Abstract
The definition of antigens for the diagnosis of human and bovine tuberculosis is a research priority. If diagnosis is to be used alongside Mycobacterium bovis BCG-based vaccination regimens, it will be necessary to have reagents that allow the discrimination of infected and vaccinated animals. A list of 42 potential M. bovis-specific antigens was prepared by comparative analysis of the genomes of M. bovis, M. avium subsp. avium, M. avium subsp. paratuberculosis, and Streptomyces coelicolor. Potential antigens were tested by applying them in a high-throughput peptide-based screening system to M. bovis-infected and BCG-vaccinated cattle and to cattle without prior exposure to M. bovis. A response hierarchy of antigens was established by comparing responses in infected animals. Three antigens (Mb2555, Mb2890, and Mb3895) were selected for further study, as they were strongly recognized in experimentally infected animals but with low or no frequency in BCG-vaccinated and naïve cows. Interestingly, all three antigens were recognized in animals vaccinated against Johne's disease, suggesting the presences of epitopes cross-reacting with M. avium subsp. paratuberculosis antigens. Eight peptides from the three antigens studied in detail were identified as immunodominant and were characterized in terms of major histocompatibility complex class II restriction element usage and shown to be restricted through both DR and DQ molecules. Reasons for antigenic cross-reactivity with M. avium subsp. paratuberculosis and refinement of the in silico strategy to predict such cross-reactivity from the primary protein sequence will be discussed. Evaluation of the peptides identified from the three dominant antigens by use of larger field studies is now a priority.
For 20 years, bovine tuberculosis (BTB) has been spreading in Great Britain and the number of detected cases of BTB continues to rise exponentially (7). The current BTB control program involves regular skin testing using the single intradermal comparative tuberculin test followed by compulsory slaughter of cattle with a positive result. Vaccination remains an additional strategy for the control of BTB, but the only currently available vaccine, Mycobacterium bovis bacillus Calmette-Guérin (BCG), sensitizes vaccinated individuals to tuberculin (2, 3). The next generation of vaccines against both human and bovine tuberculosis is likely to be based on those vaccines that can enhance preexisting immunity conferred by BCG and so will require the development of diagnostic reagents that can distinguish vaccination from infection (1, 8, 9).
The availability of pathogen genome sequences has provided a boon to researchers searching for new diagnostic markers of infection, since all of the encoded proteins and hence potential antigens are revealed. However, with an organism such as M. bovis, which contains ∼4,000 protein-encoding genes, it is not feasible to screen all proteins for their antigenicity; a biologically relevant filter must be used to sift the genome information to a manageable protein subset (13, 14). One subset that would have obvious diagnostic potential is proteins that are present only in the pathogen and not in related species; with the wealth of genome information that is now available for mycobacteria, we are in a position to define such potentially species-specific proteins for members of the M. tuberculosis complex (3-7).
We herein describe the application of comparative genomics to define novel antigens of Mycobacterium bovis. We have also assessed the potential of these antigens as diagnostic markers in cattle both experimentally and naturally infected with M. bovis, in cattle vaccinated with BCG, and in naïve animals. We report the identification of a subset of eight peptides from over 70 genes that are likely to be useful for the diagnosis of tuberculosis across a wise spectrum of genetic backgrounds in both cattle and humans.
MATERIALS AND METHODS
Iterative genome screen.
At the time the in silico genome analysis was performed, the complete sequence of M. bovis was available, with incomplete versions of the M. avium subsp. avium (http://www.tigr.org/) and M. avium subsp. paratuberculosis (http://www.cbc.umn.edu/ResearchProjects/AGAC/Mptb/Mptbhome.html) genomes. We used StandAlone TBLASTN to compare all proteins in M. bovis to those in M. avium subsp. avium, identifying M. bovis proteins that showed no or poor identity against M. avium subsp. avium. Theseproteins were then rechecked against the M. avium subsp. avium genome by using TBLASTN to ensure that no potential coding sequences had been missed. This list was further refined by screening the proteins against the six-frame translation of the M. avium subsp. paratuberculosis genome and also the fully annotated Streptomyces coelicolor sequence as a means to screen against related soil actinobacteria with the public NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST).
Antigens and peptides.
Bovine tuberculin (purified protein derivative [PPD]-B) and avian tuberculin (PPD-A) were supplied by the Tuberculin Production Unit at the Veterinary Laboratories Agency, Weybridge, Surrey, United Kingdom, and were used to stimulate whole blood at 10 μg/ml. Staphylococcal enterotoxin B (16) was included as a positive control at 5 μg/ml.
