Abstract
In a screen for proteins that interact with Jak2, we identified a previously uncharacterized 70-kDa protein and cloned the corresponding cDNA. The predicated sequence indicates that p70 contains an SH3 domain and a C-terminal domain with similarities to the catalytic motif of phosphoglycerate mutase. p70 transcripts were found in all tissues examined. Similarly, when an antibody raised against a C-terminal peptide to analyze p70 protein expression was used, all murine tissues examined were found to express p70. To investigate the in vivo role of p70, we generated a p70-deficient mouse strain. Mice lacking p70 are viable, develop normally, and do not display any obvious abnormalities. No differences were detected in various hematological parameters, including bone marrow colony-forming ability, in response to cytokines that utilize Jak2. In addition, no impairment in B- and T-cell development and proliferative ability was detected.
A variety of cytokine receptors recruit and mediate the activation of Jak2, including the receptor for erythropoietin (Epo) (7, 13). The essential role that Jak2 plays in the function of these receptor complexes is illustrated by the loss of their function in Jak2-deficient cells (15, 16). The activation of Jak2 results in the tyrosine phosphorylation of multiple sites on the receptor chains and the subsequent recruitment of a variety of proteins to the receptor complex (5, 20, 24). The proteins recruited to the receptor complex are hypothesized to be critical for the subsequent cellular responses (see, for example, reference 4). However, in the case of the receptor for Epo, mice have been derived in which the distal half of the receptor cytoplasmic domain has been deleted and in which the single remaining tyrosine has been mutated to a phenylalanine (27). Remarkably, these mice are relatively normal, and, specifically, the Epo receptor retains its ability to support erythroid lineage expansion and differentiation. In the absence of evidence that receptor tyrosines are required for recruitment of essential signaling proteins to the receptor complex, we have focused on the possibility that sites of tyrosine phosphorylation on Jak2 may play essential roles in recruiting signaling proteins (10, 21, 27).
The activation of Jak2 initially requires transphosphorylation of a critical tyrosine in the activation loop, Y1007, comparable to most tyrosine kinases (6). Following activation of kinase activity, there are a number of additional tyrosine residues that are phosphorylated. One of the major sites of autophosphorylation within murine Jak2 is Y966 (T. Matsuda and J. N. Ihle, unpublished data). In assessing the potential role for this site, we initially sought to determine which of the known signaling proteins would bind to the site by affinity isolation. Strikingly, a number of proteins known to be involved in signal transduction bound a tyrosine-phosphorylated peptide derived from the amino acid sequence surrounding Jak2 Y966. Among these proteins were phospholipase (PLC)-γ1, PLC-γ2, the phosphatidylinositol (PI) 3-kinase adapter subunits p85α and p85β, the PI 3-kinase catalytic subunit p110δ, SHC, and Stat5a and -b. The binding was unique to Jak2-Y966 among the phosphorylation sites examined and specifically required the phosphorylation of Y966. Because of the unique properties of Y966, we used a large-scale affinity purification approach to identify novel proteins that would be recruited into complexes at this site and identified a novel protein of 70 kDa. Here we report the properties of p70, demonstrate that mice lacking p70 are not detectably altered relative to wild-type mice, and show that p70 is not required for the responses of cytokines that utilize Jak2.
MATERIALS AND METHODS
Peptide pulldown assay.
DA3 cells (11) growing in media containing 10% fetal calf serum (FCS) (HyClone) and 5 ng of interleukin 3 (IL-3)/ml were washed twice with phosphate-buffered saline (PBS) and were incubated overnight in media containing 0.5% FCS. Unstimulated cells or cells treated for 10 min with 50 ng of IL-3/ml (106/assay) were lysed in buffer containing 50 mM Tris, pH 8.0, 50 mM NaCl, 5 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5 μg of leupeptin/ml, 0.1 μg of aprotinin/ml, 100 μg of phenylmethylsulfonyl fluoride (PMSF)/ml, and 1 mM Na3VO4. Following removal of the nuclei by centrifugation at 10,000 × g, cytoplasmic lysates were incubated with 20 μl of Sepharose beads that had been conjugated by glutaraldehyde either with the peptide TSQICKGMEYLGTKR or the peptide TSQICKGMEpYLGTKR. The beads were incubated with the cytoplasmic lysates for 20 min at 4°C, washed five times with buffer containing 20 mM Tris, pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 100 μM PMSF, and 1 mM Na3VO4. Bound proteins were eluted with Laemmli sample buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by Western blot analysis as described below.
Isolation of p70 protein.
Fifty liters of DA3 cells was grown to a confluence of 2 × 106 cells/ml in RPMI medium (Gibco-Life Technologies) containing 10% FCS and 5 ng of IL-3/ml. Following two washes in PBS to remove growth factor, cells were cultured overnight in RPMI medium containing 1% FCS, pelleted, and lysed in ice-cold lysis buffer (see above). Nuclei were removed by centrifugation at 10,000 × g. Approximately 100 ml (15 mg/ml) of lysate was applied to a Q-Sepharose column (2.5 by 10 cm) previously equilibrated in ice-cold buffer containing 50 mM Tris, pH 8.0, 50 mM NaCl, 5 mM EDTA, 100 μg of PMSF/ml, and 1 mM Na3VO4. The column was washed extensively in lysis buffer, and bound proteins were step eluted with lysis buffer containing 150 mM NaCl. The eluate was passed sequentially over a Sepharose column to which had been conjugated the peptide TSQICKGMEYLGTKR (1 by 2 cm), and a Sepharose column to which had been conjugated the tyrosine-phosphorylated peptide TSQICKGMEpYLGTKR (1 by 1 cm). The latter column was washed extensively with load buffer and distilled water, and bound proteins were eluted with 2% acetic acid. Following removal of the solvent by lyophilization, the eluted material was suspended in Laemmli sample buffer, resolved by SDS-PAGE, and detected by Coomassie blue staining. The protein band representing p70 was extracted from the polyacrylamide gel, and the peptide sequence of p70 protein was obtained as described earlier (23).
