signaling pathway in a variety of genetic screens (10, 14, 16, 56, 57). Recently, we and others identified a mammalian counterpart of Drosophila Mastermind, MAML1 (26, 41, 55), and showed that it (i) encodes a nuclear protein that binds to the ankyrin repeat domain of Notch1, (ii) forms a multiprotein DNA-binding complex with the intracellular domain of all four Notch receptors and CSL, and (iii) functions as a coactivator to induce transcription of the Notch target gene HES1. Further, we found that truncation mutants of MAML1 lacking either the N-terminal domain (required for Notch binding) or the transcriptional activation domain in the C terminus functioned as dominant-negative inhibitors of Notch-induced transcriptional activation of HES1. Taken together, the genetic studies of Drosophila Mastermind and the biochemical studies of mammalian MAML1 suggested a model wherein Mastermind stabilizes the Notch/CSL complex on the HES1 promoter and activates transcription through an unknown mechanism. In the fly, mastermind appears to be required for Notch signaling, but this has not been established for mammalian cells.
We report here the identification of two more members of the Mastermind-like gene family. NCBI database searching by using the N-terminal basic domain of MAML1 identified two human genes encoding proteins with significant amino acid homology in this region (Fig. 1). MAML2, located on human chromosome 11q22, shares 60% amino acid sequence identity with MAML1 in the basic domain but only 21% identify for the whole sequence. MAML3, on human chromosome 4q28, is 50% identical to MAML1 in the basic region, but again only 33% identical throughout. As determined by Northern blot, all three MAML genes appear to be expressed widely in human adult tissues.
Although the database searches did not reveal the presence of known functional motifs, several interesting structural features of the MAML proteins were noted. We have previously shown that MAML1 may undergo a posttranslational modification, since it migrates at an apparent molecular mass that is ∼20 kDa larger when produced in mammalian cells than when produced by in vitro translation. MAML3 also appears to be expressed as two bands differing by 20 kDa, which again suggests a modification. Preliminary studies indicate that MAML3 is phosphorylated and that treatment with lambda phosphatase collapses the upper band into the lower band (L. Wu and J. Griffin, unpublished results). The sites of phosphorylation, the kinase(s), and any functional significance of this phosphorylation are currently unknown. Recently, Jeffries et al. reported that MAML1 is also phosphorylated, suggesting that phosphorylation may be a general modification of this family (21). Since many proteins that are found in nuclear bodies are sumoylated (54), we previously looked for evidence of sumoylation of MAML1 (Wu and Griffin, unpublished). Western blotting with anti-SUMO-1 was inconclusive, and K→R mutations of two consensus sumoylation sites (LK 217QE and IK 298TE) failed to change the migration of the protein. It will be of interest in the future to identify any modifications of the MAML proteins, particularly if they alter function.
Another the structural feature of interest for the MAML family is the presence of polynucleotide repeats, most often CAG, leading to polyglutamine (polyQ) tracts. The longest polyQ tracts of MAML1 to -3 consist of 4, 31, and 21 glutamines, respectively. Similar polyQ stretches are commonly seen in transcription factors and other nuclear proteins. In some polyQ proteins, expansion of the polyQ tract leads to protein instability and the formation of nuclear inclusions in neurons, leading to neurotoxicity. Although there is no evidence that any of the MAML family undergoes such expansions, we have previously shown that MAML1 associates with ataxin-3, the spinocerebellar ataxia type 3 disease protein, in the nuclear inclusion bodies formed when ataxin-3 has an expanded polyQ tract (7). This observation is consistent with previous studies showing that several proteins with polyQ tracts accumulate in the nuclear inclusion bodies associated with a number of different trinucleotide repeat diseases. Since MAML1 interacts with ataxin-3, it will be of interest to also determine whether MAML2 or -3 also associate with ataxin-3 or other trinucleotide repeat disease proteins.