Multirod peptide synthesis was used to generate 20-mer peptides spanning the length of 42 proteins, with each peptide overlapping its neighbor by 12 residues (Chiron Mimotopes, Clayton, Australia). These were dissolved in Hanks balanced salt solution (Gibco) and then combined into 73 pools of between 8 and 12 peptides from each protein and used at a concentration of between 16 and 25 μg/ml of each peptide in a pool or 25 μg/ml individually. Peptides from some antigens were combined into up to four pools because of the length of the protein. Peptides from the ESAT-6 and CFP-10 proteins were synthesized, quality assessed, and formulated into a peptide cocktail as previously described (17).
IFN-γ enzyme-linked immunosorbent assay.
A gamma interferon (IFN-γ) enzyme-linked immunosorbent assay was performed as previously described (5). In addition, IFN-γ concentration (ng/ml) was quantified from optical density at 450 nm (OD450) values by use of a standard curve constructed using titrations of bovine recombinant IFN-γ (Serotec, Kidlington, Oxford, United Kingdom). A response was considered positive if the OD450 value was greater than 0.1 after subtraction of the negative control value, equivalent to 6.7 pg/ml of IFN-γ.
IFN-γ ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were performed and enumerated as previously described (17). For blocking of bovine leucocyte antigen (BoLA) class II processing, monoclonal antibodies raised against BoLA-DR and BoLA-DQ were purchased from VMRD, Inc., Pullman, Wash. Each antibody was preincubated with peripheral blood mononuclear cells (PBMC) in the ELISPOT assay plate at 5 μg/ml for 1 h at 37°C, after which the relevant peptide was added to the well and plates were incubated as normal for the IFN-γ ELISPOT assay.
Depletion of CD4+ cells from PBMC.
PBMC (1 × 107) were suspended in 100 μl of phosphate-buffered saline containing 0.1% bovine serum albumin (depletion buffer; Sigma) and incubated with 100 μl of CC30 (provided by C. J. Howard, Institute for Animal Health, Compton, United Kingdom) bovine anti-CD4+ antibody on ice for 1 h. Cells were then washed in wash buffer. Anti-pan mouse immunoglobulin G beads (150 μl; Dynal, Wirral, United Kingdom) were washed in wash buffer before incubation with cells on ice for 30 min. Afterwards, tubes containing cells and beads were placed in a magnetic particle concentrator for 2 min. The remaining liquid containing unbound cells was removed, washed, and used in the ELISPOT assay as normal. Efficiency of depletion was assessed by staining whole and depleted PBMC with fluorescein isothiocyanate-conjugated anti-CD4+ and phycoerythrin-conjugated anti-CD8+ monoclonal antibodies (Serotec, Kidlington, Oxford, United Kingdom) and by flow cytometric analysis using a CyAn cytometer (Dako Cytomation, Fort Collins, Colo.).
Cattle.
Uninfected control animals (ca. 6 months old) were obtained from herds free of BTB (10 animals) and tested for the absence of an in vitro IFN-γ response to PPD-B. A further 10 animals from BTB-free herds were vaccinated with 106 CFU of BCG Pasteur by the subcutaneous route, and heparinized blood samples were collected 3 weeks later.
Fourteen calves from BTB-free herds were infected with between 1 and 1,000 CFU of a field strain of M. bovis (AF2122/97) by the intratracheal route. Active infection was confirmed by skin test positivity (between 12 and 16 weeks postinfection), the presence of visible lesions in lungs and lymph nodes postmortem, and positive bacteriological culture of M. bovis from tissues. Blood samples were obtained 18 weeks after infection.
Blood samples were also obtained by the State Veterinary Service from naturally infected, tuberculin skin test-positive reactors within herds known to have BTB. Disease was confirmed as for experimentally infected animals.
Four calves were vaccinated subcutaneously at 2 to 3 months of age with 2 × 108 CFU of irradiated M. paratuberculosis reference strain ATCC 19698 in an oil adjuvant (Montanide ISA 775; SEPPIC). Blood samples were obtained 7 to 14 months postimmunization.
All animal experiments were carried out in compliance with the respective Animal Ethics Committees.
RESULTS
A list of 42 M. bovis genes encoding putative specific proteins was defined on the basis of iterative genome screens. These proteins encompassed a wide range of putative functions, molecular weights, and subcellular locations, and sets of overlapping synthetic peptides covering the complete sequences of these 42 proteins were prepared and formulated into 73 pools of 8 to 12 peptides (Table 1). These pools were then tested for their ability to induce IFN-γ responses in vitro in M. bovis-infected, BCG-vaccinated, and naïve cattle. All infected cattle gave positive responses to the staphylococcal enterotoxin B control and PPD-B (mean, 106.7 pg/ml; standard deviation [SD], 15.8 pg/ml). Infected cattle also responded to PPD-A (mean, 42.8 pg/ml; SD, 26.3 pg/ml) and the combined ESAT-6 and CFP-10 peptide pool (mean, 96.8 pg/ml; SD, 26.1 pg/ml). All BCG-vaccinated animals and 4 of the 10 naïve animals gave positive responses to avian tuberculin.
