Abstract
Human mitochondrial 12S rRNA A1555G mutation has been found to be associated with deafness. However, putative nuclear modifier gene(s) has been proposed to regulate the phenotypic expression of this mutation. In yeast cells, mutant alleles of MSS1, encoding a mitochondrial GTP-binding protein, manifest a respiratory-deficient phenotype only when coupled with mitochondrial 15S rRNA PR454 mutation corresponding to human A1555G mutation. This suggests that an MSS1-like modifier gene may influence the phenotypic expression of the A1555G mutation. We report here the identification and characterization of human MSS1 homolog, GTPBP3, the first identified vertebrate gene related to mitochondrial tRNA modification. The Gtpbp3 is the mitochondrial GTPase evolutionarily conserved from bacteria to mammals. Functional conservation of this protein is supported by the observation that isolated human GTPBP3 cDNA can complement the respiratory-deficient phenotype of yeast mss1 cells carrying PR454 mutation. GTPBP3 is ubiquitously expressed in various tissues as multiple transcripts, but with a markedly elevated expression in tissues of high metabolic rates. We showed that Gtpbp3 localizes in mitochondrion. These observations suggest that the human GTPBP3 is a structural and functional homolog of yeast MSS1. Thus, allelic variants in GTPBP3 could, if they exist, modulate the phenotypic manifestation of human mitochondrial A1555G mutation.
A variety of mitochondrial DNA (mtDNA) mutations have been found to be associated with many clinical abnormalities, including neuromuscular disorders, hearing loss, and diabetes (18, 45, 48). However, the nuclear background influences the phenotypic expression of pathogenic mtDNA mutations associated with human diseases. For example, different nuclear backgrounds confer a marked advantage to either the A3242G mutation in the tRNALeu(UUR) gene or wild-type mitochondrial genome (14, 50). Another example is that the nuclear background plays a determining role in biochemical phenotype of the deafness-associated A1555G mutation (23, 25). This mutation, which results from the A-to-G transition at position 1555 in the mitochondrial 12S rRNA gene, has been found to be associated with aminoglycoside-induced deafness and nonsyndromic deafness in families of various ethnic backgrounds (19, 30, 42). In the absence of aminoglycosides, the A1555G mutation produces a clinical phenotype that ranges from severe congenital deafness, through moderate progressive hearing loss of later onset (17), to completely normal hearing (17, 42). More-severe biochemical defects were observed in the mutant lymphoblastoid cell lines derived from symptomatic individuals from an Arab-Israeli family than from those of cell lines derived from asymptomatic individuals in the same family (23). These genetic and biochemical data strongly point out that the A1555G mutation is a primary factor underlying the development of deafness (23, 25). However, the nuclear modifier gene(s), or aminoglycoside antibiotics, play a synergistic role in aggravating the hearing impairment associated with the A1555G mutation (8, 19, 23, 24, 25, 42). The product of modifier nuclear gene(s), which may functionally interact with the mutated 12S rRNA, influences the phenotypic manifestation of the A1555G mutation by enhancing or suppressing the effect of the mutation (23). Despite the great efforts made last 10 years, as yet such nuclear modifier genes still remain to be identified (9).
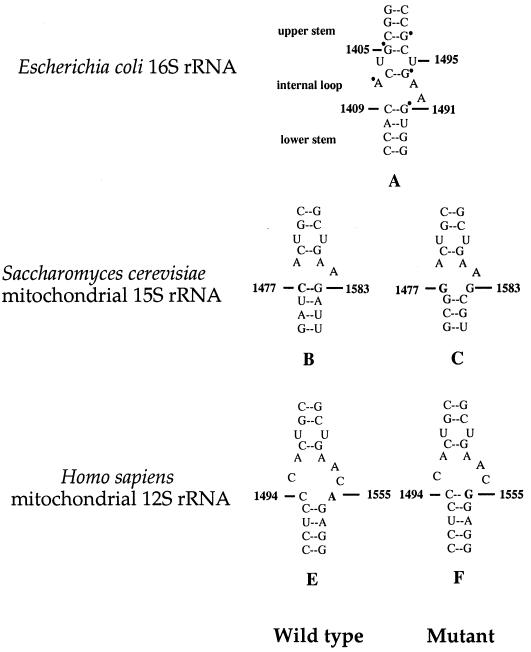
As shown in Fig. 1, the A1555G mutation is located in the region of small rRNA highly conserved from bacteria to mammals (41). The corresponding region in Escherichia coli forms an essential part of the decoding site of the ribosome (52) and is crucial for subunit association either by RNA-protein or RNA-RNA interaction (53). The same region of the bacterial small rRNA is also known to bind aminoglycoside antibiotics (39), and mutations within this region conferring antibiotic resistance have been isolated in bacteria (13, 22) and yeast mitochondria (36, 49). In fact, the new G-C pair in the human mitochondrial 12S rRNA created by the A1555G mutation facilitates the binding of aminoglycoside (27), which accounts for aminoglycosides induced hearing loss in the individuals carrying this mutation (19, 30, 42).
FIG. 1.
Secondary structure of decoding site of small rRNAs. The A-site of E. coli 16S rRNA oligonucleotide showing the DMS footprints, observed in the presence of the aminoglycosides neomycin, paromomycin (22, 39), is marked with a dot (in structure A). The corresponding regions of S. cerevisiae mitochondrial 15S rRNA and human mitochondrial 12S rRNA are shown as the wild-type versions (structures B and E) and in the versions containing the PR454 mutation (structure C) and A1555G mutation (structure F), respectively.
With the aim of identifying nuclear modifier genes, the yeast Saccharomyces cerevisiae has been used as a model organism to isolate the nuclear mutations that are involved in the phenotypic manifestation of the A1555G mutation. Interestingly, it was reported that the mutations in MSS1 or MTO1, encoding highly conserved mitochondrial proteins, expressed their respiratory-deficient phenotypes only when their mtDNA carry the PR454 mutation corresponding to the deafness-associated A1555G mutation (11, 12). In the PR454 background, mss1 or mto1 mutants fail to synthesize subunit 1 of cytochrome oxidase, thereby leading to a respiratory-deficient phenotype (11, 12). These observations strongly indicate that Mss1p or Mto1p affects the phenotypic expression of the PR454 mutation by functionally interacting with the region of the PR454 in mitochondrial 15S rRNA. In E. coli, the products of mnmE (homolog of MSS1) and gidA (homolog of MTO1) have been shown to be involved in the biosynthesis of the hypermodified nucleoside 5-methyl-aminnomethy-2-thio-uridine (mnm5s2U34) (5, 7). This modified nucleotide, found in the wobble position of several bacterial tRNAs specific for glutamate, lysine, arginine, leucine, and glutamine, has a pivotal role in the structure and function of tRNAs, including structural stabilization, aminoacylation, and codon recognition at the decoding site of small rRNA (5, 7).
