Abstract
Centromeres require specialized nucleosomes; however, the mechanism of localization is unknown. Drosophila sp. centromeric nucleosomes contain the Cid H3-like protein. We have devised a strategy for identifying elements within Cid responsible for its localization to centromeres. By expressing Cid from divergent Drosophila species fused to green fluorescent protein in Drosophila melanogaster cells, we found that D. bipectinata Cid fails to localize to centromeres. Cid chimeras consisting of the D. bipectinata histone fold domain (HFD) replaced with segments from D. melanogaster identified loop I of the HFD as being critical for targeting to centromeres. Conversely, substitution of D. bipectinata loop I into D. melanogaster abolished centromeric targeting. In either case, loop I was the only segment capable of conferring targeting. Within loop I, we identified residues that are critical for targeting. Most mutations of conserved residues abolished targeting, and length reductions were deleterious. Taken together with the fact that H3 loop I makes numerous contacts with DNA and with the adaptive evolution of Cid, our results point to the importance of DNA specificity for targeting. We suggest that the process of deposition of (Cid.H4)2 tetramers allows for discriminating contacts to be made between loop I and DNA, providing the specificity needed for targeting.
The centromere is the locus responsible for poleward movement of the chromosomes during cell division. All eukaryotic centromeres are characterized by specialized nucleosomes containing an H3-like histone (CenH3) in place of canonical H3 (6, 36). In the absence of CenH3, centromere function and localization of other kinetochore components are abolished (2, 5, 14, 27, 35). Overexpression of human CenH3 (CENP-A) leads to its mislocalization to chromosome arms where it recruits kinetochore components (42). Therefore, CenH3-containing nucleosomes form the basis for organizing the kinetochore.
Canonical nucleosomes consist of 146 bp of DNA wrapped around an octamer of two each of the core histones H2A, H2B, H3, and H4 and are the fundamental packaging unit of chromatin (45). The first step in chromatin assembly is deposition of an (H3.H4)2 tetramer, which by itself organizes 120 bp of DNA (8, 11, 24). Subsequent deposition of two H2A.H2B dimers completes a nucleosome core particle (45). CenH3 replaces canonical H3 in nucleosomes both in vivo and in vitro (1, 2, 5, 18, 25, 29, 33, 34, 35, 47). CenH3 and H3 do not heterodimerize in the same nucleosome (1, 3, 33) and are therefore probably found as separate tetramers with histone H4 before incorporation into chromatin. Maintenance of CenH3 chromatin depends upon targeting of (CenH3.H4)2 tetramers to centromeres, a process that is poorly understood.
CenH3s differ from canonical H3 in that they have diverged histone fold domains (HFDs), noncanonical N-terminal tails, and slightly longer loop I (LI) regions (12, 36). Differences between CenH3s and canonical H3 appear to specify centromeric targeting of CenH3s. Deletion analysis showed that the HFD and not the N-terminal tail of CENP-A targeted centromeres in a human cell line (33, 37). However, no single targeting element could be mapped by swapping amino acids between CENP-A and canonical H3 (33, 37). For Saccharomyces cerevisiae CenH3 (Cse4), targeting was not specifically assayed, although residues throughout the HFD were similarly required for centromere function (9, 16). Since these regions were predicted to contact the DNA, it was proposed that DNA specificity was important for centromeric targeting of CenH3 (9, 16, 33, 37). However, this proposal appears to be contradicted by the lack of DNA sequence conservation at centromeres (6, 12, 36) and by the existence of human neocentromeres, known to be devoid of the α-satellite DNA found at canonical human centromeres (7, 18, 19, 30, 43).
Recent evolutionary studies of Drosophila and Arabidopsis CenH3s support a role for DNA specificity in localizing CenH3-containing tetramers (12, 21, 39). In Drosophila, both the N-terminal tail and LI of the Cid HFD were found to be evolving adaptively (21). LI is known to form a DNA-binding domain together with LII of H4 (20), suggesting that LI mediates DNA specificity in Cid-containing chromatin, presumably at the level of targeting of (Cid.H4)2 tetramers. Given the lack of mechanistic insight into the process of CenH3 targeting to centromeres, we have investigated the targeting determinants of Cid.
We reasoned that amino acid swaps between H3 and CenH3 did not reveal a targeting element because, although they are expected to have similar structures (20, 29, 47), they perform different functions and are therefore evolving under different selective constraints. H3-containing nucleosomes mediate essential processes that impact chromatin, such as transcription and gene silencing (15), whereas CenH3-containing nucleosomes are required for centromere function and maintenance. In addition, (H3.H4)2 tetramers are assembled onto the bulk of genomic DNA, whereas (CenH3.H4)2 tetramers only need to be incorporated into a small subset of the genomic DNA sequences found at their respective centromeres. These differences in DNA-binding requirements may be driving the rapid, adaptive evolution of CenH3s (12, 21, 39). In contrast, H3 is highly conserved. Furthermore, assembly of canonical H3 and CenH3s use different processes, with most H3 being deposited in a replication-coupled manner and CenH3 deposition occurring in a replication-independent manner throughout the cell cycle (1, 32). The interpretation of swap experiments between canonical H3 and CenH3 is further complicated by the possible dimerization of chimeric proteins with endogenous partners.