TABLE 1.
Proteins identified from genome screen as potential specific antigens from M. bovis with minimal homology to environmental species
| Type of protein | M. bovis protein (Mb) | H37Rv protein (Rv) | Length (aa)a | E valueb vs:
|
||
|---|---|---|---|---|---|---|
| M. avium | M. paratuberculosis | S. coelicolor | ||||
| Transport/membrane proteins | 0454c | 0446c | 256 | 4.1 | 1.1 | None |
| 0864 | 0841 | 80 | 5.2 | 5.2 | 5.2 | |
| 3024 | 2999 | 321 | 5.7 | 4.4 | None | |
| 3881 | 3851 | 94 | 5.2 | 5.2 | 6.8 | |
| Intermediary metabolism and respiration proteins | 1580 | 1555 | 125 | None | None | None |
| 3162 | 3138 | 362 | 5.8 | 5.8 | None | |
| Other proteins | 2854c | 2830c | 71 | 5.2 | 3.1 | 2.3 |
| 1991 | 1956 | 149 | 3.5 | None | 1.2 | |
| Conserved hypothetical proteins | 0101 | 0098 | 183 | 4 | 4 | None |
| 0103 | 0100 | 78 | 4 | 4 | None | |
| 0614c | 0598C | 137 | None | 4.9 | None | |
| 0676c | 0657c | 51 | 6.9 | 6.9 | None | |
| 0679c | 0660c | 81 | None | None | None | |
| 0680c | 0661c | 145 | None | None | None | |
| 0990c | 0965c | 139 | 3.9 | 1 | 6.6 | |
| 1133c | 1103c | 106 | None | None | None | |
| 1750c | 1721c | 75 | 5.2 | 5.2 | None | |
| 1869c | 1838c | 131 | 3.4 | 1.5 | None | |
| 2579c | 2549c | 131 | 4.4 | 5.7 | 5.7 | |
| 2781c | 2760c | 89 | 4 | 4 | None | |
| 2840c | 2816c | 113 | 4 | 6.8 | None | |
| 2854c | 2830c | 71 | 5.2 | 3.1 | 2.3 | |
| 2890 | 2865 | 93 | 45 | 1.4 | 1.4 | |
| 2973c | 2949c | 199 | 6.1 | 6.1 | None | |
| 3210 | 3188 | 115 | 5.2 | None | 0.043 | |
| 3711c | 3686c | 110 | 3.1 | 1.4 | 3.1 | |
| 3895 | 3865 | 103 | None | None | None | |
| Hypothetical proteins | 0081c | 0078A | 197 | None | None | None |
| 0274c | 0268c | 169 | None | None | None | |
| 0306 | 0298 | 75 | None | None | None | |
| 1165 | 1134 | 78 | 5.2 | 5.2 | None | |
| 1807c | 1778c | 149 | 5.9 | 0.92 | 5.9 | |
| 2326c | 2304c | 69 | 5.2 | 6.8 | None | |
| 2459c | 2433c | 96 | 8.9 | 8.9 | 5.2 | |
| 2520 | 2492 | 250 | 6.8 | None | None | |
| 2555 | 2526 | 75 | None | None | None | |
| 2846c | 2822c | 124 | 3.8 | 3.8 | None | |
| 3022 | 2998 | 153 | 3.7 | 2.8 | 1.3 | |
| 3219c | 3196A | 66 | 8.9 | 8.9 | None | |
| 3796c | 3770c | 191 | 7.4 | 9.6 | None | |
aa, amino acids.
E value is a parameter that describes the number of hits one can expect to see just by chance when searching a database of a particular size. It decreases exponentially with the score that is assigned to a match between two sequences. The lower the E value (or the closer it is to zero), the more significant the match is.
