Abstract
We developed a multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB) to detect the genes encoding members of the family of variable surface-localized proteins of Streptococcus agalactiae (group B streptococcus [GBS]), namely, Bca (Cα), Rib, Epsilon (Epsilon/Alp1/Alp5), Alp2, Alp3, and Alp4, and the immunoglobulin A binding protein, Bac (Cβ). We used the assay to identify these genes in a collection of well-characterized GBS isolates and reference strains. The results showed that mPCR/RLB avoids the common problems of cross-reaction and nontypability associated with protein typing using antisera. It is as sensitive as, but more practical than, separate gene-specific PCRs and would be suitable for large molecular epidemiological studies of GBS.
Group B streptococcus (GBS) (Streptococcus agalactiae) is an important cause of sepsis, especially in neonates (19). In addition to capsular polysaccharide antigens, some S. agalactiae surface proteins are potential components of a protein-based GBS vaccine (16), and the use of some as carriers in polysaccharide conjugate vaccines is under investigation (5, 17). Identification of surface-localized protein antigens can facilitate studies of the epidemiology and pathogenesis of GBS infection (14).
The genes encoding the proteins Cα (bca), Alp2 and Alp3 (Cα-like 2 and 3) (alp2 and alp3), Epsilon (epsilon [alp1/alp5]), and Rib (rib) have been well studied, and their gene sequences are published in GenBank (2, 16, 18, 23). They belong to a family of surface-localized proteins containing internal tandem repeats, variation in the numbers of which causes variation in protein size and antigenicity (16, 23). The gene encoding the immunoglobulin A (IgA) binding protein, Cβ (bac), also has been well described (6, 7). In a previous study, we developed individual gene-specific PCRs for bca, rib, alp2, alp3, and bac (12), and Creti et al. have recently described a multiplex PCR method for bca, rib, alp2/alp3, alp4, and epsilon (2). In this study, we developed a multiplex PCR (mPCR)-based reverse line blot (RLB) hybridization assay and used it to identify seven protein antigen genes in several sets of well-characterized GBS isolates.
MATERIALS AND METHODS
GBS isolates and serotyping.
The GBS isolates used in this study were the following: 27 reference strains used in our previous studies (Table 1); 83 isolates provided by Nicola Jones, Oxford, United Kingdom, which had been characterized by multilocus sequence typing (MLST) (8); 58 isolates provided by Dele Davies and Shannon Manning, Michigan State University, East Lansing, MI, which also had been characterized by MLST (3); and 50 isolates provided by Catherine Lachenauer, Channing Laboratory, that previously had been used in a serological study of protein surface antigens (15).
TABLE 1.
Conventional serotyping, molecular serotype identification based on PCR/sequencing, and mPCR/RLB hybridization protein gene profile for 27 GBS reference strainsa
| Strain no.b | Strain identification | CS | MS | mPCR/RLB pgp |
|---|---|---|---|---|
| 1 | O90 (Rockefeller University)c | Ia | Ia | Epsilon |