On the basis of the above evidence, we have hypothesized that the human homolog MSS1 has a similar function to yeast MSS1, thereby playing a role in the phenotypic expression of deafness associated with the A1555G mutation. Thus, the isolation and characterization of the human homolog of MSS1 will lead to the deep understanding of pathogenetic mechanism of the human mitochondrial 12S rRNA A1555G mutation. In the present study, we identified and characterized the human homolog of yeast MSS1, GTPBP3 (GTP binding protein 3). First, we searched the NCBI human expressed sequence tag (EST) databases to identify potential ESTs that were homologous to the yeast Mss1p. Based on the two EST sequences, we cloned the human GTPBP3 cDNA and elucidated the genomic structure of this gene. Human GTPBP3 has been characterized by examining the gene expression in different tissues, subcellular location, and functional complementation of yeast mss1 mutants carrying the mitochondrial PR454 allele.
MATERIALS AND METHODS
Cell lines, culture conditions, and RNA extraction.
HepG2, a human hepatoblastoma cell line, was used for the extraction of RNA. Human osteosarcoma cell line 143BTK− (33) was used for the subcellular location experiment. Both 143BTK− and HepG2 cells were grown in the regular Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum. Total cellular RNA was isolated from the human HepG2 cells by using Trizol reagent (Invitrogen) according to the manufacturer's directions.
cDNA cloning of human GTPBP3.
Peptide sequence of S. cerevisiae Mss1p was subjected to a BLAST search of GenBank nucleotide sequence databases for the human ESTs and two human EST clones (GenBank accession no. BE 397995; nucleotides [nt] 470 to 994) derived from lymphoma and BE 408027 (nt 62 to 830) derived from choriocarcinoma were identified with the significant homology to yeast MSS1 open reading frame (ORF). For the identification of the 5′-terminal region of cDNA we used an antisense primer (5′-CGGAGCCTCTGCCCGCGCCTGGCATC-3′; nt 772 to 797), and to obtain the 3′-end cDNA region we used a sense primer (5′-TCCATCGTGTCCCCGGAGCCAGGGACCACC-3′; nt 880 to 909). The 5′ and 3′ regions of cDNA were obtained by reverse transcription-PCR (RT-PCR). First-strand cDNA for the use of PCR template was generated from 2 μg of RNA isolated from HepG2 cells by using the SMART RACE cDNA amplification kit (Clontech). Touch PCR was performed with Advantage-GC 2 PCR kit (Clontech). To construct the plasmid pGTPBP3 containing the entire coding region of GTPBP3 cDNA, RT-PCR was performed by using the high-fidelity Pfu DNA polymerase (Promega) and total RNA isolated from HepG2 cells as a template, with the primers containing the HindIII site (5′-CCCAAGCTTGCCCAGACTTGAAGCCACACAGGCAG-3′ [nt 1 to 26] and 5′-CCCAAGCTTAGATCCTCCTATCTCCATCCCAACCCG-3′ [nt 1794 to 1768]). The predominant PCR product was purified by agarose gel electrophoresis and subsequently cloned into a PCR 2.1-TOPO vector (Invitrogen). Nucleotide sequencing was done by using a dye terminator cycle sequencing kit (Perkin-Elmer) and an ABI PRISM 3700 genetic analyzer.
Sequence analysis.
The BLAST homology searches were performed with the available web-based programs of the National Center for Biotechnology Information (2). DNA and protein sequence alignments were carried out by using Seqweb program GAP (GCG). The percentages of identical and similar amino acids were calculated. The working draft sequences from bacterial artificial chromosome (BAC) clone AC010463 were assembled into a complete contiguous genomic sequence by using the BLAST program to compare these sequences with sequence of human GTPBP3 cDNA. The splice donor and acceptor site sequence of GTPBP3 genomic DNA was determined as described previously (43-44).
Northern blot analysis of GTPBP3 expression.
A 12-lane human multiple tissue RNA blot and an 8-lane human brain tissue RNA blot (Clontech) containing 2 μg of poly(A)+ RNA/lane were used for the present study. A 636-bp GTPBP3 cDNA fragment corresponding to nt 1220 to 1855 was random prime labeled with [32P]dATP and hybridized with the RNA blots according to the manufacturer's instructions. Membranes were then washed to a final stringency of 0.1X SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)- 0.1% sodium dodecyl sulfate at 65°C for 40 min. As an internal control, the human RNA blots were stripped and rehybridized with [32P]dATP-labeled pACT1 containing human actin β-subunit cDNA. The plasmid pACT1 was constructed by amplifying a 208-bp fragment of the human actin β-subunit gene (GenBank accession no. BC014861; nt 820 to 1027) by PCR and cloning it into the PCR 2.1-TOPO vector (Invitrogen).
Functional complementation.
The S. cerevisiae wild-type strain used for the present study was W301-1B (MATα ade2-1 trp1-1 his 3-11,15 leu2-3,112 ura3-1). The mss1 strains used in this study were W303ΔMSS1(PR454) (MATa ade2-1 trp1-1 his3-11,15 leu2-3,112 ura3-1 mss1::HIS3), and W303ΔMSS1(PS) (MATa ade2-1 trp1-1 his3-11,15 leu2-3,112 ura3-1 mss1::HIS3) (11). All yeast strains were grown in GYP medium (0.5% yeast extract Difco, 1% Bacto Peptone Difco, and 2% glucose) and GlyYP medium (0.5% yeast extract Difco, 1% Bacto Peptone Difco, and 2% glycerol). Minimal medium contains 0.67% Difco yeast nitrogen base without amino acids. This medium was supplemented with amino acids at 50 μg/ml or base at 25 μg/ml to complement the auxotrophic requirements.
The pDB20 was used for the expression of GTPBP3 in S. cerevisiae. The pDB20-GTPBP3 was constructed by inserting full-coding region of human GTPBP3 cDNA (pGTPBP3) at HindIII site of pDB20 (4). These constructs were transformed into the mss1 strain of S. cerevisiae by the method of Gietz and Schiestl (20). The Ura+ transformants were selected at 30°C on minimal glucose medium. The transformants were then replica plated on GYP or GlyYP plates, followed by incubation at 30°C for 4 days. Colonies growing on GlyYP medium were subjected to further analysis.
Subcellular localization of human Gtpbp3.
The coding region of human GTPBP3 cDNA lacking its natural stop codon was obtained by PCR by using the high-fidelity Pfu polymerase (Promega) and pGTPBP3 as the template. The primers 5′-CCCAAGCTTGCCCAGACTTGAAGCCACACAGGCAG (nt 1 to 26) and 5′-CCCAAGCTTGCCCACACAGAAGTCCTGGAA (nt 1533 to 1510) were used for the PCR amplification. The PCR products were digested with HindIII and cloned into pBluescript II KS(+) (Promega). After sequence determination, the inserts were subcloned into pEGFP-N1 (Clontech).
The resultant constructs were transfected into 143B cells by using SuperFect transfection reagent (Qiagen, Inc.) according to manufacturer's protocol. Transfected cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum for 30 h. The chamber slides were then washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde-PBS for 10 min, and permeabilized in 0.1% Triton X-100-PBS for 10 min. The cells were subsequently incubated with mouse anti-human COX1-Alexa Fluor 594 conjugate (10 μg/ml; Molecular Probes) and rabbit anti-GFP-immunoglobulin G-Alexa Fluor 488 conjugate (4 μg/ml; Molecular Probes) in 5% normal goat serum-PBS at 37°C for 2 h. The chamber slides were then viewed under a Zeiss confocal fluorescence microscope.
