Abstract
DNA and histone synthesis are both triggered at the beginning of S phase by cyclin/cdk2 activity. Previous studies showed that inhibition of DNA synthesis with hydroxyurea or cytosine arabinoside (AraC) triggers a concerted repression of histone synthesis, indicating that sustained histone synthesis depends on continued DNA synthesis. Here we show that ectopic expression of HIRA, the likely human ortholog of two cell cycle-regulated repressors of histone gene transcription in yeast (Hir1p and Hir2p), represses transcription of histones and that this, in turn, triggers a concerted block of DNA synthesis. Thus, in mammalian cells sustained DNA synthesis and histone synthesis are mutually dependent on each other during S phase. Although cyclin/cdk2 activity drives activation of both DNA and histone synthesis at the G1/S transition of cycling cells, concerted repression of DNA or histone synthesis in response to inhibition of either one of these is not accompanied by prolonged inhibition of cyclin A/cdk2 or E/cdk2 activity. Therefore, during S phase coupling of DNA and histone synthesis occurs, at least in part, through a mechanism that is independent of cyclin/cdk2 activity. Coupling of DNA and histone synthesis in S phase presumably contributes to the prompt and orderly assembly of newly replicated DNA into chromatin.
Progression through the cell cycle is driven by the sequential and periodic activation of cyclin/cdk complexes (23, 27). For example, entry into and progression through S phase is promoted by activation of cyclin E/cdk2 and cyclin A/cdk2, whereas entry into mitosis is triggered by activation of cyclin B/cdc2. Numerous lines of evidence demonstrate that cyclin/cdk2 activity plays a key role in initiation of S-phase events. Elevation of cyclin/cdk2 activity in G1 phase causes premature entry into S phase (9, 56, 57, 72), and inhibition of cyclin/cdk2 activity inhibits entry into and progression through S phase (20, 52, 67, 76). Initiation of S phase depends on activation of a number of biosynthetic processes, including DNA synthesis, histone synthesis, and chromatin assembly (58).
According to current models distinct S-phase processes, such as DNA synthesis and histone synthesis, are independently activated by cyclin/cdk2 at the start of S phase (17, 30). For example, activation of DNA synthesis depends on phosphorylation of Cdc6 and Cdc45. Increased histone synthesis in S phase is due to regulation at transcriptional and posttranscriptional levels. Histone gene transcription increases three- to fivefold as cells enter S phase, and this depends on phosphorylation of NPAT by cyclin E/cdk2 (30, 39, 42, 50, 74). However, posttranscriptional regulation accounts for the majority of the 35- to 50-fold increase in histone synthesis during S phase (42, 50). The processing of the immature intronless pre-mRNA to the mature mRNA requires a cleavage within the 3′ untranslated region (UTR); this occurs more efficiently in S phase (15, 19, 24, 37, 65), and the mRNA is also more stable at this time (10, 24, 25, 62). The processing of the pre-mRNA depends on a stem-loop structure within the 3′ UTR of the mRNA and a protein that binds to it, stem-loop binding protein (SLBP) (66, 70). Efficient pre-mRNA processing also requires the U7 snRNP, whose binding to a purine-rich sequence downstream of the stem-loop is facilitated by SLBP (13, 16), a heat-labile factor (HLF) (15), and novel zinc finger protein hZFP100, which interacts with the U7 snRNP and SLBP (14). Increased mRNA stability also depends on the stem-loop in the 3′ UTR and possibly SLBP, although the requirement for SLBP has not been directly demonstrated (15, 38, 53). Due to control of the translation and degradation of SLBP, its abundance varies through the unperturbed cell cycle in a manner that parallels the histone mRNA abundance, and this contributes to posttranscriptional regulation of histone synthesis through the cell cycle (66, 71). Exactly how SLBP levels are controlled through the cell cycle is unknown, although, as for activation of histone transcription, control is likely to be dependent on periodic cyclin/cdk2 activity. The activity of both HLF and the U7 snRNP has also been reported to increase as cells enter S phase (19, 26, 37, 65), although some workers have failed to observe the regulation of the latter (6).
Although the simultaneous activation of DNA synthesis and histone synthesis by cyclin/cdk2 activity at the G1/S transition ensures a certain level of coordination between them, in view of the inevitable impact of chromatin structure on the fidelity of nuclear processes, these processes are likely to be regulated in a very closely coordinated and concerted manner throughout S phase. Consistent with this idea, the size of the free histone pool in S phase in mammalian somatic cells is very small and hydroxyurea (HU)-mediated inhibition of DNA synthesis in mammalian cells triggers a rapid and concerted destabilization of histone mRNAs, indicating that sustained histone synthesis is dependent on ongoing DNA replication (4, 25, 62, 68). Presumably, these results reflect the existence of controls during S phase which ensure that the rate of production of histones matches the rate of their consumption by DNA replication and chromatin assembly. This, in turn, facilitates orderly chromatin assembly. However, the molecular mechanism underlying this coupling of DNA and histone synthesis and whether sustained DNA synthesis is likewise dependent on ongoing histone synthesis are not known. Moreover, whether cyclin/cdk2, in addition to its role in simultaneous activation of these events at the G1/S transition, also plays a direct role in coordination of these events in S phase is not known.
