Abstract
Meningitis caused by Streptococcus pneumoniae represents an important factor of morbidity and mortality in humans. In a significant number of cases, this disease is associated with specific clones of the organism, the so-called invasive pneumococcal clones. The aim of the study was to analyze 156 S. pneumoniae isolates identified as etiological agents of meningitis in Poland in the years 1997 through 2002. The isolates were characterized by multilocus sequence typing (MLST), and the results were compared with those obtained by pulsed-field gel electrophoresis (PFGE) and with the MLST data on invasive pneumococci from other countries. Eighty-nine different sequence types were found in the group of isolates, 50 of which had been known before including 19 of the major invasive clones. However, a significant fraction of the isolates possessed novel combinations of known and new MLST alleles. The majority of penicillin-nonsusceptible isolates belonged to the group of international multiresistant clones (Spain23F-1, Spain6B-2, Spain9V-3, Poland23F-16, and Poland6B-20), which underlined the importance of these in the dissemination of antimicrobial resistance. The results of the MLST analysis correlated well with the PFGE data, thus again demonstrating good congruence between the two typing methods for S. pneumoniae. The majority of the isolates (95.5%) belonged to families 1 or 2 of the surface protein PspA, confirming its potential usefulness as the vaccine antigen candidate.
Streptococcus pneumoniae (pneumococcus) is one of the most important bacterial agents causing meningitis in humans. For example, in the United States, after the wide implementation of the vaccine against Haemophilus influenzae type b, pneumococcal meningitis accounts for 47% of all cases of bacterial central nervous system infections, with the mortality rate exceeding 20% (27). Moreover, the disease results in long-term sequelae in a significant number of survivors, including both children and adults (1, 2, 31). Much higher mortality rates, due to invasive diseases caused by S. pneumoniae, and worse clinical outcomes, are characteristic for the developing countries (14).
Recently, multilocus sequence typing (MLST) (11), which allows for unambiguous strain identification and comparisons of bacterial isolates from different locales and types of infection, has been widely used to characterize S. pneumoniae populations worldwide (e.g., reference 5, 6, 10, 13, 19, or 25). Among others, these studies have led to the discrimination of pneumococcal clones commonly associated with invasive infections (30). For some of these clones, overrepresentation among invasive isolates relative to those from carriage suggests possibly increased virulence (5, 16, 26). MLST also plays an essential role in tracking the clones that have acquired antimicrobial resistance and which have spread widely as the so-called multiresistant international S. pneumoniae clones (22). The majority of these are nonsusceptible to β-lactams, which represent drugs of choice in the treatment of pneumococcal meningitis. The recently increasing prevalence of penicillin-nonsusceptible pneumococci (PNSP), observed among invasive isolates in many countries, results in treatment failures (20). Some of the multiresistant international clones have been identified also as being of potentially increased invasiveness, e.g., Spain23F-1, Spain6B-2, Spain9V-3, England14-9, and Poland6B-20 (30).
Since 1997, a laboratory-based surveillance program of bacterial meningitis has been operating in Poland, one of the activities of the National Reference Centre for Bacterial Meningitis (NRCBM). In this study, MLST was used to characterize S. pneumoniae isolates from meningitis cases collected by the NRCBM from 1997 to 2002. The MLST data were compared with those obtained by pulsed-field gel electrophoresis (PFGE) and the determined family types of the PspA surface protein, which is one of the main pneumococcal virulence factors and a vaccine candidate.
(Parts of this work were presented at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 14 to 17 September 2003, and the ISPPD-4, Helsinki, Finland, 9 to 13 May 2004.)
MATERIALS AND METHODS
Bacterial strains, serotyping, and susceptibility testing.
One hundred fifty-six S. pneumoniae isolates from the cerebrospinal fluid or blood of patients with meningeal symptoms from 49 health care centers located in 44 towns in Poland were included in the study. These were all meningitis-related pneumococcal isolates collected by the NRCBM over the period from 1997 to 2002. Serotypes, antimicrobial susceptibility, and PFGE types of 128 isolates from 1997 to 2001 had been described previously (28), and these isolates were supplemented with 28 new isolates identified in 2002. In the whole analyzed group, 19 isolates were obtained from infants below 2 years of age, 41 were from older children (2 to 14 years old), and 95 were from adults; in one case, the age was not reported. Serotypes of the pneumococci isolated in 2002 were determined on the basis of capsular swelling (the Quellung reaction) at the Statens Serum Institut (Copenhagen, Denmark), and MICs of penicillin (Sigma Chemical Company, St. Louis, Mo.) and cefotaxime (Polfa Tarchomin, Warsaw, Poland) were evaluated by the microdilution method as recommended by the Clinical and Laboratory Standards Institute, with the approved breakpoints (7).