RESULTS
Identification of human GTPBP3 cDNA, encoding a putative homolog of yeast Mss1p.
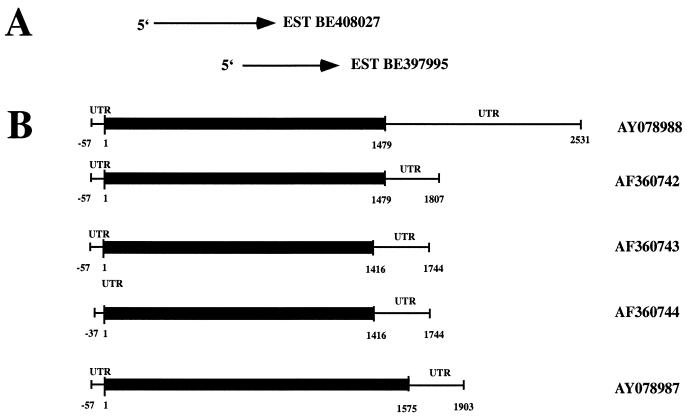
S. cerevisiae Mss1p sequence was subjected to a BLAST search of the NCBI/GenBank human EST databases. Two ESTs derived from human Burkitt lymphoma (BE 397995) and choriocarcinoma (BE 408027) were identified with significant homology to the N and C termini of yeast MSS1 ORF (Fig. 2).
FIG. 2.
Identification of human GTPBP3 cDNAs. (A) Two EST clones were used to obtain putative human GTPBP3 cDNA, which are aligned with the corresponding human GTPBP3 cDNA. (B) Structure of GTPBP3 cDNAs. The protein-coding regions of GTPBP3 are indicated by a black rectangle. Accession numbers for cDNAs and EST sequences are shown on the right.
Based on the sequence of the cDNA identified by a search of the human EST databases, we designed primers to isolate the full-length coding region and the noncoding 5′ and 3′ regions of the putative human homolog of MSS1. By using total RNA isolated from HepG2 cells as a template, a cDNA containing an ORF of 1,479 bp has been identified. This cDNA encodes 492 amino acids with a predicted molecular mass of 52,030 Da. This gene has been tentatively designated GTPBP3 (GTP-binding protein 3). To investigate the 5′ sequence of the human GTPBP3 transcript, we performed 5′-RACE (rapid amplification of cDNA ends) with total RNA isolated from HepG2 cells as a template. Through 5′-RACE, three clones were obtained. These 5′-end cDNA clones contain only one ATG codon. If we assume this ATG to be the initiation codon, the 5′-untranslated region (UTR) consists of 57 or 37 bp (Fig. 2). The 3′-RACE led to the identification of five different clones. Four clones share identical 328 bp length of 3′-UTR, whereas the 3′-UTR of the fifth clone extends an additional 713 bp. Using the primers homologous to the 5′ and 3′ ends of the cDNA, the full length of GTPBP3 cDNA of 2,588 bp has been isolated and deposited in GenBank (accession no. AY078988). In addition, four other transcripts, which are likely splicing variants with 1,864 bp (isoform II), 1,801 bp (isoform III), 1,781 bp (isoform IV), and 1,960 bp (isoform V), have been obtained, and their cDNA sequences have been deposited in GenBank (accession no. AF360742, AF0360743, AF360744, and AY078987, respectively).
Genomic structure of GTPBP3.
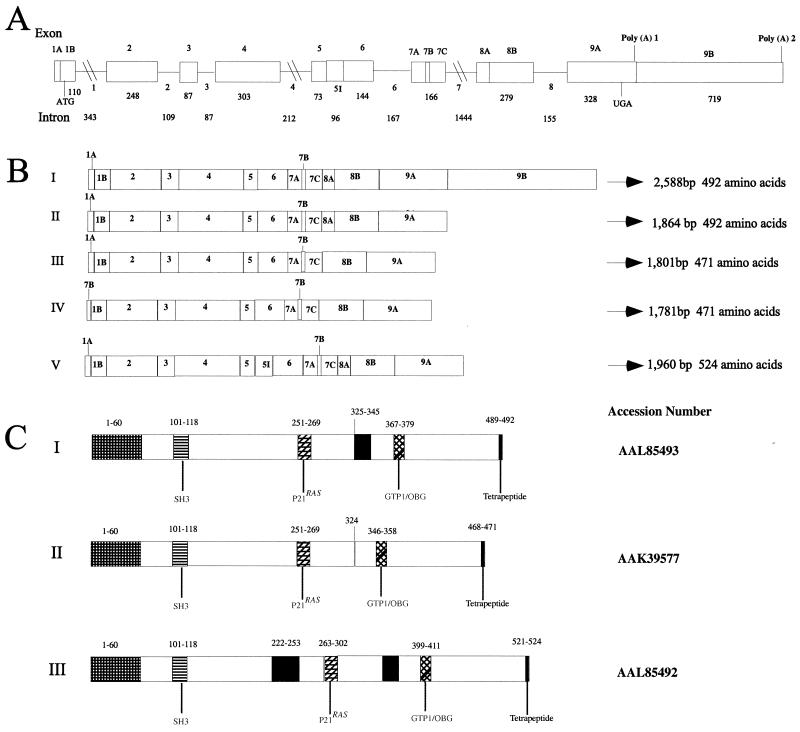
We used the GTPBP3 cDNA sequence as a query to search available the EMBO/GenBank high-throughout genome database. This search led to the identification of working-draft sequences of a BAC clone (accession no. AC010463) that contains the full length of the genomic GTPBP3 sequence. In order to define the actual intron-exon organization of the human GTPBP3-transcripted sequence, we used the BAC sequence data in conjunction with our cDNA sequence data to assemble 8-kb genomic sequences from the GTPBP3 region of chromosome 19. These data indicate that the GTPBP3 gene is composed of nine exons, which range from 73 bp to 1,027 bp in size (Fig. 3A), and eight introns varying from 87 bp (intron 3) to 1,444 bp (intron 7). The genomic organization of this GTPBP3 transcript derived from these data are summarized in Fig. 3A. In addition, two putative polyadenylation signal (AATGTAAA; nt 1843 to 1850) and (ATATAAA; nt 2543 to 2549) were observed. The intron-exon junctions are shown in Table 1. The complete nucleotide sequence of GTPBP3 has been deposited in GenBank (AF361481). Furthermore, we defined the intron-exon organization of other four transcripts by comparing these cDNA sequences with the above genomic DNA sequence data of GTPBP3. As shown in Fig. 3B, all four transcripts lack the exon 9B due to the alternative splicing. Isoform V contains intron 5, which inserts 96 bp in-frame into the coding region and produces a protein with 524 amino acids. The isoforms III and IV, which deleted 63 bp from exon 8, result in a truncated protein of 471 amino acids; however, isoform III also skipped exon 1A, which deleted 42 bp, but the addition of 22 bp from exon 7 at the 5′-UTR. The protein organization of these three isoforms is summarized in Fig. 3C. These isoforms might be of special functional interests, since they comprise in-frame insertion or deletion.
