Abstract
FCP1, a phosphatase specific for the carboxy-terminal domain of RNA polymerase II (RNAP II), was found to stimulate transcript elongation by RNAP II in vitro and in vivo. This activity is independent of and distinct from the elongation-stimulatory activity associated with transcription factor IIF (TFIIF), and the elongation effects of TFIIF and FCP1 were found to be additive. Genetic experiments resulted in the isolation of several distinct fcp1 alleles. One of these alleles was found to suppress the slow-growth phenotype associated with either the reduction of intracellular nucleotide concentrations or the inhibition of other transcription elongation factors. Importantly, this allele of fcp1 was found to be lethal when combined individually with two mutations in the second-largest subunit of RNAP II, which had been shown previously to affect transcription elongation.
The engine at the heart of the transcriptional apparatus is RNA polymerase II (RNAP II), a protein complex comprising 12 subunits that are remarkably conserved throughout eukaryotes. Crystallographic studies with Saccharomyces cerevisiae RNAP II have given us the first detailed insights, at the atomic level, of the molecular mechanism by which eukaryotic RNA polymerases transcribe DNA (14, 16). RNA polymerases cannot recognize the promoters of their target genes but rather rely on accessory proteins, known in eukaryotes as general transcription factors (GTFs) (37, 46). These factors recognize conserved core promoter elements present in most protein-coding genes and also interact directly with RNAP II, resulting in the recruitment of RNAP II to the start site of transcription.
At least two forms of RNAP II have been detected in cells. These isoforms result from the presence of a unique domain in the C-terminal region of the largest subunit of RNAP II, known as the carboxyl-terminal domain (CTD). In mammals, this domain consists of 52 repeats of the consensus heptapeptide Tyr-Ser-Pro-Thr-Ser-Pro-Ser and appears from crystallographic studies to be unstructured (14). The precise function of the CTD has been elusive, although it was speculated almost a decade ago that it constitutes a binding site for protein factors involved in transcriptional regulation and RNA processing (17, 18).
The initiation of transcription by RNAP II is a multistep process that involves separation of the DNA strands at the initiation site (promoter melting), formation of the first phosphodiester bond of the transcript, and disruption of the interactions between RNAP II and the promoter (promoter clearance). The isoform of RNAP II that associates with transcription initiation complexes contains an unphosphorylated CTD, whereas the isoform involved in transcription elongation contains a phosphorylated CTD (4, 27, 36, 62). Thus, the transition from initiation of transcription to elongation of the transcript is accompanied by extensive phosphorylation of the CTD of RNAP II.
A number of protein kinases have been implicated in the phosphorylation of the CTD during the transition from transcription initiation to elongation. Among these are the Cdk7 subunit of the GTF IIH (TFIIH) and the Cdk9 subunit of P-TEFb, a factor that regulates transcription elongation (32, 35). The Cdk7 kinase phosphorylates serine 5 of the CTD heptapeptide motif (32). And, while the specificity of human P-TEFb remains controversial, a close yeast homologue of its Cdk9 subunit, Ctk1, modifies the serine residue at position 2 of the CTD heptapeptide repeat (9).
The identification of FCP1, a protein phosphatase that appears to be dedicated to the CTD of RNAP II, supports the notion that RNAP II toggles between nonphosphorylated and phosphorylated forms during the transcription cycle. FCP1 dephosphorylates RNAP II in ternary elongation complexes (10) as well as in solution (2, 3, 6-8, 10) and, therefore, is thought to function in the recycling of RNAP II during the transcription cycle (10).
The phosphorylation pattern of the CTD undergoes precise changes during transcription; specifically, the phosphorylation of serine 5 in the CTD motif occurs between transcription initiation and promoter clearance and modification of serine 2 is found only when RNAP II is associated with the coding region of the gene (9). Biochemical experiments suggest that human FCP1 targets, with apparently similar affinities, CTDs that are phosphorylated at serine 2 (CTD-serine 2) and/or CTD-serine 5. However, studies of yeast suggest that FCP1 targets CTD-serine 2 and does not dephosphorylate CTD-serine 5 (9). Interestingly, chromatin immunoprecipitation (ChIP) experiments performed with yeast show that FCP1 remains associated with RNAP II during transcription elongation.
In the present study, we demonstrate that human FCP1 functions as an elongation factor and stimulates the rate of elongation by RNAP II. This effect is independent of the catalytic activity of FCP1. Moreover, we have isolated yeast fcp1 mutants that display synthetic phenotypes in combination with specific alleles of genes encoding other elongation factors or elongation-defective forms of RNAP II.
MATERIALS AND METHODS
Purification of FCP1 and TFIIF and its derivatives.
Recombinant FCP1 was purified by using the baculovirus expression system. SF9 cells were infected with recombinant baculovirus carrying the cDNA encoding human FCP1. Infected cells were maintained at a cell density of 106 cells/ml for 60 h at 30°C. Recombinant proteins were purified by Ni-nitrilotriacetic acid-agarose chromatography, followed by chromatography on a DEAE-cellulose column as previously described (10).
TFIIF consists of two subunits, RAP74 and RAP30. Recombinant wild-type RAP74, mutant forms of RAP74 (1-172, 1-217, and Δ137-356), wild-type RAP30, and mutant forms of RAP30 (Δ91-105 and Δ106-120) were expressed individually in Escherichia coli and purified by Ni-nitrilotriacetic acid-agarose chromatography under conditions of denaturation with 8 M urea. The individually purified polypeptides (RAP30 and RAP74) were mixed at an approximately 1:1 molar ratio under denaturing conditions (8 M urea, 0.1 M Na2HPO4, 0.01 M Tris-HCl [pH 7.9]), and the mixture was slowly dialyzed against 0.5 M urea-containing buffer. Finally, samples were dialyzed against BC500 (20 mM Tris-HCl, 0.2 mM EDTA, 10 mM β-mercaptoethanol, 10% glycerol, 500 mM KCl [pH 7.9]) and purified by gel filtration (Superdex200 fast protein liquid chromatography column; in buffer BC500). The TFIIF heterodimers were then assayed for transcription activity.
Transcription elongation assays.
In vitro transcription assays were reconstituted with highly purified, E. coli-expressed TATA-binding protein, TFIIB, TFIIE, and TFIIF, as well as highly purified, native RNAP II (18 ng) and TFIIH isolated from HeLa cells. Preinitiation complexes (PIC) were assembled on biotinylated linear DNA that contained the adenovirus major late promoter, which had been bound to streptavidin-coated Dynal beads. Specifically, the EcoRI/ScaI fragment of the plasmid pML20-47 (48) was biotinylated at the end of the EcoRI site and bound to the streptavidin beads.