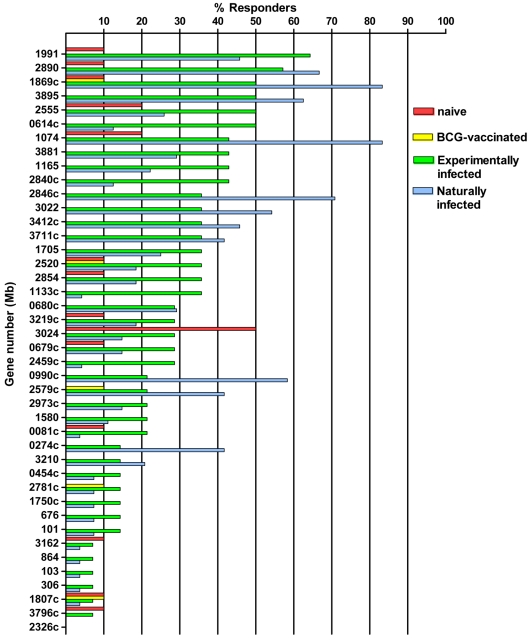
Responder frequencies against the peptide pools among the different groups are shown in Fig. 1. Among the BTB-naïve animals, 22 responses were detected to 16/73 pools; in all but one case, the responses were low level (<6.75 pg/ml IFN-γ). One peptide pool from Mb3024 elicited responses in 50% of the naïve animals tested, with one animal giving an IFN-γ concentration of 20.7 pg/ml. None of the animals responding to PPD-A recognized any of the 73 pools tested, suggesting a lack of cross-reactivity with M. avium subsp. avium antigens.
FIG. 1.
Responder frequencies following stimulation with pools containing 8 to 12 peptides from each of the 42 antigens tested (shown in Table 1) of naïve (n = 10), BCG-vaccinated (n = 10), M. bovis experimentally infected (n = 14), and M. bovis naturally infected (n = 24 to 27) animals. Where more than one peptide pool was generated for an antigen, the pool with the highest frequency is shown. A positive response was determined by the following equation: OD450 with peptides − OD450 of medium control >0.1.
Responses were less frequent among BCG-vaccinated calves, with only 4/73 pools yielding low-level IFN-γ responses. In contrast to results for naïve animals, no responses to Mb3024 were detected in this group.
Positive responses to 67/73 (91.8%) pools were seen among experimentally infected animals, and seven animals (50%) recognized ≥20 peptide pools each (median, 13.5). Table 2 shows the proportions and magnitude of responses to the most strongly recognized pools. Nine pools were recognized by ≥40% of animals tested, with a range of mean IFN-γ production of 7.71 to 18.25 pg/ml. The pools most frequently recognized by experimentally infected cattle were from Mb1991, Mb2890, Mb3895, Mb2555, Mb1869c, and Mb2846c.
TABLE 2.
Peptide pools recognized most frequently and stronglya
| Gene no. (Mb) | Pool/no. pools in gene | No. peptides in pool | Experimentally infected
|
Naturally infected
|
||
|---|---|---|---|---|---|---|
| % Responder (n = 14) | Mean IFN-γ concn (pg/ml) | % Responder (n = 22-24) | Mean IFN-γ concn (pg/ml) | |||
| 1991 | 1/2 | 9 | 64.3 | 10.7 | 45.8 | 11.8 |
| 2890b | 1/1 | 11 | 57.1 | 13.0 | 66.7 | 19.7 |
| 3895b | 1/1 | 12 | 50.0 | 18.3 | 62.5 | 12.3 |
| 2555b | 1/1 | 8 | 50.0 | 13.4 | 37.0 | 14.6 |
| 1869c | 2/2 | 7 | 50.0 | 10.3 | 83.3 | 18.3 |
| 2846c | 2/2 | 7 | 50.0 | 7.7 | 29.2 | 16.0 |
| 1074 | 2/3 | 12 | 42.9 | 15.6 | 83.3 | 25.9 |
| 1165 | 1/1 | 9 | 42.9 | 10.1 | 22.2 | 8.1 |
| 3881 | 1/1 | 12 | 42.9 | 8.9 | 29.2 | 9.0 |
| 3412c | 2/3 | 12 | 35.7 | 13.1 | 45.8 | 21.1 |
| 2846c | 1/2 | 7 | 35.7 | 5.7 | 70.1 | 24.5 |
| 3711c | 1/1 | 13 | 35.7 | 7.0 | 41.7 | 10.7 |
| 2579c | 2/2 | 7 | 35.7 | 4.4 | 41.7 | 16.3 |
| 0274c | 2/2 | 10 | 14.3 | 9.0 | 41.7 | 10.2 |
| 3022 | 2/2 | 9 | 14.3 | 5.9 | 54.2 | 12.8 |
| 0990c | 1/2 | 8 | 7.1 | 6.4 | 58.3 | 16.4 |
| 1074 | 1/3 | 12 | 7.1 | 4.7 | 58.3 | 14.8 |
Recognized in >40% of animals tested in either group.
These pools were the most frequently or strongly recognized and were resynthesized for further analysis.
Among field reactor cattle, 54 (74.0%) pools elicited a positive response in at least one animal and five animals gave responses to ≥20 pools (median, 4). Thirteen pools elicited a positive response in ≥40% of naturally infected cattle, with a range of IFN-γ production from 8.1 to 25.89 pg/ml (Table 2). The antigens most frequently recognized by naturally infected cattle were Mb1869c, Mb2846c, Mb1074, and Mb2890.