Here we examine CenH3 targeting by making swaps within a single lineage, Drosophila. Given the rapid evolution of Cid (12, 21) we predict that Cid from a diverged Drosophila species will not have the ability to target D. melanogaster centromeres. Such a diverged Cid would be a suitable recipient in swap experiments aimed at mapping targeting determinants. By using this approach, we have delineated a single targeting element, and it corresponds to LI. We show that LI is both necessary and sufficient for centromeric targeting and identify single amino acids within LI that are essential for targeting. (Cid.H4)2 tetramers may be targeted to centromeres by a LI mediated discrimination step during their assembly.
MATERIALS AND METHODS
Constructs.
All coding regions were amplified by PCR and cloned into two separate plasmid vectors for expression from either an hsp70 heat shock (HS) or Cid promoter as C-terminal green fluorescent protein (GFP) fusion proteins with a 6-amino-acid linker (QRPVAT) between the last amino acid and GFP. Proteins expressed from the vector with a Cid promoter (13) also contained an N-terminal myc tag (MEQKLISEEDLSR) flanked by a BglII restriction enzyme site encoding amino acids RS. The vector with an HS promoter (13) was modified to contain bases TAGAACA immediately before the ATG start site. This allowed higher expression levels from the HS promoter than had been observed before (1, 13). PCR primers contained XbaI/BglII sites (5′; HS and Cid vectors, respectively), NotI sites (3′), and coding sequence, starting from the first codon after the ATG for full-length Cid proteins (22). To delete the N-terminal tail, restriction enzyme sites were 5′ of R119 of D. melanogaster Cid (CidMel) or the equivalent amino acid of other H3-like proteins. Swap proteins and mutants were constructed by two consecutive PCR steps. First, two products were produced, by using either the 5′ or 3′ primer for the entire coding region and either a downstream or an upstream primer spanning the region of the swap junction or mutation. These two PCR products were combined and used as a template for the second PCR step with 5′ and 3′ primers for the entire coding region followed by cloning using restriction sites outlined above. All constructs were verified by sequencing.
Cell culture and immunostaining.
D. melanogaster Kc167 cells were cultured and transfected with 10 μg of DNA as described previously (13). For inducible constructs, HS was for 1 h at 37°C, 16 to 24 h after transfection. Recovery was for 2 to 6 h as indicated. For the Cid promoter, cells were processed 24 h after transfection. Immunostaining was carried out as described with an anti-Cid antibody and Texas red secondary antibody (13). The anti-Cid polyclonal antibody recognizes an 18-amino-acid peptide present in the N-terminal tail of D. melanogaster, D. simulans, and D. erecta Cid proteins (13). Images were analyzed using Delta Vision software (Applied Precision).
All transfections were done at least three times. Transfection efficiency varied from 50 to 90%. Data from transfections were only used if CidMel-GFP (full length or HFD) showed high transfection levels in a parallel experiment, thus controlling for transfection efficiency of different populations of Kc cells. Constructs that were scored as displaying no targeting had no detectable GFP at the centromere for at least 1,000 cells examined. For constructs scored as targeting centromeres, Cid-GFP expressed from either the Cid or HS promoters could typically be detected at the centromere in 250 to 800 cells out of every 1,000, with intensities up to 100-fold above background for the HS promoter. It should be noted that expression from the Cid promoter is cell cycle specific (13) and at low levels compared to the high expression level from the HS promoter. Although some proteins were targeted when expressed from the HS promoter but not from the Cid promoter, we never observed the converse, i.e., targeting when expressed from the Cid promoter but not from the HS promoter. The reproducible absence of green dots at the centromeres in all cells examined when a fusion protein was expressed from the Cid promoter is likely to reflect reduced targeting efficiency rather than variation in expression levels, because all open reading frames could be produced at high levels from the HS promoter irrespective of targeting, as evidenced by green fluorescence throughout the entire nucleus.
RESULTS
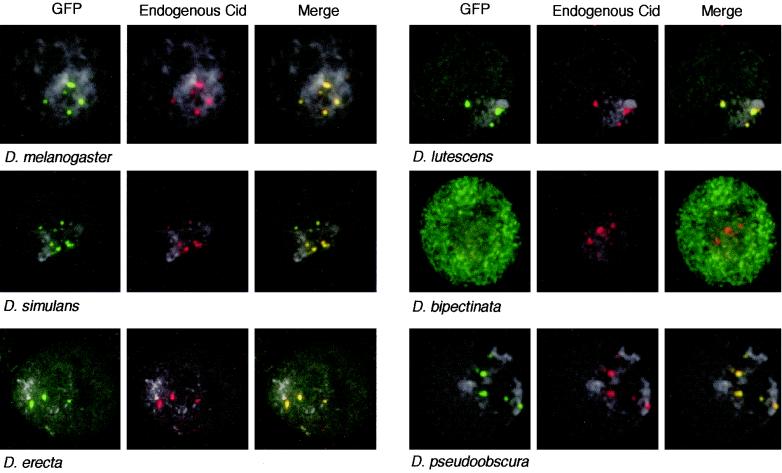
When CidMel is transiently expressed as a C-terminal GFP fusion protein in Drosophila cultured cells, it appears as green dots that overlap with endogenous Cid (immunostained in red) at centromeres (13) (Fig. 1, D. melanogaster). Given the rapid, adaptive evolution of Cid (21, 22), we expected that more diverged Cids may have lost the ability to target D. melanogaster centromeres. To find such a Drosophila Cid that does not target D. melanogaster centromeres, we expressed Cid-GFP from D. simulans (CidSim), D. erecta (CidEre), D. lutescens (CidLut), D. bipectinata (CidBip), and D. pseudoobscura (CidPse) in D. melanogaster cells. These species are 2.5 to 36 million years diverged from D. melanogaster, and their Cids range from 97 to 58% identical to CidMel at the amino acid level in the HFD (Table 1) (22). CidSim, CidEre, CidLut, and CidPse-GFP fusion proteins targeted D. melanogaster centromeres such that they could be visualized as green dots overlapping with endogenous CidMel (red dots) (Fig. 1). However, one of the more distantly related Cids, CidBip, had lost the ability to target D. melanogaster centromeres, even when it was present at high levels in the nucleus (Fig. 1, D. bipectinata).