Each of the MAML proteins localize to nuclear bodies, although some staining is also seen diffusely throughout the nucleus. MAML1 colocalizes but does not coimmunoprecipitate with PML (promyelocytic leukemia) protein in PML bodies (ND10, PML oncogenic domains) (55). MAML2 and -3 colocalize with MAML1, suggesting that they also are found predominantly in PML bodies, although this was not directly tested in these studies. However, MAML1 also was detected in a small number of nuclear bodies that lacked costaining for PML, suggesting that there is incomplete overlap of the subcellular localization of these proteins. The functions of PML bodies and other nuclear bodies remain largely unknown (33, 58). PML bodies are characterized by the accumulation of a number of transcriptional regulators in addition to PML, including p53, Sp100, SUMO-1, HAUSP(USP7), CBP, and BLM. PML bodies have been further implicated in aspects of transcriptional regulation, as storage depots, or as sites of protein catabolism. Recently, kinases localized to PML bodies have been identified, such as a nuclear body p53 kinase and homeodomain-interacting protein kinase-2. It will be of interest to determine whether the phosphorylation of MAML3, discussed above, might also occur within nuclear bodies or perhaps regulate the traffic of MAML3 into nuclear bodies. At this time, it is not known if the fraction of MAML proteins within PML bodies is “active,” i.e., involved in interactions with Notch and CSL. We note, however, that overexpression of each of the MAML proteins causes intracellular Notch1 and CSL to relocalize into nuclear bodies but that accumulation of CSL in these structures requires the simultaneous expression of both ICN1 and MAML, suggesting that it is the whole complex that accumulates in nuclear bodies and not each protein individually. Recently, Jeffries et al. showed that in ICN1-transformed RKE cells, ICN1 is detected in two different, high-molecular-weight complexes in the nucleus (21). The largest complex contains ICN1, CSL, and MAML1, and evidence was presented that MAML1 tethers ICN1 to CSL, without directly binding to CSL itself. Thus, additional proteins may be involved in this interaction, or it is possible that an ICN1/MAML1 complex has a higher affinity for CSL than does ICN1 alone. Interestingly, ICN1-ΔRAM (ICN1 lacking the binding site for CSL) could still form a complex with CSL in the nucleus in the presence, but not in the absence, of MAML1. Thus, ICN1-ΔRAM may signal in a CSL-dependent manner in the presence of MAML1, even though it fails to coprecipitate with CSL. These studies were recently complemented by Fryer et al., who demonstrated that chromatin-dependent transactivation of a synthetic promoter containing multiple CSL binding sites upstream of a TATA-containing minimal core promoter element required addition of MAML1 to ICN1 and CSL. A proximal TAD1 (aa 75 to 301) was identified as a likely high-affinity binding site for p300/CBP that was necessary for in vitro chromatin-dependent transcription. The previously identified C-terminal TAD (“TAD2”) (55) was not needed for in vitro transcription but was confirmed to be essential for in vivo function as a coactivator. Further studies will be necessary to determine whether MAML2 and MAML3 also function to recruit p300/CBP and thereby direct histone acetylation.
Despite the fairly limited amino acid homology between MAML1, -2, and -3, each was found in these studies (i) to have a C-terminal transcriptional activation domain, (ii) to bind Notch1, -2, -3, and -4, and (iii) to form a multiprotein complex with CSL by coimmunoprecipitation, provided an ICN protein was present. However, at a functional level, MAML3 could be readily distinguished from MAML1 and MAML2. In U20S cells, both MAML1 and MAML2 functioned as effective coactivators for ICN1 to -4 in the transactivation of a HES1 promoter construct. MAML3, however, functioned well with ICN4 but minimally with ICN1 to -3. Differences were also observed between MAML3 and MAML1 (or MAML2) when U20S cells were stimulated with either Jagged2 or Delta1 ligands to activate endogenous Notch receptors. Although MAML1 and MAML2 functioned as coactivators, MAML3 had minimal effects which could be due to a failure of the Jagged2 or Delta1 ligands used here to activate Notch4.