To shed light on control of histone synthesis in S phase, we have studied human HIRA, a likely regulator of histone gene expression. Based on sequence analysis, HIRA is the best candidate to be an ortholog (functional equivalent) of two regulators of histone gene transcription in Saccharomyces cerevisiae, Hir1p and Hir2p (35). The N-terminal half of HIRA is related to Hir1p, and the C-terminal half is related to Hir2p. Thus, the HIRA gene might combine the functions of the two yeast genes. In wild-type yeast and mammalian cells, histones are transcribed periodically throughout the cell cycle, with expression peaking in S phase. In yeast, Hir1p and Hir2p repress transcription of histones H2A and H2B in G1 phase and, through their ability to recruit components of the SWI/SNF chromatin-remodeling complex, activate expression in S phase (12, 61, 64). In addition, Hir1p and Hir2p are implicated in control of nucleosome assembly. Mutations in Hir genes contribute to defects in gene silencing at telomeres and silent mating loci, and Hir1p and Hir2p physically interact with the chromatin assembly factor Asf1p (29, 33, 59). Hir1p localizes to yeast centromeres and, together with chromatin assembly factor I (CAF-I), is required for formation of proper chromatin structure at these loci (60). In contrast, little is known concerning the role of the human HIRA protein. Consistent with HIRA being a regulator of cell cycle progression, like Hir1p and Hir2p, we showed previously that HIRA is a nuclear cyclin/cdk2 substrate that is phosphorylated in S phase of the cell cycle (21). Moreover, we showed that ectopic expression of HIRA inhibited DNA synthesis and arrested cells in S phase of the cell cycle (21). De Lucia and coworkers showed that HIRA is phosphorylated during mitosis and that phosphorylation is accompanied by release from nuclear chromatin (11). HIRA has been shown to interact with several proteins implicated in control of transcription and nuclear structure, including Pax3 and histones (36, 40). Recently, HIRA in Xenopus laevis egg extracts was shown to be necessary for DNA replication-independent nucleosome assembly, suggesting that, like Hir1p and Hir2p, vertebrate HIRA may have dual functions in control of cell cycle-regulated histone transcription and chromatin assembly (54).
Although results presented here support the notion that HIRA, like Hir1p and Hir2p, is a regulator of histone gene expression, the major conclusion of this work concerns concerted regulation of DNA synthesis and histone synthesis in S phase. We show that the previously observed HIRA-induced S-phase arrest is a secondary consequence of HIRA-induced repression of histone gene transcription. Therefore, completion of DNA synthesis in S phase requires sustained histone gene expression. Interestingly, HIRA-induced inhibition of DNA synthesis is not accompanied by repression of cyclin/cdk2 activity. Similarly we show that HU-induced inhibition of histone gene expression is not accompanied by prolonged inhibition of cyclin/cdk2 activity. Thus, in intact mammalian cells completion of DNA synthesis and histone synthesis are mutually dependent and coupled through a cdk2-independent pathway. This represents a novel level of S-phase control that might contribute to coordinated DNA synthesis, histone synthesis, and chromatin assembly.
MATERIALS AND METHODS
Cell culture and transfections.
U2OS cells were cultured and transfected as described previously (2). The cells were synchronized with HU as described elsewhere (34).
Plasmid construction.
pBS-GAPDH was a gift from Maureen Murphy. pMae DHFR WT and ΔE2F were gifts from Peggy Farnham. pAd-LacZ was a gift from Michael Xu. pECE-pRBΔcdk was a gift from Paul Hamel. pcDNA3 HA-HIRA has been described previously (21). All synthetic mutant versions of this plasmid were made by standard molecular biology techniques and confirmed by direct sequencing and/or restriction digest. cDNAs encoding the replication-dependent histones H1, H2A, H2B, H3, and H4 were obtained by PCR from human genomic DNA. Plasmids encoding these histones were generated by standard molecular biology techniques and confirmed by direct sequencing. pSP64T β-globinSLBP was a gift from William Marzluff. pcDNA3 HA-SLBP was made from this by a standard PCR-based approach and was confirmed by direct sequencing. pcDNA3 HA-SLBP RR/KK and pcDNA3 HA-SLBP QKQ were made by standard mutagenesis techniques. pAd HA-HIRA(421-729) was made with the Adeasy adenovirus construction kit (Qbiogene) by using standard molecular biology techniques and by following the manufacturer's instructions. Details of all cloning methods are available upon request.
Immunological techniques.
Purified rabbit polyclonal anti-SLBP was a gift from William Marzluff. Anti-cyclin A (C160) was a gift from Ed Harlow. Anti-hemagglutinin (HA; 12CA5) was purchased from Roche. Anti-cdk2 (M2), anti-cyclin E (HE11), anti-cyclin A (BF683), and anti-PCNA (PC10) were purchased from Santa Cruz Biotechnology. Anti-pRB (245) was purchased from Pharmingen. Purified rabbit polyclonal anti-NPAT has been described previously (39). Anti-HIRA monoclonal antibodies (WC15, -19, -117, and -119) have been described previously (21). The anti-HDAC4 antibody will be described in a future publication (G. D. Kao and T. J. Yen, unpublished data). Immunoprecipitation, Western blotting, and immunofluorescence were performed as described previously (21).
Nuclear runoff transcription assay.
The nuclear runoff transcription assay was performed as described elsewhere. Briefly, 24 h after infection with the appropriate adenovirus, cells were harvested and intact nuclei were isolated in Tween lysis buffer A (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% Tween 20). Nuclear runoff transcription was performed, and total RNA was isolated with TriZol (Gibco).
Collection of CD19+ transfected cells by using magnetic beads.
Cells transiently transfected with a plasmid encoding CD19 were collected by using anti-CD19-coated magnetic beads (Dynal) according to the manufacturer's instructions.
MNase assay.
The micrococcal nuclease (MNase) assay was performed as described elsewhere (73). Briefly, 24 h after infection with the appropriate adenovirus, cells were harvested and permeabilized for 2 min in 0.004% lysolecithin (Sigma) and then treated for 5 min with the amounts of MNase (Roche) indicated in the legend for Fig. 5. Total DNA was isolated by phenol-chloroform extraction and ethanol precipitation and separated on 0.6% Tris-acetate EDTA-agarose gels and visualized with ethidium bromide.