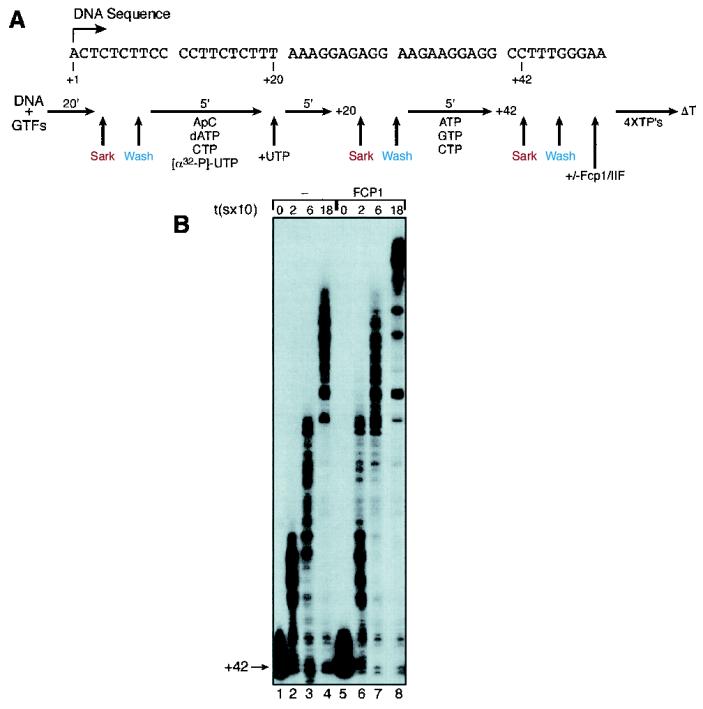
PIC were formed by incubating, for 20 min at 30°C, the GTFs, RNAP II, and the DNA template (100 ng, bound to streptavidin beads) in transcription buffer (13 mM HEPES, 5.2 mM MgCl2, 2% polyethylene glycol, 500 ng of poly[dG-dC], 45 μg of bovine serum albumin [BSA; DNase and RNase free], 10 mM dithiothreitol, 10 mM [NH4]2SO4, 0.12 mM phenylmethylsulfonyl fluoride, 0.06 mM EDTA) that contained 60 mM KCl (Txn60). Active PIC were selected by washing the beads with 0.05% Sarkosyl (in Txn60 buffer), and the resulting preparation was equilibrated with Txn60 buffer. The PIC were then incubated with RNasin (16 U) and a mixture of nucleoside triphosphates (NTPs) (NTPmix1: 0.9 mM adenylyl-(3′→5′)-cytidine (ApC), 20 μM dATP, 0.165 μM CTP, and 0.3 μM [α-32P]UTP) for 5 min at 30°C and chased with 1 μM UTP for 5 min to produce the +20 complex (see Fig. 1A). These stalled +20 complexes were washed with 0.05% Sarkosyl (in Txn60 buffer), equilibrated with Txn60 buffer, and then incubated with 2.5 μM ATP, GTP, and CTP for 5 min to produce the +42 complex (see Fig. 1A). The +42 complexes were washed with 0.05% Sarkosyl (in either Txn500 [as described above, but containing 500 mM KCl] or Txn100, as indicated in the figure legends) and equilibrated with Txn60. This +42 complex was used as starting material for the transcription elongation assays.
FIG. 1.
Effect of FCP1 on transcription elongation by RNAP II in a reconstituted system. (A) Template DNA sequence and experimental scheme for the isolation of stalled +42 complexes (see Materials and Methods for details). (B) Ternary +42 complexes (washed with Txn100 and equilibrated with Txn60) were incubated alone (lanes 1 to 4) or in the presence of FCP1 (60 ng; lanes 5 to 8) for 10 min at 30°C and then chased with 30 μM concentrations of each of the four XTPs (ATP, GTP, CTP, and UTP). Reactions were stopped at various time points (0, 20, 60, and 180 s, respectively), and then reaction products were subjected to phenol-chloroform extraction and ethanol precipitation, after which were analyzed by electrophoresis on polyacrylamide-urea gels followed by autoradiography.
Elongation reactions were performed for 10 min at 30°C by incubating the +42 complexes with FCP1 and/or TFIIF (as indicated in the figure legends). The complexes were then chased with a mixture of all four XTPs (at a concentration of 30 μM each) for various periods of time, as indicated in the figure legends. Reactions were stopped with stop buffer (0.3 M sodium acetate, 1 mM EDTA, 0.1% sodium dodecyl sulfate, 200 μg of glycogen/ml), and the reaction products were subjected to phenol-CHCl3 extraction and ethanol precipitation. Transcription elongation products were analyzed by electrophoresis on 7% acrylamide-8 M urea gels, and the gels were subjected to autoradiography.
Media and genetic manipulations.
Yeast strain constructions and other genetic manipulations were performed by standard methods (51). All yeast media were prepared as described previously (22).
Isolation of fcp1 alleles that confer sensitivity to 6-AU (6-AUs) and/or temperature (ts−).
fcp1 alleles were generated by error-prone-PCR-mediated mutagenesis. The DNA fragment encoding amino acids (aa) 29 to 445 was mutagenized in the presence of 10 μM Mn2+ with one of the deoxynucleotides under limiting conditions (30 μM). The resulting PCR products were transformed into the yeast strain YKC001 with a linear centromeric plasmid that contains the TRP1 marker and a gapped form of the FCP1 gene that lacks the region encoding aa 29 to 445. TRP+ transformants were isolated, purified, and replica plated to 5-fluoroorotic acid (5-FOA) plates to cure the plasmid-borne wild-type FCP1 allele. To identify colonies defective for growth at 38°C, the 5-FOA-resistant colonies were replica plated to SC plates lacking Trp (SC−Trp plates) and incubated at 30 and 38°C, and temperature-sensitive (ts) colonies were isolated. To determine whether the ts phenotype was linked to the fcp1 allele, plasmid DNA from fcp1 ts mutants was isolated and rescored by using the plasmid shuffle system described above. Strains that are defective for growth in the presence of 6-AU were scored on SC medium containing 300 μg of 6-azauracil (6-AU)/ml at 30°C.
Mutations that conferred 6-AUs or ts phenotypes were defined by sequencing the fcp1 gene. Sequencing the mutant fcp1-As42 allele revealed that the isoleucine at position 17 was mutated to valine and that the leucine at position 125 was changed to proline. Allele fcp1-8-13 encodes a valine-to-alanine change at aa 399, allele fcp1-9-11 encodes a glycine-to-aspartic acid change at aa 232, and fcp1-9-19 encodes a proline-to-leucine change at aa 60, a glycine-to-valine change at aa 162, and a serine-to-leucine change at aa 308.
Assay for 6-AU sensitivity.
Test strains containing an fcp1 allele that conferred 6-AU sensitivity were transformed with p186, a centromeric vector plasmid with a URA3 marker gene, and selected colonies were grown overnight by using a synthetic medium that lacked tryptophan and uracil (SC−Trp−Ura). Cultures were diluted to an optical density at 600 nm of approximately 0.2, followed by fivefold serial dilutions. The diluted cultures were spotted onto agar plates that contained SC−Trp−Ura without or with 6-AU (50 μg/ml). The plates were incubated for 3 days at 30°C and monitored for growth.
Yeast strains.