From the results obtained with experimentally infected cattle, pools of peptides from three of the antigens were selected and constituent peptides were resynthesized in bulk to allow mapping of the dominant regions within each antigen. Pools were selected on the basis of responder frequency in experimentally infected animals, as well as specificity and magnitude of response. The antigens selected were Mb2555, Mb2890, and Mb3895. These three antigens were also selected because no responses to the relevant pools were detected with BCG-vaccinated calves. Although the pool from Mb1991 was frequently recognized, the magnitudes of response were low.
The specificities of these peptide pools were further investigated using cattle experimentally vaccinated with irradiated M. avium subsp. paratuberculosis. As the amino acid sequences had also been screened against the M. avium subsp. paratuberculosis genome, the results were anticipated to be similar to those observed with BCG-vaccinated cattle or naïve cattle. However, unexpectedly all three peptide pools were strongly recognized by all four animals vaccinated against Johne's disease (data not shown).
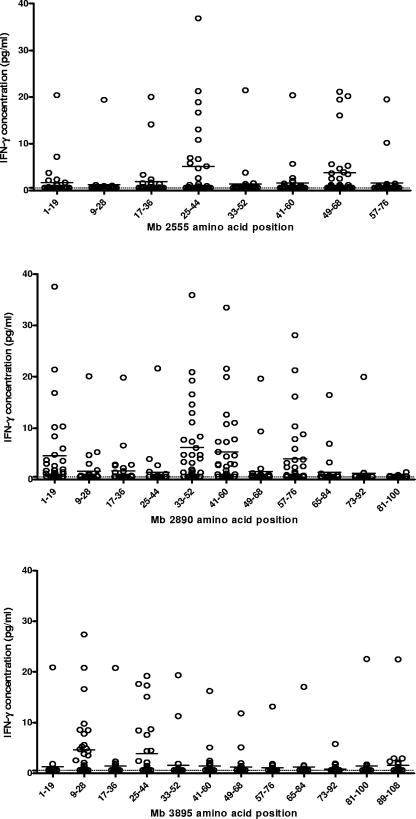
Next, responses of individual constituent peptides of the three pools were determined for M. bovis-infected cattle (Fig. 2) to define epitopic determinants. Two peptides from Mb2555 (residues 25 to 44 and 49 to 68) gave a combined sensitivity of 41.9% (mean IFN-γ concentration, 12.23 pg/ml), compared with 48.3% (13.4 pg/ml) if all eight peptides covering the protein were used. Four of the 11 peptides from Mb2890 (residues 1 to 19, 33 to 52, 41 to 60, and 57 to 76) gave sensitivity of 58.1%, compared with 64.5% for the complete pool (mean IFN-γ concentration, 12.88 pg/ml). Of the 12 peptides spanning the length of Mb3895, a combination of 2 peptides (residues 9 to 28 and 25 to 44) gave 41.9% sensitivity, compared with 51.6% for all 12 peptides. The combination of eight peptides from the three pools produced a sensitivity of 63%.
FIG. 2.
IFN-γ production in response to individual peptides from Mb2555, Mb2890, and Mb3895, tested with 11 M. bovis-infected cattle exposed in the field and 21 cattle infected with M. bovis experimentally. The horizontal bars show the mean concentration of IFN-γ produced for each peptide. The dotted line represents the threshold for positivity in the assay (3.6 pg/ml).
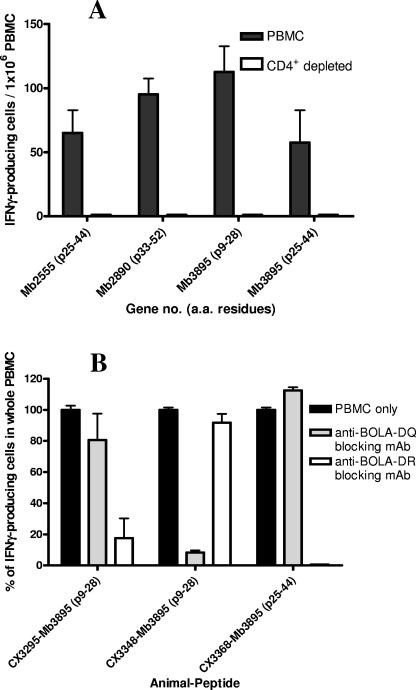
In order to characterize the peptides further with respect to responding T-cell subset, CD4+ T cells were depleted from PBMC from one experimentally infected animal (CX3368) and tested with the IFN-γ ELISPOT assay. Magnetic depletion of CD4+ PBMC left less than 1.5% CD4+ cells as demonstrated by flow cytometry (data not shown). Depletion of CD4+ T cells resulted in abrogation of all IFN-γ production for each of the four peptides tested (Mb2555, residues 25 to 55; Mb2890, residues 33 to 52; Mb3895, residues 9 to 28; and Mb3895, residues 25 to 44), as shown in Fig. 3A. The BoLA class II elements restricting the recognition of the peptides were determined next.