The ankyrin repeats of Notch receptors are essential for all known Notch functions (1). One function for the ankyrin repeat domain in the activation of transcription is the recruitment of coactivators, since certain mutations in the ankyrin repeat domain (ANK) that produce loss-of-function phenotypes abolish transcriptional activation (43). We have previously shown that MAML1 interacts with the ankyrin repeats of Notch1; therefore, we analyzed possible quantitative differences in binding between the ankyrin repeats of Notch1 and MAML proteins. We found that the interaction of MAML3 with the ankyrin repeats of Notch1 was much weaker than that with MAML1 and MAML2 as determined by mammalian two-hybrid assays (Fig. 3). Consistent with these data, we found that MAML3 was less efficient as an coactivator of ANK-induced HES-1 transcription (Fig. 7). Therefore, these MAML proteins have differential ability to interact with ANK of Notch1, and this might at least partially contribute to the differential activity of HES-1 activation that we observed. To gain insight into the mechanism of specificity of these MAML proteins and Notch receptors, we created two domain-swap fusion proteins, MM3/1 and MM1/3, in which the basic domains of MAML1 and MAML3 were exchanged. MM3/1 (MAML1 with the basic domain replaced by that of MAML3) showed weak binding to ANK and an inability to activate ANK-induced and Jagged2-induced HES-1 transcription in U20S cells, findings similar to the properties of MAML3 (Fig. 8D and 9A and B). MM1/3 (MAML3 with the basic domain replaced by that of MAML1) showed strong binding to ANK similar to MAML1, However, MM1/3 only weakly activated ANK-induced or Jagged2-induced HES-1 transcription (Fig. 8D and 9A and B). The overall profiles of MM3/1 and MM1/3 as coactivators for different Notch receptors were found to be similar to those of MAML3 and MAML1 to certain extent, respectively, although MM3/1 and MM1/3 could not restore the levels of activities achieved by MAML1 and MAML3. These results suggest that the basic domain is important for binding of MAML to ICN but are not sufficient to determine the ability of MAML to activate HES-1 promoter and that other domains of MAML proteins might also be crucial. Recently, Fryer et al. showed that a central MAML1 activation domain (TAD1) recruits CBP/p300 to promote nucleosome acetylation by the Notch transcriptional complex and activates transcription in vitro, and another C-terminal activation domain (TAD2) enhances phosphorylation of CBP/p300 and ICN and is required for Notch induced transcription in vivo (11). The three MAML sequences are relatively divergent outside of the basic domain regions, and it is possible that the TAD1 and TAD2 in MAML2 and MAML3 could be different from MAML1 in terms of functions and interactions with other factors, including CBP/p300. Since the locations of the TADs have not been mapped in the MAML2 and MAML3, it is possible that the fusion proteins described here, MM1/3 and MM3/1, could have defects in one or both of the TADs, and further studies will be important to compare p300 binding and other functions among the three coactivators. However, we demonstrate that one feature that distinguishes MAML3 from MAML1 is the ability to interact effectively with the ankyrin repeats of Notch1, and we propose that this contributes to the reduced activity of MAML3 as a coactivator for ICN1.
Overall, the fact that the biological functions of the MAML proteins in the tissue culture assays are different suggests that their in vivo functions may not be redundant. Further, it suggests that the complement of MAML proteins expressed by an individual cell could modify the outcome of Notch signaling, and thereby contribute to biological diversity.
To further explore this possibility, we looked more carefully at expression of mouse Maml genes by using in situ hybridization. The results indicate that there are a number of spatial and temporal differences between expression of Maml1 and Maml2 in the developing mouse spinal cord. For example, although Maml1 is expressed highly in the dorsal spinal cord of the embryo, Maml2 is expressed more highly in the ventral spinal cord. Notch1 is expressed in both locations, and Hes1 is expressed primarily in the dorsal spinal cord. Unfortunately, we could not visualize Maml3 in the nervous system by in situ hybridization, so it is not possible to comment at this point on whether Maml3 also shows significant variability of expression at a suborgan level. The in situ results with Maml1 and Maml2, however, coupled with the finding that MAML3 is biochemically distinct from MAML1 and MAML2, further supports the hypothesis that the three MAML proteins have nonoverlapping functions in vivo.
Taken together, our studies identified a family of Mastermind-like coactivators for Notch signaling. The three members—MAML1, MAML2, and MAML3—exhibit differential functional activities and expression patterns. It is highly likely that different functions and differential expression of these MAML genes will increase the potential diversity of signals from individual Notch receptors and ligands in distinct cell types.
Acknowledgments
This work was supported in part by NIH RO1CA36167 (J.D.G.) and a Scholar Award of the American Society of Hematology (L.W.).
