Abstract
Although CLOCK/BMAL1 heterodimers have been implicated in transcriptional regulation of several rhythmic genes in vitro through E-box sequence elements, little is known about how the circadian clock regulates rhythmic genes with diverse phases in vivo. The gene nocturnin is rhythmically transcribed in Xenopus retinal photoreceptor cells, which contain endogenous circadian clocks. Transcription of nocturnin peaks in these cells in the middle of the night, while CLOCK/BMAL1 activity peaks during the early morning. We have identified a novel protein-binding motif within the nocturnin promoter, which we designated the nocturnin element (NE). Although the NE sequence closely resembles an E-box, our data show that it functions as a cyclic AMP response element (CRE) by binding CREB. Furthermore, phosphorylated CREB (P-CREB) levels are rhythmic in Xenopus photoreceptors, with a phase similar to that of nocturnin transcription. Our results suggest that P-CREB controls the rhythmic regulation of nocturnin transcription and perhaps that of other night phase genes. The NE may be an evolutionary intermediate between the E-box and CRE sequences, both of which seem to be involved in the circadian control of transcription, but have evolved to drive transcription with different phases in these clock-containing cells.
Most organisms undergo rhythmic changes on a daily basis to synchronize diverse behavioral and physiological events to the dramatic changes in the natural environment. Many of these changes are controlled by endogenous circadian clocks that control events such as cell signaling, gene expression, hormone production, neuronal activity, and many aspects of behavior. Recent work has provided insight into many details of the molecular mechanisms that make up the circadian clock (reviewed in references 53 and 67), but how the circadian oscillatory mechanism is translated into such diverse rhythmic biological processes is not yet well understood.
One thing that is clear, however, is that rhythmic control of gene expression is an important feature of circadian clock control of rhythmic processes. Transcriptional control is important both within the central clock mechanism itself and for conveying circadian regulation to a myriad of rhythmic outputs. Recent genome-wide microarray analyses have revealed that the number of genes under circadian control is vast and that the resulting mRNA rhythms exhibit many different amplitudes, phases, and waveforms (1, 11, 15, 24, 27, 34, 42, 51, 56, 59, 63). The transcriptional (and posttranscriptional) mechanisms that underlie these different expression patterns are generally not known.
In animals, the best understood examples are those genes that are directly regulated by the basic helix-loop-helix-PAS proteins CLOCK and BMAL1. Heterodimers of these proteins activate transcription of genes that are components of the central clock, such as the period (per) and cryptochrome (cry) genes, through a conserved E-box element (reviewed in references 53 and 67). This results in the transcription of these genes with a characteristic phase (which varies in different tissues and species), and this transcriptional activation is subsequently repressed by the protein products of these genes, resulting in the negative feedback loop that makes up a portion of the central clock mechanism. In addition to this role in the central oscillator, CLOCK and BMAL1 have also been reported to activate clock-controlled genes, such as those for arginine vasopressin (avp), D-element-binding protein (dbp), and arylalkylamine N-acetyltransferase (aa-nat) by binding to E-box elements (9, 10, 33, 54).
Another transcriptional pathway that has been implicated in rhythmic regulation is the cyclic AMP (cAMP) response element (CRE)/CRE-binding protein (CREB) pathway (49, 50), although specific target genes have not been identified in clock-containing cells. CREB is a stimulus-induced transcription factor that can be activated by a large number of environmental changes (reviewed in references 57 and 66). The activity of the Drosophila CREB2 exhibits a 24-h rhythm which is abolished in per mutant flies, and a mutation in Drosophila CREB2 shortens the circadian locomotor rhythm and dampens the oscillation of per (5). In the mouse suprachiasmatic nucleus (SCN), levels of phosphorylated CREB (P-CREB) oscillate as a function of circadian time (CT), with a peak at mid to late subjective night (49), but the role of this oscillation is not known. Interestingly, the consensus sequence of the CRE is similar to the E-box sequence, and it has been suggested that there is an evolutionary relationship between these sequences (reviewed in reference 38).
The Xenopus retina contains a fully functional circadian clock localized within the photoreceptor layer (6, 7, 31). As in other vertebrates, many aspects of retinal physiology are under circadian control (reviewed in reference 3), including visual sensitivity (41), retinomotor activity (52), melatonin synthesis and release (6, 8), and gene expression (20-22). Xenopus homologs of the clock genes, including Clock, bmal1, per 1, per 2, cry 1, and cry 2, are expressed in retinal photoreceptor cells, and the phases of peak expression of these genes resemble that of the mammalian SCN, with per 1 and per 2 expression peaking in early day and that of bmal1 peaking in early night (71-73; unpublished data).
nocturnin is a novel clock-controlled gene in Xenopus retina with sequence similarity to the Saccharomyces cerevisiae transcriptional coactivator-deadenylase CCR4, and its mRNA levels exhibit a high-amplitude circadian rhythm with a mid-night peak (20, 22). Nuclear run-on assays demonstrated that this rhythm of nocturnin mRNA levels is controlled at the level of transcription (20). Previous analyses of the regulatory elements contained within this gene have shown that a novel protein-binding motif called photoreceptor conserved element II (PCE) controls the appropriate spatial expression of the nocturnin gene, resulting in specific expression in retinal photoreceptor cells (39). In this paper, we have investigated the regulatory mechanism that drives the rhythmic transcription of the nocturnin gene and have identified a novel protein-binding motif designated the nocturnin element (NE), which is similar to, but distinct from, both per-E-box (PE) and CRE consensus sequences. Our results demonstrate that CREB, but not CLOCK/BMAL1, binds to the NE. We also show that the retinal photoreceptor cells have a high-amplitude rhythm of CREB phosphorylation with a nighttime peak. The phase of this rhythm, coupled with our biochemical data, suggests that the rhythmicity of the nocturnin gene is regulated by P-CREB.
MATERIALS AND METHODS
Animals.
Xenopus laevis was purchased from Nasco (Fort Atkinson, Wis.) and maintained under conditions of 12 h of light and 12 h of darkness (LD). The care and use of Xenopus complied with all relevant federal and institutional guidelines.
In situ hybridization.
The eyes were dissected from adult frogs at Zeitgeber time (ZT) 2 and ZT 14 (ZT 0 is defined as the lights-on time, and ZT 12 is defined as the lights-off time). Eyecups (including the retina, pigment epithelium, choroid, and sclera) were prepared and then fixed overnight in 4% paraformaldehyde in phosphate-buffered saline at 4°C. The tissues were cryoprotected in 30% sucrose in phosphate-buffered saline for 2 to 4 h at 4°C and then embedded in Tissue-Tek O.C.T. compound (Ted Pella, Redding, Calif.), and cryosections (12-μm thick) were prepared. Digoxigenin-labeled antisense T7 and sense T3 probes were prepared from the cDNA clone of nocturnin. In situ hybridization was carried out as described in reference 72.
EMSAs and supershift assays.
Xenopus retinal nuclear extracts were prepared and electrophoretic mobility shift assays (EMSAs) were performed as described previously (39). Protein concentrations in nuclear extracts were determined before the performance of EMSAs to ensure equal loadings. The oligonucleotides (Life Technologies, Inc., and MWG Inc.) used as probes in the EMSAs are listed in Fig. 2B and 4. Briefly, 4 to 6 μg of retinal nuclear extract was incubated with the 5′-end-labeled probe in a total volume of 15 to 20 μl (39). For competition assays, excess unlabeled oligonucleotides were added as indicated in the figures and incubated for 20 to 30 min at room temperature prior to the addition of radiolabeled probes.
FIG. 2.