| 2 | H36B (ATCC 12401; NCTC 8187)c | Ib | Ib | AB |
| 3 | 18 RS 21 (NCTC 11079)c | II | II | R |
| 4 | M 781 (ATCC BAA-22)c | III | III | R |
| 5 | 3139c | IV | IV | Epsilon |
| 6 | CJB 111 (ATCC BAA-23)c | V | V | Alp3 |
| 7 | SS 1214c | VI | VI | Epsilon |
| 8 | 7271c | VII | VII | Alp3 |
| 9 | JM9 130013c | VIII | VIII | Alp3 |
| 10 | A909 (NCTC 11078)c | Ia | Ia | AB |
| 11 | BM110c | III | III | R |
| 12 | NZRM 908 (NCDC SS 615)c | Ia | Ia | Alp2 |
| 13 | NZRM 909 (NCDC SS 618)c | Ib | Ib | AB |
| 14 | NZRM 910 (NCDC SS 700)c | Ia | Ia | AB |
| 15 | NZRM 911 (NCDC SS 619)c | II | II | R |
| 16 | NZRM 912 (NCDC SS 620)c | III | III | Alp2 |
| 17 | NZRM 2832 (Prague 1/82; ATCC 49446)c | IV | IV | Epsilon |
| 18 | NZRM 2833 (Prague 10/84; ATCC 49447)c | V | V | Alp3 |
| 19 | NZRM 2834 (Prague 118754)c | VI | VI | AB |
| 20 | NZRM 2217 (Prague 25/60; ATCC 49448)d,e | NT | II | Alp4 |
| 21 | O90 (ATCC 12400)f | Ia | Ia | Alp2 |
| 22 | A909 (NCTC 11078)f | Ia | Ia | AB |
| 23 | 335 (NCTC 12906)f | Ia | Ia | Epsilon |
| 24 | 70339 (NCTC 12907)f | Ia | Ia | ARB |
| 25 | 65604 (3)f | III | III | R |
| 26 | 15626/81 (3)f | IV | IV | AB |
| 27 | Pattison (NCTC 9828)f | NT | II | Alp4 |
Abbreviations: CS, conventional serotype; MS, molecular serotype (serotype-specific PCR and/or partial cps sequencing; mPCR/RLB, reverse line blot assay, following multiplex PCR (all results were identical between individual PCR and mPCR/RLB); NT, nonserotypeable; A, Bca (Cα), R, Rib; B, Bac (Cβ); Epsilon, Epsilon/Alp1/Alp5; pgp, protein gene profile.
Strain numbers correspond to lane numbers in Fig. 1.
Eleven GBS reference strains from Lawrence Paoletti and Catherine Lachenauer, Channing Laboratory, Boston, Mass.
Nine GBS reference strains from Diana Martin, Streptococcus Reference Laboratory, at the Institute of Environmental Science and Research Limited, Wellington, New Zealand.
This isolate was the atypical reference strain Prague 25/60, which was nonserotypeable using capsular polysaccharide antisera but was assigned to molecular serotype II. It expresses a protein that is antigenically similar to R (and is used to generate R antiserum), encoded by the gene alp4 (12).
Seven GBS reference strains from Johan Maeland, Department of Microbiology, School of Medicine, Norwegian University of Science and Technology, Trondheim, Norway.
Results of capsular polysaccharide serotyping were provided by the donor laboratories (1, 13).
DNA preparations.
DNA was prepared from GBS cultures as described previously (13).
Molecular serotype/serosubtype and protein gene identification.
Capsular polysaccharide (cps) molecular serotype/serosubtype identification, based on PCR/sequencing of variable regions of the cps gene cluster, and protein gene profile, based on single target protein gene PCR, were performed on all isolates used in this study, as described previously (1, 11, 13).
Oligonucleotide design.
We designed two specific PCR primers and two specific probes for each of the seven GBS protein genes, using published GenBank sequences. We also designed one GBS-specific primer pair and two GBS-specific probes (9) (Table 2).
TABLE 2.