Genotypes of yeast strains are shown in Table 1. Yeast strain YKC001 is a meiotic segregant of strain JA830 (2). The viability of YKC001 was maintained by the centromeric plasmid p186 FCP1, which contains FCP1 and URA3. Transformation of YKC001 with the centromeric plasmids p185 FCP1 (FCP1-TRP1), p185 fcp1As42, p185 fcp1-8-13, p185 fcp1-9-11, and p185 fcp1-9-19, followed by counter selection of the URA3 marker with 5-FOA plates (SC−Trp containing 5-FOA) generated strains YKC002 (FCP1), YKC003 (fcp1-As42), YKC004 (fcp1-8-13), YKC005 (fcp1-9-11), and YKC006 (fcp1-9-19), respectively.
TABLE 1.
Genotypes of the strains used
| Strain | Genotype |
|---|---|
| JA830 | MATa/MATα can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp11/trp1-1 ura3/ura3 ade2-1/ade2-1 FCP1/fcp1::leu2 |
| YKC001 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [FCP1 URA3 CEN] |
| YKC002 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [FCP1 TRP1 CEN] |
| YKC003 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [fcp1-As42 TRP1 CEN] |
| YKC004 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [fcp1-8-13, TRP1 CEN] |
| YKC005 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [fcp1-9-11 TRP1, CEN] |
| YKC006 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [fcp1-9-19 TRP1 CEN] |
| YMH200 | MATaura3-52 leu2-3,112 his3 trp1-1 ade2-1 ydr1::HIS3 [CEN URA3 YDR1] |
| YKC200 | MATahis3-11,15 leu2-3,112 trp1-1 ura3 ade2-1 fcp1::LEU2 [FCP1 URA CEN] |
| YKC262 | his3 leu2-3,112 trp1-1 ura3 fcp1::LEU2 spt4::HIS3 [FCP1 URA CEN] |
| YKC263 | his3 leu2-3,112 trp1-1 ura3 fcp1::LEU2 spt4::HIS3 [fcp1-As42 TRP CEN] |
| YKC264 | his3 leu2-3,112 trp1-1 ura3 fcp1::LEU2 spt4::HIS3 [fcp1-8-13 TRP CEN] |
| YKC265 | his3 leu2-3,112 trp1-1 ura3 fcp1::LEU2 spt4::HIS3 [fcp1-9-11 TRP CEN] |
| YKC267 | his3 leu2-3,112 trp1-1 ura3 fcp1::LEU2 spt4::HIS3 [fcp1-9-19 TRP CEN] |
| FY1114 | MATα his4-912δ lys2-128δ leu2-3,112 trp1-1 ura3-52 his3 spt4::HIS3 |
| Z103 | MATα ura3-52 his3Δ200 leu2-3,112 rpb2Δ297::HIS3 [pRP2-7L(LEU2 CEN rpb2-7)] |
| Z106 | MATα ura3-52 his3Δ200 leu2-3,112 rpb2Δ297::HIS3 [pRP2-10L(LEU2, CEN, rpb2-10)], |
| FR7 | ura3 his leu2-3,112 rpb2Δ297::HIS3 fcp1::LEU2 [pRP2-7L(LEU2 CEN rpb2-7)] [FCP1 URA CEN] |
| FR10 | ura3 his leu2-3,112 rpb2Δ297::HIS3 fcp1::LEU2 [pRP2-10L(LEU2 CEN rpb2-7)] [FCP1 URA CEN] |
To generate an fcp1Δ spt4Δ double-knockout strain, strain YKC262, a MATa fcp1::LEU2 [FCP1-URA3] segregant derived from a cross between strains YMH200 and YKC001, was mated with strain FY114. This second cross yielded a segregant that grew on SC plates lacking histidine, uracil, and leucine and that was sensitive to 5-FOA (YKC262). Transformation of YKC262 with plasmids p185 FCP1, p185 fcp1As42, p185 fcp1-8-13, p185 fcp1-9-11, and p185 fcp1-9-19, followed by counter-selection of the URA3 marker on 5-FOA plates, generated strains YKC263 (FCP1), YKC264 (fcp1-As42), YKC265 (fcp1-8-13), YKC267(fcp1-9-11), and YKC268 (fcp1-9-19), respectively. To determine whether the 6-AU sensitivity of YKC264 was dependent on the genomic deletion of spt4, yeast strains YKC263 to YKC268 were transformed with pBM25, a URA3-centromeric plasmid that contains the wild-type SPT4 gene (33). To generate a strain that carries an rpb2 mutation encoding an altered form of the second-largest subunit of RNAP II with an fcp1 knockout, YKC200 was mated with either strain Z103 or Z106 (26, 49, 50). From this cross, we isolated haploid progeny that were either 5-FOA sensitive (strain FR7) or that grew on SC medium containing Trp (strain FR10). Transformation of FR7 with plasmids p185 FCP1, p185 fcp1As42, p185 fcp1-8-13, p185 fcp1-9-11, and p185 fcp1-9-19, followed by counter-selection of the URA3 marker with 5-FOA, generated strains FR71 (FCP1), FR72 (fcp1-As42), FR73 (fcp1-8-13), FR74 (fcp1-9-11), and FR75 (fcp1-9-19), respectively. Transformation of FR10 with plasmids p185 FCP1, p185 fcp1As42, p185 fcp1-8-13, p185 fcp1-9-11, and p185 fcp1-9-19, followed by counter-selection of the URA3 marker on 5-FOA plates generated strains FR101 (FCP1), FR102 (fcp1-As42), FR103 (fcp1-8-13), FR104 (fcp1-9-11), and FR105 (fcp1-9-19), respectively.
RESULTS
FCP1 affects elongation by RNAP II in a reconstituted transcription system.
To expand our previous observations, which suggested that FCP1 affects the efficiency of elongation by RNAP II independent of its catalytic activity (10), we reconstituted transcription assays in vitro with purified factors. These assays are designed so that RNAP II could be “walked” to specific sites on the DNA template. The approach used is described in the legend for Fig. 1A and is similar to the approach used previously to analyze the fate of transcription factors during the transcription cycle (61). In brief, transcription initiation complexes were formed on a bead-bound DNA template that contained the adenovirus major late promoter. Sequences downstream of the transcription start site were modified to allow the stalling of RNAP II by nucleotide starvation at defined sites. Stalled complexes were washed to remove nucleotides and free transcription factors, and then a new mixture of nucleotides was added to resume elongation in the presence or absence of transcription elongation factors. Aliquots of the reaction mixture were withdrawn at various time points, and the rate of elongation was measured by analyzing the sizes of the RNAs on denaturing polyacrylamide gels.
RNAP II elongation complexes were stalled 42 nucleotides downstream of the transcription start site (+42; Fig. 1B, lane 1). These stalled complexes were washed as described in the legend for Fig. 1A and then chased in the presence of FCP1. Under these conditions approximately 70% of the complexes stalled at +42 were able to be chased to larger products in a time-dependent manner (Fig. 1B, lanes 2 to 4). When stalled complexes were supplemented with recombinant, highly purified FCP1, two effects were observed. First, the amount of complex released from stalling was increased modestly, from 70 to approximately 90% (Fig. 1B, compare lanes 2 to 4 with lanes 6 to 8). Second, and most importantly, the rate of elongation was stimulated. For example, after 20 s of elongation, the RNA products produced in the presence of FCP1 were larger than those observed in the absence of FCP1. In fact, these longer transcripts appeared to be similar in length to those observed after 1 min of elongation in the absence of FCP1 (compare lane 6 with lanes 2 and 3). Moreover, after 1 min of chase, the elongation products formed in the presence of FCP1 were similar in size to those observed after 3 min of elongation in the absence of FCP1 (compare lane 7 with lane 4). Full-length products were observed only in the presence of FCP1 after 3 min of chase. Taken together, these results show an approximately threefold stimulation in the rate of transcription elongation upon addition of FCP1. The effects observed were specific for FCP1, as the addition of an equivalent amount of BSA was without effect (Fig. 2).