FIG. 3.
(A) Effect of depletion of CD4+ cells from PBMC on IFN-γ responses to peptide. Responses to peptides from each of the three antigens identified were shown to be CD4 restricted through BoLA class II. (B) Inhibition of IFN-γ production from CD4+ T cells by blocking of BoLA class II processing by monoclonal antibodies against class II subtypes DQ and DR. a.a., amino acid; p, peptide; mAb, monoclonal antibody.
The BoLA complex consists of one DR gene pair and up to two DQ gene pairs per haplotype, and in order to determine the restriction elements of the responses demonstrated to individual peptides, IFN-γ production in the presence of monoclonal antibodies blocking bovine DQ and DR molecules was determined. The two dominant peptides from Mb3895 were tested, and response to the peptide at amino acid position 25 to 44 was shown to be entirely restricted through BoLA-DR, with the monoclonal antibody against this moiety giving complete abrogation of IFN-γ production (Fig. 3B). The peptide at position 9 to 28 in Mb3895 was tested with two different animals and was found to be restricted through a different class II molecule in each animal (Fig. 3B). In animal CX3295, peptide 9 to 28 was recognized in the context of BoLA-DQ, as blocking of BoLA-DQ resulted in the reduction of IFN-γ-producing cells by 80%. In contrast, the same peptide was recognized coupled to BoLA-DR in animal CX3348, as addition of a monoclonal antibody against BoLA-DR resulted in a decline by 90% of IFN-γ-producing cells relative to whole PBMC.
DISCUSSION
Identifying potential new diagnostic antigens and vaccine candidates from the thousands of protein-encoding genes contained in the M. bovis genome can be rationalized by the application of comparative genomics. Using this method, 42 genes were short listed and synthesized as peptides, arranged into 73 pools of peptides for initial testing in groups of naïve, vaccinated, and infected cattle.
Screening of pools in these animals revealed three proteins with potential as diagnostic reagents, based on little or no response in M. bovis-naïve and BCG-vaccinated cattle and strong IFN-γ induction in M. bovis-infected cattle. We found that the hierarchies of recognition varied considerably between naturally and experimentally infected cattle, with much higher IFN-γ concentrations and responder frequencies seen among animals infected in the field. There are a number of possible explanations for this. First, the time since infection is unknown for animals infected in the field, so that the immune responses observed with such animals may be more advanced than for experimentally infected animals that were sampled at 18 weeks postinfection. Also, in the field, a higher accumulation of doses after repeated exposure may stimulate a higher and broader immune response. Finally, the same strain of M. bovis (AF2122/97) was always administered in our experimental model, while several other strains that may differ in ability to stimulate IFN-γ-mediated immune responses were isolated from the British herd. These findings highlight the importance of using naturally infected subjects to confirm the findings from animal models of infection. As a natural host of Mycobacterium bovis infection, cattle are an ideal subject for this purpose.
Mapping of responses to pools at the individual peptide level from three of the most immunogenic antigens in our screen allowed the number of peptides required for maximum sensitivity to be reduced from 31 to 8, giving a combined sensitivity of 63% and apparent specificity of 100% in relation to the BCG-vaccinated animals tested here. While this is clearly lower than the reported sensitivity of antigens such as ESAT-6 and CFP-10 (4, 17), a small proportion of cattle infected with M. bovis fail to demonstrate a response to ESAT-6 and CFP-10 and these peptides may be useful in these cases (12).
Despite the low reactivity of the peptide pools representing Mb2555, Mb2890, and Mb3895 in BCG-vaccinated and naïve cattle, all three antigens were cross-reactive in cows vaccinated with irradiated M. avium subsp. paratuberculosis, despite their apparent lack of homology with M. avium subsp. paratuberculosis genes (Table 1). This paradox was resolved by searching the M. avium subsp. paratuberculosis and M. avium subsp. avium genomes for sequences homologous to the short individual 20-mer peptide sequences of Mb2555, Mb2890, and Mb3895. This approach revealed numerous small regions of identity with unrelated M. avium subsp. paratuberculosis proteins (Table 3 shows results for Mb2890) but not with M. avium subsp. avium proteins. These were not detected when we used the whole amino acid sequence as the query. However, the potential of these peptides may still be realized. In the case of Mb2890, for example, only one peptide (p57 to p76) out of the four M. bovis immunodominant peptides described in Fig. 2 showed any significant homology with the sequence of M. avium subsp. paratuberculosis antigens (Table 3). The other three dominant peptides, which showed no such homology, could still be used for M. bovis-specific diagnosis. The diagnosis of Johne's disease (a major burden on the farming industry) is often hampered by interference induced by skin testing for BTB; therefore, this approach also has the potential to overcome difficulties in distinguishing animals infected with M. bovis from those infected with M. avium subsp. paratuberculosis that are otherwise identical (6, 11).