Oligonucleotide primers used in this study
| Primera | Target | Tm (°C)b | GenBank accession no. | Sequencec |
|---|---|---|---|---|
| cfbSb | cfb | 59.93 | X72754 | 326 ATG ATG TAT CTA TCT GGA ACT CTA GTG 352 |
| Sag59PSd | cfb | 58.55 | X72754 | 368 TTT TCA CCA GCT GTA TTA GAA GTA 391 |
| Sag190PAd | cfb | 59.74 | X72754 | 522 GTT CCC TGA ACA TTA TCT TTG AT 500 |
| cfbAb | cfb | 59.74 | X72754 | 585 CGC AAT GAA GTC TTT AAT TTT TC 563 |
| bcaSb | bca | 60.45 | M97256 | 313 GCT TAC ATA GAT TTA TAT GAT GTA AAA TTA GG 344 |
| bcaSp | bca | 57.62 | M97256 | 370 GTT TTA GAA CAA GGT TTT ACA GC 392 |
| bcaAp | bca | 58.60 | M97256 | 579 CT TAT CCC TCA AGG TTG TTG 560 |
| bcaAb | bca | 58.91 | M97256 | 637 CAG TAC GAC TTT CTT CCG TC 618 |
| alp23Sb | alp2/alp3 | 59.89 | AF208158 | 1254 CAT GGA AGT GAC AAT TAT GAA AG 1276 |
| alp2Sp | alp2 | 59.24 | AF208158 | 1366 CTT CCG CCA GAT AAA ATT AAG 1386 |
| alp2Ap | alp2 | 58.25 | AF208158 | 1576 CTG TTG ACT TAT CTG GAT AGG TC 1554 |
| alp2Ab | alp2 | 60.81 | AF208158 | 1603 CCA CTG TAA CTT TTA CAG GAA CTT C 1579 |
| Alp23Sb | alp2/alp3 | 59.89 | AF291065 | 1534 CAT GGA AGT GAC AAT TAT GAA AG 1556 |
| alp3Sp | alp3 | 57.11 | AF291065 | 1643 GTT CTT CCG CTT AAG GAT AG 1662 |
| alp3Ap | alp3 | 60.17 | AF291065 | 1711 CGG TGT TTC TCC TAC TTT GAC 1691 |
| alp3Ab | alp3/rib | 59.99 | AF291065 | 1847 CTT TTG AAC CAT CTG GGT AAG 1827 |
| ribSb | rib | 63.88 | U58333 | 219 AGA TAC TGT GTT TGC AGC TGA AGT AA 244 |
| ribSp | rib | 59.07 | U58333 | 256 G CTG TTA CGT TAA ACA CAA ATA TG 279 |
| ribAp | rib | 57.92 | U58333 | 448 C AAC AGT AGT CAA TTC AGA AGG 427 |
| ribAb | rib | 57.16 | U58333 | 577 CTA TTT TAT CTC TCA AAG CTGAAG 554 |
| alp4Sb | alp4 | 61.47 | AJ488912 | 158 TGT TAG CAG CTG AAG TAG TTG AAG 181 |
| alp4Sp | alp4 | 63.70 | AJ488912 | 187 GCT GCA ACA TTA AAT ACT GCC AT 209 |
| alp4Ap | alp4 | 60.62 | AJ488912 | 325 TGT AAT AAA TAG CAG TGT ATC CCG 302 |
| alp4Ab | alp4 | 60.05 | AJ488912 | 350 GCA TAA ACT TTT GAA CCT TGT G 329 |
| epsilonSb | epsilon | 64.75 | U33554 | 304 CTG TGT TTG CAG CTG AGG TG 323 |
| epsilonSp | epsilon | 58.30 | U33554 | 452 CCC TTC TAA TTA TTC AGC AAA C 473 |
| epsilonAp | epsilon | 60.91 | U33554 | 685 GGA TCA TTT GCA TTT TCA ATT AC 663 |
| epsilonAb | epsilon | 62.52 | U33554 | 719 CA GTA CAT CTT TTC GAC TAT CAT CG 695 |
| GBS1360Sb | bac | 65.82 | X59771 | 1337 AAG GCT ATG AGT GAG AGC TTG GAG 1360 |
| GBS1716Sp | bac | 58.32 | X59771 | 1697 AAA AGT GAT TCG AAG ACG AC 1716 |
| GBS1716Ap | bac | 59.28 | X59771 | 1735 AAG ATC ACT GAA GTC CAA CG 1716 |
| GBS1937Ab | bac | 64.80 | X59771 | 1960 CTG CTC TGG TGT TTT AGG AAC TTG 1937 |
Suffixes: “b” indicates primers 5′ labeled with biotin; “p” indicates probes 5′ labeled with amine.
The primer Tm values are those provided by the primer synthesizer (Sigma-Aldrich).
Numbers represent the numbered base positions at which primer sequences start and finish (numbering start points 1 refer to the start point 1 of the corresponding gene GenBank accession number).