FIG. 3.
Structure of human GTPBP3 gene and its products. (A) Diagram of human GTPBP3 gene structure. Boxes represent exons, with the positions of initiation and termination codons indicated. Two putative poly(A) sites are indicated. The scale shown applies only to exons; entire region spans more than 6 kb of genomic DNA. The nine exons and eight introns are numbered, and their sizes are indicated below in base pairs. (B) Schematic representation of five GTPBP3 transcripts. The transcripts result from the alternative splicing of exons 1A, 7B, 8A, and 9B and intron 5. “I” was original identified by genomic prediction. “II to IV” result from the deletion of exons, and “V” is due to the retention of intron 5. (C) Scheme for the structure of Gtpbp3 and its isoforms. Numbers gives the position of residues in proteins in relation to the first methionine. The accession numbers are indicated in the left. The functional domains are shaded with different patterns. The N-terminal targeting sequence comprises the residues at positions 1 to 60. The region shaded gray is the 21-residue deletion at positions 325 to 345 in the isoform II, whereas the region shaded black is an insertion of 32 amino acids at position 222 in the isoform III.
TABLE 1.
Splice donor and acceptor for human GTPBP3 transcripta
| Exon | Splice donor sequence (intron size [bp]) | Splice acceptor sequence | Exon |
|---|---|---|---|
| GGGCCCCCTGCCCAG | 1 | ||
| 1 | CTCGCAGGTGGGGCT (343) | CGGTTCCAGGTTGTG | 2 |
| 2 | TTCCCAGGTGAGGGT (109) | TCACATTAGGTCCCC | 3 |
| 3 | GCCTTGGGTGAGTTG (87) | TTCCTGCAGGCAGCG | 4 |
| 4 | CACCAAAGCAAGTCC (212) | CCCCGCCAGGCTCTG | 5 |
| 5 | GAGCAAGGTGGGTCT (96) | CACCCACAGCCGACA | 6 |
| 6 | CTGCTCAGTGAGTAG (167) | CTCCCCCAGGTCGGA | 7 |
| 7 | GGGAGAGGTGGGCGG (1,444) | CTCCCGCAGGCTAGA | 8 |
| 8 | CTGCAGTGTGAGCCC (155) | CTCTTCCAGGTGTGGG | 9 |
Functional splice donor and acceptor sequences are indicated in boldface.
Human Gtpbp3 is a conserved mitochondrial GTP-binding protein.
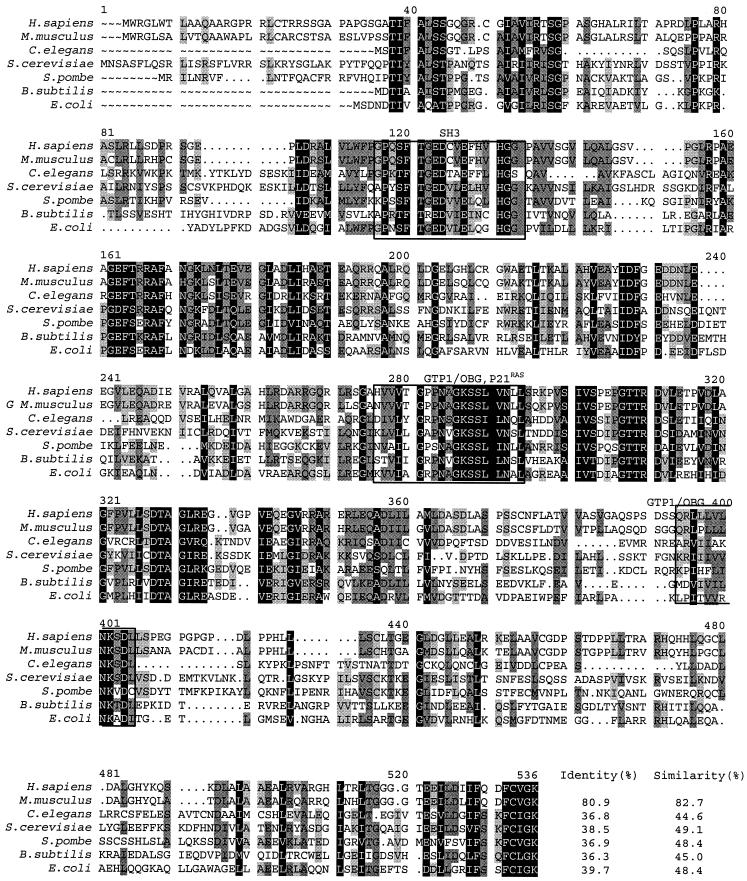
The protein alignments of human GTPBP3 with other identified MSS1 homologs, including S. cerevisiae (12), Mus musculus, Schizosaccharomyces pombe, Caenorhabditis elegans, and E. coli (1), and Bacillus subtilis (47) revealed an extensive conservation of amino acid sequences (Fig. 4). In particular, the identities of the overall predicated amino acid sequences of human GTPBP3 with homologs of S. cerevisiae, M. musculus, C. elegans, S. pombe, B. subtilis, and E. coli were 39, 81, 37, 37, 36, and 40%, respectively, whereas the similarities were 49, 82, 45, 48, 45, and 48%, respectively.
FIG. 4.
Sequence alignment of human Gtpbp3 with its homologs. The alignment was generated by using the GCG program. The organisms and corresponding accession numbers used for this analysis were as follows: M. musculus GTPBP3 (AAK35216), S. cerevisiae Mss1p (P32559), E. coli THDF (AAA62057), S. pombe Mss1p (CAB60697), C. elegans (T21994), and B. subtilis THDF (CAA44403). Numbers gives the positions of residues in proteins in relation to the first methionine of the S. cerevisiae Mss1p. Three highly conserved domains—SH3, GTP1/OBG, and P21Ras—are boxed. Amino acid residues shaded dark are identical; those shaded dark gray or light gray are similar in at least five residues or four residues of seven homologs, respectively. The percent sequence identities and similarities between Gtpbp3 and its homologs are indicated beside the alignment at the bottom of the figure.
Like the homolog of S. cerevisiae, the product of human GTPBP3 contains the typical features of mitochondrial target presequence with regularly spaced basic, hydroxylated, and hydrophobic residues (28). The MitoProt program (http//www.mips.biochem.mpg.de/proj/medgen/mitop) predicts with a high probability that human Gtpbp3 localizes to mitochondrion. This protein belongs to a family of GTPase, which is involved in tRNA modification (7, 10). In addition, this protein contains three highly conserved domains: two GTP-binding domains: GTP1/OBG (6) and the p21Ras-like domain (3), as well as the SH3 domain (40). The GTP-binding domains of human GTPBP3 contain the p21Ras motif at positions 251-HVVVTGPPNAGKSSLVNLL-269 and the GTP1/OBG GTP-binding motif, which resides at positions 367-QRLLLVLNKSDL-378. The SH3 domain of this protein localizes at positions 101-GPQSFTGEDCVEFHVHGG-118, which is involved in protein-protein interactions (40). From these data, the high sequence conservation between human Gtpbp3 and S. cerevisiae Mto1p suggests that they appear to be structural homolog and leads us to test whether the human protein can complement the characterized function of yeast protein.