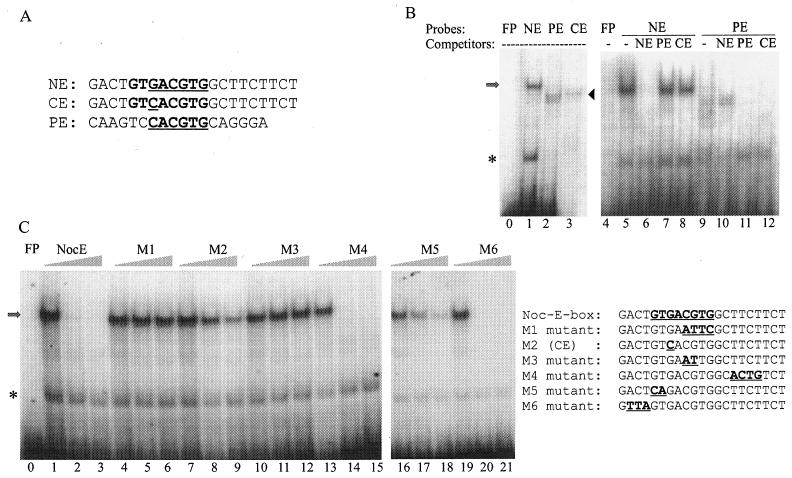
NE sequence as defined by EMSAs. (A) Sequence alignment of the NE, CE, and PE. (B) EMSAs were performed with a nuclear extract mixture isolated from Xenopus retina at ZTs 2 and 14. The identities of the radiolabeled probes and 100-fold-molar-excess cold competitors are given at the tops of the lanes. The arrow and arrowhead mark the positions of specific DNA-protein complexes revealed when NE and PE, respectively, were used as probes. The asterisk indicates nonspecific binding activities. (C) The NE was used as the probe in all lanes. Identities of the competitors are given at the tops of the groups of three lanes. Each group contains 0×, 50×, and 200× concentrations of cold competitors along with the radiolabeled NE probe. The sequences of the probe and competitors are shown to the right of the gels. Note that M2 is identical to CE. Lanes 0 and 4 in panel B and lane 0 in panel C show controls of free probes (FP) without retinal nuclear extracts. Each gel is a representative example of gels from three repeated experiments.
FIG. 4.
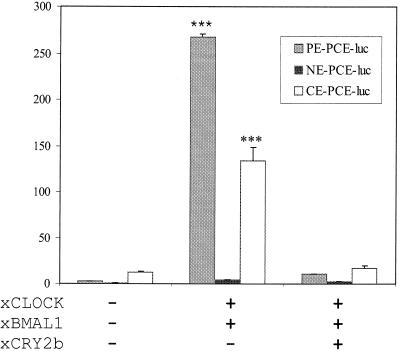
Effects of xCLOCK and xBMAL1 on PE, NE, and CE promoter activities in transiently transfected Cos 7 cells. The sequences of the different E-box-like elements shown are listed in the legend for Fig. 2A. The null-renilla vectors were cotransfected as internal controls. Sample values are means ± standard errors of determinations with four samples. All results were normalized to the relative luciferase activity in cells transfected with the PE-PCE-luc reporter gene only, whose activity was assigned a value of 1. ***, P < 0.001 (the value is significantly different from that for the respective control [reporter only]) by analysis of variance by the Tukey-Kramer multiple-comparison test. These data are representative of results from three independent experiments.
To identify the individual transcription factor(s) involved in the DNA-protein complex formation, supershift assays were performed. Antibodies against CREB-1 (Santa Cruz Biotechnology, Inc.), phospho-CREB (Ser 133; Cell Signaling Technology), and Xenopus CLOCK (xCLOCK; a gift of Gregory Cahill, University of Houston) were coincubated with the nuclear extracts for 1 to 4 h at room temperature prior to the addition of probes. Samples were electrophoresed at a constant voltage of 210 V at 4°C for 2 h. Dry gels were exposed to film for 1 to 24 h at room temperature.
Plasmid construction.
Complementary oligonucleotides (Life Technologies, Inc., and MWG Inc.) containing three repeats of PCE were synthesized flanked with restriction enzyme-cutting sites (SacI and HindIII [underlined]): 5′-CCGAGCTCGGGGTCAGACAGGCTTATAGGCAGACAGGCTTATATCTGGCAGACAGGCTTATATCTGCCAAGCTTCC-3′ (39). Three repeats of PCE served as a basal promoter for transcription initiation. The sequence of the PE is from the mouse per promoter (positions −156 to −139) (32), and the sequence of the NE is from the Xenopus nocturnin promoter (positions −104 to −95) (39). Standard recombinant DNA techniques were used to assemble the PE-PCE-luciferase (PE-PCE-luc), NE-PCE-luc, and consensus E-box-PCE-luc (CE-PCE-luc) reporter constructs. Briefly, three repeats of the different elements (sequences are listed in Fig. 2A) flanked with restriction enzyme cutting sites (5′-XhoI and SacI-3′) were linked to PCE sequences at the SacI site and then together subcloned upstream of the firefly luc reporter in pGL2-Basic vector (Promega) at the XhoI and HindIII sites.
The cDNA clones of xClock, Xenopus bmal1 (xbmal1), and Xenopus cry 2b (xcry 2b) (71-73; unpublished data) were subcloned downstream of the human cytomegalovirus (CMV) promoter in the pVAX-1 vector (Invitrogen). All constructs were confirmed by sequencing. pCMV-PKA, pCMV-CREB, and pCMV-K-CREB vectors were obtained from Clontech Laboratories. pCMV-PKA contains the gene encoding the catalytic subunit of protein kinase A (PKA) driven by the CMV promoter (25, 43). pCMV-K-CREB expresses a mutant form of the human CREB protein that contains mutations in its DNA-binding domain (65).
Transient transfections.
Cos 7 and NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum and maintained at 37°C in a water-saturated atmosphere containing 95% air and 5% CO2. For transfection, the cells were distributed into 12-well plates and cultured overnight until they were ∼70% confluent. Both 3T3 cells and Cos 7 were transiently transfected with DNA plasmids by lipofection with Lipofectamine reagent (Life Technologies, Inc.).
Thymidine kinase (TK)-renilla vectors (Promega, Madison, Wis.) were originally cotransfected into the Cos 7 cells as internal controls for estimation of transfection efficiency. However, the results showed that TK-renilla itself was activated 5- to 10-fold in the presence of xCLOCK and xBMAL1 (data not shown). Therefore, pRL-null plasmid vector (promoterless renilla; Promega) was used as the internal control and it yielded much more consistent results than TK-renilla under various conditions (reference 4 and data not shown).
For each transfection in Cos 7 cells, 250 ng of firefly luc reporter vector, 10 ng of null-renilla luc (with or without 250 ng each of xClock, xbmal1, and xcry 2b), and pEGFP-C2 vector (used for DNA compensation) were incubated with OPTI-MEM-Lipofectamine solution (Life Technologies, Inc.) for 0.5 h. 3T3 cells were transfected with 250 ng of firefly luc reporter vector and 10 ng of null-renilla luc (with or without the addition of 250 ng of CREB, 1 μg of PKA, and 250 ng of K-CREB). Cells were then exposed to the DNA-liposome complexes in OPTI-MEM (Life Technologies, Inc.) for 5 h and transferred to Dulbecco's modified Eagle medium for another 16 to 20 h. The activities of the firefly and renilla luc genes were measured with a Dual-luciferase assay kit (Promega).
Immunohistochemistry.
Xenopus eyes were removed from adult frogs, and eyecups (containing sclera, choroid, retinal pigment epithelium, and neural retina) were transferred to culture medium at 21°C. The culture medium was a defined medium of balanced salts, amino acids, and 100 μM 5-hydroxy-trptophan and was equilibrated to pH 7.4 with 95% O2-5% CO2 (8, 23). At ZT 12, half of the samples were maintained under conditions of LD and the other half were transferred to constant darkness (DD). On the second day of culture, eyecups were collected at appropriate times and sections were prepared as described above for in situ hybridization.