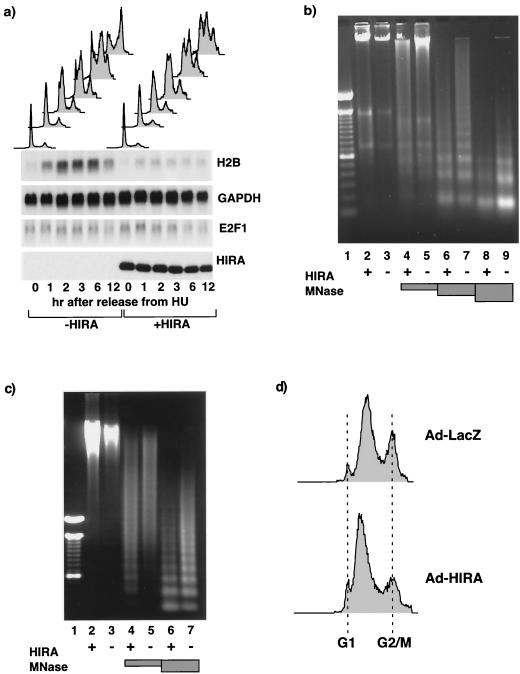
FIG. 5.
HIRA-induced histone gene repression precedes S-phase arrest and generates incompletely assembled chromatin. (a) U2OS cells were transiently transected with pcDNA3 or pcDNA3 HA-HIRA WT as indicated together with pCMV-CD19. The cells were arrested at the G1/S transition with HU and then released into S phase. Cells were harvested at the indicated times afterwards, and the cell cycle distribution of the CD19+ cells (top) and abundances of histone H2B, GAPDH, and E2F1 mRNAs (indicated at the right) in the CD19+ cells were determined. Total-cell lysates were Western blotted with anti-HA (HIRA). (b) U2OS cells were infected with an adenovirus expressing LacZ (lanes 3, 5, 7, and 9) or HIRA(421-729) (lanes 2, 4, 6, and 8). Twenty-four hours later the cells were permeabilized and treated without MNase or with 0.25, 2.5, or 25 Worthington units of MNase/ml (shaded boxes). Genomic DNA was purified, fractionated by 0.6% agarose gel electrophoresis, and visualized with ethidium bromide. The size markers in lane 1 are a 100-bp ladder. (c) U2OS cells were infected with an adenovirus expressing LacZ or HIRA(421-729). After 4 h 2 mM HU was added to the cells, and 20 h later it was washed off and the cells were allowed to progress into S phase for 4 h. The cells were harvested, permeabilized, and treated without MNase or with 0.25 or 2.5 Worthington units of MNase/ml (shaded boxes). Genomic DNA was purified, fractionated by 0.6% agarose gel electrophoresis, and visualized with ethidium bromide. The size markers in lane 1 are a 100-bp ladder. (d) Samples from cells harvested for panel c were also processed for FACS analysis to determine the cell cycle distribution. Ad, adenovirus.
Other techniques.
In vitro kinase assays (2), fluorescence-activated cell sorter (FACS) analysis of CD19+ transfected cells (2), luciferase assays (46), and Northern blotting (49) have been described elsewhere.
RESULTS
We showed previously that ectopic expression of human HIRA in U2OS cells triggered S-phase arrest (21). Since human HIRA is a likely orthologue of yeast Hir1p and Hir2p, two repressors of histone expression, we hypothesized that ectopic expression of HIRA represses histone gene expression and that this, in turn, triggers S-phase arrest. To test whether HIRA is able to repress histone gene expression, human U2OS cells were transiently transfected with a plasmid encoding wild-type HA-tagged HIRA. RNA was prepared from the transfected cells and Northern blotted to determine the abundance of histone mRNAs. As shown in Fig. 1a, ectopic expression of HIRA profoundly repressed the steady-state abundance of mRNA encoding each histone subtype. Moreover, this repression was specific since there was no effect on the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. Of note, the human genome contains multiple copies of each histone subtype (http://genome.nhgri.nih.gov/histones/index.html) (3). For example, there are 16 copies of histone H2A clustered on the short arm of chromosome 6. Most of these are expressed specifically in S phase of the cell cycle (replication dependent), and the DNA coding sequences are greater than 80% identical. However, there are also a number of histone genes that are expressed outside of S phase (replacement variants). These replacement variants have greater sequence divergence at the DNA level (the H2A Z gene is only 40 to 50% identical to the replication-dependent H2A genes), and the mRNAs are significantly larger due to a longer 3′ UTR and poly(A) tail. Under the conditions used here and with replication-dependent histones as probes, only the replication-dependent histones are detectable (W. F. Marzluff, personal communication). We focused on these histones because we reasoned that their repression would be most likely to trigger S-phase arrest.
FIG. 1.
Ectopic expression of HIRA represses histone gene transcription. (a) U2OS cells were transiently transfected with pcDNA3 or pcDNA3 HA-HIRA as indicated together with pCMV-CD19. Transfected CD19+ cells were collected, and total RNA was prepared and Northern blotted to determine abundances of the indicated mRNAs. EtBr, ethidium bromide. (b) U2OS cells were transiently transfected with pcDNA3 (lanes 1 to 4), pcDNA3 HA-HIRA (lane 5 to 8), or pcDNA3 HA-HIRA plus pcDNA3 HA-SLBP (lanes 9 to 12) together with pCMV-CD19. Thirty-six hours later 10 μg of actinomycin D/ml was added, and at the indicated times afterwards the CD19+ transfected cells were collected and the abundances of histone and GAPDH mRNAs were determined by Northern blotting. Total-cell lysates were Western blotted with anti-HA. (c) The histone mRNA signals from panel b were quantitated on a phosphorimager and expressed as a percentages of control (100% = signal intensity at time zero). (d) U2OS cells were infected with an adenovirus encoding LacZ (lanes 1 and 2) or HIRA(421-729) (lanes 3 and 4). Twenty-four hours later nuclei were harvested and the rate of histone H2B transcription was determined in a runoff assay. 32P-labeled mRNA (5 × 106 ethanol-precipitable cpm per membrane) was used to probe a membrane slot blotted with pcDNA3Δ (lanes 2 and 4) or pcDNA3ΔH2B (lanes 1 and 3). (e) The signals in panel d were quantitated on a phosphorimager, corrected for the background hybridization on vector alone, and expressed as arbitrary units.