Functional complementation of the yeast mss1 mutant by GTPBP3 cDNA.
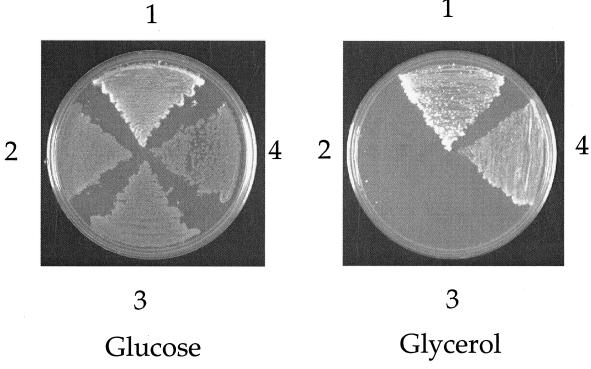
To examine the functional conservation of GTPBP3, we tested its ability to complement the respiratory defect of the yeast mss1 mutants carrying the PR454 allele of mitochondrial 15S rRNA gene. The entire coding region of GTPBP3 cDNA was cloned into an expression vector pDB20 (4). The pDB20 carries a URA3 marker gene of yeast and is the ADH1 promoter-based yeast expression vector. The resulting construct was transformed into a yeast mss1 strain, and the resultant Ura+ transformants were tested for their growth on glycerol-rich medium. As shown in Fig. 5, the growth defect of the mss1 mutant on glycerol medium was restored by the GTPBP3 cDNA. This strongly suggested that GTPBP3 is able to functionally complement the yeast mss1 mutation carrying the PR454 allele of mitochondrial genome.
FIG. 5.
Overexpression of human GTPBP3 suppresses the defective growth of the yeast mss1 mutant on glycerol medium. The yeast strains were spread on glucose- and glycerol-rich media and then incubated at 30°C for 4 days. Sector 1, wild-type haploid strain W303-1B; sector 2, W303ΔMSS1(PR); sector 3, W303ΔMSS1(PR) transformed with vector pDB20; sector 4, W303ΔMSS1(PR) with the introduction of pDB20-GTPBP3.
GTPBP3 is expressed in multiple tissues.
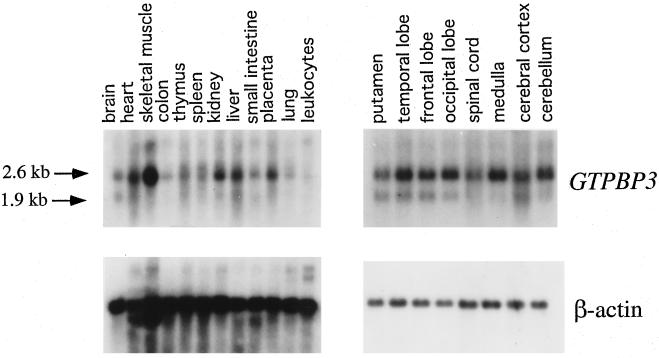
To investigate the tissue-specific expression of GTPBP3, a 32P-labeled 636 bp of GTPBP3 cDNA probe was hybridized with RNA blots of multiple human tissues. Figure 6 shows that this GTPBP3 probe detects two sets of transcripts with ca. 2.6 and 1.9 kb, which are in a good agreement with the length of 2.6 and 1.9 kb of cDNA. GTPBP3 is ubiquitously expressed, but there are significant variations in steady-state GTPBP3 mRNA levels among tissues. Compared with the tissues of a high metabolic rate, such as skeletal muscle, kidney, liver, and heart, the expression of GTPBP3 in colon, lung, and leukocytes appears to be very low. GTPBP3 is expressed moderately in the brain, thymus, spleen, and small intestine. In the brain, the expression of GTPBP3 in the putamen, spinal cord is lower than in other parts of brain. Significant differences in the ratio of two sets of transcripts were observed in different tissues. Generally, the 2.6-kb transcript is expressed at much higher level than the 1.9-kb transcripts in all tissues. In particular, the 2.6-kb transcript is highly abundant in the skeletal muscle; medulla and cerebellum, but the 1.9-kb transcript is almost undetectable in these tissues.
FIG. 6.
Multiple-tissue Northern blot analysis of GTPBP3 expression. Human 12-lane multiple-tissue blot and 8-lane brain MTN blot I (Clontech) containing 2 μg of poly(A)+ RNA per lane was hybridized with a 32P-labeled fragment containing human 636-bp GTPBP3 cDNA probe according to the manufacturer's protocol. The blot was then stripped and rehybridized with 32P-labeled human β-actin probe as a control.
Subcellular location of the product of human GTPBP3.
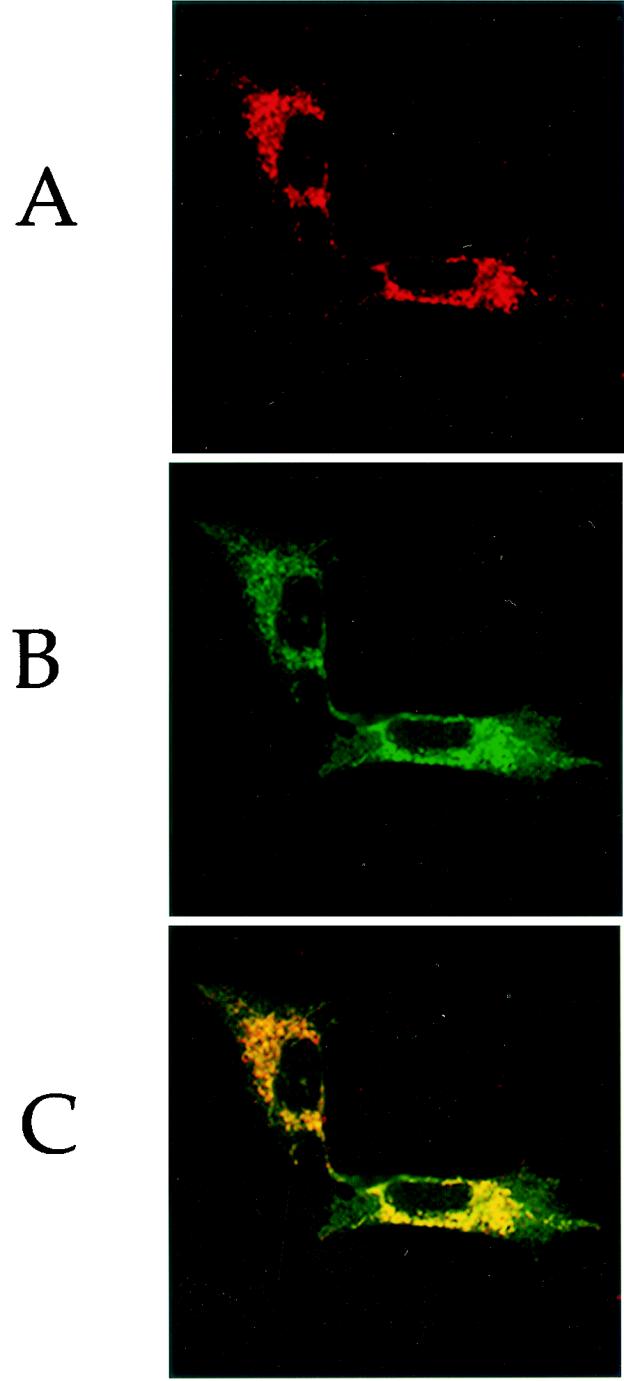
To determine the subcellular localization of the human GTPBP3 protein, a human GTPBP3 cDNA lacking its natural stop codon was obtained by PCR with the human GTPBP3 cDNA clone pGTPBP3 as the template. After the sequence determination, the insert was subsequently subcloned into the expression vector pEGFP-N1 (Clontech). This construct was then transiently transfected into the human 143B cells, immunostained with the antibodies and analyzed with a confocal microscope system. Figure 7 shows the immunofluorescence pattern of transfected 143B cells double labeled with rabbit monoclonal antibody specific for the GFP and mouse monoclonal antibody to COX1, a subunit of cytochrome c oxidase (COX) complex in the mitochondrial inner membrane. A typical mitochondrially staining pattern was observed with both antibodies (Fig. 7A and B) and superimposition of the two panels showed complete overlap of the two patterns (Fig. 7C). These data demonstrated that the product of human GTPBP3 localizes exclusively to mitochondria.