Cloning of p70 cDNA.
The peptides obtained from microsequencing p70 were used to screen a database of expressed sequence tags (ESTs), identifying an EST that coded for three of the five peptides (GenBank accession no. AA190028). The EST fragment was used as a probe to screen a randomly primed mouse brain cDNA library (Stratagene). Positive clones (4) representing partial fragments of p70 cDNA were isolated in a screen of 7.5 × 105 individual clones conducted according to standard techniques, and cDNA inserts were subcloned into pBluescript and subjected to sequence analysis. Overlapping cDNA fragments were pieced together to generate a full-length cDNA.
Rabbit polyclonal antibodies.
A peptide corresponding to the predicted C terminus of p70 (CPTGGFNWRETLLQE) was synthesized, conjugated to glutaraldehyde-activated keyhole limpet hemocyanin, and used to immunize two rabbits (Rockland). Before use, antibodies were purified by affinity chromatography over a peptide column.
Transfections, immunoprecipitations and Western blots.
COS7 cell transfections were performed with Polyfect (Qiagen) according to the manufacturer's instructions. Cell lysates were made by lysing cells in ice-cold lysis buffer (see above), clarified by centrifugation, rotated at 4°C with specific antibody for 2 h, and incubated 1 h with 20 μl of protein A Sepharose. Beads were washed five times with cold wash buffer (20 mM Tris, pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 100 μM PMSF, and 1 mM Na3VO4), and bound protein was eluted with Laemmli sample buffer and separated by SDS-PAGE. Protein was transferred to nitrocellulose using a semidry transfer apparatus (Bio-Rad), blocked with 3% bovine serum albumin in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.1% Tween 20), incubated at 23°C for 1 h with specific antibody, washed with TBST, incubated for 1 h with horseradish peroxidase-conjugated secondary antibody (Amersham), and developed according to the manufacturer's instructions.
Generation of p70-deficient mice.
p70 genomic clones were isolated from a W9.5 embryonic stem cell BAC library (Incyte Genomics, St. Louis, Mo.) by screening with [α-32P]dCTP-labeled EST no. AA198028 as a probe. A restriction enzyme map of the p70 locus was determined using BAC clones and murine genomic DNA. A 10-kb fragment of p70 genomic sequence containing two exons encoding amino acid residues 9 to 88 was subcloned into pBluescript. A 2.2-kb SphI fragment containing the two exons was replaced with a cDNA encoding the neomycin resistance gene driven by the thymidine kinase promoter oriented in the direction opposite to that of p70 transcription. A cDNA encoding the diphtheria toxin gene driven by the thymidine kinase promoter (25) to be used in negative selection was then placed 3′ of the p70 homology region to complete the targeting construct. p70-deficient mice were generated essentially as previously described (22). Briefly, 129/SvJ E14 ES cells were electroporated with 20 μg of SalI-linearized targeting vector and selected with 350 μg of Geneticin (Gibco-BRL)/ml. Four hundred fifty G418-resistant clones were isolated, expanded, and analyzed by Southern analysis for correct integration of the mutation, by probing with a BamHI genomic fragment 3′ to the region of homology within the targeting construct. Five clones heterozygous for the induced mutation were injected into C57BL/6 blastocyts, which were then implanted into pseudopregnant females. Male chimeras from three clones successfully passed the induced mutation through the germ line. Heterozygote agouti mice were interbred to produce wild-type and p70-deficient littermates on a mixed 129/SvJ/BL/6 background. Genotype was determined by PCR using the following primers: wild-type allele, CGGTGAGTAAGAATCCGCTCAGTA and CACATGCTGCCTGAACGCTTCTT (300 bp); and knockout allele, CGGTGAGTAAGAATCCGCTCAGTA and GCATCGCCTTCTATCGCCTTCTTG (150 bp). Animals were housed under specific-pathogen-free conditions under institutional guidelines.
DNA and RNA analysis.
Genomic DNA was isolated by lysing tissue in buffer (100 mM Tris, pH 8.0, 5 mM EDTA, 0.2% SDS, 200 mM NaCl, and 250 μg of proteinase K/ml), incubating 12 h at 55°C, extracting with phenol-chloroform, and precipitating with ethyl alcohol. After restriction enzyme digestion, DNA was separated by agarose gel electrophoresis and transferred to positively charged nylon (Amersham) for hybridization with labeled probes (Rediprime; Amersham) according to standard techniques. RNA was prepared by extracting tissue with Trizol according to the manufacturer's instructions (Gibco BRL), separated by formaldehyde gel electrophoresis, and analyzed as described above. A Northern blot of murine poly(A)+ RNA (Clontech) was probed with a radiolabeled fragment of p70 cDNA corresponding to nucleotides encoding amino acid residues 469 to 501.
Histology.
For organ histological analysis, tissues were fixed overnight in 10% buffered formalin (Fisher Scientific). Fixed tissues were embedded in paraffin, sectioned (10 μm), and stained with hematoxylin and eosin.
FACS analysis and cell purification.
Whole spleens, thymi, or lymph nodes were dissected from littermates and crushed through a 70-μm-pore-size nylon mesh in PBS containing 2% FCS. Bone marrow was obtained by flushing femurs and tibias with the same buffer. Red blood cells were lysed by addition of buffer (pH 7.2) containing 150 mM NH4Cl, 1 mM KHCO3, and 0.1 mM EDTA; debris was removed by straining; and cells were labeled with fluorescein isothiocyanate (FITC)-conjugated antibodies as indicated. Routine fluorescence-activated cell sorter (FACS) analysis was performed with a FACSCalibur (Becton Dickinson). For proliferation assays, B and T cells were purified to >98% purity by labeling with FITC-conjugated Thy1.2 antibody (Pharmingen) and phycoerythrin (PE)-conjugated B220 antibody (Pharmingen), and sorting with a MoFlo Cell Sorter (Cytomation). In some instances, B cells were purified by complement-mediated lysis utilizing anti-Thy1.2 monoclonal antibody and a mixture of guinea pig and rabbit complement (Cederlane) according to the instructions of the manufacturer.