MLST, PFGE, PspA families, and data analysis.
Total DNAs of isolates were obtained with the Genomic DNA Prep Plus kit (A & A Biotechnology, Gdynia, Poland) and used subsequently as PCR templates. The MLST analysis was performed on aroE, gdh, gki, recP, spi, xpt, and ddl housekeeping gene loci as originally proposed by Enright and Spratt (11). The Web database (www.mlst.net) was used for assigning allele numbers for particular loci and, on the basis of the resulting allelic profiles, the sequence types (STs) of isolates. To assure data quality, all the single-, double-, and triple-locus variants (SLVs, DLVs, and TLVs, respectively) and the novel allelic combinations without more than four matches to known STs were subjected to the repeated typing. The novel allelic variants and profiles were reported to the MLST database. The eBURST analysis (12), available at www.mlst.net, was used to estimate the relationships among the isolates and to construct a population snapshot. The eBURST was based on the group or clonal complex definition, according to which its members share six out of seven MLST loci. The PFGE analysis of the isolates from 2002, including genomic DNA preparation, its digestion with the SmaI restriction enzyme (MBI Fermentas, Vilnius, Lithuania), and electrophoretic separation of the resulting fragments, was performed according to Lefévre et al. (21). For classification of the isolates, those with identical patterns were considered representatives of a single PFGE type, designated with an Arabic number, and isolates that differed by one to three bands were identified as subtypes of the same type, marked by additional lower case letters (e.g., 1a or 1b). The PspA families 1 and 2 were distinguished by PCR, using specific primers and reaction conditions reported by Coral et al. (8). The diversity indexes and confidence intervals (CI) were calculated by the approach described by Grundmann et al. (15). For the statistical data evaluation, the chi-square test and the Fisher exact test were used with 95% confidence intervals.
RESULTS
MLST of pneumococci.
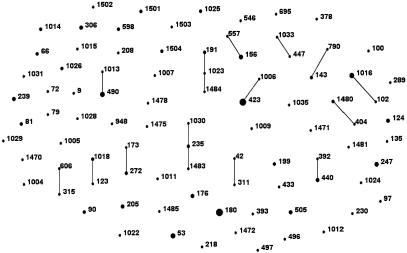
In the studied group of 156 S. pneumoniae isolates, all the MLST loci were polymorphic, and the number of alleles varied from 18 for aroE to 27 for ddl (an average of 22.85 alleles per locus). Altogether, 89 different allelic profiles were identified (Table 1), 50 of which (present in 105 isolates [67.3%]) specified STs previously reported to the MLST database. The allelic profiles of the remaining isolates consisted either of novel combinations of known alleles (27 allelic profiles; STs 598, 606, 1004 to 1009, 1011 to 1016, 1018, 1022 to 1026, 1028 to 1031, 1033, 1470, and 1480) or contained new allelic variants (12 profiles; STs 1471, 1472, 1475, 1478, 1481, 1483 to 1485, 1502, and 1503). The eBURST analysis (Fig. 1) revealed the presence of 15 clonal complexes and 56 singletons that contained 61 (39%) and 95 (61%) isolates, respectively.
TABLE 1.