FIG. 7.
Human Gtpbp3 is a mitochondrial protein. 143B cells were transiently transfected with pEGFP-N1-GTPBP3. (A) Intracellular distribution of GTPBP3-GFP fusion protein was visualized by using the GFP antibody conjugated with Alexa Fluor 488. (B) The expression of the COX1 subunit in the same cell was assessed by the successive application of monoclonal antibody against human COX1 conjugated with Alexa Fluor 594. (C) A superimposition of panels A and B shows the complete overlap of the two staining patterns.
DISCUSSION
In the present study, we identified the full-length cDNA sequence and elucidated the general genomic structure of human GTPBP3 gene. The GTPBP3 is the first identified vertebrate gene related to the mitochondrial tRNA modification. Alignment of the product of human GTPBP3 with yeast Mss1p, as well as with other homologs, including mouse, S. pombe, E. coli, and C. elegans homologs, shows a high amino acid sequence conservation, as well as a similarity in size. In particular, the amino acid sequence identity between human Gtpbp3 and S. cerevisiae Mss1p (11) is 39%, whereas the identity between human Gtpbp3 and E. coli 50K GTPase protein (1), encoded by mnmE, is 40%. Human Gtpbp3, similar to all identified homologs, is a multiple-domain GTP-binding protein, as shown in Fig. 3C. The first 80 amino acids of N-terminal sequence is a characteristic mitochondrial targeting sequences (28). The MitoProt program predicts that the probability for human Gtpbp3 to export into mitochondria is 0.985, suggesting that the Gtppb3 has a highly mitochondrial targeting signal. In addition, this program also predicts that the cleaved site for the maturation of this protein localizes at the residue 90. Immunofluorescence analysis of 143B cells expressing the GTPBP3-GFP fusion protein demonstrated the colocalization of the GTPBP3 protein with the mitochondrial protein COX1. This strongly indicated that this protein functions in the mitochondria. As shown in Table 2, this protein carries three highly conserved domains from bacteria to mammal: two GTP-binding domains GTP1/OBG and p21Ras-related domain, as well as one SH3 domain. The SH3 domain of this protein, like that of other proteins (40), may mediate protein interaction by binding certain proline-rich amino acid sequences. The GTP-binding domains of Gtpbp3, including p21Ras and GTP1/OBG (3, 6), similar to those of 50K GTPase protein in E. coli (10), may be very important for its GTPase activity. In addition, the C-terminal domain of Gtpbp3 contains the tetrapeptide motif C-V-G-K at the extreme C terminus. This C-terminal cysteine might be involved in membrane association of Gtpbp3 as it occurs with other GTPases carrying this residue (6), and might also be involved in a putative tRNA-modifying activity of this protein (32). This evolutionary conservation between distantly related species suggests that human GTPBP3 and its homologs possess a fundamental cellular role.
TABLE 2.
Similarity and identity of the domains in Gtpbp3 (H. Sapiens)
| Domain and organism | Similarity (%) | Identity (%) |
|---|---|---|
| SH3 | ||
| M. musculus | 100 | 100 |
| C. elegans | 65 | 65 |
| S. cerevisiae | 76 | 76 |
| S. pombe | 71 | 71 |
| B. subtilis | 59 | 53 |
| E. coli | 67 | 67 |
| p21Ras | ||
| M. musculus | 95 | 95 |
| C. elegans | 67 | 56 |
| S. cerevisiae | 67 | 67 |
| S. pombe | 79 | 68 |
| B. subtilis | 71 | 65 |
| E. coli | 78 | 72 |
| GTP1/OBG | ||
| M. musculus | 83 | 83 |
| C. elegans | 73 | 55 |
| S. cerevisiae | 83 | 58 |
| S. pombe | 67 | 50 |
| B. subtilis | 70 | 50 |
| E. coli | 60 | 40 |
GTPBP3 homologs of other species have been shown to be involved in the tRNA modification, thus playing an important role in the translation process. In particular, the E. coli mnmE mutants (homolog of GTPBP3) are defective in the biosynthesis of the hypermodified nucleoside 5-methyl-aminnomethy-2-thiouridine (mnm5s2U34) (5, 7). This modified nucleotide, found in the wobble position of several bacterial tRNAs specific for glutamate, lysine, arginine, leucine, and glutamine, has a pivotal role in the function of tRNAs, including both tRNA identity and the codon recognition specificity (7). It has been shown that the defect in modification of this nucleotide led to the deficient aminoacylation of these tRNAs (35), as well as to an alteration in codon-anticodon interaction (26), consequently affecting the efficiency and accuracy of translation (26, 46). The synthesis of mnm5s2U34 is a complicated process and has been proposed to occur in multiple steps in bacteria (7). The E. coli mnmE (homolog of MSS1) product was shown to be responsible for the first step of the modification in position 5 of uracil (46). The localization of mitochondrion and the degree of amino acid conservation between human Gtpbp3 and this group of proteins apparently indicate that the Gtpbp3 is involved in the mitochondrial tRNA modification.