The species-specific primers have been previously published (9). All other primers were designed specifically for this study.
Primer design.
The primers were designed to have similar physical characteristics in order to allow simultaneous amplification in a multiplex reaction. Their lengths were between 20 and 32 bp, melting temperatures between 57.2°C and 65.8°C, and amplicon sizes in the range of 193 to 624 bp (Table 2). The specificities of primer sequences were determined by comparison with GenBank, using FASTA, to ensure that there would be minimal risk of cross-hybridization with other regions of the GBS genome or unrelated genes. All multiplex PCR primers were 5′ biotinylated to allow detection of hybridization with a streptavidin peroxidase substrate.
Probe design.
To allow optimal hybridization under the common conditions, probes were also designed to have similar physical characteristics: lengths between 20 and 24 bp and melting temperatures between 57.1°C and 63.7°C (Table 2). FASTA searches were performed to compare their sequences with those in GenBank. All probes have a 5′ amine group to facilitate covalent linkage to the nylon membrane and allow membranes to be stripped and reused repeatedly.
Multiplex PCR system.
The 25-μl eight-primer-pair mPCR mixture was prepared as follows: 5 μl template DNA, 0.25 μl each of all forward (50 pmol/μl) and reverse (50 pmol/μl) primers, 1.25 μl deoxynucleoside triphosphates (2.5 mM of each deoxynucleoside triphosphate), 2.5 μl 10× PCR buffer, 3 μl 25 mM (4.5 mM final) MgCl2, 0.1 μl QIAGEN hotstart Taq polymerase (5 units/μl); also, water was added to 25 μl. The PCR program was performed according to the QIAGEN Hotstart Taq polymerase kit instructions: 95°C for 15 min, 1 cycle, 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, 35 cycles, 72°C for 10 min, 1 cycle, and 22°C hold. A total of 8 μl of each PCR product was separated by electrophoresis on a 1.5% agarose gel to confirm successful amplification (13). The remaining PCR products were used for RLB hybridization (5 μl of PCR product for each).
RLB hybridization.
The RLB hybridization assay was based on the method described previously (21, 22), except that the hybridization temperature was 60°C, the conjugate used was streptavidin-peroxidase (Roche Diagnostics Co.) diluted 1:4,000 in 2× SSPE (SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.5% sodium dodecyl sulfate, and the time of exposure to X-ray film (Hyperfilm; Amersham) was 7 min. RLB results were regarded as positive when both probes for a particular gene gave positive results. To optimize hybridization conditions, the probes were tested at several twofold dilutions, starting at a concentration of 0.6 pM, and the final labeling concentration was generally 0.2 to 0.4 pM. In our laboratory, membranes labeled with probes have been stripped and reused at least 20 times without any significant loss of signal (data not shown).
The sensitivity and specificity of mPCR/RLB were compared with those of single-protein-gene-specific PCR (1, 2, 12).
Database similarity search and sequence comparison.
Databases were searched for sequence similarity using the FASTA program in the SeqSearch program group. Sequences were compared using the BESTFIT and GAP programs in the Comparison program group. All programs are provided in WebANGIS, Australian National Genomic Information Service, third version.
RESULTS
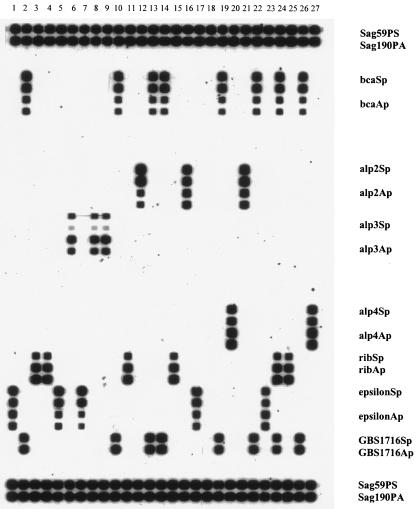
The RLB results for the 27 reference strains are shown in Fig. 1. Multiplex PCR products from all isolates hybridized with the control GBS-specific probes and, with one exception, with one or more sets of protein gene-specific probes. The exception was strain no. 18, which failed to hybridize with any protein gene probes on the membrane shown in Fig. 1. However, on repeat testing, it hybridized with both alp3 probes. Strain no. 24 was the only exception to the rule that each strain hybridized with only one of the six sets of probes representing the variable surface-localized protein family (represented by Cα, R, and Alp, etc.); it hybridized with both bca (Cα) and rib (Rib). All eight isolates that hybridized with Cα gene (bca) probes also hybridized with Cβ gene (bac) probes.