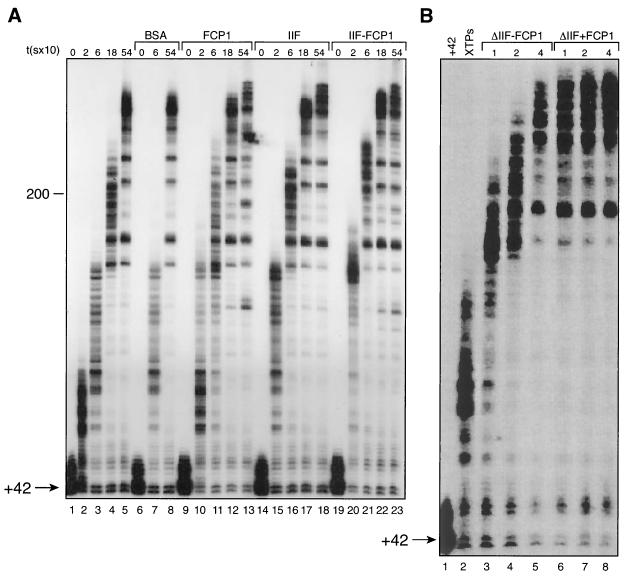
FIG. 2.
Transcription elongation by FCP1 in the presence and absence of TFIIF. (A) Ternary +42 complexes were isolated as described in the legend to Fig. 1, washed with Txn100, equilibrated with Txn60, and incubated for 10 min at 30°C alone (lanes 1 to 5), with FCP1 (60 ng; lanes 9 to 13), with TFIIF (1.5 ng; lanes 15 to 18), or with FCP1 and TFIIF (lanes 19 to 23). Reaction mixtures were chased with 30 μM concentrations of each of the four XTPs, and reactions were stopped at various time points (0, 20, 60, 180, and 540 s, respectively). BSA (60 ng) was used as a control protein (lanes 6 to 8). (B) Ternary +42 complexes (washed with Txn500 and equilibrated with Txn60) were incubated with various concentrations of TFIIF (0.38, 0.75, and 1.5 ng in lanes 3, 4, and 5, respectively) alone or in the presence of an additional 60 ng of FCP1 (lanes 6 to 8) for 10 min at 30°C. The reaction mixtures were then chased with 30 μM concentrations of each of the four XTPs, and reactions were stopped after 1 min.
We next compared the effect of FCP1 on elongation with that of TFIIF, a known elongation factor that interacts with FCP1 (2, 8, 10). As in the above experiments, RNAP II complexes that had been stalled at +42 were supplemented with FCP1 or TFIIF or both. FCP1 and TFIIF were both able to stimulate elongation rates on their own (Fig. 2A, compare lanes 2 to 5 with lanes 10 to 13 and 15 to 18); however, the patterns of paused complexes at early elongation time points for the two factors differed. For example, TFIIF appeared to be more efficient at stimulating elongation at early time points. After 20 and 60 s of elongation, although the lengths of the growing RNAs appeared relatively similar with both TFIIF and FCP1, the number of paused complexes proximal to the promoter was decreased in the presence of TFIIF (compare lanes 15 and 16 with lanes 10 and 11). At later time points (3 and 9 min), we again observed that the lengths of the elongation products produced in the presence of either FCP1 or TFIIF appeared to be similar; however, the intermediate elongation complexes were modestly decreased in the presence of TFIIF (compare lanes 12 and 13 with lanes 17 and 18). In addition, TFIIF failed to increase the number of stalled complexes released from the +42 site upon nucleotide addition (compare lanes 15 to 18 with lanes 10 to 13 and 2 to 5). The addition of both FCP1 and TFIIF to the stalled complexes resulted in an increase in the elongation rates observed when each of the factors was added individually; moreover, the efficiency of elongation was clearly enhanced. For example, after 20 s of elongation in the presence of FCP1 and TFIIF, the number of paused intermediates was quite small compared to the numbers observed when each factor was added individually. In addition, the bulk of the RNA appeared to be in relatively homogeneous elongation complexes. After 1 min of elongation in the presence of FCP1 and TFIIF, although the elongation complexes appeared to be heterogeneous, the bulk of the RNA was larger than the RNA species observed when TFIIF and FCP1 were added separately (compare lane 21 with lanes 11 and 16).
To expand these elongation studies, we analyzed the effect of adding various amounts of TFIIF in the presence and absence of saturating amounts of FCP1. Reaction mixtures were chased for 1 min. The results of this experiment demonstrate that, in the presence of excess FCP1, TFIIF is most efficient at stimulating elongation (Fig. 2B).
FCP1 can affect transcription elongation in the absence of TFIIF.
To further characterize the function of FCP1 during elongation by RNAP II and to study the dependence of FCP1 on TFIIF, a series of TFIIF mutant proteins were used in elongation studies.