The human GTPBP3, similar to other nuclear genes coding for mitochondrial proteins, such as frataxin (34), SURF1 (51), MTO1 (37), and SCO2 (31), is predominantly expressed in the tissues with a high metabolic rate, including skeletal muscle, liver, and heart tissues, as well as brain tissue. Two sets of transcripts of 2.6 and 1.9 kb have been detected by Northern analysis. The 2.6-kb transcript is expressed at a much higher level than the 1.9-kb transcripts in all tissues. These observations suggest that the 2.6-kb mRNA is more stable than the 1.9-kb mRNA. By using RT-PCR, molecular cloning, and cDNA sequencing, five different cDNAs have been identified. Among these, a cDNA of 2,588 bp is consistent with the 2.6-kb transcript identified by Northern blot analysis, while four other transcripts of 1,864 bp (isoform II), 1,801 bp (isoform III), 1,781 bp (isoform IV), and 1,960 bp (isoform V) are in good agreement with the 1.9-kb transcripts. As shown in Fig. 2, the 3′-UTRs of the four isoforms are 712 bp shorter than that of GTPBP3 cDNA with 2,588 bp. Apparently, the shorted UTRs of these isoforms results from the alternative poly(A) site selection. Different 3′-UTRs have been shown to be associated with differential stability or translatability of various mRNA products (15). In addition, the 5′-UTR of isoform IV contains 37 bp, whereas that of other cDNAs has 57 bp. This difference in the 5′-UTR may be implicated in regulating transcription efficiency as reported in other genes (21). The alternative splicing also leads to the in-frame insertion and deletion in this gene. The isoform V, resulting from the retention of intron 5, produces a protein with 524 amino acids, whereas the truncated protein of 471 amino acids that is translated from isoform III and IV is due to the partial deletion of exons 8. It has been shown that different protein isoforms may regulate or interact with distinct sets of target (29). One example is that the splicing variants of the Wilms' tumor 1 gene encode proteins that have opposite effects on tumorigenicity (38). It is likely that different isoforms of GTPBP3 have different functions in various tissues. Further characterization of these isoforms is now being carried out in this laboratory.
The A1555G mutation in the 12S rRNA gene is one of the most common causes of nonsyndromic deafness and aminoglycoside-induced deafness (17, 18). Although there are common features among the patients carrying the A1555G mutation, specifically a high-frequency, symmetrical sensorineural hearing loss, the severity of hearing impairment varies greatly from severe congenital deafness to normal hearing (17, 42). An overall decrease in the rate of mitochondrial protein labeling was observed in the mutant lymphoblastoid cell lines derived from Arab-Israeli family, compared with that in control cell lines (23). However, a more severe mitochondrial dysfunction was observed in the lymphoblastoid cell lines derived from symptomatic individuals than those from asymptomatic individuals carrying the A1555G mutation (23). In contrast, under a constant nuclear background, there were very similar biochemical defects, including the reduction in the rate of mitochondrial protein synthesis between transmitochondrial cell lines derived from symptomatic and asymptomatic individuals (25). These genetic and biochemical evidences firmly suggest that the A1555G mutation is a primary factor underlying the development of deafness (23, 25), but the nuclear background determines the penetrance of the A1555G mutation in the Arab-Israeli family (23, 25). Very likely, the biochemical defect associated with the A1555G mutation primarily results from the fact that the hypermodified tRNAs, synthesized by the participation of GTPBP3, are less efficient for the decoding of the codon ending in G than A, as proposed in the yeast mitochondrial PR454 mutation (7). As a result, this mutation may affect the efficiency and accuracy of codon-anticodon interaction, thereby leading to a defect in the mitochondrial protein synthesis (23, 25). In yeast cells, mutations in MSS1 are proposed to cause the defect in the synthesis of mnm5s2U34 and consequently reduced mitochondrial tRNA modification. Apparently, these unmodified tRNAs do not function accurately with the altered mitochondrial ribosomes, thereby aggravating the mitochondrial translational defect associated with the PR454 mutation (11, 12). The finding that the GTPBP3 can functionally complement the respiratory deficient phenotype of yeast mss1 allele carrying the PR454 mutation suggests that the products of yeast MSS1 and human GTPBP3 have a similar function in mitochondrial tRNA modification. By functionally interacting with the decoding region of small rRNA, particularly in the site of A1555G, the product of GTPBP3 may regulate the translational efficiency and accuracy of codon-anticodon base pairings in the mitochondrial ribosomes. Thus, the defect in the expression or mutation(s) of GTPBP3, as described in the yeast MSS1 (11, 12), could act as a nuclear modifier factor and then contribute to the phenotypic variability of A1555G mutation by enhancing or suppressing the phenotypic manifestation of the A1555G mutation. To further understand the role of GTPBP3 in the phenotypic expression of the A1555G mutation, it certainly is of interest to determine whether there is the polymorphism(s) and/or mutation(s) in the GTPBP3 in the asymptomatic or symptomatic individuals from the Arab-Israeli family and other pedigrees carrying the A1555G mutation.
Acknowledgments
This work was supported by Public Health Service grants DC04958 and DC05230 from the National Institute on Deafness and Other Communication Disorders and by research grant awards from the United Mitochondrial Disease Foundation and the Deafness Research Foundation.
We thank Alex Tzagoloff at Columbia University for the yeast strains, Leonard Guarente at Massachusetts Institute of Technology for the pDB20 vector, and You-Hai Xu for the HepG2 cell line. We are grateful to Chuck Loftice and Debbie Baker for technical and clerical support.
REFERENCES
- 1.Alam, K. Y., and D. P. Clark. 1991. Molecular cloning and sequence of the THDF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J. Bacteriol. 173:6018-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbacid, M. 1987. RAS genes. Annu. Rev. Biochem. 56:779-827. [DOI] [PubMed] [Google Scholar]
- 4.Becker, D. M., J. D. Fikes, and L. Guarente. 1991. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl. Acad. Sci. USA 88:1968-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björk, G. R. 1996. Stable RNA modification, p. 861-886. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, B. K. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 6.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125-132. [DOI] [PubMed] [Google Scholar]
- 7.Brégeon, D., V. Colot, M. Miroslav, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 15:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bu, X., H. Y.Yang, M. Shohat, and J. I. Rotter. 1993. Two locus mitochondrial and nuclear gene models for mitochondrial disorders. Genet. Epidemiol. 9:27-44. [DOI] [PubMed] [Google Scholar]
- 9.Bykhovskaya, Y., X. Estivill, K. Taylor, T. Hang, M. Hamon, R. A. Casano, H. Yang, J. I. Rotter, M. Shohat, and N. Fischel-Ghodsian. 2000. Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am. J. Hum. Genet. 66:1905-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabedo, H., F. Macian, M. Villarroya, J. C. Escudero, M. Martinez-Vicente, E. Knecht, and M. E. Armengod. 1999. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 18:7063-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colby, G., M. Wu, and A. Tzagoloff. 1998. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 273:27945-27952. [DOI] [PubMed] [Google Scholar]