TABLE 3.
BLAST alignments of peptides from Mb2890 against M. avium subsp. paratuberculosis genomea
| Peptide sequence | Homology with M. paratuberculosis | Protein | Length of homolog (aa)b | Identity (%) |
|---|---|---|---|---|
| MRILPISTIKGKLNEFVDAV | No | NA | ||
| IKGKLNEFVDAVSSTQDQIT | Yes | MAP2191 | 15 | 60 |
| SigA | 12 | 75 | ||
| VDAVSSTQDQITITKNGAPA | Yes | MAP3094c | 15 | 47 |
| SigA | 12 | 75 | ||
| MmpS3 | 13 | 61 | ||
| DQITITKNGAPAAVLVGADE | Yes | MAP4037c | 15 | 60 |
| MAP3094c | 17 | 47 | ||
| MAP2984 | 7 | 100 | ||
| GAPAAVLVGADEWESLQETL | No | NA | ||
| GADEWESLQETLYWLAQPGI | No | NA | ||
| QETLYWLAQPGIRESIAEAD | Yes | MAP2324C | 10 | 70 |
| Yes | MAP3063 | 10 | 80 | |
| QPGIRESIAEADADIASGRT | Yes | MAP3034 | 17 | 64 |
| EmbR_2 | 12 | 66 | ||
| MAP2372c | 15 | 66 | ||
| MAP3898 | 13 | 69 | ||
| MAP1543 | 20 | 50 | ||
| AEADADIASGRTYGEDEIRA | Yes | MAP3898 | 13 | 69 |
| MAP3034 | 9 | 77 | ||
| EmbR_2 | 13 | 53 | ||
| InfB | 6 | 100 | ||
| SGRTYGEDEIRAEFGVPRRP | Yes | OtsA | 10 | 80 |
| MAP0023 | 35 | 37 | ||
| GRTYGEDEIRAEFGVPRRPH | Yes | OtsA | 10 | 80 |
| CmaA1 | 15 | 60 |
Showing homologs identified with their length and identity (number of identical amino acids in matching peptide sequence).
aa, amino acids; NA, not applicable.
This study has confirmed that bioinformatics can prioritize target genes for subsequent immunological analysis but that one cannot yet predict immunogenicity or T-cell cross-reactivity (and thereby specificity) and highlights that this approach has to remain an iterative process between in silico prediction and experimental in vivo and in vitro immunology. Based on the data presented here, we propose a refinement of the in silico antigen prioritization protocol. First, potential M. bovis specific antigens should be identified in silico by using full-length sequences as queries, followed by a second in silico search for short regions of homology within the selected proteins by rescreening the protein in, for example, 20-mer sections. In this way, peptides with a high degree of homology with proteins from other organisms, e.g., >50%, could be eliminated from the analysis, and peptide pools consisting of peptides with no or low homology to potentially cross-reactive proteins could be prepared for immunological analysis.
Considering the highly polymorphic BoLA class II complex and the relatively outbred nature of the United Kingdom herd, peptides for the diagnosis of bovine tuberculosis need to be highly promiscuous to give maximum sensitivity. The most immunogenic peptides identified in this study have been shown to be restricted through both DR and DQ molecules, suggesting likely widespread recognition, with presentation of one peptide mediated through both DR and DQ molecules in different animals. This phenomenon of recognition of a peptide through multiple major histocompatibility complex haplotypes has been seen previously in response to a vaccine against foot-and-mouth disease virus, where one peptide was recognized in the context of four BoLA haplotypes (15). It has previously been demonstrated that bioinformatic methods for identifying peptides that are immunogenic in humans also predict epitopes that are promiscuously recognized in cattle (18) and that antigens identified as immunogenic in cattle are also diagnostically useful for M. tuberculosis infection in humans (10). Therefore, it is likely that the peptides identified in this study will have considerable potential for the diagnosis of human tuberculosis, as the coding sequences of the eight most immunogenic peptides in M. tuberculosis are identical.
In conclusion, the combination of bioinformatics and high-throughput screening in a relevant host has allowed efficient prioritization of 42 candidate antigens from the 4,000 protein-coding genes in the M. bovis genome and identification of a subset of eight peptides with the greatest diagnostic potential. Their further evaluation through larger field studies is now a priority.
Acknowledgments
This study was funded by the Department for Environment, Food, and Rural Affairs, United Kingdom.