Slides with retinal sections were blocked with CBT blocking solution (0.25% Carageenan lambda σ, 1% bovine serum albumin, 0.3% Triton X-100) and 0.1% sodium azide in TBS (50 mM Tris base, 150 mM NaCl; pH 7.5) for 1 to 2 h at room temperature. After being blocked, sections were incubated with the P-CREB primary antibody (1:50 dilution; Cell Signaling Technology) in a humidified chamber overnight; they were then washed three times for 10 min each time with washing solution (WS) consisting of 1× TBS and 0.3% Triton X-100. For fluorescent visualization of the P-CREB antibody, Cy3-conjugated donkey anti-rabbit immunoglobulin G antibody (heavy plus light chains) (1:1,500 dilution; Jackson ImmunoResearch Laboratories, Inc.) was incubated with the sections for 1 h at room temperature and then the sections were washed three times with WS for 10 min each time. Sections were rinsed with water and mounted with Fluoromount G (Electron Microscopy Sciences). Images were obtained on an Olympus IX 70 inverted microscope and digitized with the OlymPix camera system. Data analyses were performed in a blind manner: all pictures were taken from slides with covered labels and with the same exposure times. Five sections per retina from two animals were examined for each time point.
RESULTS
Cellular localization of nocturnin transcripts.
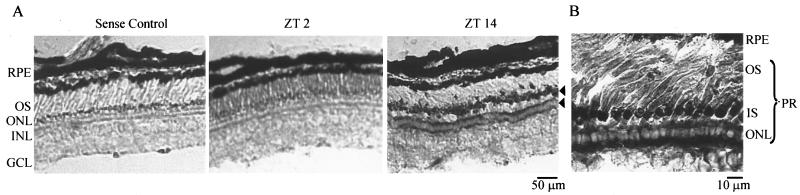
Nocturnin is a retina-specific gene in Xenopus laevis, and its mRNA levels exhibit a high-amplitude circadian rhythm in both the LD and DD cycles (20). It was previously shown by in situ hybridization with radiolabeled probes that nocturnin was specifically expressed in the photoreceptor layer within the retina (22). To more carefully determine cellular localization of nocturnin transcripts, in situ hybridization with digoxigenin-labeled nocturnin probes was performed on Xenopus retinas that had been dissected and fixed at ZTs 2 and 14. Consistent with the radioactive in situ results (22), we detected intense staining in photoreceptor cells at ZT 14 (Fig. 1A). nocturnin mRNA could be seen only at night (ZT 14) and not at ZT 2 (Fig. 1A), as was previously demonstrated by Northern blot experiments (20). With higher magnification, it is clear that the staining is present in the perinuclear region and inner segments of rod and cone photoreceptor cells (Fig. 1B), verifying that both rods and cones synthesize nocturnin mRNA rhythmically.
FIG. 1.
Temporal and spatial analyses of the nocturnin mRNA expression in Xenopus retina by in situ hybridization. Xenopus eyes were dissected and fixed at ZTs 2 and 14, and 12-μm-thick cryosections were prepared. (A) In situ hybridization was done with a digoxigenin-labeled sense probe at ZT 14 (left) and antisense probes at ZT 2 (middle) and ZT 14 (right). Arrowheads point to the heavy in situ labeling in the inner segments and the perinuclear regions of the photoreceptor cells. (B) A higher-magnification image of the in situ staining at ZT 14 is shown. RPE, retinal pigment epithelium; PR, photoreceptor cells; OS, outer segments; IS, inner segments; ONL, outer nuclear layer (photoreceptor cell nuclei); INL, inner nuclear layer; GCL, ganglion cell layer.
Specificity of the NE-binding activity.
By comparing the nocturnin gene 5′-end-flanking sequence with known protein-binding motifs, we have identified several putative protein-binding sites. One is PCE, which is present as three perfect repeats in the proximal nocturnin promoter (positions −77 to −18) and is sufficient and necessary to direct a reporter gene specifically to retinal photoreceptor cells (39). Another is the NE, an E-box-like element also in the proximal nocturnin promoter (positions −104 to −95). No other E-box sequences were found within ∼8,000 bp of nocturnin 5′-end-flanking sequence. The NE itself is not sufficient to direct reporter genes to photoreceptor cells (data not shown), and deletion of the NE did not affect the spatial expression of a reporter gene (39). However, since E-boxes have been implicated in rhythmic transcription in other genes, we wondered whether the NE might contribute to nocturnin's rhythmic transcription.
Alignment of the core sequences of the NE and PE (Fig. 2A) shows that they differ at one position in the core sequence (the NE is GACGTG, while the PE is CACGTG) as well as throughout the flanking sequences. This first position has previously been reported to be less well conserved among E-boxes of this class (60). A chimeric sequence with the conserved E-box core sequence from the PE (CACGTG) but with the NE flanking sequences was constructed and designated the CE (Fig. 2A). To determine whether these sequences recognize a similar transcription factor(s), EMSAs were performed with retinal nuclear extracts (a mixture of extracts isolated at ZTs 2 and 14). Lane 1 in Fig. 2B shows that one prominent shifted band was detected with the NE probe. The band at the bottom is due to nonspecific binding activities (data not shown). Double bands were detected with both the PE and CE probes (lanes 2 and 3) and show a mobility distinct from that seen with the NE probe. Although the sequence of the CE probe is different at only one base pair from that of the NE probe, the shifted bands with the CE probe exhibit a pattern like that of the PE probe (lanes 2 and 3), indicating that the core E-box sequence is critical for generating these shifted bands.
The addition of a 100-fold molar excess of cold oligonucleotides identical to the radiolabeled NE probe inhibited the NE-binding activity (Fig. 2B, lane 6). However, the same amounts of nonlabeled PE and CE competitors had no effect on the shifted band (lanes 7 and 8), suggesting that this binding activity is specific to the NE. When the PE was used as a probe, an excess of nonlabeled NE competitor had no effect on the PE-shifted band (lane 10), but the same amount of excess cold CE competitor inhibited the shift as effectively as the PE competitor. These results indicate that the specific DNA-binding activity observed with the NE probe is different than that seen with the PE and CE.
To determine the sequence motif necessary for this NE-specific binding, further competition assays were conducted with the retinal nuclear extract mixture (Fig. 2C). The addition of excessive unlabeled oligonucleotides identical to the radiolabeled NE probe resulted in a dosage-dependent inhibition of the DNA-binding activity (lanes 1 to 3). The binding was not inhibited by competition with oligonucleotides that were mutated in any of the underlined portion of the 5′-GTGACGTG-3′ sequence (M1, shown in Fig. 2C in lanes 4 to 6, and M3, shown in lanes 10 to 12), demonstrating that this sequence is required for the DNA-protein interaction. Addition of excess M2(CE, shown in Fig. 2C in lanes 7 to 9) or M5 (lanes 16 to 18) inhibits the binding in a dose-dependent manner although less effectively than addition of the NE itself, suggesting that the underlined part of the sequence 5′-GTGACGTG-3′ is also important for specific binding. The competitors with mutations on either side of these sequences (M4 and M6) resulted in a potent dosage-dependent inhibition (lanes 13 to 15 and 19 to 21) similar to that seen with the use of a competitor identical to the probe. These results indicate that these sequences are not critical for binding. Therefore, the protein-binding element NE is defined as GTGACGTG.
xCLOCK recognizes the PE but not the NE.