Proliferation assays.
B or T cells (105) were placed in individual wells of a round-bottomed 96-well plate in RPMI media containing 10% FCS, 10 mM HEPES, pH 7.0, 2 mM glutamine, 1 mM sodium pyruvate (Gibco BRL), 50 μM β-mercaptoethanol (Gibco BRL), 0.1 mM nonessential amino acids (Gibco BRL), and 10 μg of gentamicin (Gibco BRL)/ml. B-cell mitogens were at the following concentrations: immunoglobulin M (IgM), 10 μg/ml; IL-4, 100 ng/ml; anti-CD40, 1 μg/ml; and lipopolysaccharide, 10 μg/ml. For T-cell proliferation assays, a solution of 10 μg of the indicated antibody/ml was used to coat the bottom of each well by incubation at 37°C for 2 h. IL-2 at 25 ng/ml was included where indicated. Each assay was conducted in triplicate, and the data presented are representative of three individual experiments.
Blood cell counts and colony assays.
To analyze peripheral blood, samples were taken from the orbital sinus using heparinized microhematocrit capillary tubes (Fisher). Twenty microliters was analyzed by a MASCOT Hemavet 3700R counter (CDC Technologies), scoring for white blood cells and red blood cells. Hematocrits were calculated by a Hemavet counter based on red blood cell counts and mean corpuscular volume. To assay bone marrow progenitor colony-forming ability, bone marrow was flushed as described above, cells were plated, and colonies were scored as previously described (27).
RESULTS
Identification and cloning of p70.
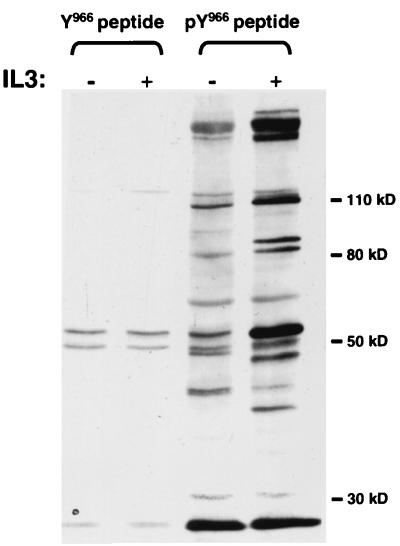
Jak2 is activated in the context of a number of cytokine receptors (12). As a consequence, it is tyrosine phosphorylated at 10 predominant sites (Matsuda and Ihle, unpublished data). Among these sites, murine Jak2-Y966 was identified as a potentially important site based on mutational analysis and on the ability of a phosphopeptide containing this site to bind a number of proteins known to be involved in signal transduction. These proteins include such molecules as Stat5a and -b, PLC-γ1, PLC-γ2, the PI 3-kinase adapter subunits p85α and -β, the PI 3-kinase catalytic subunit p110δ, and SHC (Fig. 1). To identify novel proteins that might be recruited to this site, a large-scale affinity column approach was used as detailed in Materials and Methods and the affinity-purified proteins were sequenced. Protein microsequencing of a 70-kDa protein identified five peptides, three of which were found to be encoded in the sequence of a cDNA for an uncharacterized EST (GenBank accession no. AA190028). To clone the complete p70 cDNA, one of the ESTs was used to isolate four overlapping cDNA fragments from a murine library. The predicted protein from the compiled cDNA sequences would contain 603 amino acids with a calculated molecular mass of 67.5 kDa and included all five peptides identified by microsequencing (Fig. 2A ).
FIG. 1.
Peptide pulldown assay with Jak2-derived peptides. Cells were starved of growth factor overnight and were either left untreated (−) or treated with IL-3 for 5 min (+). Lysates were prepared and incubated with Sepharose beads to which nonphosphorylated Y966 peptide (TSQICKGMEYLGTKR) or tyrosine phosphorylated pY966 peptide (TSQICKGMEpYLGTKR) was conjugated, as outlined in Materials and Methods. Bound proteins were separated by SDS-PAGE, and tyrosine-phosphorylated proteins were analyzed by Western blotting with antiphosphotyrosine antibodies.
FIG. 2.
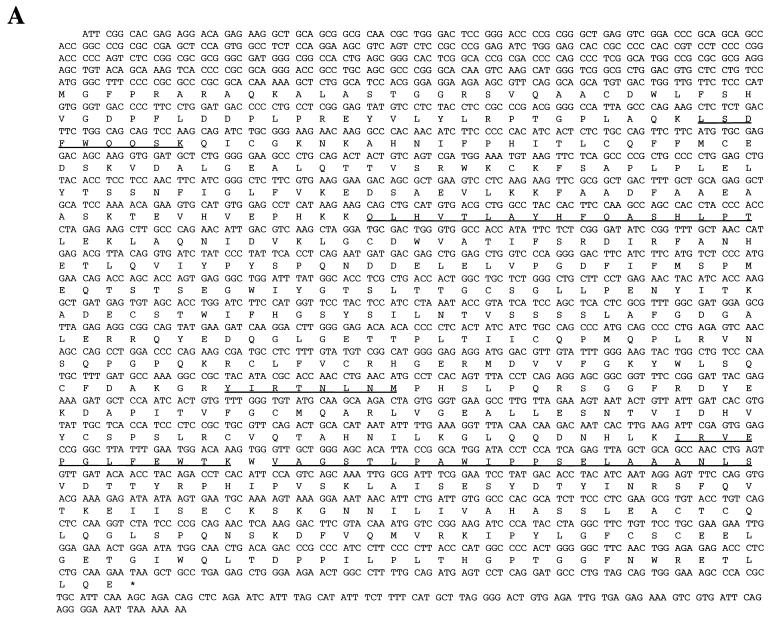
The p70 gene. (A) Nucleotide sequence and predicted amino acid sequence of p70 open reading frame. Underlined amino acids indicate peptides obtained from microsequencing-purified p70 protein, as described in Materials and Methods. (B) Alignment of C-terminal region of p70 with a consensus sequence built by the Protein Family (Pfam) database for members of the phosphoglycerate mutase family (1). The two arginines, two histidines, and glutamic acid predicted to be involved in catalytic activity are highlighted. Identical residues are indicated with two dots; chemically similar residues are indicated with one dot. (C) Peptide competition analysis. Proteins precipitated from lysates of DA3 cells by anti-p70 peptide antibody were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was probed with anti-p70 peptide antibody. Prior to immunoprecipitation, lysates were incubated for 15 min with the following: lane 1, no peptide; lane 2, antigenic peptide; and lane 3, unrelated peptide. Number of kilodaltons is given on right. (D) Interaction of p70 with phosphorylated Jak2 Y966 peptide. DA3 cytoplasmic lysates were subjected to immunoprecipitation with anti-p70 peptide antibodies (lane 1) or affinity pulldown assays utilizing nonphosphorylated Y966 peptide conjugated to Sepharose (lane 2) or phosphorylated Y966 peptide conjugated to Sepharose (lane 3). Blotting antibody was anti-p70 peptide antibody.