- 12.Decoster, E., A. Vassal, and G. Faye. 1993. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 232:79-88. [DOI] [PubMed] [Google Scholar]
- 13.De Stasio, E. A., and A. E. Dahlberg. 1990. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16S ribosomal RNA. J. Mol. Biol. 212:127-133. [DOI] [PubMed] [Google Scholar]
- 14.Dunbar, D. R., P. A. Moonie, H. T. Jacobs, and I. J. Holt. 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl. Acad. Sci. USA 92:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwalds-Gilbert, G., K. L. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 19:2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elseviers, D., L. A. Petrullo, and P. J. Gallagher. 1984. Novel Escherichia coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 12:3521-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estivill, X., N. Govea, A. Barcelo, E. Perello, C. Badenas, E. Romero, L. Moral, R. Scozzari, L. D'Urbano, M. Zeviani, and A. Torroni. 1998. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am. J. Hum. Genet. 62:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischel-Ghodsian, N. 1999. Mitochondrial deafness mutations reviewed. Hum. Mut. 13:261-270. [DOI] [PubMed] [Google Scholar]
- 19.Fischel-Ghodsian, N., T. R. Prezant, X. Bu, and S. Oztas. 1993. Mitochondrial ribosomal RNA gene mutation in a patient with sporadic aminoglycoside ototoxicity. Am. J. Otolaryngol. 4:399-403. [DOI] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and R. H. Schiestl. 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7:253-263. [DOI] [PubMed] [Google Scholar]
- 21.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 22.Gregory, S. T., and A. E. Dahlberg. 1995. Nonsense suppressor and antisuppressor mutations at the 1409-1491 base pair in the decoding region of Escherichia coli 16S rRNA. Nucleic Acids Res. 23:4234-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan, M. X., N. Fischel-Ghodsian, and G. Attardi. 1996. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 6:963-971. [DOI] [PubMed] [Google Scholar]
- 24.Guan, M. X., N. Fischel-Ghodsian, and G. Attardi. 2000. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum. Mol. Genet. 9:1787-1793. [DOI] [PubMed] [Google Scholar]
- 25.Guan, M. X., N. Fischel-Ghodsian, and G. Attardi. 2001. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 10:573-580. [DOI] [PubMed] [Google Scholar]
- 26.Hagervall, T. G., and G. R. Björk. 1984. Undermodification in the first position of the anticodon of supG-tRNA reduces translational efficiency. Mol. Gen. Genet. 196:194-200. [DOI] [PubMed] [Google Scholar]
- 27.Hamasaki, K., and R. R. Rando. 1997. Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism, which causes aminoglycoside-induced deafness. Biochemistry 36:12323-12328. [DOI] [PubMed] [Google Scholar]
- 28.Hartl, F. U., and W. Neupert. 1990. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science 247:930-938. [DOI] [PubMed] [Google Scholar]
- 29.Hsu, T., J. A. Gogos, S. A. Kirsh, and F. C. Kafatos. 1992. Multiple zinc finger forms resulting from developmentally regulated alternative splicing of a transcription factor gene. Science 257:1946-1950. [DOI] [PubMed] [Google Scholar]
- 30.Hutchin, T., I. Haworth, K. Higashi, N. Fischel-Ghodsian, M. Stoneking, C. Saha-Arnos, and G. Cortopassi. 1993. A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nucleic Acids Res. 21:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaksch, M., I. Ogilvie, J. Yao, G. Kortenhaus, H. G. Bresser, K. D. Gerbitz, and E. A. Shoubridge. 2000. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 9:795-801. [DOI] [PubMed] [Google Scholar]
- 32.Kealey, J. T., and D. V. Santi. 1991. Identification of the catalytic nucleophile of tRNA (m5U54) methyltransferase. Biochemistry 30:9724-9728. [DOI] [PubMed] [Google Scholar]
- 33.King, M. P., and G. Attardi. 1989. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246:500-503. [DOI] [PubMed] [Google Scholar]
- 34.Koutnikova, H., V. Campuzano, F. Foury, P. Dolle, O. Cazzalini, and M. Koenig. 1997. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 35.Krüger, M. K., and M. A. Sørensen. 1998. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 284:609-620. [DOI] [PubMed] [Google Scholar]
- 36.Li, M., A. Tzagoloff, K. Underbrink-Lyon, and N. C. Martin. 1982. Identification of the paromomycin-resistance mutation in the 15S rRNA gene of yeast mitochondria. J. Biol. Chem. 257:5921-5928. [PubMed] [Google Scholar]
- 37.Li, X., R. Li, X. Lin, and M. X. Guan. 2002. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of the deafness-associated mitochondrial 12S rRNA A1555G mutation. J. Biol. Chem. 277:27256-27264. [DOI] [PubMed] [Google Scholar]
- 38.Menke, A. L., N. Riteco, R. C. van Ham, C. de Bruyne, F. J. Rauscher, A. J. van der Eb III, and A. G. Jochemsen. 1996. Wilms' tumor 1 splice variants have opposite effects on the tumorigenicity of adenovirus-transformed baby-rat kidney cells. Oncogene 12:537-546. [PubMed] [Google Scholar]
- 39.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 40.Musacchio, A., M. Saraste, and M. Wilmanns. 1994. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat. Struct. Biol. 1:546-551. [DOI] [PubMed] [Google Scholar]
- 41.Neefs, J. M., Y. Van de Peer, P. De Rijik, A. Goris, and R. De Wachter. 1991. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 19(Suppl.):1987-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prezant, T. R., J. V. Agapian, M. C. Bohlman, X. Bu, S. Oztas, W. Q. Qiu, K. S. Arnos, G. A. Cortopassi, L. Jaber, J. I. Rotter, M. Shohat, and N. Fischel-Ghodsian. 1993. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4:289-294. [DOI] [PubMed] [Google Scholar]
- 43.Rogan, P. K., B. M. Faux, and T. D. Schneider. 1998. Information analysis of human splice site mutations. Hum. Mut. 12:153-171. [DOI] [PubMed] [Google Scholar]
- 44.Schneider,T. D. 1997. Information content of individual genetic sequences. J. Theor. Biol. 189:427-441. [DOI] [PubMed] [Google Scholar]
- 45.Schon, E. A., E. Bonilla, and S. DiMauro. 1997. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr. 29:131-149. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan, M. A., J. F. Cannon, F. H. Webb, and R. M. Bock. 1985. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J. Bacteriol. 161:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trach, K., and J. M. Hoch. 1989. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171:1362-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 49.Weiss-Brummer, B., and A. Huttenhofer. 1989. The paromomycin resistance mutation (PARR454) in the 15S rRNA gene of the yeast Saccharomyces cerevisiae is involved in ribosomal frameshifting. Mol. Gen. Genet. 217:362-369. [DOI] [PubMed] [Google Scholar]
- 50.Yoneda, M., A. Chomyn, A. Martinuzzi, O.Hurko, and G. Attardi. 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyelopathy. Proc. Natl. Acad. Sci. USA 89:11164-11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, Z., J. Yao, T. Johns, K. Fu, I. De Bie, C. Macmillan, A. P. Cuthbert, R. F. Newbold, J. Wang, M. Chevrette, G. K. Brown, R. M. Brown, and E. A. Shoubridge. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20:337-343. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann, R. A., C. L. Thomas, and J. Wower. 1990. Structure and function of rRNA in the decoding domain and at the peptidyltransferase center, p. 331-347. In W. E. Hill, P. B. Moore, A. Dahlberg, D. Schlessinger, R. A. Garrett, and J. R. Warner (ed.), The ribosome: structure, function and evolution. American Society for Microbiology, Washington, D.C.
- 53.Zwieb, C., D. K. Jemiolo, W. F. Jacob, R. Wagner, and A. E. Dahlberg. 1986. Characterization of a collection of deletion mutants at the 3′-end of 16S ribosomal RNA of Escherichia coli. Mol. Gen. Genet. 203:256-264. [DOI] [PubMed] [Google Scholar]