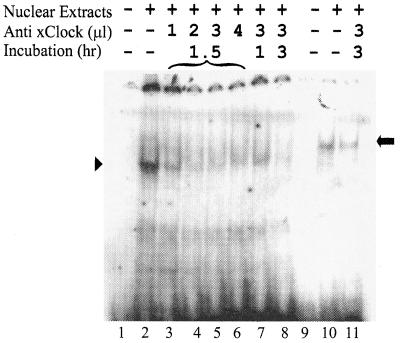
It is known that the heterodimer of the CLOCK and BMAL1 proteins bind the PE and can transactivate reporter genes in vitro (13, 18). To test whether xCLOCK is a component of the specific NE- or PE-shifted complexes, EMSAs in the presence of anti-xCLOCK antibody were conducted. The xCLOCK antibody specifically binds the DNA-binding domain of the xCLOCK protein; therefore, the addition of xCLOCK antibody should disrupt any DNA-CLOCK complexes. Coincubation of the xCLOCK antibody with the nuclear extract prior to the addition of the PE probe resulted in a dose- and time-dependent inhibition of the DNA-protein interactions (Fig. 3, lanes 2 to 8), verifying that these bands contain CLOCK, as expected. However, the xCLOCK antibody had no effect on the shifted band when the NE was used as the probe (lanes 10 to 11), suggesting that the specific binding to the NE is not due to CLOCK and BMAL heterodimers.
FIG. 3.
EMSAs in the presence of the xCLOCK antibody. Shifted complexes are compared after the use of the PE (lanes 1 to 8) and NE (lanes 9 to 11) as probes. The amounts of antibody and the incubation periods are labeled at the tops of the lanes. Lanes 1 and 9 are the free probes without nuclear extracts. The arrowhead and arrow mark the positions of the specific DNA-protein complexs with the PE and NE probes, respectively.
xCLOCK and xBMAL1 transactivate PE-PCE-luc but not the NE-PCE-luc reporter gene in transfected Cos 7 cells.
Transient transfections in Cos 7 cells were performed to examine whether the E-box-binding activities detected in the EMSAs are functional. Previous data demonstrated that the PCE-binding proteins are enriched in retina only (39) and that PCE itself without the E-box enhancer element is not sufficient to activate transcription in the presence or absence of xCLOCK and xBMAL1 in transfected Cos 7 cells (data not shown). Therefore, PCE was used as a minimal promoter to test the effects of the NE, CE, and PE in these transfection assays.
The ability of xCLOCK and xBMAL1 to activate transcription through binding to the different promoter elements was examined. Expression constructs containing xCLOCK and xBMAL1 were cotransfected with the NE-, CE-, or PE-PCE-luc reporter constructs into Cos 7 cells. xCLOCK and xBMAL1 markedly increased the luciferase activity in Cos 7 cells transfected with the PE-PCE-luc reporter construct but not with NE-PCE-luc (Fig. 4), consistent with the EMSA results (Fig. 3). One base pair mutation of G to C in the 5′ end of the E-box core sequence (converting NE to CE) restored the activation (Fig. 4), even though the flanking sequences are identical to those of NE-PCE-luc (Fig. 2A). The increase of CE-PCE-luc activity is less than that of PE-PCE-luc, which may indicate some role for the flanking sequences. An expression plasmid containing xcry was cotransfected into the Cos 7 cells along with xCLOCK and xBMAL1, which resulted in the expected complete repression of the transactivation activities by xCLOCK and xBMAL1 (37, 70), suggesting that this activation is specific. These data indicate that within the core sequence of the PE, the CACGTG at the 5′ end is critical for the specificity of DNA binding and activation by CLOCK/BMAL1. It also confirms the result of the EMSAs, suggesting that xCLOCK does not bind the NE. If the NE is necessary for the rhythmic expression of nocturnin, this regulation must occur through some non-CLOCK mechanism.
CREB binds to the NE and P-CREB transactivates NE-PCE-luc reporter.
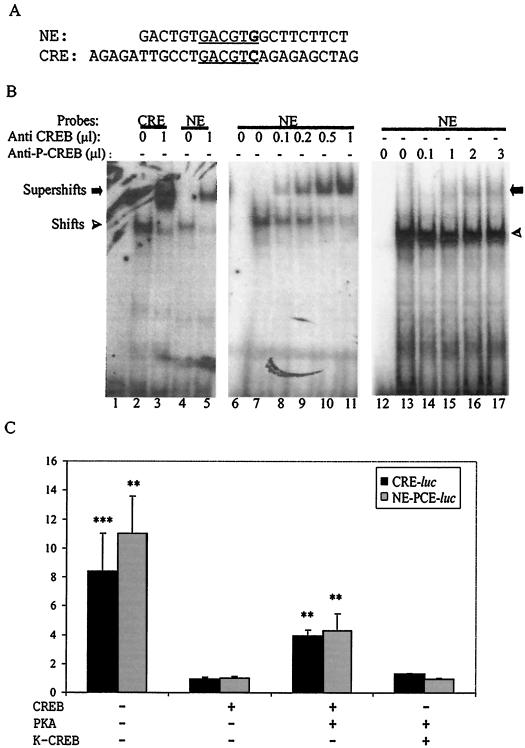
The NE sequence is also very similar to a CRE (Fig. 5A). It is known that CREB homodimers bind CREs and that after phosphorylation, activated P-CREB can induce transcription of downstream genes (57). We tested whether the complex bound to the NE contained the CREB protein. EMSAs with retinal nuclear extract and a control consensus CRE probe resulted in a shift similar to that seen with the NE probe (compare lanes 2 and 4, Fig. 5B). Two antibodies, anti-CREB and anti-P-CREB, were used to examine whether CREB is a component of the NE-binding protein complex. Coincubation of the anti-CREB antibody with retinal nuclear extracts before the addition of the probes resulted in the formation of similar, more slowly migrating bands with both the NE and CRE probes (Fig. 5B, lanes 3 and 5). The intensity of the NE and CRE supershift bands increased with increasing amounts of the anti-CREB antibody (lanes 6 to 11). These data indicate that the NE-bound complex contains CREB. Coincubation of the anti-P-CREB antibody also resulted in supershifting the band of the NE-binding protein complex, indicating that the active form of CREB can also bind to this element (lanes 12 to 17). The supershifted bands with anti-P-CREB antibodies are much weaker than those produced by the anti-CREB antibodies. This is probably due to the low concentration of P-CREB, since normally only a small portion of the CREB in cell nuclei is in the phosphorylated form (57).
FIG. 5.
CREB binds to NE and activates transcription of a downstream reporter gene. (A) Sequence alignment of the CRE and NE. (B) CREB recognizes the NE in EMSAs. EMSAs to detect supershifts were performed in the presence of anti-CREB and anti-P-CREB antibodies. The identities of the probes and the amounts of antibody and incubation periods are labeled at the tops of the lanes. (C) P-CREB transactivated both CRE-luc and NE-PCE-luc reporter genes in transiently transfected NIH 3T3 cells. CRE-luc and NE-PCE-luc reporter genes were cotransfected with CREB, CREB plus PKA, or K-CREB plus PKA into 3T3 cells. Sample values are means ± standard errors of results for eight samples. All results were normalized to the relative luciferase activities in cells transfected with the CRE-luc reporter gene plus CREB, which was assigned a value of 1.0. ***, P < 0.001; **, P < 0.01 (the value was significantly different from that for the respective control [CREB only]).
Transient-transfection assays of NIH 3T3 cells were performed to examine whether P-CREB would activate the transcription of the reporter gene through the CRE or NE enhancer elements. The transfection procedure alone results in some activation of the CRE- and NE-PCE-luc reporters, most likely due to the transient endogenous CREB phosphorylation that results from the medium change during the transfection procedure (Fig. 5C). When an expression plasmid containing CREB under the control of a strong CMV promoter was cotransfected into the cells, strong repression of the transactivation by the endogenous signals was observed, since the overexpressed CREB should be largely in the nonphosphorylated form. When plasmids expressing a constitutively active form of PKA and CREB were cotransfected together, both the CRE-luc and NE-PCE-luc reporter genes were induced four- to fivefold. This transactivation is prevented when a mutant form of CREB (K-CREB), which blocks CREB-induced transcription (65), was coexpressed with PKA into the 3T3 cells (Fig. 5C). Taken together with the results of the binding studies, these data indicate that the NE is acting like a CRE, in that it binds CREB and minimal promoters containing this element are transcriptionally activated by P-CREB.
P-CREB exhibits a circadian pattern in retinal photoreceptor cells.