The protein sequence of p70 reveals two previously defined protein domains. Residues 211 to 271 are predicted to contain an SH3 domain, a domain that interacts with protein sequences containing prolines and hydrophobic residues (17, 26). A second region in the carboxyl-terminal portion is 46% similar to the catalytic domain motif found in members of the bis-phosphogylcerate mutase family. Members of this family are structurally related enzymes that catalyze the transfer of phosphogroups from small molecules such as phosphoglycerate and fructose-2,6-phosphate (1, 18). Modeling of the catalytic domain of family members reveals a hydrophobic core surrounding a constellation of active site residues (2, 9), including two conserved histidines, two conserved arginines, and a glutamic acid (14). Five residues of murine p70, Arg-344, His-345, Arg-427, Glu-455, and His-530, are present in an orientation consistent with the existence of an evolutionary relationship between p70 and members of the bis-phosphoglycerate mutase family (Fig. 2B).
Screening of databases resulted in the identification of both human and Drosophila homologues of murine p70, and, importantly, both genes contained the SH3 and potential catalytic domain of the bis-phosphoglycerate mutase family.
Biological characterization of p70.
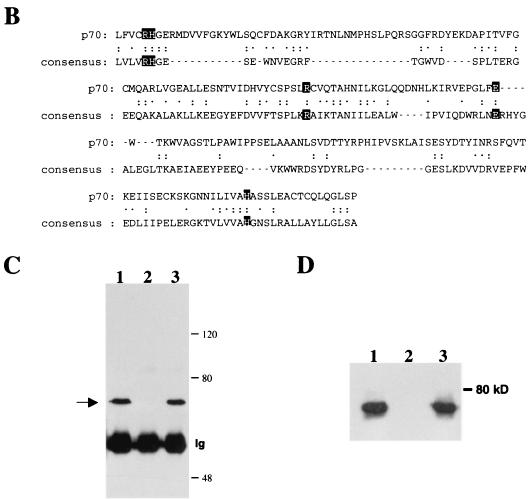
To initially characterize the biological functions of p70, an antiserum against a C-terminal peptide sequence was generated. As illustrated (Fig. 2C), this antibody recognizes a protein of 70 kDa and this recognition was blocked by the immunizing peptide but not by an unrelated peptide. Moreover, this antiserum detected a 70-kDa protein that was specifically isolated by the Jak2 Y966 phosphopeptide affinity column and was not obtained from a peptide affinity column that contained the nonphosphorylated peptide (Fig. 2D). To further assess the ability of p70 to interact with Jak2, COS7 cells were cotransfected with expression vectors for Jak2 and a C-terminal Flag-tagged version of p70. As illustrated in Fig. 3, p70 was readily detected in the immunoprecipitates of Jak2. Conversely, immunoprecipitation of p70 resulted in the coimmunoprecipitation of Jak2. However, in spite of the readily detectable association of the two proteins in cotransfection experiments, we have been unable to demonstrate the coimmunoprecipitation of the endogenous proteins under a variety of conditions resulting in the activation of Jak2.
FIG. 3.
Coimmunoprecipitation of Jak2 and p70. Cos cells were cotransfected as indicated, and lysates were subjected to immunoprecipitation (IP)/Western blot analysis with the indicated antibody.
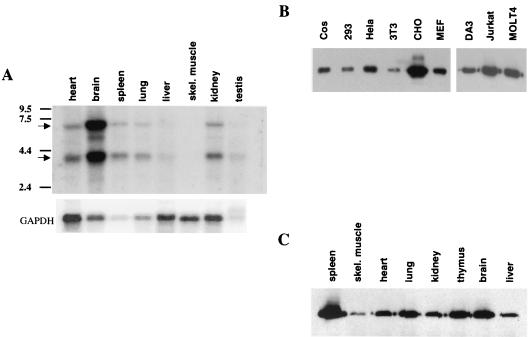
To begin to obtain insights into the potential biological function of p70, the pattern of expression of p70 was determined. As illustrated in Fig. 4A, there are two transcripts of 4 and 6 kb that contain p70 sequences. Both of these transcripts are larger than the compiled cDNA sequence, suggesting that additional 5′ and/or 3′ untranslated regions exist. By Northern analysis, p70 transcripts are present in a variety of tissues. Consistent with this, a broad pattern of p70 protein expression was detected in a variety of cell lines as well as various mouse tissues.