Serotypes, STs, allelic profiles, PFGE types and PspA families of 156 Polish CSF pneumococcal isolates
| Serotype | STa,b | Allelic profilea,c | Comment | No. of isolates | PFGE typesd | PspAe |
|---|---|---|---|---|---|---|
| 1 | *306 | 12-8-13-5-16-4-20 | 3 | 47a, b | 1 | |
| 1031 | 16-31-4-34-6-4-94 | 1 | 98 | 1 | ||
| 2 | 1504 | 2-13-4-1-6-147-14 | 2 | 57, 58 | 2 | |
| 3 | *180 | 7-15-2-10-6-1-22 | 11 | 1 | 2 | |
| 505 | 46-8-2-10-6-1-22 | DLV of ST180 | 3 | 2 | 1 | |
| 378 | 13-9-15-14-10-16-19 | 1 | 84 | 2 | ||
| 1024 | 10-5-28-11-17-111-17 | 1 | 77 | 2 | ||
| 4 | *205 | 10-5-4-5-13-10-18 | 3 | 41 a, b | 2/NT | |
| *247 | 16-13-4-5-6-10-14 | 4 | 39, 42 a, b | 2 | ||
| *695 | 16-13-4-4-6-113-18 | 1 | 41b | 2 | ||
| 1022 | 8-8-4-1-15-1-20 | 1 | 80 | 2 | ||
| 5 | *289 | 16-12-9-1-41-33-33 | Columbia5-19 | 1 | 63 | 1 |
| 6A | 208 | 10-13-2-1-6-19-14 | 1 | 28 | 2 | |
| 490 | 2-13-9-1-6-19-14 | 5 | 16, 17, 18, 79, 81 | 2 | ||
| 1014 | 2-10-1-43-6-31-6 | 2 | 26, 99 | 2 | ||
| 6B | *90 | 5-6-1-2-6-3-4 | Spain6B-2 | 2 | 76 a, b | NT |
| 135 | 7-5-4-12-6-20-46 | 1 | 100 | 2 | ||
| *176 | 7-13-8-6-10-6-14 | 3 | 13, 14, 73 | 1 | ||
| *315 | 20-28-1-1-15-14-14 | Poland6B-20 | 1 | 12 | 2 | |
| 606 | 16-28-1-1-15-14-14 | Poland6B-20 (SLV of ST315) | 1 | 12 | 2 | |
| 497 | 7-25-4-2-48-20-28 | 1 | 15 | 1 | ||
| 948 | 1-8-9-1-10-3-6 | 1 | 89 | 2 | ||
| 1013 | 2-8-9-1-6-19-14 | 1 | 11 | 2 | ||
| 7F | *191 | 8-9-2-1-6-1-17 | 2 | 7, 8 | 2 | |
| 1023 | 8-9-2-5-6-1-17 | 1 | 6 | 2 | ||
| 1484 | 66-9-2-5-6-1-17 | 1 | 6 | 2 | ||
| 8 | *53 | 2-5-1-11-16-3-14 | 5 | 24, 27, 71 | 1 | |
| 404 | 7-9-15-11-42-1-70 | 1 | 85 | 2 | ||
| 1480 | 7-9-15-11-93-1-70 | 2 | 23a, b | 2 | ||
| 9N | 66 | 2-8-2-4-6-1-1 | 1 | 40 | 2 | |
| 1008 | 1-5-4-32-25-1-8 | 1 | 70 | 1 | ||
| 9V | *156 | 7-11-10-1-6-8-1 | Spain9V-3 | 2 | 38b, c | 2 |
| 557 | 7-11-10-1-6-58-1 | Spain9V-3 (SLV of ST156) | 1 | 38a | 2 | |
| 239 | 15-17-4-16-6-19-17 | 3 | 37 | 1 | ||
| 10A | 97 | 5-7-4-2-10-1-27 | 1 | 56 | 1 | |
| 598 | 2-12-2-16-27-28-14 | 1 | 55a | NT | ||
| 10B | 598 | 2-12-2-16-27-28-14 | 1 | 55b | 2 | |
| 11A | 1012 | 2-5-29-18-42-3-18 | 1 | 65 | 2 | |
| 11B | 1009 | 1-5-6-1-17-16-11 | 1 | 66 | 2 | |
| 12F | *218 | 10-20-14-1-6-1-29 | 1 | 72 | NT | |
| 14 | *9 | 1-5-4-5-5-1-8 | England14-9 | 1 | 93 | 1 |
| *124 | 7-5-1-8-14-11-14 | 3 | 29, 31 | 2 | ||
| 143 | 7-5-10-18-6-8-1 | 2 | 30c | 2 | ||
| *156 | 7-11-10-1-6-8-1 | Spain9V-3-14 | 2 | 30b, 38d | 2 | |