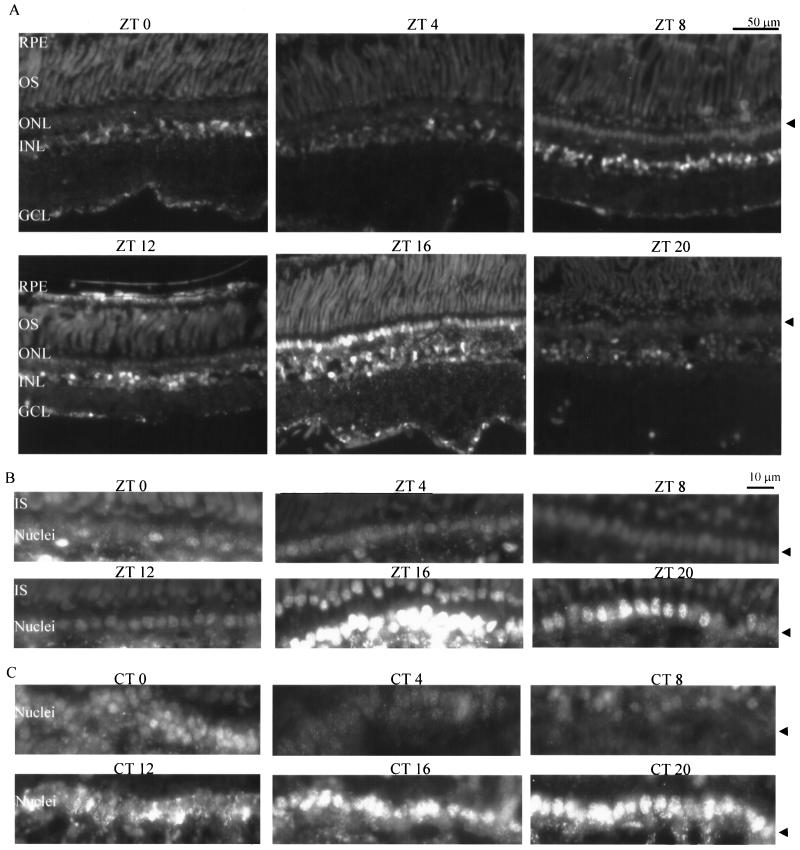
If CREB plays a role in driving the rhythmic transcription of nocturnin, one of the possible mechanisms by which this is achieved is that CREB itself is phosphorylated rhythmically. Therefore, we examined the temporal and spatial pattern of P-CREB in the Xenopus retina. Eyecups were collected every 4 h over a 24-h cycle under both LD and DD conditions. Sections of retinas were labeled immunohistochemically to detect the presence of P-CREB. The results demonstrated that P-CREB could be seen in the photoreceptor cells, inner nuclear layer, and ganglion cell layer (Fig. 6A). Comparison of the time points showed that a robust rhythm of P-CREB was present in the photoreceptor cells under both LD (Fig. 6A and B) and DD (Fig. 6C) conditions. Maximal expression of P-CREB in the nuclei of photoreceptor cells was observed at around ZT 16 under LD conditions, and its expression levels dropped markedly during the day (Fig. 6A and B). Under DD conditions, some signal was seen in the perinuclear regions of the photoreceptor cells at CT 12 (CT 0 defines the beginning of subjective day), and high levels of P-CREB were observed in the nuclei of photoreceptor cells at both CTs 16 and 20 (Fig. 6C). Some variation of P-CREB staining was also detected in other retinal cell types, but no consistent rhythm was observed. Western blot experiments using whole retinal extracts did not demonstrate an obvious circadian rhythm, most likely due to the nonrhythmic levels of P-CREB in nonphotoreceptor cell types (data not shown).
FIG. 6.
Circadian oscillations of P-CREB in Xenopus retina. Retina sections were collected every 4 h during a 24-h LD (A and B) or DD (C) cycle. The retina sections were immunolabeled to detect the expression of P-CREB. (A) Fluorescent images of retina sections at different times of day during an LD cycle. The arrow points to the layer of photoreceptor cell nuclei (ONL). (B and C) Higher-magnification images of the photoreceptor nuclei at different times in the LD (B) and DD (C) cycles show that P-CREB levels fluctuate in a circadian rhythm. CT 0 defines the beginning of subjective day, and CT 12 defines the beginning of subjective night. Five sections were analyzed for each time point. Sections stained with secondary antibody alone (without primary antibody) showed no signal (not shown).
DISCUSSION
Our results suggest that circadian expression of nocturnin is driven by rhythms in levels of P-CREB, which activates transcription through binding to the NE. Our data demonstrate that CREB isolated from retinal nuclear extracts binds the NE sequence within the proximal nocturnin promoter in vitro and that P-CREB can activate transcription of a reporter gene from a minimal promoter containing the NE in transiently transfected cells. Within the retina, we show that P-CREB levels cycle as a function of CT in retinal photoreceptor cells, and the phase of this rhythm correlates well with the rhythmic transcription pattern of the nocturnin gene.
This nighttime phase of peak P-CREB levels suggests that the retinal clock can use this transcriptional activator as a mechanism to drive rhythmic gene expression that is out of phase with the rhythm in CLOCK/BMAL1 activity. This phase consideration is significant, because CLOCK/BMAL1 dimers, acting through E-boxes, have been implicated in the activation of many rhythmic genes, both within the central clock mechanism (per and cry genes) and downstream of the clock. The strongly rhythmic activity of CLOCK/BMAL1 is a critical feature of the central clock mechanism, and therefore, downstream genes directly regulated by this dimer would be predicted to have phases similar to that of the per and cry genes. This is the case for two of the direct targets of the CLOCK/BMAL1 heterodimer, the gene encoding neuropeptide arginine vasopressin (33) and the gene encoding the transcription factor DBP (54); in the mouse SCN, both genes are transcribed rhythmically, with phases similar to those of the per and cry genes (peaking in early day).
In Xenopus retina, the per 1 gene is expressed such that peak mRNA levels occur during the first few hours of the day (73), suggesting that the transactivation activity of CLOCK/BMAL1 is highest during late night or early day. This is inconsistent with a role for this heterodimer in directly activating transcription of genes like nocturnin that peak during the night, and indeed, our data demonstrate that the CLOCK/BMAL1 dimer does not bind or transactivate the nocturnin promoter through the E-box-like NE. In addition to nocturnin, there may well be other night-phase genes that are controlled by the clock via the rhythms of P-CREB in the photoreceptors.
However, a clear-cut role for CLOCK/BMAL1 in dictating a per-like phase of transcriptional activation to E-box-containing genes is not obvious in all cases. Examination of the promoters of several other rhythmic genes with variable phases has revealed the presence of E-box-like elements. In most cases, these E-box sequences have been shown to be capable of activating downstream reporter genes in heterologous systems when CLOCK and BMAL1 are present, but in most cases, the relevance of this has not been tested in vivo. In some cases, these genes are expressed with phases very different phases from those of per and cry genes, and it is not clear how a direct role for CLOCK/BMAL1 can be simply reconciled. For example, the aa-nat gene in the chicken pineal is expressed with a peak in mid-night (ZT 18) (10) while chicken per 2 has a peak in mid-day in this tissue (14). Yet this gene contains an E-box that can be regulated by CLOCK/BMAL1 in transiently transfected cells (10). One possible explanation is that in this case and others like it, the E-box may not be regulated by CLOCK/BMAL1 but by other closely related heterodimers (for example, MOP4/BMAL1 [10]) that have distinct temporal patterns of activity.