FIG. 1.
Targeting of Cid-GFP from different Drosophila species to D. melanogaster centromeres. Cid-GFP proteins were induced from the HS promoter (4-h recovery). A typical transfected nucleus is shown for each construct. Cid-GFP proteins (panels at left; green) from D. melanogaster, D. simulans, D. erecta, D. lutescens, and D. pseudoobscura are present at D. melanogaster centromeres (middle panels; stained red by anti-Cid antibody). The merge of the Cid-GFP and endogenous Cid panels is shown in the panels at right (overlap in yellow). DNA is stained by DAPI (4′,6′-diamidino-2-phenylindole) and shown in light grey. CidBip-GFP is never present at D. melanogaster centromeres (red), even when high levels of GFP are found diffusely in the nucleus.
TABLE 1.
Conservation of amino acids between the HFD of H3 and different CenH3s
| HFD | % Amino acid identify with:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CidMel | CidSim | CidEre | CidLut | CidBip | CidPse | CENP-A | Cse4p | HCP3 | |
| CidMel | 100 | 97 | 87 | 86 | 59 | 58 | 46 | 38 | 35 |
| H3 | 42 | 40 | 40 | 38 | 39 | 41 | 63 | 67 | 58 |
We further analyzed the behavior of CidBip-GFP compared to CidMel-GFP in D. melanogaster cells. CidMel-GFP targeting of centromeres was observed in both interphase and mitotic cells after expression at low levels (Fig. 2A and C). At high expression levels, CidMel-GFP localized both to centromeres and euchromatin (Fig. 2B and D). In contrast, CidBip-GFP was never observed at D. melanogaster centromeres, but there were two kinds of interphase patterns. One is a euchromatic pattern (Fig. 2E), where no localization is seen at the heterochromatic chromocenter, which contains the centromeres (1). The other pattern shows CidBip-GFP distributed uniformly throughout the nucleus (Fig. 2F).
FIG. 2.
Expression patterns of full-length CidMel-GFP (A to D) and CidBip-GFP (E to H) proteins. Cid-GFP fusion proteins were transiently expressed from the HS promoter (panels at left). Both endogenous and expressed CidMel are stained red by anti-Cid antibody (center panels). DAPI is grey. Typical images for interphase and mitotic nuclei are shown (merged for GFP and antibody in the panels at right). For mitotic figures, cells were allowed to recover either 2 or 6 h after HS to allow visualization of Cid-GFP expressed during G2 or S phase of the cell cycle (1).
Time course analysis revealed that the two different patterns are found in populations of cells at different stages of the cell cycle. GFP fusion proteins were induced for 1 h by an HS pulse. Kc cells show a very regular cell cycle, including a G2 phase of ∼5 h (1). Therefore, mitotic chromosomes seen 2 h after HS must have been in G2 phase at the time of the induction, whereas mitotic chromosomes seen 6 h after HS must have been from cells that were induced during S phase. A pulse of CidMel-GFP targeted it to centromeres both in S-phase and G2 cells (Fig. 2C and D). CidBip-GFP was not incorporated into chromatin in G2 phase cells (Fig. 2G) but could be visualized in euchromatin where the protein had been induced during S phase (Fig. 2H). We conclude that CidBip-GFP is competent for assembly into D. melanogaster chromosomes in S phase, but not into centromeres. CidMel must therefore contain a centromeric targeting element that CidBip does not.
The HFD of CENP-A has been shown to target it to human centromeres (33, 37). We found that the HFD of CidMel similarly targets it to D. melanogaster centromeres (Fig. 3A). Just like the full-length CidBip, the HFD of CidBip did not localize to D. melanogaster centromeres (Fig. 3A). Furthermore, a chimera of the CidMel N-terminal tail and the CidBip HFD also did not localize to D. melanogaster centromeres, and conversely, a chimera of the CidBip N-terminal tail and CidMel HFD did localize (not shown). Therefore, targeting determinants map to the HFD. The HFD of CidBip has 59% amino acid identity to the CidMel HFD (Table 1) (22) with differences spread throughout the domain. To select candidate targeting determinants, we compared the HFDs of Cids that are localized to D. melanogaster centromeres (CidMel and CidLut) to the CidBip HFD that is not localized (Fig. 3B). We concentrated on amino acids that are unique to CidBip. Three regions of the CidMel HFD (N-terminal region [NR], LI, and C-terminal region [CR]) were chosen for swap experiments with the CidBip HFD. These regions encompass all but four of the amino acids that are unique to CidBip HFD compared to both CidMel and CidLut.
FIG. 3.