We thank David H. Rowitch for helping with in situ hybridization experiments and Jingxuan Liu for technical assistance.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Aster, J., W. Pear, R. Hasserjian, H. Erba, F. Davi, B. Luo, M. Scott, D. Baltimore, and J. Sklar. 1994. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp. Quant. Biol. 59:125-136. [DOI] [PubMed] [Google Scholar]
- 3.Beatus, P., and U. Lendahl. 1998. Notch and neurogenesis. J. Neurosci. Res. 54:125-136. [DOI] [PubMed] [Google Scholar]
- 4.Berset, T., E. F. Hoier, G. Battu, S. Canevascini, and A. Hajnal. 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during Caenorhabditis elegans vulval development. Science 291:1055-1058. [DOI] [PubMed] [Google Scholar]
- 5.Bettler, D., S. Pearson, and B. Yedvobnick. 1996. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics 143:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruckner, K., L. Perez, H. Clausen, and S. Cohen. 2000. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406:411-415. [DOI] [PubMed] [Google Scholar]
- 7.Chai, Y., L. Wu, J. D. Griffin, and H. L. Paulson. 2001. The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J. Biol. Chem. 276:44889-44897. [DOI] [PubMed] [Google Scholar]
- 8.Davis, R. L., and D. L. Turner. 2001. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20:8342-8357. [DOI] [PubMed] [Google Scholar]
- 9.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 10.Fortini, M. E., and S. Artavanis-Tsakonas. 1994. The suppressor of hairless protein participates in notch receptor signaling. Cell 79:273-282. [DOI] [PubMed] [Google Scholar]
- 11.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallahan, D., C. Jhappan, G. Robinson, L. Hennighausen, R. Sharp, E. Kordon, R. Callahan, G. Merlino, and G. H. Smith. 1996. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56:1775-1785. [PubMed] [Google Scholar]
- 13.Giraldez, A. J., L. Perez, and S. M. Cohen. 2002. A naturally occurring alternative product of the mastermind locus that represses notch signalling. Mech. Dev. 115:101-105. [DOI] [PubMed] [Google Scholar]
- 14.Go, M. J., and S. Artavanis-Tsakonas. 1998. A genetic screen for novel components of the notch signaling pathway during Drosophila bristle development. Genetics 150:211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, M., L. Y. Jan, and Y. N. Jan. 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17:27-41. [DOI] [PubMed] [Google Scholar]
- 16.Helms, W., H. Lee, M. Ammerman, A. L. Parks, M. A. T. Muskavitch, and B. Yedvobnick. 1999. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev. Biol. 215:358-374. [DOI] [PubMed] [Google Scholar]
- 17.Hofelmayr, H., L. J. Strobl, C. Stein, G. Laux, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 1999. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J. Virol. 73:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iso, T., V. Sartorelli, G. Chung, T. Shichinohe, L. Kedes, and Y. Hamamori. 2001. Herp, a new primary target of notch regulated by ligand binding. Mol. Cell. Biol. 21:6071-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 21.Jeffries, S., D. J. Robbins, and A. J. Capobianco. 2002. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol. Cell. Biol. 22:3927-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joutel, A., C. Corpechot, A. Ducros, K. Vahedi, H. Chabriat, P. Mouton, S. Alamowitch, V. Domenga, M. Cecillion, E. Marechal, J. Maciazek, C. Vayssiere, C. Cruaud, E. A. Cabanis, M. M. Ruchoux, J. Weissenbach, J. F. Bach, M. G. Bousser, and E. Tournier-Lasserve. 1996. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383:707-710. [DOI] [PubMed] [Google Scholar]
- 23.Joutel, A., C. Corpechot, A. Ducros, K. Vahedi, H. Chabriat, P. Mouton, S. Alamowitch, V. Domenga, M. Cecillion, E. Marechal, J. Maciazek, C. Vayssiere, C. Cruaud, E. A. Cabanis, M. M. Ruchoux, J. Weissenbach, J. F. Bach, M. G. Bousser, and E. Tournier-Lasserve. 1997. Notch3 mutations in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), a mendelian condition causing stroke and vascular dementia. Ann. N. Y. Acad. Sci. 826:213-217. [DOI] [PubMed] [Google Scholar]
- 24.Kageyama, R., and T. Ohtsuka. 1999. The Notch-Hes pathway in mammalian neural development. Cell Res. 9:179-188. [DOI] [PubMed] [Google Scholar]
- 25.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitagawa, M., T. Oyama, T. Kawashima, B. Yedvobnick, A. Kumar, K. Matsuno, and K. Harigaya. 2001. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 21:4337-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojika, S., and J. D. Griffin. 2001. Notch receptors and hematopoiesis. Exp. Hematol. 29:1041-1052. [DOI] [PubMed] [Google Scholar]
- 28.Krebs, L. T., Y. Xue, C. R. Norton, J. R. Shutter, M. Maguire, J. P. Sundberg, D. Gallahan, V. Closson, J. Kitajewski, R. Callahan, G. H. Smith, K. L. Stark, and T. Gridley. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343-1352. [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda, K., S. Tani, K. Tamura, S. Minoguchi, H. Kurooka, and T. Honjo. 1999. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274:7238-7244. [DOI] [PubMed] [Google Scholar]