FIG. 4.
p70 expression analysis. (A) Northern blots containing poly(A)+ RNA from multiple murine tissues were probed with a p70 cDNA probe (upper panel) or a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (lower panel). Positions of p70 mRNA and GAPDH mRNA are indicated, as are positions of standard markers, in kilobases. (B) Lysates prepared from the indicated cell lines were subjected to immunoprecipitation/Western blot analysis utilizing anti-p70 peptide antibody. (C) Lysates prepared from the indicated murine tissues were subjected to immunoprecipitation/Western blot analysis utilizing anti-p70 peptide antibody.
Derivation of p70-deficient mice.
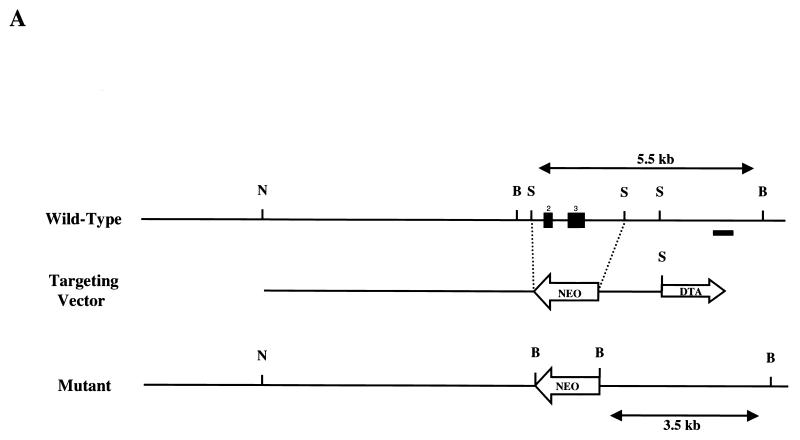
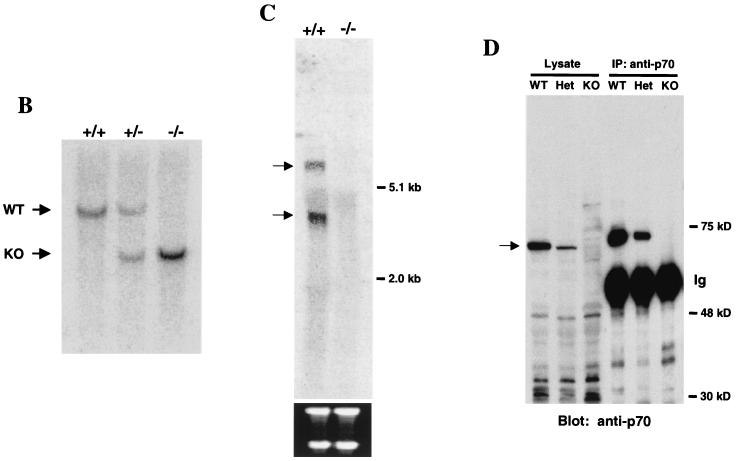
To explore the in vivo function of the p70 gene, p70-deficient mice were generated by gene targeting (3). Genomic p70 clones were isolated from a W9.5 ES cell BAC genomic library, and four partially overlapping clones were obtained. These clones did not contain the most amino terminal residues of p70 but did contain an exon encoding amino acid residues 9 to 26, designated as exon 2. As indicated in Fig. 5A, a targeting vector was constructed that would remove two coding exons (amino acids 9 to 88) and replace them with a neomycin resistance gene. The targeting vector was introduced into E14 ES cells, and five correctly targeted, independent clones were obtained. Three independent heterozygous ES cell lines were used to generate chimeric mice that transmitted the mutated allele through the germ line. Mice heterozygous for the induced mutation were interbred to produce homozygous mutant mice. Transmission of the mutated allele was confirmed by Southern analysis (Fig. 5B). Northern analysis with a probe downstream of the deleted exons was utilized to confirm the absence of p70 mRNA in mice homozygous for the mutation (Fig. 5C). Finally, anti-p70 antibody was utilized to confirm the absence of p70 protein or an amino-truncated protein in homozygous mice (Fig. 5D).
FIG. 5.
p70 targeting. (A) Structure of the p70 targeting construct. The two filled boxes represent putative exons 2 and 3. NheI (N), BamHI (B), and SphI (S) sites are indicated, as is the location of the probe utilized in Southern analysis to distinguish wild-type and targeted alleles (black bar). Neo, neomycin resistance cassette; DTA, diphtheria toxin cassette. (B) Southern blot of BamHI-digested genomic DNA from +/+, +/−, and −/− mice probed with the external probe illustrated in Fig. 4A. Bands generated by the wild-type (+/+), heterozygote (+/−), and targeted (−/−) alleles are indicated. WT, wild type; KO, knockout. (C) Northern analysis of RNA isolated from wild-type (+/+) or p70-deficient (−/−) thymi. The blot was probed with a p70 cDNA probe that was 3′ to the targeted exons as indicated in Materials and Methods. The bottom panel indicates the 28S and 18S RNAs as loading controls. (D) Immunoprecipitation (IP)/Western blot analysis of levels of p70 protein in splenic lysates prepared from +/+ (WT), +/− (Het), and −/− (KO) littermates.
Analysis of p70-deficient mice.
Interbreeding of p70 heterozygote mice produced litters with the expected Mendelian ratio of +/+, +/−, and −/− mice (1:2:1). At a gross level, p70-deficient mice were indistinguishable from their wild-type and p70 heterozygote littermates, developed normally, appeared healthy, did not succumb to any unusual pathology, and survived with a comparable longevity. Similarly, male and female p70−/− mice did not display any defects in their reproductive abilities. Histological examination of various organs revealed no differences between wild-type and p70-deficient mice. In addition, the architectures of the thymus, spleen, and lymph nodes were indistinguishable from those in wild-type mice. Blood serum chemistry (levels of albumin, alanine aminotransferase, amylase, bilirubin, creatinine, cholesterol, glucose, globulin, and cations) was not significantly different from that of wild-type mice. Additionally, wild-type and knockout mice that were starved overnight and administered glucose displayed similar responses to intraperitoneal glucose tolerance tests.