LI is necessary and sufficient for targeting of Cid to D. melanogaster centromeres. (A) CidMel HFD-GFP localizes to D. melanogaster centromeres, whereas CidBip HFD-GFP does not. Representative fields of transfected cells are shown. The secondary structure of the Cid HFD is represented by an orange (CidMel) or blue (CidBip) line, with α-helices shown as boxes. (B) Alignment of the HFD of Drosophila H3 with the HFD of CidMel (M) (orange), CidLut (L), and CidBip (B) (blue). Hyphens denote gaps in the alignment. The secondary structure of H3 HFD (20) is represented by a line above the sequence, with α-helices shown as boxes. Regions that were swapped between CidMel HFD and CidBip HFD are underlined in the CidBip HFD sequence and marked NR (NR of the HFD), LI, and CR (CR of the HFD). Note that the alignment cannot accurately identify the beginning and the end of LI in the Cid HFD (22). (C) Typical cells transfected with CidBip HFD-GFP with the NR (HFDBip NRMel), LI (HFDBip LIMel), or CR (HFDBip CRMel) segments of CidMel substituted for their native counterparts. The secondary structure for the HFD of each construct is shown schematically underneath the image with segments from D. bipectinata and D. melanogaster colored blue and orange, respectively. (D) Typical cells transfected with CidMel HFD-GFP with substitutions of the NR (HFDMel NRBip), LI (HFDMel LIBip) or CR (HFDMel CRBip) regions of CidBip. The secondary structure for the HFD of each construct is shown schematically as in panel C.
We substituted the NR, LI, or CR regions of CidBip with those of CidMel and assayed centromeric targeting in D. melanogaster cells. Strikingly, we found that a single segment of CidMel, LI, was sufficient to target Cid to D. melanogaster centromeres when introduced into CidBip (Fig. 3C). Neither the CidMel NR nor the CR targeted Cid to centromeres when introduced into CidBip HFD (Fig. 3C). Furthermore, replacing LI of CidMel with that of CidBip disrupted CidMel HFD centromeric targeting (Fig. 3D), whereas the NR or CR regions of CidBip HFD had no effect on centromeric targeting of CidMel HFD. These data show that LI is both necessary and sufficient for centromeric targeting of Cid.
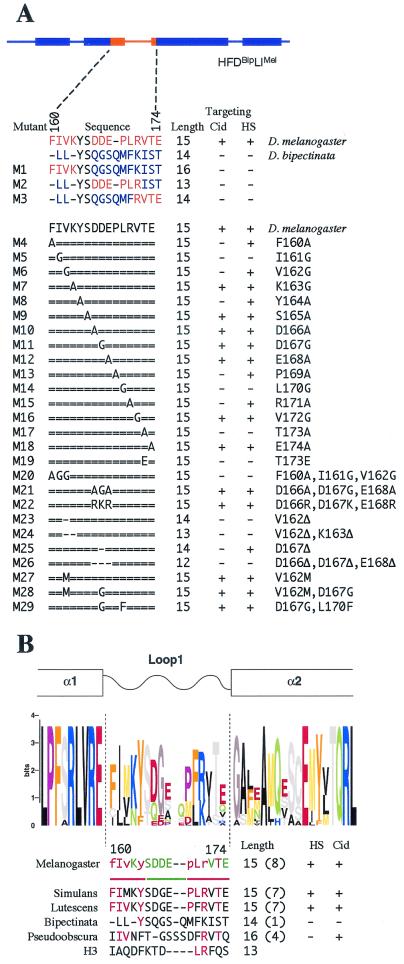
LI of CidMel is 1 amino acid longer than that of CidBip and differs from it in 12 of the remaining 14 amino acids (Fig. 4A). To identify critical amino acids for centromere targeting of CidMel LI, we performed a mutational analysis. Mutations were introduced into the CidBip HFD with LI of CidMel (HFDBip LIMel), because this construct localizes to centromeres yet is not expected to dimerize with endogenous CidMel. Shorter segments of CidMel LI did not target Cid to D. melanogaster centromeres, suggesting that multiple contacts within LI are required for targeting (Fig. 4A; M1, M2, and M3 mutants). To identify key targeting residues, we introduced single-amino-acid scanning mutations to alanine or glycine throughout D. melanogaster LI (Fig. 4A, M4 to M18). The results are displayed together with a sequence Logos representation of Drosophila LI in Fig. 4B.
FIG. 4.
Targeting of LI mutants of HFDBip LIMel-GFP. (A) The sequence of LI for each construct is shown, with hyphens indicating gaps in the alignment (22). Amino acids unique to D. melanogaster or D. bipectinata LI are shown in orange and blue, respectively. Equal signs indicate amino acids identical to D. melanogaster LI. For point mutants the amino acid changes in D. melanogaster LI are shown to the right. Numbering is relative to amino acids in D. melanogaster full-length Cid. The number of amino acids in LI (compared to 13 of LI asarbitrarily defined in histone H3) is indicated (length). Targeting is indicated by a plus sign, and the lack thereof is indicated by a minus sign for both the Cid and HS promoter constructs. (B) Single point mutations to alanine or glycine within D. melanogaster LI that affect targeting are aligned with the logos representation for Drosophila LI (22). The logos representation was generated from an alignment of Cid from a large collection of Drosophila species (22). In this graphical representation, the height of each amino acid indicates the level of conservation across Drosophila species, and amino acids with similar properties are similarly colored. Point mutations that abolish targeting are shown in red, whereas those that allow targeting are shown in green. The lack of targeting at low levels only (when expressed from the Cid promoter) is indicated by lowercase red letters. The red lines indicate more conserved amino acids that are required for targeting, whereas the green line denotes the hypervariable region. The sequence of LI from different species of Drosophila as well as H3 is shown. Hyphens denote gaps in the alignment. Amino acids that are involved in targeting by D. melanogaster LI are shown in red in the other LI regions. The LI length relative to 13 amino acids in H3 is shown (Length), with the number of amino acids conserved with those that are crucial for targeting for D. melanogaster (red) in parentheses. Targeting of different LI regions when inserted into the D. bipectinata HFD is indicated by a plus sign (targeting) or minus sign (no targeting) for the Cid and HS promoters.