The abundance of histone mRNAs is regulated at transcriptional and posttranscriptional levels (50). If HIRA is an ortholog of Hir1p and Hir2p and represses histone gene transcription, then the observed histone mRNA repression should be due to decreased transcription rather than decreased mRNA stability. To test whether HIRA altered the half-life of histone mRNAs, U2OS cells were transiently transfected with a plasmid encoding HA-HIRA and then treated with actinomycin D to block new transcription. RNA was harvested at various times, and the abundance of histone mRNAs was determined by Northern blotting. As shown in Fig. 1b and c, although the steady-state mRNA abundance was lower in the presence of HIRA, there was no significant difference in the rates of decay of the mRNA with and without HIRA. The observed decay of the mRNA was not an indirect consequence of inhibition of DNA replication by actinomycin D because, under these experimental conditions, the extent of inhibition of DNA synthesis after 30 min of actinomycin D treatment was only between 6 and 18% in different experiments, as determined by measuring 5′ bromodeoxyuridine incorporation (data not shown). In contrast, when HIRA was coexpressed with SLBP, the half-life of the mRNA was greatly extended. This observation is consistent with the fact that SLBP binds to the stem-loop in the 3′ UTR of the histone mRNA, which is known to regulate mRNA stability. Indeed it has been proposed previously that SLBP is the regulator of histone mRNA stability (15). Taken together, the evidence indicates that HIRA does not promote degradation of histone mRNAs. Next we asked whether expression of HIRA decreased transcription of histone genes. To answer this question, U2OS cells were infected with an adenovirus directing the expression of LacZ or HIRA(421-729) (a fragment of HIRA which efficiently decreases mRNA abundance [see Fig. 4]). Intact nuclei were harvested and used in transcription runoff assays to determine the rate of ongoing histone H2B transcription. As shown in Fig. 1d and e, ectopic expression of HIRA(421-729), but not LacZ, caused a decrease in histone H2B transcription. Ectopic expression of HIRA(421-729) similarly repressed transcription of histones H1, H2A, and H3 (data not shown).
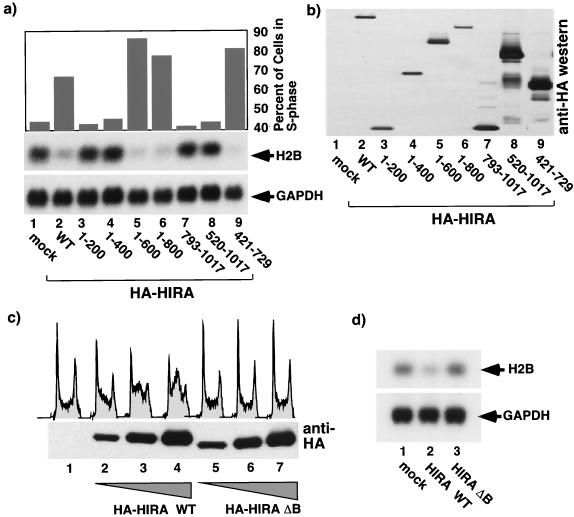
FIG. 4.
S-phase arrest and histone mRNA repression depend on the evolutionarily conserved B domain of HIRA. (a) U2OS cells were transiently transfected with a plasmid encoding wild-type (WT) HA-HIRA (lane 2) or a series of deletion mutants (lanes 3 to 9) or the empty vector (pcDNA3; lane 1) together with pCMV-CD19. The cells were harvested, and cell cycle distribution and abundances of histone H2B and GAPDH mRNAs in the transfected CD19+ cells were determined. (b) Total-cell lysates were Western blotted with anti-HA. (c) U2OS cells were transiently transfected with pcDNA3 (lane 1), 3, 10, or 28 μg of pcDNA3 HA-HIRA WT (lanes 2 to 4), or 3, 10, or 28 μg of pcDNA3 HA-HIRAΔB (lanes 5 to 7) together with pCMV-CD19. The cells were harvested, the cell cycle distribution of the CD19+ transfected cells was determined, and an aliquot of total-cell lysate was Western blotted with anti-HA. (d) U2OS cells were transiently transfected with pcDNA3 (lane 1), pcDNA3 HA-HIRA WT (lane 2), or pcDNA3 HA-HIRA ΔB (lane 3) together with pCMV-CD19. Transfected CD19+ cells were collected, and total RNA was Northern blotted to determine expression of GAPDH and histone H2B mRNAs.
Consistent with the finding that ectopically expressed HIRA repressed histone gene expression at the transcriptional level, endogenous HIRA and at least one molecule implicated in repression of transcription, HDAC4, colocalized in nuclear foci in primary WI38 cells (Fig. 2a to c) (45, 69). Similar foci were observed with three out of four monoclonal antibodies to HIRA (WC19, WC117, and WC119) but not with control antibodies (data not shown). Interestingly, only 10 to 30% of the nuclei contained detectable HIRA foci (Fig. 2d and e). Therefore we tested whether the appearance of HIRA foci was cell cycle regulated by using NPAT staining as a marker of cell cycle position. NPAT, a regulator of histone gene expression, is localized in subnuclear structures called Cajal bodies, and the number and localization of NPAT-containing Cajal bodies vary in the cell cycle (39, 74). In primary human fibroblasts in mid- and late G1, there are two NPAT-containing Cajal bodies associated with histone gene clusters on chromosome 6. During S phase, two additional foci are recruited to histone gene clusters on chromosome 1. Four foci are maintained until metaphase, when they disappear. Soon after reentry into G1, two NPAT-containing foci reform on chromosome 6. Thus, the number of NPAT-containing foci can be used as a general measure of cell cycle position. As shown in Fig. 3a to c cells with uncondensed chromatin containing HIRA foci generally contained zero or two NPAT-containing foci. Moreover, those rare cells that contained four NPAT-containing foci and HIRA foci generally contained the fewest and least-intense HIRA foci (data not shown). Conversely, cells lacking HIRA foci frequently contained four NPAT-containing foci. As summarized in Fig. 3j, approximately 85% of cells with HIRA foci had zero or two NPAT dots (probably G1 phase). In contrast, of the total cell population (visualized by DAPI [4′,6′-diamidino-2-phenylindole] staining) approximately 55% had zero or two NPAT dots. This proportion is comparable to the proportion of G1-phase cells determined by FACS analysis (data not shown).