Because of the relatively high expression of p70 in hematopoietic tissues, various hematological parameters were examined. There were no significant differences between wild-type and mutant mice in the number of red cells, hemoglobin levels, or hematocrits. Similarly, there were no significant effects on the numbers and morphology of peripheral blood leukocytes, including neutrophils, eosinophils, basophils, monocytes, and lymphocytes. The recovery of bone marrow cells was normal, and in colony assays of bone marrow hematopoietic progenitors, there were no significant alterations in the frequency and morphology of colonies in response to a variety of cytokines (Table 1). Importantly, the response to cytokines that are known to require Jak2, including Epo and IL-3, was not detectably affected by the absence of p70.
TABLE 1.
In vitro colony formation of bone marrow hematopoietic progenitors from wild-type and p70-deficient littermates in response to various cytokinesa
| Colony type | Cytokine | No. of colonies formed from p70 genotype
|
|
|---|---|---|---|
| +/+ | −/− | ||
| CFU-E | Epo | 121 ± 9 | 100 ± 13 |
| BFU-E | Epo, IL-3 | 27 ± 5 | 25 ± 5 |
| CFU-G | G-CSF | 39 ± 3 | 37 ± 16 |
| CFU-GM | GM-CSF | 27 ± 7 | 33 ± 8 |
| CFU-Mix | IL-3 | 43 ± 12 | 41 ± 10 |
| CFC-Mix | IL-3, IL-6, SCF, Epo | 12 ± 2 | 16 ± 3 |
| CFU-Eos | IL-5 | 37 ± 9 | 29 ± 5 |
| CFU-Meg | Tpo | 11 ± 2 | 13 ± 5 |
| CFU-Mix | SCF | 32 ± 5 | 37 ± 4 |
| Pre-B | IL-7 | 13 ± 4 | 10 ± 2 |
Bone marrow cells from four animals of each genotype were prepared for colony assay.
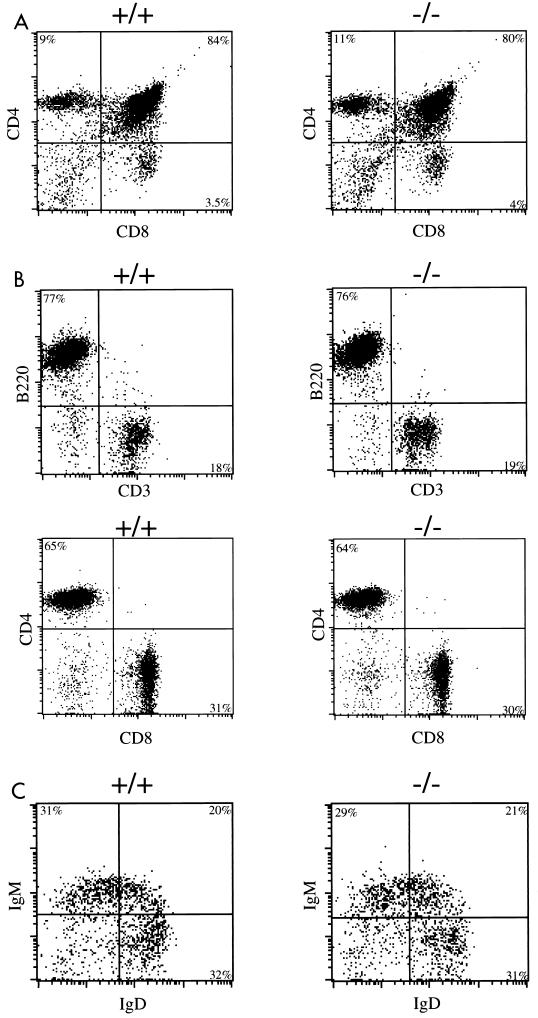
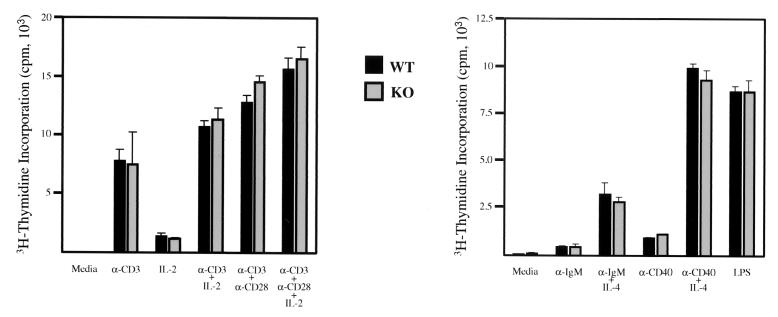
With regard to the lymphoid lineages, we observed normal numbers of pro-B (IgM− B220low, CD43+, and CD25−), pre-B (IgM−, B220+, CD43−, and CD25+), and mature B (IgM+, B220+, CD43−, and CD25−) cells in the bone marrow of p70-deficient mice (data not shown) (8). Similarly, levels of bone marrow CD117+, Ter119+, Gr1+, and Mac1+ cells were normal in the knockout mice. Flow cytometry analysis of developing thymocytes revealed no difference in numbers and proportions of double-negative (CD4− CD8−), double-positive (CD4+ CD8+), and single-positive (CD4+ CD8− or CD4− CD8+) (Fig. 6A) cells, indicating that T-cell development is unaffected by p70 deficiency. Peripheral T-cell numbers, as measured by anti-Thy1.2 or anti-CD3 reactivity, were likewise unaffected by the absence of p70, as were the levels and ratio of CD4 or CD8 single-positive T cells (Fig. 6B). To assess whether the deletion of p70 affected mature B-cell development, we analyzed by flow cytometry B-cell populations in the spleen and lymph nodes by using antibodies to B220, CD19, IgM, and IgD. There was no significant difference in numbers and maturation of peripheral B cells between +/+ and −/− mice (Fig. 6C). We then analyzed the proliferative ability of peripheral lymphocytes when stimulated by different agents. B and T cells were purified by FACS and were cultured in the presence of various mitogenic agents. Figure 7 demonstrates the absence of a significant difference between the ability of wild-type and p70-deficient B and T cells to respond to different stimuli.