- 30.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275:17211-17220. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., I. D. Krantz, Y. Deng, A. Genin, A. B. Banta, C. C. Collins, M. Qi, B. J. Trask, W. L. Kuo, J. Cochran, T. Costa, M. E. Pierpont, E. B. Rand, D. A. Piccoli, L. Hood, and N. B. Spinner. 1997. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16:243-251. [DOI] [PubMed] [Google Scholar]
- 32.Lowell, S., P. Jones, I. Le Roux, J. Dunne, and F. M. Watt. 2000. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10:491-500. [DOI] [PubMed] [Google Scholar]
- 33.Matera, A. G. 1999. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9:302-309. [DOI] [PubMed] [Google Scholar]
- 34.Miele, L., and B. Osborne. 1999. Arbiter of differentiation and death: Notch signaling meets apoptosis. J. Cell Physiol. 181:393-409. [DOI] [PubMed] [Google Scholar]
- 35.Morimura, T., R. Goitsuka, Y. Zhang, I. Saito, M. Reth, and D. Kitamura. 2000. Cell cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 275:36523-36531. [DOI] [PubMed] [Google Scholar]
- 36.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 37.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21:7761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panin, V. M., and K. D. Irvine. 1998. Modulators of Notch signaling. Semin. Cell Dev. Biol. 9:6-17. [DOI] [PubMed] [Google Scholar]
- 39.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T-cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petcherski, A., and J. Kimble. 2000. LAG-3 is a putative trancriptional activator in the Caenorhabditis elegans Notch pathway. Nature 405:364-368. [DOI] [PubMed] [Google Scholar]
- 41.Petcherski, A. G., and J. Kimble. 2000. Mastermind is a putative activator for Notch. Curr. Biol. 10:R471-R473. [DOI] [PubMed] [Google Scholar]
- 42.Rangarajan, A., C. Talora, R. Okuyama, M. Nicolas, C. Mammucari, H. Oh, J. C. Aster, S. Krishna, D. Metzger, P. Chambon, L. Miele, M. Aguet, F. Radtke, and G. P. Dotto. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roehl, H., M. Bosenberg, R. Blelloch, and J. Kimble. 1996. Roles of the RAM and ANK domains in signaling by the Caenorhabditis elegans GLP-1 receptor. EMBO J. 15:7002-7012. [PMC free article] [PubMed] [Google Scholar]
- 44.Ronchini, C., and A. J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell. Biol. 21:5925-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder, T., and U. Just. 2000. mNotch1 signaling reduces proliferation of myeloid progenitor cells by altering cell-cycle kinetics. Exp. Hematol. 28:1206-1213. [DOI] [PubMed] [Google Scholar]
- 46.Shawber, C., D. Nofziger, J. J. Hsieh, C. Lindsell, O. Bogler, D. Hayward, and G. Weinmaster. 1996. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122:3765-3773. [DOI] [PubMed] [Google Scholar]
- 47.Smoller, D., C. Friedel, A. Schmid, D. Bettler, L. Lam, and B. Yedvobnick. 1990. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 4:1688-1700. [DOI] [PubMed] [Google Scholar]
- 48.Strobl, L. J., H. Hofelmayr, C. Stein, G. Marschall, M. Brielmeier, G. Laux, G. W. Bornkamm, and U. Zimber-Strobl. 1997. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-Jκ. Immunobiology 198:299-306. [DOI] [PubMed] [Google Scholar]
- 49.Taniguchi, Y., T. Furukawa, T. Tun, H. Han, and T. Honjo. 1998. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 18:644-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varnum-Finney, B., L. Wu, M. Yu, C. Brashem-Stein, S. Staats, D. Flowers, J. D. Griffin, and I. D. Bernstein. 2000. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci. 113(Pt. 23):4313-4318. [DOI] [PubMed] [Google Scholar]
- 51.Wang, S., and B. A. Barres. 2000. Up a notch: instructing gliogenesis. Neuron 27:197-200. [DOI] [PubMed] [Google Scholar]
- 52.Weinmaster, G. 2000. Notch signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10:363-369. [DOI] [PubMed] [Google Scholar]
- 53.Weinmaster, G. 1998. Notch signaling: direct or what? Curr. Opin. Genet. Dev. 8:436-442. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, V. G., and D. Rangasamy. 2001. Intracellular targeting of proteins by sumoylation. Exp. Cell Res. 271:57-65. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26:484-489. [DOI] [PubMed] [Google Scholar]
- 56.Xu, T., and S. Artavanis-Tsakonas. 1990. Deltex, a locus interacting with the neurogenic genes, Notch, Delta, and mastermind in Drosophila melanogaster. Genetics 126:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, T., I. Rebay, R. J. Fleming, T. N. Scottgale, and S. Artavanis-Tsakonas. 1990. The Notch locus and the genetic circuitry involved in early Drosophila neurogenesis. Genes Dev. 4:464-475. [DOI] [PubMed] [Google Scholar]
- 58.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]