Three point mutations (M5, M14, and M17 [Fig. 4A]) each abolished centromeric targeting. Five other point mutations (M4, M6, M8, M13, and M15) resulted in weak targeting, because Cid HFD-GFP could only be detected at the centromere when expressed at high levels from the HS promoter. Interestingly, these detrimental mutations cluster towards either end of LI (Fig. 4B, red lines), whereas amino acids in the center of LI were insensitive to mutation (Fig. 4A, M9, M10, M11, and M12; Fig. 4B, green line). In fact, targeting was unaffected when three central acidic amino acids were mutated to neutral residues (Fig. 4A, M21), or even to basic amino acids (Fig. 4A, M22). An alignment of Cid LI from different Drosophila species (22) supports this functional characterization of central, hypervariable amino acids flanked by more conserved regions.
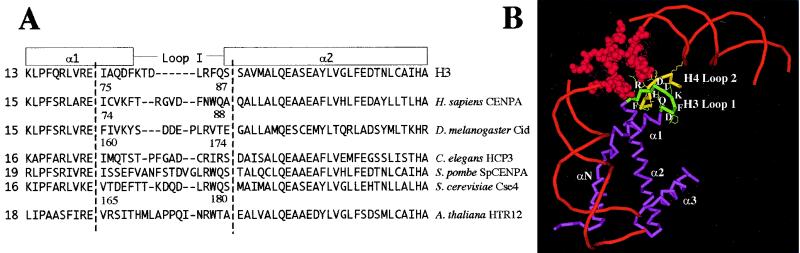
A hypervariable region flanked by more-conserved amino acids may be a hallmark of LI in CenH3 histones in general. For example, CENP-A targeting is unaffected by replacement of the central four hypervariable amino acids (T79, R80, G81, and V82) by only two (KT) (33) Fig. 5A), whereas deletion of three central hypervariable amino acids of Cse4 (K172, D173, and Q174) similarly does not affect viability in a null background (9, 16) (Fig. 5A). In contrast, changing conserved residues causes severe effects: point mutation of W86 of CENP-A revealed a requirement for an aromatic amino acid for targeting, and mutation of the respective W178 in Cse4 to either Y, D, or E was lethal (16, 33) (Fig. 5A).
FIG. 5.
LI regions of CenH3s differ in sequence and length from LI of canonical H3. (A) Alignment of LI and part of the adjacent α1- and α2-helices from a selection of known CenH3 proteins (5, 13, 35, 37, 38, 39). The secondary structure for H3 is shown schematically above the sequence (20). Dashed lines mark the extent of the LI region for CenH3, found to be critical for targeting of Cid in this study. The length of LI relative to 13 amino acids in H3, is shown next to the sequence. Hyphens denote gaps. Numbers of amino acids are relative to each starting methionine. (B) Schematic of a partial canonical nucleosome (20). LI of H3 (green) forms a DNA-binding domain with LII of H4 (yellow). Protein is represented by wire backbone traces (purple for H3), and the DNA is shown in red. Secondary structures are labeled, and side chains are indicated for the loops.
We note that position 173 of all Cids contains only threonine or serine (22). Mutation of this threonine of CidMel LI to alanine abolished targeting (Fig. 4B, M17), consistent with the possibility that phosphorylation may play a role in LI function. Targeting was also abolished by mutation of this threonine to a glutamic acid (Fig. 4A, M19), which mimics constitutive phosphorylation (Fig. 4A).
All CenH3s have a longer LI region than that of canonical H3 (Fig. 5B). We found that shortening D. melanogaster LI by one amino acid compromised centromeric targeting of HFDBip LIMel (Fig. 4A, M23 and M25). Shortening LI by two or more amino acids abolished all targeting (Fig. 4A, M24 and M26), even when the deletion was within the hypervariable region where mutation of the same amino acids had no effect on targeting (Fig. 4A, compare M26 with M21 and M22). The added length of the LI peptide backbone compared to that of H3 appears to be necessary to ensure targeting, rather than any specific interactions of the acidic side chains of D166, D167, and E168. In contrast, the Cse4 LI was found to tolerate a three-amino-acid deletion (16). In this case, LI is not the primary targeting determinant, but rather Cse4 is targeted by CBF3 binding to its cognate centromeric DNA element, CDEIII (28), and so LI may be evolving under relaxed constraints. We conclude that the crucial targeting function of CidMel LI is mediated by several amino acids located at both ends of the loop and that a minimum length of LI appears to be required for targeting.