FIG. 2.
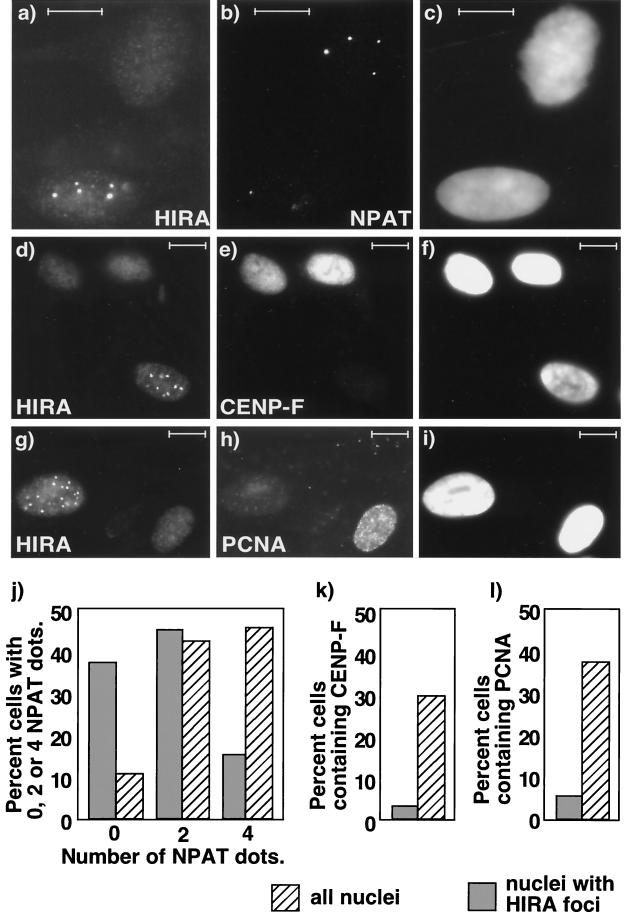
Endogenous HIRA forms subnuclear foci that contain HDAC4 in a subpopulation of primary cells. Primary human WI38 cells grown on coverslips were stained with anti-HIRA (a and d), anti-HDAC4 antibodies (b), and DAPI (c and e) to visualize the DNA. Bars, 10 μM.
FIG. 3.
Endogenous HIRA forms subnuclear foci in G1 phase of the cell cycle. Primary human WI38 cells were stained with antibodies to HIRA (a, d, and g), NPAT (b), CENP-F (e), and PCNA (h) and with DAPI (c, f, and i) to visualize the DNA. (j) The NPAT-containing foci in 60 randomly selected DAPI-stained nuclei (hatched bars) and 60 nuclei containing HIRA foci (shaded bars) were counted, and the numbers of cells with zero, two, and four foci were expressed as percentages of the total. (k) The cells staining positive for CENP-F-containing foci in 100 randomly selected DAPI-stained nuclei (hatched bars) and 100 nuclei containing HIRA foci (shaded bars) were counted, and the numbers of positive and negative cells were expressed as percentages of the total. (l) Same as panel k except that cells stained with antibodies to PCNA were counted. Bars, 10 μM.
To verify that HIRA foci were absent from S- and G2-phase cells, we double stained cells with antibodies to HIRA and two other markers of cell cycle position, PCNA and CENP-F. PCNA is expressed only in S-phase cells, whereas CENP-F is expressed only in cells in G2 phase and M phase (28, 43). As expected based on the results obtained with anti-NPAT antibodies, CENP-F and PCNA were virtually absent from cells containing HIRA foci (Fig. 3d to i, k, and l). We also observed that cells with HIRA foci tended to stain less brightly with DAPI (Fig. 2d and e), which is also consistent with them having a 2n DNA content (39). Taken together, the results based on double staining with NPAT, PCNA, and CENP-F antibodies show that HIRA foci are absent from S- and G2-phase WI38 cells. This indicates the cells with HIRA foci are in G1 or G0 phase or they are senescent. Ongoing experiments are designed to differentiate between these possibilities and determine the function of the HIRA foci.
Previously we reported that in human U2OS cells (an osteosarcoma cell line) HIRA was present diffusely throughout the entire nucleus (21). The staining patterns reported for each cell line were observed with multiple monoclonal antibodies to HIRA but not control antibodies of the same subclass(immunoglobulin G1; data not shown). The reason for the difference is not known at this point but may relate to the cell type or the transformed (U2OS) versus primary (WI38) nature of the cells. Unfortunately, the fact that HIRA is present as a diffuse nuclear stain in U2OS cells prevented us from asking whether HIRA and HDAC4 colocalize in these cells. Despite this, the colocalization of HIRA with HDAC4 in primary WI38 cells is consistent with these two proteins having a shared function in human cells. Since HDAC4 activity is linked to transcriptional repression, this, in turn, is consistent with the results for U2OS cells that implicate HIRA in repression of histone gene transcription (45, 69).