| 790 | 7-5-10-18-6-58-1 | 1 | 30a | 2 | ||
| 15B/C | *199 | 8-13-14-4-17-4-14 | 2 | 49 | 2 | |
| 1025 | 10-13-2-8-17-16-8 | 2 | 48a, b | 2 | ||
| 1502 | 7-2-1-8-102-1-163 | 1 | 59 | 1 | ||
| 16F | 1004 | 1-2-15-5-17-16-1 | 1 | 67 | 2 | |
| 17F | 123 | 7-2-40-1-10-1-45 | 1 | 60 | 2 | |
| 392 | 7-5-1-1-6-31-14 | 1 | 61 | 2 | ||
| 18C | *102 | 5-13-11-4-15-12-19 | 1 | 92 | 2 | |
| 1016 | 5-13-11-4-15-1-19 | SLV of ST102 | 5 | 52, 53, 54a, b | 2 | |
| 496 | 42-35-29-36-9-39-18 | 1 | 102 | 1 | ||
| 1472 | 1-96-9-1-9-14-6 | 1 | 101 | 2 | ||
| 19A | 1007 | 1-5-4-5-15-1-18 | 1 | 36 | 1 | |
| 19F | 79 | 2-27-2-4-9-4-31 | 1 | 35 | 2 | |
| *423 | 1-5-4-12-5-3-8 | 10 | 34a, b | 1 | ||
| 1006 | 1-5-1-12-5-3-8 | 1 | 34c | 2 | ||
| 1029 | 13-16-19-5-6-20-26 | 1 | 33 | 2 | ||
| 1471 | 1-5-4-12-5-26-159 | 1 | 32 | 1 | ||
| 20 | 235 | 15-8-8-18-15-1-31 | 2 | 43, 45 | 1 | |
| 1026 | 10-16-32-1-15-28-31 | 2 | 44 | 1 | ||
| 1030 | 15-8-8-18-15-1-9 | 1 | 46 | 1 | ||
| 1483 | 65-8-8-18-15-1-31 | 1 | 94 | 1 | ||
| 22A | 1485 | 67-2-40-11-10-1-45 | 1 | 22 | 2 | |
| 433 | 1-1-4-1-18-58-17 | 1 | 20 | NT | ||
| 1018 | 7-2-40-1-17-1-45 | 2 | 19, 25 | 2 | ||
| 22F | 1470 | 1-2-19-66-15-1-11 | 1 | 21 | 2 | |
| 23A | 42 | 1-8-9-9-6-4-6 | 1 | 78 | 2 | |
| 1011 | 1-8-9-2-6-4-72 | 1 | 74 | 1 | ||
| 23F | *81 | 4-4-2-4-4-1-1 | Spain23F-1 | 2 | 3 | 2 |
| 173 | 7-13-8-1-10-6-36 | Poland23F-16 | 1 | 4 | 1 | |
| 272 | 7-13-8-1-10-6-37 | Poland23F-16 (SLV of ST173) | 2 | 82a, b | 1 | |
| 311 | 1-8-9-1-6-4-6 | 1 | 90 | 1 | ||
| 440 | 7-5-1-1-13-31-14 | 2 | 5a, 9 | 2 | ||
| 24F | 72 | 2-13-2-4-9-4-1 | 1 | 10 | 2 | |
| 230 | 12-19-2-17-6-22-14 | 1 | 88 | 1 | ||
| 27 | 1475 | 2-17-107-24-9-105-14 | 1 | 69 | NT | |
| 28F | 546 | 16-44-1-16-9-11-17 | 1 | 97 | 1 | |
| 1503 | 2-44-1-16-106-11-17 | 1 | 97 | 1 | ||
| 31 | 1005 | 1-2-29-1-46-14-8 | 1 | 64 | 1 | |
| 33F | 100 | 5-12-29-12-9-39-18 | 1 | 62 | 1 | |
| 34 | 1015 | 2-25-2-16-6-28-14 | 1 | 81 | 2 | |
| 1481 | 7-12-53-1-101-1-8 | 1 | 50 | 2 | ||
| 1501 | 7-8-1-70-101-146-8 | 2 | 51 | 2 | ||
| 35F | 1028 | 13-8-1-8-6-1-14 | 1 | 83 | 2 | |
| 37 | 447 | 29-33-19-1-36-22-31 | 1 | 96 | 1 | |
| 1033 | 29-33-19-1-13-22-31 | 1 | 87 | 1 | ||
| 38 | 393 | 10-43-41-18-13-49-6 | 1 | 103 | 1 | |
| 39 | 1478 | 7-5-92-16-103-1-31 | 1 | 68 | 1 | |
| Rough | 66 | 2-8-2-4-6-1-1 | 1 | 86 | 2 | |
| 440 | 7-5-1-1-13-31-14 | 1 | 5b | 1 |
New STs and alleles in bold.