In some other examples, genes that are in phase with per mRNA rhythms have been shown to be regulated by a non-CLOCK/BMAL1 mechanism. In the rat, the aa-nat gene undergoes a high-amplitude peak of expression during the night in both the pineal and retina. The mammalian retina contains an endogenous clock (62), but the mammalian pineal rhythms are driven by rhythmic norepinephrine input from the circadian clock in the SCN (35). In both these tissues, the per mRNA rhythms in rats have peaks with phases similar to that of the peak of aa-nat message (61), and the rat aa-nat gene contains a perfect consensus E-box in the first intron (9). However, a portion of intronic sequence containing this E-box was capable of activating transcription of a downstream reporter gene in a CLOCK/BMAL1-dependent manner when it was transfected into retinal cells but not in pineal cells, although the rat pineal is known to express CLOCK and BMAL1 (46). Data from transgenic rats indicate that within the pineal, the aa-nat mRNA rhythms can be driven by P-CREB (resulting from increases in cAMP following norepinephrine stimulation) through two equally important CREs (9). Thus, it appears that these two tissues use distinct transcriptional mechanisms and distinct enhancer elements to generate the rhythms of aa-nat message, even though in both cases, CLOCK/BMAL1 activity appears to be present. Interestingly, in the rat pineal the per 1 gene (but not the per 2 gene) also seems to be primarily regulated by CREB rather than by CLOCK/BMAL1 (61, 64). Therefore, the use of P-CREB to drive these rhythms may be a reflection of the fact that this tissue does not contain a robust endogenous clock.
Other evidence for CRE involvement in rhythms.
In addition to the data obtained with the rat aa-nat gene, there are several other lines of evidence that implicate a role for CREB, via the CRE, in controlling circadian-rhythm-relevant gene transcription. Within vertebrates, CREB phosphorylation has been studied largely in the context of acute induction of genes by light and other phase-shifting stimuli, and evidence suggests that it may be involved in the entrainment mechanism. Exposure of hamsters to light during the night results in rapid phosphorylation of CREB at Ser 133 and Ser 142 within the SCN (17, 19, 50). This phosphorylation response to light is gated by the circadian clock and happens only at times within the circadian cycle when light causes phase shifts in locomotor activity. Likewise, light pulses during the subjective night, but not during the subjective day, triggered a marked increase in CRE-mediated reporter gene expression in transgenic mouse SCN (49).
Among P-CREB's potential clock-related targets are the per genes. Light pulses or other resetting stimuli result in acute induction of per 1 and per 2 in mammalian systems, and this appears to be mediated by P-CREB, since CREB phosphorylation temporally precedes or coincides with the acute per inductions (2, 44, 58, 64, 68, 69, 74). Furthermore, mice carrying a mutant form of CREB that cannot be phosphorylated at Ser 142 showed severely diminished induction of per after a light pulse and their ability to entrain to light is compromised (17). The presence of CRE-like sequences within the human and mouse per 1 gene-flanking sequences indicates that P-CREB's role in the acute induction may be direct (32).
Our data suggest that the daily rhythm of nocturnin transcription is regulated by P-CREB within the clock-containing photoreceptor cells. Although there are suggestions from other systems that CREB phosphorylation may contribute to rhythmic gene expression (in addition to its better-known acute effects), very little is known about the mechanism or the targeted genes, particularly in cells that contain endogenous clocks. In Drosophila melanogaster, a promoter containing a CRE has been shown to drive cycling of a reporter gene, and mutation in the CRE element significantly reduced the amplitude of the rhythm (5). Likewise, in transgenic mice, a promoter containing a CRE drives transcriptional cycling of a reporter gene in the mouse SCN (but not other brain regions) both in cyclic light and under constant conditions, with peak expression occurring between late subjective night and mid subjective day (49). Although the identities of the target genes are not known, these data suggest that rhythmic P-CREB may be used by the clock to drive the rhythmic transcription of genes within clock-containing tissues like the SCN and Xenopus photoreceptors.
How does the circadian clock generate CREB phosphorylation rhythms?
Phosphorylation of CREB can result from activation of many different signaling pathways (reviewed in reference 66), and the number of steps between the Xenopus photoreceptor circadian clock mechanism and rhythmic CREB phosphorylation is not known. Activation of PKA via increases in intracellular cAMP levels is one of the well-studied signaling pathways that result in phosphorylation of CREB. There are several examples of tissues that exhibit circadian rhythms of cAMP levels, including the previously mentioned mammalian pineal gland, in which daily changes in cAMP levels result from the rhythmic norepinephrine input from the SCN. As mentioned above, these cAMP oscillations produce the rhythmic phosphorylation of CREB that is important for stimulating transcription of the aa-nat and per 1 genes (64). Among tissues that contain robust endogenous circadian clocks, circadian rhythms of cAMP have also been demonstrated in chicken pineal (47, 48) and chicken retina (P. M. Iuvone, personal communication). In hamster retina, cAMP levels are also rhythmic, but its levels were examined only under cyclic light conditions (16). However, within the Xenopus retina, although changing levels of cAMP are clearly involved in the phase-shifting effects of dopamine, it is not clear whether cAMP levels fluctuate with a circadian rhythm (28-30). It is known that dopamine from inner retinal neurons acts through D2-like dopamine receptors on photoreceptors (8, 45) to reset the clock. Since D2-like dopamine receptors cause decreases in cAMP levels, normal rhythms of dopamine may contribute to a cAMP rhythm in photoreceptors.
Alternatively, CREB phosphorylation can also be mediated by other pathways, including calcium/calmodulin-dependent kinase IIα (CaMKIIα) and mitogen-activated protein (MAP) kinases (66). Although there is no data on the rhythmicity of these kinases in Xenopus retina, there is a recent report that the activities of Erk MAP kinase and CamKIIα are rhythmic in chick photoreceptor cells (36), the chick pineal (55), and the mouse SCN (50). In addition, Erk MAP kinase activity also exhibits clear circadian rhythms in bullfrog retina, but this rhythm is confined to a population of amacrine cells in the inner nuclear layer (26).
Evolutionary relationship of the CRE and E-box.
Our results indicate that xCLOCK/BMAL1 binding is very selective for the 6-bp consensus E-box sequence. The presence of a G in the first position of the NE, instead of the consensus C, drastically reduces the affinity of the CLOCK/BMAL1 dimer for the sequence. Generation of the single-base-pair change (replacing the G with the C) within the context of the NE flanking sequence restores CLOCK/BMAL1 binding to the element in EMSAs and also restores the ability of this element to mediate CLOCK/BMAL1 transcriptional activation. Thus, a single-base-pair change within this sequence effectively causes a switch between a functional E-box and a functional CRE. The close consensus sequence relationship of these two elements has sparked previous speculation that these elements may be evolutionarily related, particularly since they both have clock-related functions (38). Indeed, the NE within the nocturnin gene appears to be a sequence hybrid of these two elements, with only a single-base-pair change from either the CE or the consensus CRE.
This easy conversion from the CRE to E-box also suggests that the sequences flanking the 6-bp E-box are not critical for normal CLOCK/BMAL1 function (since the CACGTG within the NE-flanking sequence functioned nearly as well as the real PE sequence, at least in the context of the biochemical and transient-transfection assays). This is consistent with the fact that flanking sequences do not seem to be well conserved among different E-box sequences from different species, from different per genes, or indeed even different E-boxes within a single per gene (reviewed in reference 38). Despite this lack of obvious conservation, the sequences surrounding the PE in Drosophila do seem to have some impact on proper temporal and spatial expression patterns, although the exact role of these sequences is unclear (12, 38, 40).
Conclusions.
Although the E-box and the CRE have emerged from many studies as key elements in circadian transcriptional regulation, it is clear that our current understanding of the circadian control of transcription is still quite superficial. Many circadian-rhythm-controlled genes have both of these elements, often in several copies, and very little is known about whether these elements interact with one another. Nothing is known about what makes a simple 6-bp sequence (which should occur by chance in the genome approximately every 4,000 bases) into a real E-box that confers circadian cycling of transcription. Finally, it seems obvious that despite the nearly ubiquitous presence of E-boxes and CREs in the rhythmic genes that have been examined to date, there must be other means of driving rhythmic gene transcription. Since some of the known rhythmic genes themselves encode transcription factors, one can imagine that further cascades of gene expression may ultimately produce a wide range of subtle control, including many different phases, amplitudes, and waveforms.