This study would not have been possible without the contribution of the State Veterinary Service and, in particular, Linda Farrant, in identifying naturally M. bovis-infected, tuberculin-positive cattle. We also express our appreciation to the staff of the Animal Service Unit at VLA and, in particular, Derek Clifford for their dedication to the welfare of test animals.
K.E., P.C., S.G., H.M., M.G., K.W., S.M., G.H., and M.V. have no conflicts.
REFERENCES
- 1.Brennan, M. J. 2005. The tuberculosis vaccine challenge. Tuberculosis (Edinburgh) 85:7-12. [DOI] [PubMed] [Google Scholar]
- 2.Buddle, B. M., G. W. de Lisle, A. Pfeffer, and F. E. Aldwell. 1995. Immunological responses and protection against Mycobacterium bovis in calves vaccinated with a low dose of BCG. Vaccine 13:1123-1130. [DOI] [PubMed] [Google Scholar]
- 3.Buddle, B. M., D. Keen, A. Thomson, G. Jowett, A. R. McCarthy, J. Heslop, G. W. De Lisle, J. L. Stanford, and F. E. Aldwell. 1995. Protection of cattle from bovine tuberculosis by vaccination with BCG by the respiratory or subcutaneous route, but not by vaccination with killed Mycobacterium vaccae. Res. Vet. Sci. 59:10-16. [DOI] [PubMed] [Google Scholar]
- 4.Buddle, B. M., N. A. Parlane, D. L. Keen, F. E. Aldwell, J. M. Pollock, K. Lightbody, and P. Andersen. 1999. Differentiation between Mycobacterium bovis BCG-vaccinated and M. bovis-infected cattle by using recombinant mycobacterial antigens. Clin. Diagn. Lab. Immunol. 6:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockle, P. J., S. V. Gordon, A. Lalvani, B. M. Buddle, R. G. Hewinson, and H. M. Vordermeier. 2002. Identification of novel Mycobacterium tuberculosis antigens with potential as diagnostic reagents or subunit vaccine candidates by comparative genomics. Infect. Immun. 70:6996-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coussens, P. M. 2004. Model for immune responses to Mycobacterium avium subspecies paratuberculosis in cattle. Infect. Immun. 72:3089-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert, M., A. Mitchell, D. Bourn, J. Mawdsley, R. Clifton-Hadley, and W. Wint. 2005. Cattle movements and bovine tuberculosis in Great Britain. Nature 435:491-496. [DOI] [PubMed] [Google Scholar]
- 8.Glyn Hewinson, R. 2005. TB vaccines for the world. Tuberculosis (Edinburgh) 85:1-6. [DOI] [PubMed] [Google Scholar]
- 9.Krebs, J. 1997. Bovine tuberculosis in cattle and badgers. Ministry of Agriculture, Fisheries, and Food Publications, London, United Kingdom.
- 10.Liu, X. Q., D. Dosanjh, H. Varia, K. Ewer, P. Cockle, G. Pasvol, and A. Lalvani. 2004. Evaluation of T-cell responses to novel RD1- and RD2-encoded Mycobacterium tuberculosis gene products for specific detection of human tuberculosis infection. Infect. Immun. 72:2574-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackintosh, C. G., R. E. Labes, and J. F. Griffin. 2005. The effect of Johne's vaccination on tuberculin testing in farmed red deer (Cervus elaphus). N. Z. Vet. J. 53:216-222. [DOI] [PubMed] [Google Scholar]
- 12.Pollock, J. M., R. M. Girvin, K. A. Lightbody, R. A. Clements, S. D. Neill, B. M. Buddle, and P. Andersen. 2000. Assessment of defined antigens for the diagnosis of bovine tuberculosis in skin test-reactor cattle. Vet. Rec. 146:659-665. [DOI] [PubMed] [Google Scholar]
- 13.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Lierop, M. J., P. R. Nilsson, J. P. Wagenaar, J. M. Van Noort, J. D. Campbell, E. J. Glass, I. Joosten, and E. J. Hensen. 1995. The influence of MHC polymorphism on the selection of T-cell determinants of FMDV in cattle. Immunology 84:79-85. [PMC free article] [PubMed] [Google Scholar]
- 16.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 17.Vordermeier, H. M., A. Whelan, P. J. Cockle, L. Farrant, N. Palmer, and R. G. Hewinson. 2001. Use of synthetic peptides derived from the antigens ESAT-6 and CFP-10 for differential diagnosis of bovine tuberculosis in cattle. Clin. Diagn. Lab. Immunol. 8:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vordermeier, M., A. O. Whelan, and R. G. Hewinson. 2003. Recognition of mycobacterial epitopes by T cells across mammalian species and use of a program that predicts human HLA-DR binding peptides to predict bovine epitopes. Infect. Immun. 71:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]