The representatives of major invasive clones (26, 30) are marked by an asterisk; STs associated with increased virulence (5, 16, 26) are underlined.
In the order aroE-gdh-gki-recP-spi-xpt-ddl.
The numbering of PFGE types according to Skoczynska and Hryniewicz (28).
NT, not typeable.
FIG. 1.
Population snapshot of 156 pneumococcal isolates from meningitis in Poland in the years 1997 to 2002, as revealed by the eBURST analysis.
ST180 was the most prevalent ST among the isolates (11 isolates [7%]), followed by ST423 (10 isolates), ST53, ST490, and ST1016 (5 isolates each), and ST156 and ST247 (4 isolates each). Of the remaining STs, 7 were represented by three isolates each (STs 124, 176, 205, 239, 306, 440, and 505), 16 were identified in two isolates each, and 59 were identified in single isolates. Among the STs with multiple isolates, no specific geographic distribution in the country was observed. ST180 and its DLV, ST505, were recovered primarily from adults, with the exception of a single isolate from a 9-year-old patient. The ST180/ST505 isolates were thus overrepresented in adults (statistical significance: χ2 test, P = 0.009; Fisher exact test, P = 0.009).
Of the STs observed frequently in the invasive disease in different countries (30), 19 STs characteristic for 57 isolates were identified in this study (Table 1). Considering also their five SLVs (66 isolates altogether), the clones constituted 42.3% of the whole studied group. Of these, six clones with 16 isolates (10.3%) were those with the confirmed overrepresentation in invasive infections compared to carriage (STs 9, 124, 156, 191, 205, and 306) (5, 16, 26). Almost all the STs with multiple isolates were identified as invasive clones, with the only exception being ST490. Among the 65 STs of the remaining 90 isolates, many had not been previously observed.
Congruence of STs with pneumococcal serotypes and PFGE types.
Thirty-nine different serotypes occurred within the analyzed group of pneumococci (Table 1). A good congruence between serotypes and STs was found, and almost every ST was associated with one serotype, with the exception of ST156 (serotypes 9V and 14) and ST598 (10A and 10B), which could have resulted from capsule switching. On the other hand, numerous serotypes (1, 3, 4, 6A, 6B, 8, 9N, 9V, 10A, 14, 15B/C, 17F, 18C, 19F, 20, 22A, 23F, 24F, 28F, and 34) contained isolates of unrelated STs. The PFGE analysis (Table 1) revealed the presence of 119 different banding patterns that were classified into 102 PFGE types, with 13 types split further into 30 subtypes altogether. In general, there was a high concordance level between MLST and PFGE classifications, with the clear majority of STs corresponding to separate PFGE types, and vice versa. However, 12 STs encompassed different PFGE types (STs 53, 124, 176, 191, 235, 247, 440, 490, 1014, 1016, 1018, and 1504), and, occasionally, the same PFGE type characterized isolates of different, though usually related, STs (PFGE types 6, 12, 30, 41, and 97). The diversity indexes for the investigated group of S. pneumoniae isolates were 95.67% (CI, 94.72 to 96.61%) in the case of serotyping, 98.52% (CI, 97.88 to 99.16%) for MLST, and 99.13% (CI, 98.55 to 99.72%) for PFGE.
PNSP among the meningitis-related isolates.