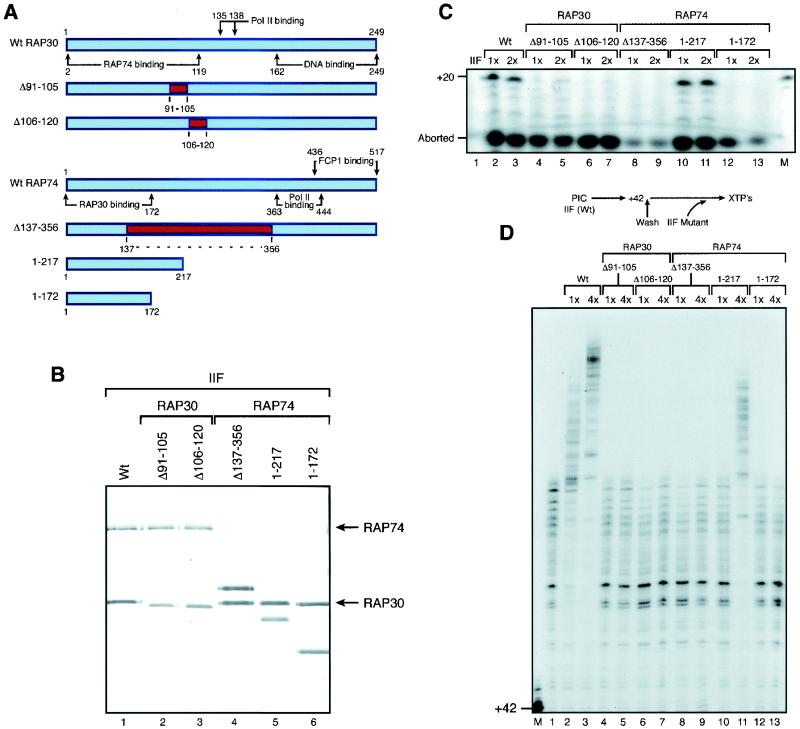
TFIIF has diverse functions in the transcription cycle. This multifaceted factor participates in (i) the delivery of RNAP II to promoter sequences (47, 57), (ii) promoter clearance (13, 55), (iii) elongation (5, 15), and (iv) the recycling of RNAP II. The recycling function is thought to be accomplished through TFIIF-mediated stimulation of the FCP1-CTD phosphatase activity (10). Distinct domains in the RAP30 and RAP74 subunits of TFIIF perform these diverse functions. Our goal was to isolate TFIIF mutants that are defective in the stimulation of elongation but that retain the other functions required for transcription. To this end, we obtained a variety of RAP30 (55) and RAP74 (28, 29, 57) deletion mutants that had been previously characterized by others, expressed the mutant polypeptides in bacteria, and purified the recombinant proteins and used them to reconstitute TFIIF activities in in vitro transcription assays (Fig. 3A and B). The mutant versions of TFIIF were analyzed for their ability to function during transcription initiation, promoter clearance, and elongation. All but one of the mutants analyzed were capable of mediating transcription initiation, as measured in an abortive-initiation assay; the one mutant that was impaired in this function had a large internal deletion in RAP74 (Δ137-356) (Fig. 3C). In the same assay, we analyzed the ability of the mutant TFIIF proteins to mediate promoter clearance by generating a 20-nucleotide transcript. This assay revealed that the two RAP30 mutants with internal deletions, Δ91-105 and Δ106-120, were severely impaired, as reported previously (55). It is interesting that one TFIIF mutant that contained a RAP74 subunit with a deletion of 300 COOH-terminal amino acids was able to function in promoter clearance (Fig. 3C); however, the removal of an additional 45 aa from the C-terminal end impaired this activity (Fig. 3C). To analyze the functions of the mutant TFIIF proteins during elongation, transcription initiation complexes were formed by using wild-type TFIIF and elongation complexes stalled at the +42 site were produced. The complexes were washed extensively with transcription buffer containing 0.5 M KCl and Sarkosyl so that any wild-type TFIIF associated with the stalled elongation complex was removed (data not shown). The complexes were then washed with transcription buffer and allowed to reinitiate elongation in the presence of the various TFIIF mutants. The efficiency of elongation was analyzed as described above. This analysis revealed that TFIIF mutants defective in promoter clearance were also defective in stimulating elongation (Fig. 3D).
FIG. 3.
Characterization TFIIF mutants used for FCP1 interaction and elongation assays. (A) Schematic diagram of the TFIIF mutants used in these experiments. Wt, wild type. (B) Silver stain of the various TFIIF mutant proteins (heterodimers), which had been purified through the gel filtration step. (C) Functions of the various TFIIF mutants in transcription initiation. PIC were assembled as described for Fig. 1 with all of the GTFs and with two different amounts of either wild-type TFIIF or mutant versions of TFIIFs (0.2 and 0.8 nmol) in transcription buffer (as described in Materials and Methods) and incubated at 30°C for 30 min. NTPmix1 (0.9 mM ApC, 20 μM dATP, 0.165 μM CTP, and 0.3 μM [α-32P]UTP) was added, and the reaction mixture was incubated for 5 min at 30°C and chased with cold UTP for an additional 5 min (see Materials and Methods) as described for Fig. 1. Reactions were stopped by adding stop buffer, and the reaction products were subjected to phenol-chloroform extraction and ethanol precipitation and then were separated by electrophoresis on a polyacrylamide-urea gel and visualized by autoradiography. (D) Functions of the various TFIIF mutants in elongation. Stalled ternary +42 complexes were isolated as described for Fig. 1 (by using wild-type TFIIF in the PIC), washed with 0.05% Sarkosyl, washed twice with Txn500, and equilibrated with Txn60. Isolated +42 complexes were then incubated alone (lane 1) or with two different concentrations (0.2 and 0.8 nmol) of either wild-type (lanes 2 and 3, respectively) or mutant (lanes 4 to 13) TFIIFs for 10 min at 30°C. The reaction mixtures were then chased with 30 μM concentrations of each of the four XTPs for 1 min. Products were analyzed as described for panel C.
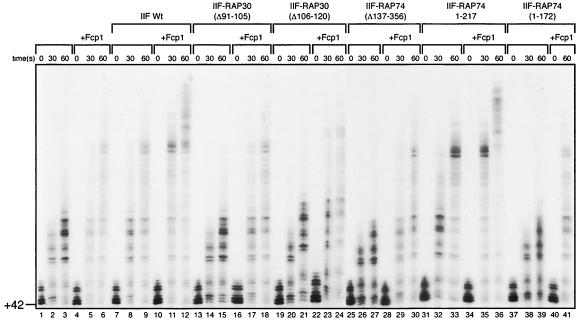
The ability of FCP1 to stimulate transcription elongation in the presence of the various TFIIF mutants was analyzed as described in the legend for Fig. 3D, except that elongation was allowed to proceed for 30 and 60 s (Fig. 4). Consistent with the results presented above, FCP1 was able to stimulate elongation whether or not TFIIF was present in the reaction mixture and addition of the TFIIF mutant proteins did not affect the ability of FCP1 to stimulate elongation. Moreover, the TFIIF mutant that lacked the RAP74 C-terminal sequences, which include the previously mapped FCP1 binding site (RAP74 Δ218-517), was able to stimulate elongation. More importantly, in the presence of FCP1, the elongation-stimulating activity of the TFIIF C-terminal mutant was also further activated and was similar to activities observed with wild-type TFIIF and FCP1. Thus, we conclude that TFIIF and FCP1 have independent functions during elongation by RNAP II. In agreement with our previous findings, we also observed that the elongation-stimulatory activity of FCP1 is independent of FCP1-associated phosphatase activity (data not shown) (10).
FIG. 4.
Kinetics of transcription elongation with FCP1 in the presence and absence of various TFIIF mutants. Ternary +42 complexes were isolated as described for Fig. 3D and incubated for 10 min at 30°C under the following conditions: alone (lanes 1 to 3), with FCP1 alone (60 ng; lanes 4 to 6), with wild-type (Wt) TFIIF (8 pmol) in the absence of FCP1 (lanes 7 to 9), with wild-type TFIIF in the presence of FCP1 (lanes 10 to 12), with mutant TFIIFs (40 pmol) in the absence of FCP1, and with mutant TFIIFs in the presence of FCP1 (60 ng) (lanes 13 to 41). The reactions were then chased with 30 μM concentrations of each of the four XTPs and stopped at various time points. The reaction products were analyzed as described for Fig. 3C.
Isolation of fcp1 alleles that implicate the FCP1 protein in RNAP II elongation in vivo.
In an attempt to study the FCP1 elongation effect in vivo, we used error-prone PCR to generate altered forms of yeast Fcp1. To avoid the TFIIF interaction domain, which was previously mapped to the COOH-terminal region of FCP1 (2, 3), the mutation sites were limited to the NH2 terminus of Fcp1. We first used a plasmid-shuffling technique to test the mutant fcp1 alleles for their ability to complement growth in the absence of the essential wild-type FCP1 gene. The viable strains that contained fcp1 alleles were then tested for several phenotypes, including heat and cold sensitivity and 6-AU sensitivity. The nucleotide analog 6-AU causes the depletion of the cellular nucleotide pool and diminishes the growth of a yeast strain when transcription elongation is compromised (30, 41).