FIG. 1.
Results of mPCR/RLB hybridization for 27 reference strains. Lanes 1 to 27 represent the 27 reference strains, in the same order as in Table 1; the 20 labeled probes are in the same order as in Table 2. Duplicate rows for the same probe represent different probe concentrations (0.2 or 0.4 pM, respectively). Probe targets in rows from top to bottom are as follows: rows 1 and 2, GBS species specific; 3 and 4, blank; 5 to 8, Cα or Bca protein gene (protein gene profile code A [see Table 1, footnote a]); 9 to 12, blank; 13 to 16, alp2; 17 to 20, alp3; 21 to 24, blank; 25 to 28, alp4; 29 to 31, rib (protein gene profile code R); 32 to 35, epsilon/alp1/alp5; 36 and 37, Cβ or Bac protein gene (protein gene profile code B); 38 and 39, blank; 40 and 41, GBS species-specific gene. PCR product of strain no. 18 hybridized with GBS-specific probes but with none of the protein gene probes. However, on retesting, its PCR product hybridized with alp3 probes.
Comparison of mPCR/RLB and single-gene-specific PCR.
To confirm the sensitivity and specificity, we compared the results of mPCR/RLB with those of single-gene-specific PCR. For all 27 reference strains (Table 1) and 191 isolates studied (Table 3), the results of mPCR/RLB were the same as those of single-gene-specific PCR.
TABLE 3.
The relationship between protein gene profiles and capsular polysaccharide synthesis gene MS for 191 GBS isolates
| MS (na) | No. of isolates with protein antigen gene profileb
|
||||||
|---|---|---|---|---|---|---|---|
| A | AB | R | Epsilon | Alp2 | Alp3 | Alp4 | |
| Ia (18) | 1 | 17 | |||||
| Ib (16) | 1 | 15 | |||||
| II (7) | 2 | 1 | 3 | 1 | |||
| III (96) | 6 | 85 | 4 | 1 | |||
| IV (1) | 1 | ||||||
| V (13) | 3 | 10 | |||||
| VI (20) | 19 | 1 | |||||
| VII (2) | 2 | ||||||
| VIII (18) | 18 | ||||||
| Total (191) | 29 | 16 | 88 | 23 | 4 | 31 | |
n, number of isolates.
Protein antigen gene profile codes are the following: A, bca positive; B, bac positive; R, rib positive; Alp2, alp2 positive; Alp3, alp3 positive; Alp4, alp4 positive; Epsilon, epsilon (= epsilon/alp1/alp5) positive.
Comparison of mPCR/RLB with conventional protein serotyping.
Of the 50 isolates for which protein subtyping had been done previously, using antisera, 48 had reacted with two or more antisera (15), indicating a high degree of cross-reactivity. By contrast, mPCR/RLB showed complete specificity, with no cross-reactions between different gene probes (Table 3). However, one isolate (no. 24) contained two different protein genes (in addition to bac).
DISCUSSION
In this study, we developed an mPCR/RLB method to identify 6 GBS surface protein genes and the IgA binding protein, Cβ, and used it to further characterize 27 reference strains and 191 isolates. With the exception of a single reference strain (no. 24 [70339] [NCTC 12907]), which contained both bca and rib, all isolates contained only one of the six members—rib, bca, epsilon/alp1/alp5, alp2, alp3, or alp4—of the variable surface protein gene family. The Cβ gene (bac) was present with one or other of these genes (usually bca) in 8 of 27 reference strains and 16 of 191 isolates (most of which belonged to molecular serotype [MS] Ib).