Eighteen (11.5%) penicillin-nonsusceptible isolates were present among the studied pneumococci, the majority of which (15 isolates) were resistant to this compound (MICs, ≥2 μg/ml). Among these PNSP, seven isolates were intermediate and five were resistant to cefotaxime. Usually, the PNSP either represented the international multiresistant clones (22), such as Spain23F-1 (ST81, two isolates), Spain6B-2 (ST90, two isolates), Poland23F-16 (ST173 and ST272, three isolates), Poland6B-20 (ST315 and ST606, two isolates), and Spain9V-3 (ST156 and ST557, five isolates), or were related to them (ST143 and ST790). The Spain9V-3 and related clones (STs 143 and 790) showed an increased frequency of isolation from meningitis patients over time. Four isolates were recovered between 1997 and 2001, while the other four representatives of this group were identified in 2002 (χ2 test, P = 0.015; the Fisher exact test, P = 0.035). The members of the Poland23F-16 clone, with high-level penicillin resistance (MICs, 4 to 8 μg/ml), were recovered exclusively from young children (two patients 6 months old and one patient 24 months old). Two isolates with an intermediate susceptibility to penicillin (MIC, 0.12 μg/ml) belonged to Poland6B-20 (25). The remaining single PNSP isolate, unrelated to the international resistant clones, was classified into ST230 and serotype 24F.
PspA families.
Fifty-nine (37.8%) isolates harbored the gene encoding PspA of family 1, and 90 isolates (57.7%) represented PspA family 2. Seven isolates (4.5%) could not be ascribed to either family due to the reproducible lack of PCR amplification using standard primers. Out of the STs characteristic for the major invasive clones (30) and their SLVs found in this study, 5 clones (20 isolates) contained PspA belonging to family 1, 11 clones (41 isolates) obtained PspA belong to family 2, and 2 clones (3 isolates) were untypeable. Thus, some tendency of association of family 2 with invasive clones was observed, but without statistical significance (χ2 test, P = 0.15; the Fisher exact test, P = 0.17). Out of the international multiresistant clones, three were of the PspA family 1 (Columbia5-19, England14-9, and Poland23F-16), three were of the family 2 (Poland6B-20, Spain9V-3, and Spain23F-1), and one was untypeable (Spain6B-2).
DISCUSSION
Being responsible for 20.9% of the reported cases, S. pneumoniae represents the third important factor in bacterial meningitis in Poland, following Neisseria meningitidis (40.9%) and H. influenzae (26.4%) (29). Its lower incidence than H. influenzae is most probably due to the poor implementation of anti-Haemophilus influenzae type b vaccination in the country. Results of the study reported here demonstrate clearly the high clonal heterogeneity of the meningitis-associated pneumococci in Poland. Until now, it was rare for MLST analyses to be focused exclusively on such pneumococcal isolates, as in many studies, the meningitis-related isolates were investigated together with isolates from other types of invasive infections (5, 6, 11, 13, 19). Data similar to ours were obtained in a study on Spanish cerebrospinal fluid S. pneumoniae isolates, recovered in the period from 1997 to 1998. Sixty-five STs among 106 isolates were found (10), and the MLST diversity index calculated by us from these data was virtually the same (98.53%; CI, 97.86 to 99.2%) as the one in our analysis. However, only 14 STs (42, *53, 72, *81, *90, 97, 100, 123, 135, *156, *180, *218, 235, and *247; the asterisks mark STs associated with the major invasive clones) (30) were common between the isolates from Spain and Poland.
The frequent occurrence of a given clone in invasive pneumococcal disease either may be a result of its increased virulence potential or may reflect its ubiquitous occurrence in the bacterial population, as deduced from comparisons made between invasive and carriage isolates (5, 16, 26). In our study, almost half of the analyzed group (42.3%) belonged to the clones commonly recovered from invasive infections (30) and their closely related variants, and they included the vast majority of the STs with multiple isolates. Most of these clones were those that, in general, are widely spread in pneumococcal populations and are probably not of higher virulence. The clones considered to be more virulent on the basis of their overrepresentation among isolates from the invasive disease (5, 16, 26) constituted only 10% of the studied group. The remaining isolates were characterized by high clonal diversity and, usually, a particular ST specified only one or two isolates. The only relatively more prevalent clone that has not been associated with the aforementioned invasive clones, ST490 (five isolates), belonged to serotype 6A, which is characteristic rather for carriage isolates (16). Therefore, the structure of the meningitis-related pneumococci in Poland can be described as consisting in one part of clones that have spread successfully on the continental or even global scale (commonly occurring clones and clones with increased virulence) and in the other of highly diverse strains out of which many have been recovered only locally. A similar structure characterized the corresponding group of isolates from Spain (10), where the major invasive clones with their SLVs constituted 41.5% of the isolates, and clones of the possibly increased virulence, 10.4%.