FIG. 6.
Flow cytometry analysis of T and B cells. Calculated percentages are displayed within each quadrant. (A) Thymocytes from wild-type and p70-deficient mice were stained with Cyc-conjugated anti-CD4 and FITC-conjugated anti-CD8. (B) Splenocytes from wild-type and p70-deficient mice were stained with Cyc-conjugated anti-B220 and FITC-conjugated anti-CD3 antibodies (top pair). Alternatively, splenocytes were stained with PE-conjugated anti-Thy1.2, Cyc-conjugated anti-CD4, and FITC-conjugated anti-CD8. The CD4/CD8 profile of splenocytes gated on Thy 1.2-positive cells is displayed (bottom panel). (C) Splenocytes from +/+ or −/− mice were stained with FITC-conjugated anti-IgD, PE-conjugated anti-IgM, and Cyc-conjugated anti-B220. The IgM/IgD profile of splenocytes gated on B220-positive cells is displayed. Shown are representative FACS analyses of at least six mice of each genotype.
FIG. 7.
T- and B-cell proliferation assays. FACS-purified splenic T cells (left) or FACS-purified splenic B cells (right) were cultured in the presence of the indicated stimuli for 48 h. Cell proliferation was measured by adding 1 μCi of [3H]thymidine to the cultures. Standard deviations for triplicate samples are indicated by the error bars. These data are representative of three T-cell and B-cell proliferation assays. LPS, lipopolysaccharide; WT, wild type; KO, knockout.
DISCUSSION
The studies presented here are part of a general effort to identify proteins that are recruited to cytokine receptor complexes through the direct, or indirect, interaction with tyrosine-phosphorylated Jak2. As illustrated, a novel 70-kDa protein was identified that specifically binds to a peptide containing one of the major sites of autophosphorylation of murine Jak2, Tyr-966, and that coimmunoprecipitates with Jak2 in transfected cells. The basis for the interaction with Jak2 has not been precisely defined. In particular, although binding is specific for the phosphorylated peptide, binding occurs in the absence of any known phosphotyrosine binding domains. It is therefore likely that an adapter protein is involved in the binding. For example, a catalytic subunit of PI 3-kinase, p110δ, is also isolated from the Y966 phosphopeptide affinity column through its association with the PI 3-kinase adapter subunits p85α and -β (S.-T. Jou and Ihle, unpublished data). Studies are in progress to assess the potential for p70 to interact with several known adapter proteins.
As illustrated, interaction with Jak2 can also be shown to occur in cells in which the two proteins are overexpressed. However, numerous attempts to demonstrate coimmunoprecipitation of endogenous proteins under conditions of activation of Jak2 have failed. It is possible that the fraction of Jak2 activated in these situations, which would be limited by the receptor numbers, is too low to allow detection of the proteins. Alternatively, it is possible that overexpression gives rise to associations that are not physiologically evident. Similarly, isolation on affinity columns may be occurring at protein concentrations that are not present in a physiologically relevant receptor complex. In this regard it is important to note that, although Stat5a and Stat5b bind specifically to the tyrosine-phosphorylated mJak2-Y966 peptide, the recruitment and activation of Stat5a and -b in the context of the Epo receptor require specific receptor tyrosines (19). Since it was not possible to strictly rule in, or out, a role for p70 in Jak2 activity based on these biochemical approaches, we also utilized a genetic approach.
Beyond a potential role in signaling from cytokine receptors utilizing Jak2, the properties of p70 provide few additional clues regarding its physiological role. The gene is conserved, as evidenced by the existence of homologues in other species. However, in these species, p70 is similarly an uncharacterized gene product of an EST and consequently nothing is known regarding its function. The gene is widely expressed in mouse tissues, which does not allow a focus for function on a particular lineage of cells or tissue. The protein contains an SH3 domain that would implicate it in signal transduction. The protein also contains a domain with homology to the catalytic domain of members of the bis-phosphoglycerate mutase family of enzymes. However, we have been unable to demonstrate a catalytic function for this region of p70. Members of the phosphoglycerate mutase family of enzymes are not known to have protein interaction domains, such as an SH3 domain. The function of the SH3 domain of p70 might be either to target it to a specific molecular complex or to enable its interaction with regulatory proteins.
One important approach to defining gene function is to determine the consequences of eliminating the gene in the general context of mouse development. As illustrated here, we were able to develop a strain of mice in which the p70 gene contained an internal deletion that resulted in a protein null phenotype. Extensive characterization of these mice has failed to identify any phenotypic characteristics that are different from those of wild-type mice. Most specifically, none of the receptor systems that utilize Jak2 that we examined were affected by the absence of p70, demonstrating the lack of a nonredundant, essential role in signaling. The absence of any phenotypic alterations in such cases can often be attributed to functional redundancy between family members.
Acknowledgments
The technical assistance of Linda Synder, Kristen Rothammer, and Sandra Moore is gratefully acknowledged. We thank John Raucci for his expert assistance in generating p70-deficient mice and the personnel of the Animal Resources Center for care and monitoring of animals. The support of Neena Carpino, Mahnaz Paktinat, Richard Cross, and Virginia Valentine for FACS and karyotypic analysis is gratefully appreciated. Special thanks to past and present members of the Ihle lab for fruitful discussions.