Our study confirmed previously reported relationships between cps serotypes and surface protein gene profiles (10, 12, 16). For example, most serotype III isolates (our molecular serosubtypes [msst] III-1 and III-2) were associated with rib (20), and msst III-3 with alp2 (16). Serotype Ib was associated with bca and bac (14), and serotypes V and VIII with alp3 (16). However, since the relationship was not absolute, different combinations of cps serotype and protein gene profile identified many serovariants, which will be useful in epidemiological studies and in formulation of conjugate vaccines (14, 16).
Most of the 50 isolates which had been subtyped using antisera against surface proteins express “laddering” patterns in Western blots with two or more antisera (15). This serological cross-reactivity probably arises from conserved epitopes present within more than one surface protein. However, in our study, all strains tested possessed one and, with a single exception, only one of the six members of the surface protein antigen gene family—rib, bca, epsilon, alp2, alp3, and alp4 (14, 16). These results indicate that immunologic typing of GBS surface proteins with currently available antisera is potentially misleading and should be used with caution, especially for epidemiological studies.
Unlike the use of antisera, molecular methods of protein “typing” do not demonstrate protein expression, which is clearly important for virulence. However, since the results of existing immunological typing methods are so difficult to interpret, further investigation is needed to identify reliable markers of gene expression. Our mPCR/RLB assay is an appropriate method for epidemiological typing, especially when combined with other molecular markers, as in our three-component system (10). If combined with an immunological assay, such as a Western blot, the mPCR/RLB would clarify the interpretation of results.
The use of a polyvalent GBS conjugate vaccine, containing all of the prevalent capsular polysaccharides, has been proposed as a strategy for the prevention of GBS infections. One or more of the GBS surface proteins could be conjugated with GBS polysaccharides and independently contribute to protective immunity against the corresponding protein-containing serotypes in an effective vaccine. In this context, a rapid, reliable molecular test capable of identifying the GBS surface protein genes, such as mPCR/RLB, would facilitate cost-effective epidemiological studies of circulating GBS strains to identify protein genes profiles.
This mPCR/RLB method has a number of advantages over our previous protein gene profiling method (12). It is easy to perform and does not require the use of gels; amplification products of 43 isolates can be tested simultaneously, and therefore, the method would be suitable for high-throughput GBS typing (4). Furthermore, it can detect all seven protein genes in a single PCR and on one membrane, which can be reused more than 20 times. The assay (for 43 isolates) can be completed within one working day. Although we have not tested its sensitivity compared with culture, it has the potential to be used directly for clinical specimens (4).
Our method also has practical advantages over the multiplex PCR described by Creti et al. (2). First, it does not involve the use of gels, and the use of two probes to identify PCR products increases the sensitivity (by amplifying the signal) and specificity (the result is considered positive only if both probes are hybridized). Second, unlike Creti et al., we can distinguish alp2 from the closely related alp3 gene by RLB. This is important, because these proteins classically cross-react with typing antisera but are clearly associated with different capsular polysaccharide serotypes (alp2 with msst III-3 [3, 10] and alp3 with serotypes V and VIII [16]). Third, our mPCR/RLB method also includes the Cβ (IgA binding) protein gene, which is an important virulence factor.
Finally, because this method uses only 20 of 43 lanes in the MiniBlotter, additional targets can be added to both mPCR and RLB, such as serotype-specific cps sequences (11) or mobile genetic elements, which are included in our three-component genotyping system (10), or antibiotic resistance genes.
In conclusion, our mPCR/RLB assay is a more sensitive and specific method for the identification of GBS surface protein gene profiles than the use of antisera and more efficient and convenient than separate PCRs or multiplex PCR with gel electrophoresis. Application of PCR/RLB in future epidemiological studies and for surveillance will facilitate the appropriate formulation of candidate GBS vaccines.