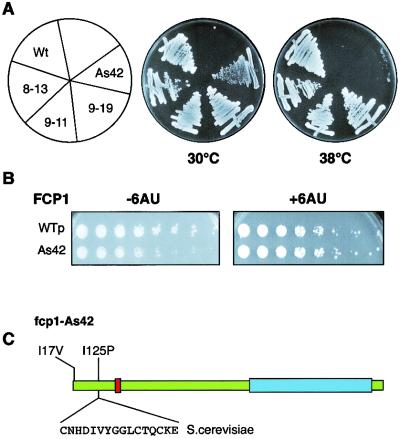
The fcp1-As42 mutant exhibited a tight ts phenotype at 38°C and grew slower than the wild-type strain at 30°C (Fig. 5A and B). When yeast cells were grown at 38°C, 6-AU was generally toxic, even in the wild-type strain. However, growth at 30°C was restored when the fcp1-As42 mutant was grown in the presence of 6-AU (Fig. 5B). In fact, the fcp1-As42 mutant grew better in 6-AU than did yeasts that carry the wild-type allele (data not shown). Thus, 6-AU suppresses the slow-growth phenotype associated with the fcp1-As42 allele.
FIG. 5.
Isolation of FCP1 alleles that implicate FCP1 in elongation by RNAP II in vivo. Mutant fcp1 alleles were generated by error-prone PCR and tested for their ability to complement growth in the absence of wild-type (Wt) genomic FCP1 by using a plasmid-shuffling technique. Viable strains containing fcp1 alleles were then tested for various phenotypes, including heat and cold sensitivity (A) and 6-AU sensitivity (B). WTp, plasmid bearing the wild-type gene. (C) Precise mutation in the fcp1-As42 allele. The sequence represents the putative Zn binding domain, and the replacement is within this domain. Red, catalytic conserved domain; blue, RAP74 interaction domain in C terminus of FCP1.
DNA sequence analyses of the fcp1 alleles revealed two encoded amino acid substitutions. One of the replacements (I125P) lies in the putative zinc binding region, whereas the other is an I17V replacement (Fig. 5C).
Synthetic phenotypes associated with certain alleles of fcp1 and spt4 or rpb2.
To further analyze the effect of FCP1 in elongation, we sought to identify putative synthetic phenotypes associated with the fcp1-As42 allele and alleles of genes that encode proteins known to affect elongation. We began the analysis by monitoring changes in the growth of strains that carry previously characterized mutations in elongation factors that confer 6-AU sensitivity. We first analyzed SPT4. SPT4 and SPT5 were initially characterized as genes that encode polypeptides that affect chromatin structure (54, 60). More recently, genetic studies have shown that certain alleles of these genes confer phenotypes that suggest defects in transcription elongation (19, 56). In human cells, the products of homologues of SPT4 and SPT5 are subunits of DSIF, a protein complex that regulates transcription elongation (56). In yeast, SPT5 is essential for viability, whereas SPT4 is not (19, 31, 54). However, upon deletion of SPT4, yeast becomes 6-AU sensitive (31).
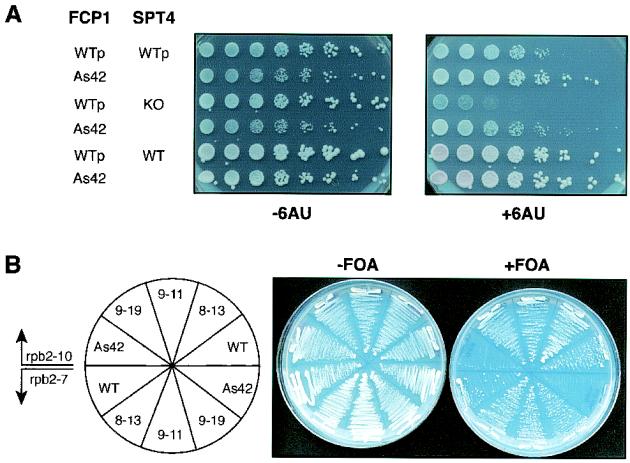
A yeast strain that lacks both SPT4 and FCP1 was constructed in the presence of a plasmid bearing the wild-type FCP1 allele on a URA3 plasmid. When fcp1-As42 replaced the wild-type FCP1 allele in an spt4 knockout background, the slow-growth phenotype associated with the fcp1-As42 allele was suppressed and the 6-AU sensitivity associated with the spt4 deletion was dampened (Fig. 6A). The 6-AU resistance phenotype did not depend on the presence of SPT4 (Fig. 6A). Similar results were obtained with PPR2, a gene that encodes the well-characterized transcription elongation factor TFIIS, which stimulates the intrinsic 3′-to-5′ RNase activity associated with RNAP II. This activity allows RNAP II to pass through “roadblocks” in the DNA template (59). Taken together, these genetic experiments suggest that FCP1 affects elongation in vivo; however, its function in elongation is different from the functions of DSIF and TFIIS. Coupling the in vivo results with the biochemical results described above, we conclude that FCP1 affects the rate of transcription elongation.
FIG. 6.
Synthetic phenotypes associated with various mutant alleles of fcp1 and either spt4 or rpb2. (A) 6-AU sensitivity assay of the fcp1-As42 allele in the absence of SPT4. Isogenic yeast strains that contained either wild-type (WT) FCP1 or mutant fcp1-As42 were analyzed for 6-AU sensitivity in the background of a genomic knockout (KO) of SPT4. Versions of the FCP1 and SPT4 alleles carried by the strains that were assayed are indicated at the left. WTp, plasmid bearing the wild-type gene. (B) fcp1-As42 is synthetically lethal when elongation is compromised, as it is when certain rpb2 alleles are present. The viability of yeast strains that contained mutant rpb2 alleles with either mutant fcp1 alleles (fcp1-As42, fcp1-9-19, fcp1-9-11, or fcp1-8-13) or wild-type FCP1 is shown. The fcp1 plasmids were shuffled into strains that contained either rpb2-7 (top) or rpb2-10 alleles (bottom), and the newly created strains were then streaked on agar plates with (+FOA; to counter-select the wild-type FCP1) or without (−FOA; control) FOA.
To further decipher the function of FCP1 during elongation, we attempted to identify synthetic phenotypes caused by the fcp1-As42 allele and alleles of rpb2, the gene that encodes the second-largest subunit of RNAP II. Two alleles of RPB2 have been relatively well characterized; a single amino acid replacement of Pro with Ser at residue 1018 (rpb2-10) results in 6-AU sensitivity and compromises the RNAP II elongation rate in vitro (30, 41), and the substitution of a Cys residue for an Arg at position 122 (rpb2-7) results in 6-AU sensitivity but does not affect the elongation rate in vitro (41). Accordingly, fcp1-As42 was shuffled into strains that bear either the rpb2-10 or rpb2-7 alleles. Strains that contained either rpb2-10 or rpb2-7 were not viable when these alleles were combined with the fcp1-As42 allele (Fig. 6B). This “synthetic lethal” phenotype observed with the rpb2 mutant alleles and fcp1-As42 was allele specific, as three other fcp1 mutant alleles did not affect viability (Fig. 6B); indeed, these three fcp1 alleles actually suppressed the slow-growth phenotype associated with rpb2-7 and rpb2-10 (data not shown). In addition, these three alleles of fcp1 (fcp1-8-13, -9-11, and -9-19) rescued the 6-AU sensitivity associated with rpb2-7 (data not shown). Finally, these alleles did not alter the 6-AU sensitivity associated with rpb2-10 (data not shown).