Acknowledgments
We thank Richard Day, Margarida Barroso, and Julie Baggs for technical advice and Gregory Cahill for providing the Xenopus CLOCK antibody. We also thank Jianhua Cang, Haisun Zhu, and other lab members for many helpful comments on the manuscript.
This work was funded by National Eye Institute grant EY11489 (C.B.G.).
REFERENCES
- 1.Akhtar, R. A., A. B. Reddy, E. S. Maywood, J. D. Clayton, V. M. King, A. G. Smith, T. W. Gant, M. H. Hastings, and C. P. Kyriacou. 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12:540-550. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, U., Z. Sun, G. Eichele, and C. Lee. 1997. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91:1055-1064. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, F. E., and C. B. Green. 2000. Symphony of rhythms in the Xenopus laevis retina. Microsc. Res. Tech. 50: 360-372. [DOI] [PubMed] [Google Scholar]
- 4.Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26, 28. [DOI] [PubMed] [Google Scholar]
- 5.Belvin, M. P., H. Zhou, and J. C. Yin. 1999. The Drosophila dCREB2 gene affects the circadian clock. Neuron 22:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besharse, J. C., and P. M. Iuvone. 1983. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature 305:133-135. [DOI] [PubMed] [Google Scholar]
- 7.Cahill, G. M., and J. C. Besharse. 1993. Circadian clock functions localized in Xenopus retinal photoreceptors. Neuron 10:573-577. [DOI] [PubMed] [Google Scholar]
- 8.Cahill, G. M., and J. C. Besharse. 1991. Resetting the circadian clock in cultured Xenopus eyecups: regulation of retinal melatonin rhythms by light and D2 dopamine receptors. J. Neurosci. 11:2959-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W., and R. Baler. 2000. The rat arylalkylamine N-acetyltransferase E-box: differential use in a master vs. a slave oscillator. Brain Res. Mol. Brain Res. 81:43-50. [DOI] [PubMed] [Google Scholar]
- 10.Chong, N. W., M. Bernard, and D. C. Klein. 2000. Characterization of the chicken serotonin N-acetyltransferase gene. Activation via clock gene heterodimer/E box interaction. J. Biol. Chem. 275:32991-32998. [DOI] [PubMed] [Google Scholar]
- 11.Claridge-Chang, A., H. Wijnen, F. Naef, C. Boothroyd, N. Rajewsky, and M. W. Young. 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32:657-671. [DOI] [PubMed] [Google Scholar]
- 12.Darlington, T. K., L. C. Lyons, P. E. Hardin, and S. A. Kay. 2000. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J. Biol. Rhythms 15:462-471. [DOI] [PubMed] [Google Scholar]
- 13.Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. L. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603. [DOI] [PubMed] [Google Scholar]
- 14.Doi, M., Y. Nakajima, T. Okano, and Y. Fukada. 2001. Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Proc. Natl. Acad. Sci. USA 98:8089-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffield, G. E., J. D. Best, B. H. Meurers, A. Bittner, J. J. Loros, and J. C. Dunlap. 2002. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 12:551-557. [DOI] [PubMed] [Google Scholar]
- 16.Faillace, M. P., M. I. Sarmiento, L. N. Siri, and R. E. Rosenstein. 1994. Diurnal variations in cyclic AMP and melatonin content of golden hamster retina. J. Neurochem. 62:1995-2000. [DOI] [PubMed] [Google Scholar]
- 17.Gau, D., T. Lemberger, C. von Gall, O. Kretz, N. Le Minh, P. Gass, W. Schmid, U. Schibler, H. W. Korf, and G. Schutz. 2002. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 34:245-253. [DOI] [PubMed] [Google Scholar]
- 18.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564-1569. [DOI] [PubMed] [Google Scholar]
- 19.Ginty, D. D., J. M. Kornhauser, M. A. Thompson, H. Bading, K. E. Mayo, J. S. Takahashi, and M. E. Greenberg. 1993. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260:238-241. [DOI] [PubMed] [Google Scholar]
- 20.Green, C. B., and J. C. Besharse. 1996. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA 93:14884-14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, C. B., and J. C. Besharse. 1994. Tryptophan hydroxylase expression is regulated by a circadian clock in Xenopus laevis retina. J. Neurochem. 62:2420-2428. [DOI] [PubMed] [Google Scholar]
- 22.Green, C. B., and J. C. Besharse. 1996. Use of a high stringency differential display screen for identification of retinal mRNAs that are regulated by a circadian clock. Brain Res. Mol. Brain Res. 37:157-165. [DOI] [PubMed] [Google Scholar]
- 23.Green, C. B., M.-Y. Liang, B. M. Steenhard, and J. C. Besharse. 1999. Ontogeny of circadian and light regulation of melatonin release in Xenopus laevis embryos. Brain Res. Dev. Brain Res. 117: 109-116. [DOI] [PubMed] [Google Scholar]
- 24.Grundschober, C., F. Delaunay, A. Puhlhofer, G. Triqueneaux, V. Laudet, T. Bartfai, and P. Nef. 2001. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J. Biol. Chem. 276:46751-46758. [DOI] [PubMed] [Google Scholar]
- 25.Hagiwara, M. 1995. Regulation of gene expression through phosphorylation of transcription factors. Seikagaku 67:1388-1391. (In Japanese.) [PubMed] [Google Scholar]
- 26.Harada, Y., K. Sanada, and Y. Fukada. 2000. Circadian activation of bullfrog retinal mitogen-activated protein kinase associates with oscillator function. J. Biol. Chem. 275:37078-37085. [DOI] [PubMed] [Google Scholar]
- 27.Harmer, S. L., J. B. Hogenesch, M. Straume, H. S. Chang, B. Han, T. Zhu, X. Wang, J. A. Kreps, and S. A. Kay. 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110-2113. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa, M., and G. M. Cahill. 1998. Cyclic AMP resets the circadian clock in cultured Xenopus retinal photoreceptor layers. J. Neurochem. 70:1523-1531. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa, M., and G. M. Cahill. 1999. Modulation of rhythmic melatonin synthesis in Xenopus retinal photoreceptors by cyclic AMP. Brain Res. 824:161-167. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa, M., and G. M. Cahill. 1999. A role for cyclic AMP in entrainment of the circadian oscillator in Xenopus retinal photoreceptors by dopamine but not by light. J. Neurochem. 72:1812-1820. [DOI] [PubMed] [Google Scholar]
- 31.Hayasaka, N., S. I. LaRue, and C. B. Green. 2002. In vivo disruption of Xenopus CLOCK in the retinal photoreceptor cells abolishes circadian melatonin rhythmicity without affecting its production levels. J. Neurosci. 22:1600-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hida, A., N. Koike, M. Hirose, M. Hattori, Y. Sakaki, and H. Tei. 2000. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics 65:224-233. [DOI] [PubMed] [Google Scholar]
- 33.Jin, X., L. P. Shearman, D. R. Weaver, M. J. Zylka, G. J. de Vries, and S. M. Reppert. 1999. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96:57-68. [DOI] [PubMed] [Google Scholar]
- 34.Kita, Y., M. Shiozawa, W. Jin, R. R. Majewski, J. C. Besharse, A. S. Greene, and H. J. Jacob. 2002. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics 12:55-65. [DOI] [PubMed] [Google Scholar]
- 35.Klein, D. C., and R. Y. Moore. 1979. Pineal N-acetyltransferase and hydroxyindole-O-methyltransferase: control by the retinohypothalamic tract and the suprachiasmatic nucleus. Brain Res. 174:245-262. [DOI] [PubMed] [Google Scholar]