Consistent with our results for the full-length Cid proteins, the LI regions of CidSim and CidLut also targeted Cid to D. melanogaster centromeres when introduced into CidBip HFD (Fig. 4A, M28 and M29). The CidBip HFD with CidPse LI localized to D. melanogaster centromeres when it was expressed at levels high enough to appear in euchromatin too (Fig. 4B). Centromeric targeting correlates with the degree of conservation of the amino acids that are critical for centromeric targeting of CidMel LI (Fig. 4B [number of critical amino acids that are conserved is shown in parentheses]). CidPse LI contains four of the eight critical amino acids and weakly targets Cid to D. melanogaster centromeres. CidBip LI contains only one of the eight amino acids and fails to target Cid to D. melanogaster centromeres. It is interesting that the CidBip N-terminal tail has undergone a nine-amino-acid repeat expansion (22). We have shown that this expansion is significantly similar to DNA minor groove-binding motifs and have suggested that these and other minor groove binding repeats in the Cid N-terminal tail facilitate centromere-specific binding (22). CidBip may rely on its N-terminal tail for targeting to D. bipectinata centromeres. In this way, there would be a relaxed constraint on LI in the D. bipectinata lineage. We conclude that the LI region is a common targeting element of CidMel, CidSim, CidLut, and CidPse for D. melanogaster centromeres, whereas CidBip may have followed a different evolutionary trajectory and lost this function.
DISCUSSION
We used Cid from different species of the Drosophila lineage to delineate a centromeric targeting element, LI. It is striking that adaptive evolution of Cid has also occurred in LI (21) and that the amino acids within this region are changing rapidly in the Drosophila lineage (22). Our study highlights the power of using an evolutionary analysis to guide ascertainment of functional protein domains.
LI was found to be one of several regions of the HFD to be required for targeting and centromere function in previous swap experiments between CenH3s and canonical H3 (9, 16, 33). These experiments did not reveal a single targeting element, likely because of different selective constraints acting on canonical H3 and CenH3s. Canonical H3 did not target centromeres in swap experiments (33, 37), perhaps because centromeric targeting signals had to compete against regulatory elements in the highly conserved histone H3. Like CENP-A LI (33), LI of CidMel does not target centromeres when introduced into canonical H3, either in the presence or absence of the H3 N-terminal tail (data not shown). CidMel LI is also insufficient to target CenH3 heterologs (Cse4 or Caenorhabditis elegans HCP3) to D. melanogaster centromeres (data not shown). This is consistent with the possibility that CenH3s are derived from multiple independent lineages where they are evolving under different selective constraints. Indeed, CenH3 heterologs cannot functionally substitute for one another: Cse4, CENP-A, and HCP3 do not target centromeres when expressed in Drosophila cells (13), and chimeric CenH3s fail to rescue centromere defects in yeast (16).
Our mutational analysis identifies a framework of amino acids within LI that are essential for targeting. These more conserved amino acids are likely to play a general role in maintaining LI structure. In the crystal structure of the nucleosome, LI of H3 forms a DNA-binding domain together with LII of H4 (10, 20) (Fig. 5B). The arginine side chain of H3 is inserted into the minor groove of the DNA (Fig. 5B, R84 of H3), and hydrogen bonds are formed between DNA phosphate groups and protein main chain and side chains. There are three hydrogen bonds between H3 LI (R84, F85, and Q86) and LII of H4. L170 and R171 of CidMel are conserved with the equivalent residues in LI of H3 (L83 and R84) (Fig. 5B). Mutation of L170 in the same position of Cse4 LI (Fig. 5B; L176 of Cse4) is lethal when combined with another point mutation that is involved in contacting H4 in canonical nucleosomes (9). Mutation of L170 in Cid LI may disrupt contact with H4 and possibly also with the DNA.
In contrast to the essential nature of conserved framework residues, mutation of the central amino acids of LI did not affect targeting. These mutationally flexible amino acids coincide with a hypervariable region of LI found in a sequence alignment of Cids from across the Drosophila lineage (22). These amino acids are changing rapidly, perhaps in response to the changing DNA satellites at centromeres (12, 21). However, the hypervariable amino acids within LI did not appear to be crucial for targeting to D. melanogaster centromeres in our assay, because LI from either CidMel or CidSim targeted D. melanogaster centromeres. Since the adaptive evolution between these species has occurred within LI of the HFD (21), it is likely that D. simulans LI mediates discrimination between D. melanogaster and D. simulans centromeric DNA (21). Discrimination might result from subtle differences in binding affinity during assembly or from a step subsequent to targeting, such as nucleosome stability. It should be noted that even a slight advantage in centromere function that would not be detected experimentally will be expected to drive to fixation within a population (12).