To test whether S-phase arrest and repression of histone transcription might depend on the same molecular activity of HIRA, we tested a panel of synthetic HIRA mutants for their ability to repress histone expression and cause S-phase arrest when transiently expressed in cells. As shown in Fig. 4a and b, there was a perfect correlation between the ability of the mutants to cause S-phase arrest and histone repression. Interestingly, we noted that all active HIRA mutants retain residues 439 to 475 of human HIRA. This stretch of amino acids in human HIRA was previously identified based on sequence comparisons as a domain (the B domain) that is conserved in HIRA orthologues from a wide range of species, from humans to yeast (32). To specifically test whether the B domain is required for HIRA activity, we made a mutant lacking only the B domain (HIRAΔB). As shown in Fig. 4c and d, HIRAΔB was unable to induce either S-phase arrest or histone repression. This suggests that HIRA-induced repression of both histone transcription and S-phase arrest results from a single conserved molecular function of HIRA that is mediated by the evolutionarily conserved B domain.
If HIRA-induced histone mRNA repression is the cause of the S-phase arrest, then histone repression should precede S-phase arrest. To test this, we analyzed the kinetics of the onset of the two events in synchronized cells. U2OS cells were transfected with a plasmid encoding wild-type HIRA and then treated with HU for 20 h. Because HU arrests cells at the G1/S transition and within S phase, this method provides an enrichment of cells at G1/S rather than perfect synchronization (Fig. 5a). The cells were then released from the arrest, and at various times afterward the cells were harvested and the cell cycle distribution and the histone mRNA abundance were determined. As shown in Fig. 5a, mock-transfected cells progressed through S phase and by 12 h were mostly in the G2/M phase of the cell cycle. Expression of histone H2B peaked in mid-S phase. HIRA-expressing cells progressed relatively normally through S phase initially and then arrested between 3 and 6 h into S phase. However, in these cells histone H2B mRNA barely increased even during the first few hours of S phase, when DNA replication was occurring. In contrast, two other genes normally expressed in S phase of the cell cycle, the E2F1 and dihydrofolate reductase genes, were unaffected by ectopic HIRA expression (Fig. 5a and data not shown). Thus, histone mRNA repression is specific and occurs before S-phase arrest.
If repression of histone synthesis is the cause of the S-phase arrest, then it might be possible to detect defects in chromatin structure that result from reduced levels of histones during chromatin assembly. To test this, cells were infected with an adenovirus directing the expression of HIRA or, as a control, LacZ. Twenty-four hours later the cells were harvested and chromatin was prepared and digested with MNase. Since MNase cleaves DNA between nucleosomes (73), perturbations in nucleosome incorporation are expected to increase MNase sensitivity. Chromatin from HIRA-expressing cells was more sensitive to MNase digestion, as indicated by the more rapid degradation of high-molecular-weight DNA in chromatin from HIRA-expressing cells (Fig. 5b, lanes 4 and 5). In addition the extended “nucleosomal ladder” was less apparent in chromatin from HIRA-expressing cells (Fig. 5b, lanes 6 and 7). The difference in MNase sensitivity between chromatin from control and HIRA-expressing cells was subtle but reproducible and comparable to the difference in MNase sensitivities of chromatin from wild-type and histone-deficient yeast (22, 31). To rule out the possibility that the heightened MNase sensitivity of chromatin from HIRA-expressing cells was a consequence of the HIRA-induced enrichment of the cells in S phase of the cell cycle, cells were again infected with adenovirus directing expression of HIRA or LacZ. Six hours after infection the cells were treated for 24 h with HU to arrest them at the G1/S transition and then released into S phase of the cell cycle for 6 h. As shown in Fig. 5d, both populations of cells had DNA contents indicative of predominantly S-phase cells. However, the chromatin from HIRA-expressing cells was, once again, more sensitive to digestion by MNase (Fig. 5c). Taken together these results indicate that chromatin from HIRA-expressing cells is more sensitive to MNase digestion. Although changes in MNase sensitivity might result from other alterations in nuclear structure, this result is entirely consistent with DNA replication occurring in the absence of proper chromatin assembly.
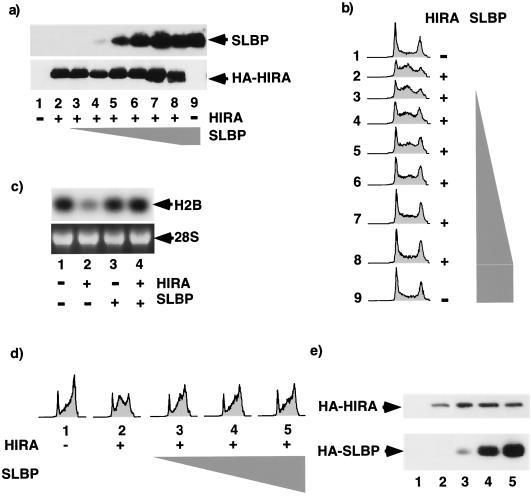
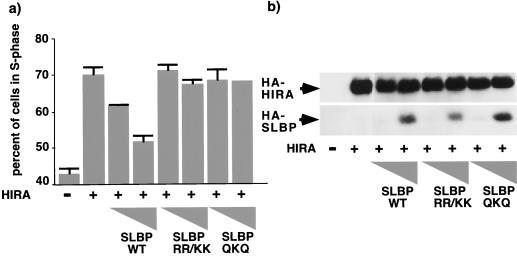
Next we reasoned that, if histone repression is the sole cause of S-phase arrest, then it should be possible to override the S-phase arrest by restoring the level of histones to normal. This was accomplished by coexpressing HIRA with SLBP, which, as shown earlier, stabilizes the residual histone mRNA and overrides HIRA-induced histone mRNA repression (Fig. 1b and c). In addition, ectopic expression of SLBP is likely to promote more-efficient processing of the pre-mRNA (66, 71). U2OS cells were transiently transfected with a plasmid encoding HIRA in the absence or presence of increasing amounts of SLBP. Cells were then harvested for FACS analysis to determine the cell cycle distribution. Expression of HIRA alone caused histone mRNA repression (Fig. 6c, lane 2) and S-phase arrest (Fig. 6b, lane 2). When SLBP was coexpressed with HIRA, it restored histone mRNA levels to normal (Fig. 6c, lane 4) and completely overcame the HIRA-induced S-phase arrest in a dose-dependent manner (Fig. 6b, lanes 3 to 8). In addition, coexpression of SLBP was able to override HIRA-induced S-phase arrest in synchronized cells that were first arrested in mimosine and then released into S phase of the cell cycle (Fig. 6d and e). Twelve hours after release from mimosine-induced arrest most of the control cells were in G2/M phase of the cell cycle. In contrast, most of the HIRA-expressing cells were arrested in S phase. Coexpression of SLBP with HIRA largely abolished the S-phase arrest. There was no effect of SLBP alone on cell cycle progression in synchronized or unsynchronized cells (Fig. 6b and data not shown). Since SLBP has not been reported to have any activity other than control of histone mRNA metabolism, it seems likely that the ability of SLBP to override the HIRA-induced S phase is a consequence of its effect on histone mRNA abundance. However, as confirmation of this we tested the ability of two subtle SLBP mutants that, as Dominski and coworkers have shown, do not bind to histone mRNAs (13) to override HIRA-induced S-phase arrest. Although both mutants were expressed equivalently to the wild-type protein, neither mutant was able to override the arrest (Fig. 7). Therefore, this result very strongly indicates that HIRA-induced S-phase arrest is a consequence of reduced histone gene expression.