Acknowledgments
We thank our colleagues for providing GBS isolates other than those isolated in our laboratory. Lawrence Paoletti and Catherine Lachenauer, Channing Laboratory, Boston, Mass., provided 11 reference strains, including nine strains of GBS serotypes (Ia to VIII) and another two reference strains (BM110 and A909); Diana Martin, Streptococcus Reference Laboratory, at the Institute of Environmental Science and Research Limited, Wellington, New Zealand, provided nine international reference GBS type strains, including strains of serotypes Ia to VI; Johan Maeland, Department of Microbiology, School of Medicine, Norwegian University of Science and Technology, Trondheim, Norway, provided seven serotype reference strains used in his previous studies; Nicola Jones, Nuffield Department of Clinical Laboratory Sciences, Institute for Molecular Medicine, John Radcliffe Hospital, Oxford OX3 9DU, United Kingdom, provided 83 GBS strains used in a previous MLST study; 58 isolates were provided by Dele Davies and Shannon Manning, Michigan State University, East Lansing, Mich., which had been used previously in a study of MLST; and 50 clinical isolates were from Catherine Lachenauer, Channing Laboratory and Division of Infectious Diseases, Department of Medicine, Brigham and Women's Hospital, Children's Hospital, Boston, MA 02115.
REFERENCES
- 1.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2004. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creti, R., F. Fabretti, G. Orefici, and C. von Hunolstein. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies, H. D., N. Jones, T. S. Whittam, S. Elsayed, N. Bisharat, and C. J. Baker. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097-1102. [DOI] [PubMed] [Google Scholar]
- 4.Gold, B. 2003. Origin and utility of the reverse dot-blot. Expert Rev. Mol. Diagn. 3:143-152. [DOI] [PubMed] [Google Scholar]
- 5.Gravekamp, C., D. L. Kasper, J. L. Michel, D. E. Kling, V. Carey, and L. C. Madoff. 1997. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infect. Immun. 65:5216-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heden, L. O., E. Frithz, and G. Lindahl. 1991. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21:1481-1490. [DOI] [PubMed] [Google Scholar]
- 7.Jerlstrom, P. G., G. S. Chhatwal, and K. N. Timmis. 1991. The IgA-binding beta antigen of the C protein complex of group B streptococci: sequence determination of its gene and detection of two binding regions. Mol. Microbiol. 5:843-849. [DOI] [PubMed] [Google Scholar]
- 8.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke, D., C. Menard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 10.Kong, F., D. Martin, G. James, and G. L. Gilbert. 2003. Towards a genotyping system for Streptococcus agalactiae (group B streptococcus): use of mobile genetic elements in Australasian invasive isolates. J. Med. Microbiol. 52:337-344. [DOI] [PubMed] [Google Scholar]
- 11.Kong, F., L. Ma, and G. L. Gilbert. 2005. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J. Med. Microbiol. 54:113-1138. [DOI] [PubMed] [Google Scholar]
- 12.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kvam, A. I., A. Efstratiou, L. Bevanger, B. D. Cookson, I. F. Marticorena, R. C. George, and J. A. Maeland. 1995. Distribution of serovariants of group B streptococci in isolates from England and Norway. J. Med. Microbiol. 42:246-250. [DOI] [PubMed] [Google Scholar]
- 15.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 16.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson, C., M. Stalhammar-Carlemalm, and G. Lindahl. 1996. Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and alpha. Infect. Immun. 64:3518-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel, J. L., L. C. Madoff, K. Olson, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89:10060-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 20.Stalhammar-Carlemalm, M., L. Stenberg, and G. Lindahl. 1993. Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, H., F. Kong, P. Jelfs, G. James, and G. L. Gilbert. 2004. Simultaneous detection and identification of common cell culture contaminant and pathogenic Mollicutes strains by reverse line blot hybridization. Appl. Environ. Microbiol. 70:1483-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wastfelt, M., M. Stalhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]