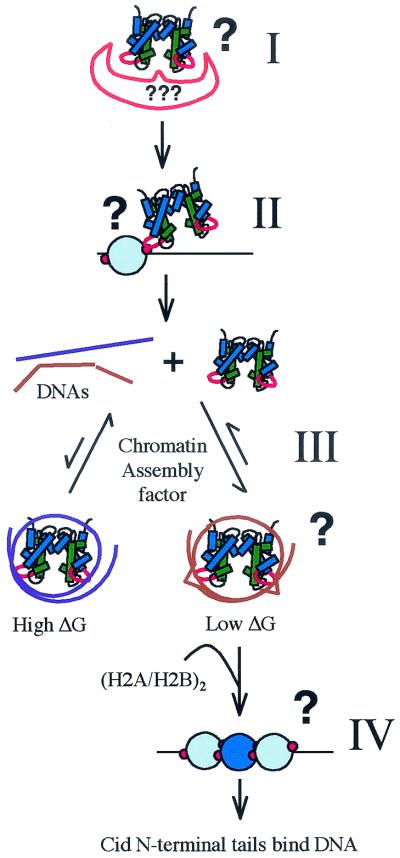
Cid LI could be a discriminating factor at any step in nucleosome assembly (Fig. 6). First, LI may dictate interaction of (Cid.H4)2 tetramers with a specialized chromatin assembly factor that targets centromeres (Fig. 6, step I). Currently there are no assembly factors that are known to act exclusively at centromeres (31, 46). Furthermore, overexpressed Cid (Fig. 2) and CENP-A (42) are incorporated into euchromatin, suggesting that (CenH3.H4)2 tetramers can be assembled outside of centromeres by general chromatin assembly factors. The best candidate for a specific assembly factor is Schizosaccharomyces pombe Mis6p, which is required for targeting of SpCENP-A (38). However, it is unclear if Mis6p directly recruits SpCENP-A, and homologs of Mis6p are not involved in targeting CENP-A or Cse4 (23, 26). The inability of CidMel LI to target CenH3 heterologs to D. melanogaster centromeres suggests that LI does not act as a simple recognition site for a centromere-specific Drosophila assembly factor. In addition, such a protein-protein interaction should evolve to an optimal conformation, leading to the conservation of LI rather than the adaptive evolution observed in multiple Drosophila lineages (21, 22).
FIG. 6.
Steps in the targeting process where LI may be involved. LI may be required for binding of the (Cid.H4)2 tetramer to a putative centromere-specific nucleosome assembly factor (step I). LI may mediate the interaction of unassembled (Cid.H4)2 tetramers with Cid chromatin at centromeres, guiding assembly of new Cid nucleosomes at the location of existing centromeres (step II). LI may be a discriminating factor in DNA specificity during the assembly of (Cid.H4)2 tetramers (step III). Assembly of (Cid.H4)2 tetramers is an equilibrium process, perhaps assisted by chromatin assembly or remodeling factors which would act to lower the activation energy for transfer of tetramers between different DNA sequences (brown and purple double helices). DNA sequences (brown) that have the lowest free energy in the complex are favored for reversible deposition of tetramers, and their assembly goes to completion. LI may act at the level of DNA specificity by dictating a deformation in the DNA (schematically shown as a kink in the DNA). The LI regions may be required to fit correctly in between existing Cid nucleosomes (step IV). Minor groove binding of the Cid N-terminal tails may complete assembly of new Cid nucleosomes and result in formation of unique higher order centromeric chromatin structure.
Instead of targeting the centromere by binding an assembly factor that localizes to centromeres, Cid LI may target centromeres by mediating an interaction between existing centromeric nucleosomes and unassembled Cid tetramers (Fig. 6, step II). Cid LI likely protrudes from both tetramers and nucleosomes, consistent with such an interaction. However, this protein-protein interaction also should evolve to an optimum and so does not explain the adaptive evolution of Drosophila LI (21, 22). Rather than mediating an interaction between an unassembled tetramer and an existing centromeric nucleosome, LI may mediate the correct fitting (32, 36) of new centromeric nucleosomes in between existing ones (Fig. 6, step IV). Such an LI-discriminating assembly mechanism would rely on contacts of LI to both proteins and DNA, consistent with coevolution of LI and centromeric satellites. LI of CidBip may thus prevent targeting of CidMel HFD by posing a structural hindrance to its inclusion within centromeric chromatin.
LI may also specify targeting by preferential assembly of (Cid.H4)2 tetramers on certain DNAs (Fig. 6, step III) (21, 33). Different DNA sequences may compete for binding of (Cid.H4)2 tetramers during chromatin assembly. Those DNA sequences that are capable of adopting a favorable conformation for interaction could then be strongly preferred during the assembly step. In an equilibrium assembly process, complexes with the lowest free energy will prevail and be available for binding of H2A.H2B dimers and subsequent chromatin maturation. A recent energetic study suggests that the histone-DNA contacts at the central one or two turns of nucleosomal DNA provide a nucleation point for wrapping DNA around the rest of the octamer (4). If assembly indeed starts at the dyad, a noncanonical LI could present a critical energetic barrier 25 bp in either direction from the central contact point. The location of LI at superhelical positions +2.5 and −2.5 of the nucleosome (20) ensures that a DNA molecule will encounter LI at an interval of 50 bp during assembly. Different DNA sequences can have a substantial free-energy difference for canonical nucleosomes (40). In the case of CenH3 nucleosomes, the free-energy differences for different DNAs may be even larger due to constraints imposed by the increased length of LI. This could result in substantial preference for one DNA over another.
Despite differences at the primary sequence level, centromeric satellites contain patterns consistent with a bias in nucleosome assembly: nucleosomal unit repeat lengths are a common occurrence (12), and there is evidence for positioning of CENP-A nucleosomes on α-satellite DNA (41). Although some sequences appear to be preferred at centromeres (30, 44), optimal conformations for assembly of (CenH3.H4)2 tetramers need not be specified by just one primary sequence (17, 44). Indeed, a periodic secondary structural feature may be required for optimal binding to LI. It is not surprising that such DNA features have escaped detection: predicting positioning strength based on DNA sequence is notoriously difficult even for canonical nucleosomes (40). Neocentromeres may represent DNA sequences with free energy of nucleosome formation higher than those of bona fide centromeres but lower than those of other sequences in the genome. Although neocentromeres do not contain α-satellite DNA, they do show a sequence bias (18, 19), consistent with DNA preference in targeting of (CENP-A.H4)2 tetramers.