FIG. 6.
HIRA-induced S-phase arrest is overridden by the histone mRNA-stabilizing protein, SLBP. (a) U2OS cells were transiently transected with pcDNA3 (lanes 1 and 9) or pcDNA3 HA-HIRA WT (lanes 2 to 8) in the absence or presence of pcDNA3 HA-SLBP (0.01, 0.1, 0.25, 0.5, 1, or 3 μg [triangle]) together with pCMV-CD19. Total-cell lysates were prepared and Western blotted with antibodies to SLBP or HA-HIRA. (b) Same as panel a except that the cell cycle distribution of CD19+ cells was determined. (c) RNA was prepared from transfected CD19+ cells and Northern blotted to determine abundance of histone H2B. (d) U2OS cells were transiently transfected with pcDNA3 or pcDNA3 HA-HIRA WT in the absence or presence of pcDNA3 HA-SLBP (3, 8, or 18 μg [triangle]) together with pCMV-CD19. The cells were arrested in G1 phase with mimosine and then released into S phase. The cell cycle distribution of the CD19+ cells was determined 12 h after release. (e) Cell lysates from panel d were Western blotted with anti-HA antibodies.
FIG. 7.
Override of HIRA-induced S-phase arrest by SLBP requires binding of SLBP to 3′ UTR of histone mRNA. (a) U2OS cells were transiently transfected with pcDNA3 or pcDNA3 HA-HIRA WT in the absence or presence of pcDNA3 HA-SLBP WT, pcDNA3 HA-SLBP RR/KK, or pcDNA3 HA-SLBP QKQ (0.2 or 1 μg [triangles]) together with pCMV-CD19. Cells were harvested, and the cell cycle distribution was determined by FACS. (b) Lysates were Western blotted with antibodies to HA.
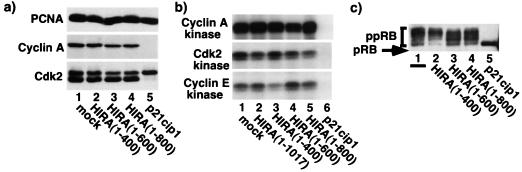
Since activation of DNA synthesis at the G1/S transition requires activation of cyclin/cdk2 activity, next we wanted to test whether inhibition of DNA synthesis due to repression of histone synthesis depends on inhibition of cyclin/cdk2 activity. To do this, we asked whether ectopic expression of wild-type HIRA (or active mutants; Fig. 4) affected a number of markers of cyclin/cdk2 activity. As shown in Fig. 8, ectopic expression of HIRA had no significant effect on expression of cdk2 or cyclin A. Similarly, HIRA did not affect cdk2-, cyclin A-, or cyclin E-associated kinase activity or pRB phosphorylation. In contrast, ectopic expression of the cyclin/cdk2 inhibitor p21cip1 inhibited cyclin/cdk2 activity and pRB phosphorylation as expected. Interestingly, ectopic expression of HIRA had no effect on other markers of normal S-phase progression, such as PCNA expression and phosphorylation of cdk2 on T160 (the faster-migrating form of cdk2 in Fig. 8a). Taken together, these results indicate that HIRA-induced inhibition of DNA synthesis is not accompanied by inhibition of cyclin/cdk2 activity.
FIG. 8.
HIRA-induced S-phase arrest is independent of changes in cyclin/cdk2 activity. (a) U2OS cells were transiently transfected with pcDNA3 (lane 1), pcDNA3 encoding the indicated HIRA deletion mutants (lanes 2 to 4), or pRC-CMVp21cip1 (lane 5) together with pCMV-CD19. The transfected CD19+ cells were collected, and lysates were prepared and Western blotted with anti-PCNA, anti-cyclin A, and anti-cdk2 as indicated. (b) Same as panel a except lysates were used to assay cdk2, cyclin A, and cyclin E by immunoprecipitation-kinase assay. (c) Same as panel a except cell lysates were Western blotted with anti-pRB.