DISCUSSION
Over the last several years, researchers have begun to appreciate the complexity of elongation by RNAP II (for reviews see references 11, 38, 42, and 52). Taken together, the results of various studies have established that transcription elongation by RNAP II is a highly regulated process (38, 42). This regulation is mediated by a number of factors that operate at different steps in the elongation process.
Some of these factors, such as NELF, inhibit elongation (56), most likely during promoter clearance. This inhibition is thought to be connected with a checkpoint that couples 5′ capping of nascent RNAs with the establishment of an elongation-competent form of RNAP II (for details see references 34 and 38). Other factors, such as P-TEFb, overcome the negative effect of NELF and also positively stimulate transcription elongation by phosphorylating the CTD of RNAP II at a step subsequent to transcription initiation (1). P-TEFb is a multifunctional factor, as it is also required for the establishment of an RNAP II elongation-competent complex (9, 38). Other factors function to stimulate transcription elongation by increasing the rate of elongation or by preventing the pausing and/or arrest of the elongation complex (43, 44, 52, 53). TFIIF, SIII, and ELL (52, 53) are among the factors known to stimulate the elongation rate directly. TFIIS stimulates a 3′-to-5′ RNase activity present in RNAP II and is required to overcome arrest (20, 21, 45).
In this study, we showed that FCP1, a phosphatase that is highly specific for the CTD of RNAP II (10), stimulates transcription elongation. The FCP1 elongation-stimulatory activity was demonstrated by using a reconstituted transcription system. All of the components of this system were defined biochemically as well as in vivo. The in vivo characterization involved analyses of the phenotypes associated with various fcp1 alleles in combination with distinct alleles of other factors previously shown to have a function in elongation. The FCP1 elongation-stimulatory activity is independent of the CTD phosphatase activity (10) (data not shown).
The in vitro transcription experiments allowed us to analyze the effect of FCP1 on elongation at a mechanistic level and to study the relation between FCP1 and TFIIF. TFIIF was shown previously to affect elongation rates (5, 15), to interact with FCP1 (8, 10), and to regulate the phosphatase activity of FCP1 (10). Here, FCP1 was shown to stimulate elongation rates independent of TFIIF. However, we also observed that the elongation-stimulatory effects of TFIIF and FCP1 are additive. Although the precise mechanism by which TFIIF and FCP1 stimulate elongation rates is unknown, we speculate that the effect is mediated through interactions with RNAP II and/or the nascent RNA which result in the alteration of the structure of the RNAP II elongation complex. Interactions with the nascent RNA could be mediated through the BRCT domain present at the C terminus of FCP1 (25, 63). It was recently suggested that the BRCT domain serves as an RNA binding domain.
Because the elongation-stimulatory effects of TFIIF and FCP1 are additive, it is likely that the two proteins stimulate elongation rates by distinct mechanisms (for example, they may induce different conformational changes in the elongation complex). Recent studies have demonstrated that FCP1 directly interacts with the Rpb4 subunit of RNAP II (24), and previous studies demonstrated that the small subunit of TFIIF, RAP30, interacts with the Rpb5 subunit of RNAP II (58). Although it is not known which subunit of RNAP II interacts with RAP74 (the largest subunit of TFIIF), DNA-protein cross-linking studies with transcriptionally competent initiation complexes indicate that RAP74 is in close proximity to RNAP II subunits Rpb1 and Rpb2 (23). Therefore, it is likely that FCP1 and TFIIF each interact directly with a distinct subunit of RNAP II.
It is also possible that TFIIF interacts with nascent RNA indirectly by binding to FCP1. However, even though TFIIF and FCP1 interact directly, this interaction appears not to be required for the elongation-stimulatory activity of these factors. This notion is supported by our experiments with a TFIIF derivative that contained a version of RAP74 from which aa 218 to 517 (which include the FCP1 binding site) had been deleted (RAP74 1-217). RAP74 1-217 was shown to interact functionally with FCP1 and RNAP II; that is, TFIIF derivatives that contained this mutant version of RAP74 increased elongation rates to levels above those observed with FCP1 alone (Fig. 4).
The biochemical observations that demonstrate a role for FCP1 during the elongation phase of the transcription cycle are further supported by experiments performed in vivo. Recent studies have established a genetic interaction between FCP1 and the elongation factor TFIIS (12). And our own results described herein, derived from the isolation of a variety of fcp1 alleles, established a genetic relationship between FCP1 and various other factors that affect elongation. For example, the fcp1-As42 allele was found to cause lethality in combination with alleles of rpb2 that confer sensitivity to 6-AU, a drug that diminishes intracellular concentrations of GTP and UTP and that is thought to slow elongation in vivo. Under wild-type conditions, elongation is an efficient process; however, reductions in the concentrations of the nucleotide pools result in an imbalance of elongation, which is reflected phenotypically as slow growth in the presence of 6-AU. Mutations in elongation factors such as DSIF and TFIIS can enhance the sensitivity of a strain to 6-AU; interestingly, 6-AU sensitivity can also be decreased when elongation factors with diminished capacity are present. Indeed, the fcp1-As42 allele was found to suppress 6-AU sensitivity. Similar results have been observed with certain alleles encoding other transcription factors, such as the chromatin-specific elongation factor FACT (39). We speculate that, because the products of these alleles impair the function of elongation factors that are required constantly to maintain efficient elongation rates, the moving RNAP II complex proceeds at a slower rate and can then tolerate a decrease in the concentration of the nucleotide pool. This “recovered balance” between the efficiency of the elongating machinery and the nucleotide pool results, phenotypically, in better growth in the presence of 6-AU.
Our biochemical analyses established that, although TFIIF and FCP1 both stimulate elongation rates, the mechanisms by which they accomplish this feat are apparently different. The concentration of TFIIF that is required for optimal stimulation of elongation is catalytic with respect to RNAP II elongation complexes. This finding is supported by previous biochemical studies that showed that TFIIF is transiently required to stimulate elongation rates and that TFIIF is released from mature elongation complexes (61). These early biochemical observations are supported by recent ChIP experiments that revealed that TFIIF is not associated with active elongation complexes (40). On the other hand, stoichiometric concentrations of FCP1, with respect to RNAP II elongation complexes, are required for optimal stimulation of elongation, and recent ChIP experiments suggest that FCP1 remains associated with the elongation complexes as the transcription reaction proceeds (9). Therefore, these results collectively imply that that there is a constant requirement for FCP1 during elongation and that the mode of action of FCP1 is different from those of TFIIF and TFIIS.