This work was supported by NRSA 5-F32-CA83269 to N.C., by Cancer Center CORE grant CA21765, by grants RO1 DK42932 and PO1 HL53749 to J.N.I., and by the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazan, J. F., R. J. Fletterick, and S. J. Pilkis. 1989. Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Proc. Natl. Acad. Sci. USA 86:9642-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capecchi, M. R. 1994. Targeted gene replacement Sci. Am. 270:52-59. [DOI] [PubMed] [Google Scholar]
- 4.Carter-Su, C., L. Rui, and M. R. Stofega. 2000. SH2-B and SIRP: JAK2 binding proteins that modulate the actions of growth hormone. Recent Prog. Horm. Res. 55:293-311. [PubMed] [Google Scholar]
- 5.Damen, J. E., L. Liu, H. Wakao, A. Miyajima, P. Rosten, A. B. Jefferson, P. W. Majerus, J. Krosl, R. K. Humphries, and G. Krystal. 1997. The role of erythropoietin receptor tyrosine phosphorylation in erythropoietin-induced proliferation. Leukemia 3(Suppl.):423-425. [PubMed] [Google Scholar]
- 6.Feng, J., B. A. Witthuhn, T. Matsuda, F. Kohlhuber, I. M. Kerr, and J. N. Ihle. 1997. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol. Cell. Biol. 17:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadina, M., D. Hilton, J. A. Johnson, A. Morinobu, A. Lighvani, Y. J. Zhou, R. Visconti, and J. J. O'Shea. 2001. Signaling by type I and II cytokine receptors: ten years after. Curr. Opin. Immunol. 13:225-268. [DOI] [PubMed] [Google Scholar]
- 8.Hardy, R. R., and K. Hayakawa. 2001. B cell developmental pathways. Annu. Rev. Immunol. 19:595-621. [DOI] [PubMed] [Google Scholar]
- 9.Hasemann, C. A., E. S. Istvan, K. Uyeda, and J. Deisenhofer. 1996. The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure 4:1017-1029. [DOI] [PubMed] [Google Scholar]
- 10.He, T. C., N. Jiang, H. Zhuang, and D. M. Wojchowski. 1995. Erythropoietin-induced recruitment of Shc via a receptor phosphotyrosine-independent, Jak2-associated pathway. J. Biol. Chem. 270:11055-11061. [DOI] [PubMed] [Google Scholar]
- 11.Ihle, J. N., and D. Askew. 1989. Origins and properties of hematopoietic growth factor-dependent cell lines. Int. J. Cell Cloning 7:68-91. [DOI] [PubMed] [Google Scholar]
- 12.Ihle, J. N. 1996. Janus kinases in cytokine signaling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:159-199. [DOI] [PubMed] [Google Scholar]
- 13.Ihle, J. N., W. Thierfelder, S. Teglund, D. Stravopodis, D. Wang, J. Feng, and E. Parganas. 1998. Signaling by the cytokine receptor superfamily. Ann. N. Y. Acad. Sci. 865:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Jedrzejas, M. J. 2000. Structure, function, and evolution of phosphoglycerate mutases: comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog. Biophys. Mol. Biol. 73:263-287. [DOI] [PubMed] [Google Scholar]
- 15.Neubauer, H., A. Cumano, M. Muller, H. Wu, U. Huffstadt, and K. Pfeffer. 1998. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93:397-409. [DOI] [PubMed] [Google Scholar]
- 16.Parganas, E., D. Wang, D. Stravopodis, D. Topham, J. C. Marine, S. Teglund, E. F. Vanin, S. Bodner, O. R. Colamonici, J. M. van Deursen, G. Grosveld, and J. N. Ihle. 1998. Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385-395. [DOI] [PubMed] [Google Scholar]
- 17.Pawson, T., and J. D. Scott. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075-2080. [DOI] [PubMed] [Google Scholar]
- 18.Pilkis, S. J., M. O. Lively, and M. R. el-Maghrabi. 1987. Active site sequence of hepatic fructose-2,6-bisphosphatase. Homology in primary structure with phosphoglycerate mutase. J. Biol. Chem. 262:12672-12675. [PubMed] [Google Scholar]
- 19.Quelle, F. W., D. Wang, T. Nosaka, W. E. Thierfelder, D. Stravopodis, Y. Weinstein, and J. N. Ihle. 1996. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol. Cell. Biol. 16:1622-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui, L., D. R. Gunter, J. Herrington, and C. Carter-Su. 2000. Differential binding to and regulation of JAK2 by the SH2 domain and N-terminal region of SH2-Bβ. Mol. Cell. Biol. 20:3168-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeshita, T., T. Arita, M. Higuchi, H. Asso, K. Endo, H. Kuroda, N. Tanaka, K. Murata, N. Ishii, and K. Sugamura. 1997. STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity 6:449-457. [DOI] [PubMed] [Google Scholar]
- 22.Van Deursen, J., A. Heerschap, F. Oerlemans, W. Ruitenbeek, P. Jap, H. ter Laak, and B. Wieringa. 1993. Skeletal muscles of mice deficient in muscle creatine kinase lack burst activity. Cell 74:621-631. [DOI] [PubMed] [Google Scholar]
- 23.Wang, R., R. Kobayashi, and J. M. Bishop. 1996. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc. Natl. Acad. Sci. USA 93:8425-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witthuhn, B. A., F. W. Quelle, O. Silvennoinen, T. Yi, B. Tang, O. Miura, and J. N. Ihle. 1993. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell 74:227-246. [DOI] [PubMed] [Google Scholar]
- 25.Yagi, T., S. Nada, N. Watanabe, H. Tamemoto, N. Kohmura, Y. Ikawa, and S. Aizawa. 1993. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal. Biochem. 214:77-86. [DOI] [PubMed] [Google Scholar]
- 26.Yu, H., J. K. Chen, S. Feng, D. C. Dalgarno, A. W. Brauer, and S. L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933-945. [DOI] [PubMed] [Google Scholar]
- 27.Zang, H., K. Sato, H. Nakajima, C. McKay, P. A. Ney, and J. N. Ihle. 2001. The distal region and receptor tyrosines of the Epo receptor are non-essential for in vivo erythropoiesis. EMBO J. 20:3156-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]