The present study underlines the primary importance of the Cid HFD and LI for centromeric targeting. We have suggested that the N-terminal tails of Cid may also bind DNA, specifically in the minor groove (22). Such an interaction may result in a unique higher-order structure of centromeric core chromatin (22). Minor groove binding of the Cid N-terminal tail is likely to occur at a step subsequent to wrapping of the DNA around the octamer (Fig. 6) and may contribute to the stability of Cid nucleosomes.
Acknowledgments
We thank the Drosophila Species Center for the various fly strains used, Harmit Malik and Kami Ahmad for stimulating discussions and comments on the manuscript, and Judy O'Brien for technical assistance.
We were supported by the Cancer Research Fund of the Damon Runyon Cancer Research Foundation, DRG-1554 (D.V.), and by the Howard Hughes Medical Institute (H.S.H. and S.H.).
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blower, B. D., and G. H. Karpen. 2001. The role of Drosophila Cid in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blower, M. D., B. A. Sullivan, and G. H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brower-Toland, B. D., C. L. Smith, R. C. Yeh, J. T. Lis, C. L. Peterson, and M. D. Wang. 2002. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA 99:1960-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature 401:547-548. [DOI] [PubMed] [Google Scholar]
- 6.Choo, K. H. A. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1:165-177. [DOI] [PubMed] [Google Scholar]
- 7.Depinet, T. W., J. L. Zackowski, W. C. Earnshaw, S. Kaffe, G. S. Sekhon, R. Stallard, B. A. Sullivan, G. H. Vance, D. L. Van Dyke, H. F. Willard, A. B. Zinn, and S. Schwartz. 1997. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum. Mol. Genet. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 8.Dong, F., and K. E. van Holde. 1991. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc. Natl. Acad. Sci. USA 88:10596-10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harp, J. M., B. L. Hanson, D. E. Timm, and G. J. Bunick. 2000. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 56:1513-1534. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, J. J., D. J. Clark, and A. P. Wolffe. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 88:6829-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff, S., K. Ahmad, and H. S. Malik. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098-1102. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19:6130-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, J. 2000. Neocentromeres and alpha satellite: a proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 9:149-154. [DOI] [PubMed] [Google Scholar]
- 18.Lo, A. W., J. M. Craig, R. Saffery, P. Kalitsis, D. V. Irvine, E. Earle, D. J. Magliano, and K. H. Choo. 2001. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo, A. W. I., D. J. Magliano, M. C. Sibson, P. Kalitsis, J. M. Craig, and K. H. A. Choo. 2001. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromeric DNA. Genome Res. 11:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Malik, H. S., and S. Henikoff. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik, H. S., D. Vermaak, and S. Henikoff. 2002. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Measday, V., D. W. Hailey, I. Pot, S. A. Givian, K. M. Hyland, G. Cagney, S. Fields, T. N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 25.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 26.Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W. C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2:463-476. [DOI] [PubMed] [Google Scholar]
- 27.Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A. A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, D. K., K. O'Day, H. L. Trong, H. Charbonneau, and R. L. Margolis. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schueler, M. G., A. W. Higgins, M. K. Rudd, K. Gustashaw, and H. F. Willard. 2001. Genomic and genetic definition of a functional human centromere. Science 294:109-115. [DOI] [PubMed] [Google Scholar]
- 31.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelby, R. D., K. Monier, and K. F. Sullivan. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein, and P. C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan, K. F. 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11:182-188. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan, K. F., M. Hechenberger, and K. Masri. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi, K., E. S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215-2219. [DOI] [PubMed] [Google Scholar]
- 39.Talbert, P. B., R. Masuelli, A. P. Tyagi, L. Comai, and S. Henikoff. 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thastrom, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 41.Vafa, O., and K. F. Sullivan. 1997. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7:897-900. [DOI] [PubMed] [Google Scholar]
- 42.Van Hooser, A. A., I. I. Ouspenski, H. C. Gregson, D. A. Starr, T. J. Yen, M. L. Goldberg, K. Yokomori, W. C. Earnshaw, K. F. Sullivan, and B. R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529-3542. [DOI] [PubMed] [Google Scholar]
- 43.Warburton, P. E., M. Dolled, R. Mahmood, A. Alonso, S. Li, K. Naritomi, T. Tohma, T. Nagai, T. Hasegawa, H. Ohashi, L. C. Govaerts, B. H. Eussen, J. O. Van Hemel, C. Lozzio, S. Schwartz, J. J. Dowhanick-Morrissette, N. B. Spinner, H. Rivera, J. A. Crolla, C. Y. Yu, and D. Warburton. 2000. Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am. J. Hum. Genet. 66:1794-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willard, H. F. 2001. Neocentromeres and human artificial chromosomes: an unnatural act. Proc. Natl. Acad. Sci. USA 98:5374-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolffe, A. P. 1998. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, Calif.
- 46.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodelling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoda, K., S. Ando, S. Morishita, K. Houmura, K. Hashimoto, K. Takeyasu, and T. Okazaki. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97:7266-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]