The prevalence of penicillin nonsusceptibility in meningitis-related pneumococci in Poland (11.5%) remained moderate over the studied period in comparison to that in many other countries, e.g., France (40.5%) or Spain (38%) (9, 10); however, the majority of PNSP isolates demonstrated the full penicillin resistance phenotype and nonsusceptibility to cefotaxime, the “third-generation” cephalosporin. In the case of central nervous system infections, the pneumococcal nonsusceptibility to β-lactams correlates with therapeutic failures (20). Pneumococci belonging to several international multiresistant clones were previously shown to circulate in Poland (25), and such clones or related strains were predominant among the meningitis-associated Polish PNSP. The only PNSP isolate unrelated to any international clone (the isolate of serotype 24F and ST230) had been previously isolated in Italy from three patients with meningitis (24) and from a bacteremic patient in Denmark (www.mlst.net) and, therefore, might represent a novel resistant epidemic pneumococcal clone, possibly with increased virulence properties.
Due to the significant variability of pneumococcal capsular polysaccharides (17), the currently available antipneumococcal vaccines based on these compounds provide protection against only a limited number of serotypes. Therefore, there is a growing interest in vaccines composed of pneumococcal protein(s), especially the surface-exposed, highly immunogenic PspA, which induces antibodies protective against pneumococcal infection in animal models (4). However, PspA is one of the most polymorphic S. pneumoniae gene products, and studies on its sequence from unrelated isolates have discriminated six distinct clades classified into three families (18). Epidemiological studies in South American countries showed that over 90% of pneumococci expressed either family 1 or 2 PspA, with approximately equal frequencies (3, 8, 23), while in Poland, family 2 seems to be predominant. These results show that PspA-based vaccines may be an important future alternative for protection against this life-threatening disease caused by greatly divergent organisms.
Modern medicinal practice significantly influences the composition of bacterial populations by the use of antimicrobials and vaccines. The predominance of penicillin-resistant pneumococci in Spain, with its high level antibiotic consumption (10), and the decline of vaccine and vaccine-related serotypes in pneumococcal invasive disease in the United States after implementation of the 7-valent conjugate vaccine (32) constitute clear examples of such impact. In contrast, the highly divergent group of pneumococci isolated from meningitis cases in Poland might be closer to a more natural state than in other countries where stronger selective pressure has been exerted on the pneumococcal population.
Acknowledgments
We thank Paweł Grzesiowski for help with statistical calculations and Anna Klarowicz for her excellent technical assistance. We acknowledge the use of the pneumococcal MLST database which is located at the Imperial College, London, United Kingdom and is funded by the Wellcome Trust, with special thanks to Angela Brueggemann for assigning numbers for new alleles and STs found in this study.
This study was partially financed by a grant from the Polish Committee for Scientific Research (3P0A 062 23), and the publication costs were covered by the Polish National Node of Global Biodiversity Information Facility.
REFERENCES
- 1.Arditi, M., E. O. Mason, Jr., J. S. Bradley, T. Q. Tan, W. J. Barson, G. E. Schutze, E. R. Wald, L. B. Givner, K. S. Kim, R. Yogev, and S. L. Kaplan. 1998. Three-year multicenter surveillance of pneumococcal meningitis in children: clinical characteristics, and outcome related to penicillin susceptibility and dexamethasone use. Pediatrics 102:1087-1097. [DOI] [PubMed] [Google Scholar]