- 36.Ko, G. Y., M. L. Ko, and S. E. Dryer. 2001. Circadian regulation of cGMP-gated cationic channels of chick retinal cones. Erk MAP kinase and Ca2+/calmodulin-dependent protein kinase II. Neuron 29:255-266. [DOI] [PubMed] [Google Scholar]
- 37.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 38.Kyriacou, C. P., and E. Rosato. 2000. Squaring up the E-box. J. Biol. Rhythms 15:483-490. [DOI] [PubMed] [Google Scholar]
- 39.Liu, X., and C. B. Green. 2001. A novel promoter element, photoreceptor conserved element II, directs photoreceptor-specific expression of nocturnin in Xenopus laevis. J. Biol. Chem. 276:15146-15154. [DOI] [PubMed] [Google Scholar]
- 40.Lyons, L. C., T. K. Darlington, H. Hao, J. Houl, S. A. Kay, and P. E. Hardin. 2000. Specific sequences outside the E-box are required for proper per expression and behavioral rescue. J. Biol. Rhythms 15:472-482. [DOI] [PubMed] [Google Scholar]
- 41.Manglapus, M., M. Parshley, K. Stewart, G. Engbretson, B. Knox, and R. Barlow. 1999. Circadian changes of ERG responses in the Xenopus laevis. Investig. Ophthalmol. Vis. Sci. 40:S610. [Google Scholar]
- 42.McDonald, M. J., and M. Rosbash. 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107:567-578. [DOI] [PubMed] [Google Scholar]
- 43.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 44.Motzkus, D., E. Maronde, U. Grunenberg, C. C. Lee, W. Forssmann, and U. Albrecht. 2000. The human PER1 gene is transcriptionally regulated by multiple signaling pathways. FEBS Lett. 486:315-319. [DOI] [PubMed] [Google Scholar]
- 45.Muresan, Z., and J. C. Besharse. 1993. D2-like dopamine receptors in amphibian retina: localization with fluorescent ligands. J. Comp. Neurol. 331:149-160. [DOI] [PubMed] [Google Scholar]
- 46.Namihira, M., S. Honma, H. Abe, Y. Tanahashi, M. Ikeda, and K. Honma. 1999. Daily variation and light responsiveness of mammalian clock gene, Clock and BMAL1, transcripts in the pineal body and different areas of brain in rats. Neurosci. Lett. 267:69-72. [DOI] [PubMed] [Google Scholar]
- 47.Nikaido, S. S., and J. S. Takahashi. 1998. Day/night differences in the stimulation of adenylate cyclase activity by calcium/calmodulin in chick pineal cell cultures: evidence for circadian regulation of cyclic AMP. J. Biol. Rhythms 13:479-493. [DOI] [PubMed] [Google Scholar]
- 48.Nikaido, S. S., and J. S. Takahashi. 1989. Twenty-four hour oscillation of cAMP in chick pineal cells: role of cAMP in the acute and circadian regulation of melatonin production. Neuron 3:609-619. [DOI] [PubMed] [Google Scholar]
- 49.Obrietan, K., S. Impey, D. Smith, J. Athos, and D. R. Storm. 1999. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J. Biol. Chem. 274:17748-17756. [DOI] [PubMed] [Google Scholar]
- 50.Obrietan, K., S. Impey, and D. R. Storm. 1998. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat. Neurosci. 1:693-700. [DOI] [PubMed] [Google Scholar]
- 51.Panda, S., M. P. Antoch, B. H. Miller, A. I. Su, A. B. Schook, M. Straume, P. G. Schultz, S. A. Kay, J. S. Takahashi, and J. B. Hogenesch. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307-320. [DOI] [PubMed] [Google Scholar]
- 52.Pierce, M. E., and J. C. Besharse. 1985. Circadian regulation of retinomotor movements. I. Interaction of melatonin and dopamine in the control of cone length. J. Gen. Physiol. 86:671-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63:647-676. [DOI] [PubMed] [Google Scholar]
- 54.Ripperger, J. A., L. P. Shearman, S. M. Reppert, and U. Schibler. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14:679-689. [PMC free article] [PubMed] [Google Scholar]
- 55.Sanada, K., Y. Hayashi, Y. Harada, T. Okano, and Y. Fukada. 2000. Role of circadian activation of mitogen-activated protein kinase in chick pineal clock oscillation. J. Neurosci. 20:986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaffer, R., J. Landgraf, M. Accerbi, V. V. Simon, M. Larson, and E. Wisman. 2001. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 58.Shearman, L., M. Zylka, D. Weaver, L. Kolakowski, and S. Reppert. 1997. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nucleus. Neuron 19:1261-1269. [DOI] [PubMed] [Google Scholar]
- 59.Storch, K., O. Lipan, I. Leykin, N. Viswanathan, F. Davis, W. Wong, and C. Weitz. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417:78-83. [DOI] [PubMed] [Google Scholar]
- 60.Swanson, H. I., W. K. Chan, and C. A. Bradfield. 1995. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 270:26292-26302. [DOI] [PubMed] [Google Scholar]
- 61.Takekida, S., L. Yan, E. S. Maywood, M. H. Hastings, and H. Okamura. 2000. Differential adrenergic regulation of the circadian expression of the clock genes Period1 and Period2 in the rat pineal gland. Eur. J. Neurosci. 12:4557-4561. [DOI] [PubMed] [Google Scholar]
- 62.Tosini, G., and M. Menaker. 1996. Circadian rhythms in cultured mammalian retina. Science 272:419-421. [DOI] [PubMed] [Google Scholar]
- 63.Ueda, H. R., A. Matsumoto, M. Kawamura, M. Iino, T. Tanimura, and S. Hashimoto. 2002. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277:14048-14052. [DOI] [PubMed] [Google Scholar]
- 64.von Gall, C., I. Schneider-Huther, M. Pfeffer, F. Dehghani, H. W. Korf, and J. H. Stehle. 2001. Clock gene protein mPER1 is rhythmically synthesized and under cAMP control in the mouse pineal organ. J. Neuroendocrinol. 13:313-316. [DOI] [PubMed] [Google Scholar]
- 65.Walton, K. M., R. P. Rehfuss, J. C. Chrivia, J. E. Lochner, and R. H. Goodman. 1992. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)- regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol. Endocrinol. 6:647-655. [DOI] [PubMed] [Google Scholar]
- 66.West, A. E., W. G. Chen, M. B. Dalva, R. E. Dolmetsch, J. M. Kornhauser, A. J. Shaywitz, M. A. Takasu, X. Tao, and M. E. Greenberg. 2001. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 98:11024-11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, J. A., and A. Sehgal. 2001. Molecular components of the circadian system in Drosophila. Annu. Rev. Physiol. 63:729-755. [DOI] [PubMed] [Google Scholar]
- 68.Yagita, K., and H. Okamura. 2000. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 465:79-82. [DOI] [PubMed] [Google Scholar]
- 69.Yagita, K., F. Tamanini, G. T. van Der Horst, and H. Okamura. 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292:278-281. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, H., and C. B. Green. 2001. A putative flavin electron transport pathway is differentially utilized in Xenopus CRY1 and CRY2. Curr. Biol. 11:1945-1949. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, H., and C. B. Green. 2001. Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors. Mol. Vis. 7:210-215. [PubMed] [Google Scholar]
- 72.Zhu, H., S. LaRue, A. Whiteley, T. D. L. Steeves, J. S. Takahashi, and C. B. Green. 2000. The Xenopus Clock gene is constitutively expressed in retinal photoreceptors. Brain Res. Mol. Brain Res. 75:303-308. [DOI] [PubMed] [Google Scholar]
- 73.Zhuang, M., Y. Wang, B. M. Steenhard, and J. C. Besharse. 2000. Differential regulation of two period genes in the Xenopus eye. Brain Res. Mol. Brain Res. 82:52-64. [DOI] [PubMed] [Google Scholar]
- 74.Zylka, M. J., L. P. Shearman, D. R. Weaver, and S. M. Reppert. 1998. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1-20. [DOI] [PubMed] [Google Scholar]