Acknowledgments
We thank M. Hampsey for discussions, J. Greenblatt for yeast strain JA830, F. Winston for strain FY1114 and plasmid FB1101pBM25, R. Young for strains Z103 and Z106, and D. Luse for plasmid pML20-47. We also thank S. J. Conaway for RAP30 mutants and Z. Burton for RAP74 mutants. We also thank members of the Reinberg laboratory for helpful suggestions and M. Hampsey and E. Friedl for comments on the manuscript.
This work was supported by a grant from the NIH (GM-37120) and the Howard Hughes Medical Institute to D.R.
S. S. Mandal and H. Cho contributed equally to this work.
REFERENCES
- 1.Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, J., R. S. Chambers, M. S. Kobor, Y. Ho, M. Cartier, D. Bolotin, B. Andrews, C. M. Kane, and J. Greenblatt. 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:14300-14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault, J., G. Pan, G. K. Dahmus, M. Cartier, N. Marshall, S. Zhang, M. E. Dahmus, and J. Greenblatt. 1998. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem. 273:27593-27601. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomew, B., M. E. Dahmus, and C. F. Meares. 1986. RNA contacts subunits IIo and IIc in HeLa RNA polymerase II transcription complexes. J. Biol. Chem. 261:14226-14231. [PubMed] [Google Scholar]
- 5.Bengal, E., O. Flores, A. Krauskopf, D. Reinberg, and Y. Aloni. 1991. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol. Cell. Biol 11:1195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, R. S., and M. E. Dahmus. 1994. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J. Biol. Chem. 269:26243-26248. [PubMed] [Google Scholar]
- 7.Chambers, R. S., and C. M. Kane. 1996. Purification and characterization of an RNA polymerase II phosphatase from yeast. J. Biol. Chem. 271:24498-244504. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, R. S., B. Q. Wang, Z. F. Burton, and M. E. Dahmus. 1995. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270:14962-14969. [DOI] [PubMed] [Google Scholar]
- 9.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, H., T. K. Kim, H. Mancebo, W. S. Lane, O. Flores, and D. Reinberg. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 12.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulombe, B., and Z. F. Burton. 1999. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63:457-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 15.Flores, O., E. Maldonado, and D. Reinberg. 1989. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J. Biol. Chem. 264:8913-8921. [PubMed] [Google Scholar]
- 16.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 17.Greenleaf, A. L. 1993. A positive addition to a negative tail's tale. Proc. Natl. Acad. Sci. USA 90:10896-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenleaf, A. L. 1993. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem. Sci. 18:117-119. [DOI] [PubMed] [Google Scholar]
- 19.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izban, M. G., and D. S. Luse. 1993. The increment of SII-facilitated transcript cleavage varies dramatically between elongation-competent and -incompetent RNA polymerase II ternary complexes. J. Biol. Chem. 268:12874-12885. [PubMed] [Google Scholar]
- 21.Izban, M. G., and D. S. Luse. 1993. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J. Biol. Chem. 268:12864-12873. [PubMed] [Google Scholar]
- 22.Kim, S., K. Cabane, M. Hampsey, and D. Reinberg. 2000. Genetic analysis of the YDR1-BUR6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol. Cell. Biol. 20:2455-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T. K., R. H. Ebright, and D. Reinberg. 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418-1422. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, M., H. Suzuki, and A. Ishihama. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22:1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobor, M. S., L. D. Simon, J. Omichinski, G. Zhong, J. Archambault, and J. Greenblatt. 2000. A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7438-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodziej, P. A., and R. A. Young. 1991. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol. Cell. Biol. 11:4669-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laybourn, P. J., and M. E. Dahmus. 1989. Transcription-dependent structural changes in the C-terminal domain of mammalian RNA polymerase subunit IIa/o. J. Biol. Chem. 264:6693-6698. [PubMed] [Google Scholar]
- 28.Lei, L., D. Ren, and Z. F. Burton. 1999. The RAP74 subunit of human transcription factor IIF has similar roles in initiation and elongation. Mol. Cell. Biol. 19:8372-8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei, L., D. Ren, A. Finkelstein, and Z. F. Burton. 1998. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol. Cell. Biol. 18:2130-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lennon, J. C., III, M. Wind, L. Saunders, M. B. Hock, and D. Reines. 1998. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD-modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, H., L. Zawel, L. Fisher, J. M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 33.Malone, E. A., J. S. Fassler, and F. Winston. 1993. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol. Gen. Genet. 237:449-459. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 35.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien, T., S. Hardin, A. Greenleaf, and J. T. Lis. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75-77. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 38.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 39.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 40.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 41.Powell, W., and D. Reines. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinberg, D., and R. G. Roeder. 1987. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J. Biol. Chem. 262:3331-3337. [PubMed] [Google Scholar]
- 44.Reines, D., P. Ghanouni, W. Gu, J. Mote, Jr., and W. Powell. 1993. Transcription elongation by RNA polymerase II: mechanism of SII activation. Cell Mol. Biol Res 39:331-338. [PubMed] [Google Scholar]
- 45.Reines, D., and J. Mote, Jr. 1993. Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl. Acad. Sci. USA 90:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 47.Ruppert, S., and R. Tjian. 1995. Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev. 9:2747-2755. [DOI] [PubMed] [Google Scholar]
- 48.Samkurashvili, I., and D. S. Luse. 1998. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol. Cell. Biol. 18:5343-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scafe, C., C. Martin, M. Nonet, S. Podos, S. Okamura, and R. A. Young. 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scafe, C., M. Nonet, and R. A. Young. 1990. RNA polymerase II mutants defective in transcription of a subset of genes. Mol. Cell. Biol. 10:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider, J. C., and L. Guarente. 1991. Vectors for expression of cloned genes in yeast: regulation, overproduction, and underproduction. Methods Enzymol. 194:373-388. [DOI] [PubMed] [Google Scholar]
- 52.Shilatifard, A. 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 12:1437-1446. [DOI] [PubMed] [Google Scholar]
- 53.Shilatifard, A. 1998. The RNA polymerase II general elongation complex. Biol. Chem. 379:27-31. [DOI] [PubMed] [Google Scholar]
- 54.Swanson, M. S., and F. Winston. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, S., R. C. Conaway, and J. W. Conaway. 1995. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc. Natl. Acad. Sci. USA 92:6042-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, B. Q., and Z. F. Burton. 1995. Functional domains of human RAP74 including a masked polymerase binding domain. J. Biol. Chem. 270:27035-27044. [DOI] [PubMed] [Google Scholar]
- 58.Wei, W., D. Dorjsuren, Y. Lin, W. Qin, T. Nomura, N. Hayashi, and S. Murakami. 2001. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J. Biol. Chem. 276:12266-12273. [DOI] [PubMed] [Google Scholar]
- 59.Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 61.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 62.Zawel, L., and D. Reinberg. 1993. Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acid Res. Mol. Biol. 44:67-108. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, X., S. Morera, P. A. Bates, P. C. Whitehead, A. I. Coffer, K. Hainbucher, R. A. Nash, M. J. Sternberg, T. Lindahl, and P. S. Freemont. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J 17:6404-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]