- 2.Bedford, H., J. de Louvois, S. Halket, C. Peckham, R. Hurley, and D. Harvey. 2001. Meningitis in infancy in England and Wales: follow up at age 5 years. BMJ 323:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandileone, M. C., A. L. Andrade, E. M. Teles, R. C. Zanella, T. I. Yara, J. L. Di Fabio, and S. K. Hollingshead. 2004. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil: towards novel pneumococcal protein vaccines. Vaccine 22:3890-3896. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., R. C. Tart, E. Swiatlo, J. P. Dillard, P. Smith, K. A. Benton, B. A. Ralph, A. Brooks-Walter, M. J. Crain, S. K. Hollingshead, and L. S. McDaniel. 1998. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 11:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, S. C., K. J. Scott, and S. M. McChlery. 2004. Serotypes and sequence types of pneumococci causing invasive disease in Scotland prior to the introduction of pneumococcal conjugate polysaccharide vaccines. J. Clin. Microbiol. 42:4449-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2005. M100-S15. Performance standards for antimicrobial susceptibility testing, 15th informational supplement. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 8.Coral, M. C. V., N. Fonseca, E. Castañeda, J. L. Di Fabio, S. K. Hollingshead, and D. E. Briles. 2001. Pneumococcal surface protein A of invasive Streptococcus pneumoniae isolates from Colombian children. Emerg. Infect. Dis. 7:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doit, C., B. Picard, C. Loukil, P. Geslin, and E. Bingen. 2000. Molecular epidemiology survey of penicillin-susceptible and -resistant Streptococcus pneumoniae recovered from patients with meningitis in France. J. Infect. Dis. 181:1971-1978. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., A. Fenoll, D. Griffiths, and B. G. Spratt. 1999. The three major Spanish clones of penicillin-resistant Streptococcus pneumoniae are the most common clones recovered in recent cases of meningitis in Spain. J. Clin. Microbiol. 37:3210-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwood, B. 1999. The epidemiology of pneumococcal infection in children in the developing world. Philos. Trans. R. Soc. Lond. B 354:777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundman, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanage, W. P., T. H. Kaijalainen, R. K. Syrjanen, K. Auranen, M. Leinonen, P. H. Makela, and B. G. Spratt. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 18.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jefferies, J. M., A. Smith, S. C. Clarke, C. Dowson, and T. J. Mitchell. 2004. Genetic analysis of diverse disease-causing pneumococci indicates high levels of diversity within serotypes and capsule switching. J. Clin. Microbiol. 42:5681-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klugman, K. P. 2004. Clinical relevance of antibiotic resistance in pneumococcal infections, p. 331-338. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, D.C.
- 21.Lefévre, J. C., G. Faucon, A. M. Sicard, and A. M. Gase. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mollerach, M., M. Regueira, L. Bonofiglio, R. Callejo, J. Pace, J. L. Di Fabio, S. Hollingshead, D. Briles, and Streptococcus pneumoniae Working Group. 2004. Invasive Streptococcus pneumoniae isolates from Argentinian children: serotypes, families of pneumococcal surface protein A (PspA) and genetic diversity. Epidemiol. Infect. 132:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 25.Sadowy, E., J. Zhou, E. Meats, M. Gniadkowski, B. G. Spratt, and W. Hryniewicz. 2003. Identification of multidrug-resistant Streptococcus pneumoniae strains isolated in Poland by multilocus sequence typing. Microb. Drug Resist. 9:81-86. [DOI] [PubMed] [Google Scholar]
- 26.Sandgren, A., K. Sjöström, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques-Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 27.Schuchat, A., K. Robinson, J. D. Wenger, L. H. Harrison, M. Farley, A. L. Reingold, L. Lefkowitz, and B. A. Perkins. 1997. Bacterial meningitis in the United States in 1995. N. Engl. J. Med. 337:970-976. [DOI] [PubMed] [Google Scholar]
- 28.Skoczyñska, A., and W. Hryniewicz. 2003. Genetic relatedness, antibiotic susceptibility, and serotype distribution of Streptococcus pneumoniae responsible for meningitis in Poland, 1997-2001. Microb. Drug Resist. 9:175-182. [DOI] [PubMed] [Google Scholar]
- 29.Skoczyñska, A., P. Kriz, H. B. Konradsen, and W. Hryniewicz. 2000. Characteristics of the major etiologic agents of bacterial meningitis isolated in Poland in 1997-1998. Microb. Drug Resist. 6:147-153. [DOI] [PubMed] [Google Scholar]
- 30.Spratt, B. G., W. P. Hanage, and A. B. Brueggemann. 2004. Evolutionary and population biology of Streptococcus pneumoniae, p. 119-135. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, D.C.
- 31.van de Beek, D., B. Schmand, J. de Gans, M. Weisfelt, H. Vaessen, J. Dankert, and M. Vermeulen. 2002. Cognitive impairment in adults with good recovery after bacterial meningitis. J. Infect. Dis. 186:1047-1052. [DOI] [PubMed] [Google Scholar]
- 32.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat for the Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]