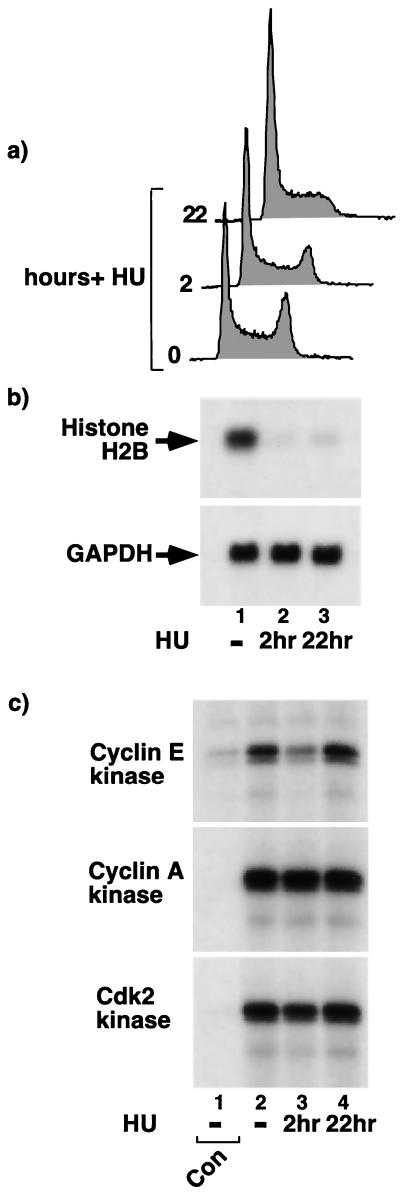
Previous studies showed that inhibition of DNA synthesis with HU triggers a concerted destabilization of histone mRNAs (4, 25, 62, 68). Although the mechanism underlying destabilization is unknown, this shows that sustained histone synthesis depends on continued DNA synthesis. Conversely, here we have shown that sustained DNA synthesis depends on continued histone synthesis. Together, these results establish that DNA synthesis and histone synthesis are dependent on each other. In view of our results showing that HIRA-induced inhibition of histone synthesis and DNA synthesis is independent of changes in cyclin/cdk2 activity, we tested whether HU-induced inhibition of DNA synthesis and histone synthesis is also independent of changes in cyclin/cdk2 activity. Asynchronously growing U2OS cells were treated without or with HU for 2 or 22 h. Within 2 h of HU treatment there was a discernible decrease in the number of cells in G2/M phase of the cell cycle (Fig. 9a). By 22 h there was a very marked decrease in the G2/M compartment and a corresponding increase in the G1 population. As expected, and as shown in Fig. 9b, HU treatment caused a marked decrease in the steady-state abundance of histone mRNAs within 2 h. Abundance of histone mRNAs remained repressed for at least 22 h. In contrast, HU had no effect on cyclin A/cdk2 activity at either 2 or 22 h after addition (Fig. 9c). Interestingly, although HU caused a transient inhibition of cyclin E/cdk2 activity at 2 h, by 22 h after HU addition cyclin E/cdk2 activity levels returned to normal. Consistent with HU transiently inhibiting cyclin E/cdk2 is a previous report showing that HU treatment for 12 h or less inhibited cdk2 (in a cdk2 immunoprecipitate) and its activator cdc25A (47), and another report showing that activity of cyclin A/cdk2 and E/cdk2 isolated from cells arrested with HU for 20 or 40 h is higher than the activity of the same complexes in G1 phase (44). The observed transient inhibition of cyclin E/cdk2 activity by HU is similar to the transient inhibition of this kinase by other forms of genotoxic stress, such as UV light and ionizing radiation (18, 41). Taken together, these results suggest that, although a transient inhibition of cyclin E/cdk2 activity might contribute to initial repression of histone mRNA abundance, inhibition of cyclin/cdk2 activity does not account for the repression of histone gene expression caused by prolonged HU treatment. After prolonged treatment with HU, DNA synthesis and histone synthesis are both repressed but cyclin/cdk2 activity is elevated.
FIG. 9.
HU-induced histone mRNA repression is independent of changes in cyclin/cdk2 activity. (a) U2OS cells were treated with 2 mM HU for 0, 2, or 22 h as indicated and then harvested and processed for FACS analysis to determine the cell cycle distribution of the cells. (b) Cells were treated as in panel a, and total cellular RNA was prepared and Northern blotted to determine the abundances of the histone H2B and GAPDH mRNAs. (c) Cells were treated as in panel a, lysates were prepared and processed for immunoprecipitation-kinase assay with antibodies to cyclin E (HE111; top), cyclin A (C160; middle), and cdk2 (M2; bottom) as indicated. In each case, lane 1 is derived from extract immunoprecipitated with an appropriate negative-control antibody (mouse 9E10 for HE111 and C160 and purified rabbit anti-mouse polyclonal for M2).
DISCUSSION
These results provide the first experimental evidence to suggest that human HIRA, like Hir1p and Hir2p in yeast, is a cell cycle-regulated repressor of histone gene transcription (Fig. 1 to 3). First, the fact that ectopic expression of HIRA represses histone gene transcription is obviously consistent with the idea that it normally functions to repress transcription of these genes in vivo. Second, the observation that HIRA colocalizes with a known transcriptional repressor, HDAC4, in G1 phase of the cell cycle is also consistent with HIRA being a transcriptional repressor molecule. The fact that HIRA activity specifically required the evolutionarily conserved B domain (Fig. 4) is consistent with the idea that transcriptional repression and S-phase arrest depend on the interaction of HIRA with a conserved HIRA effector protein (32). However, while these data are consistent with HIRA being a repressor of histone gene transcription in vivo, they by no means prove this, and future studies will address this issue in more detail. A major question concerns the role of the HIRA/HDAC4 foci. Although these foci are consistent with a shared role of HIRA and HDAC4 in transcription repression, in G1-phase cells these foci do not colocalize with NPAT-containing foci, which are thought to be located at the histone gene cluster on chromosome 6 (39, 74). This clearly argues against a direct role for these foci in repression of histone gene transcription. The visible HIRA/HDAC4 foci might represent sites of assembly of active repressor complexes that subsequently relocalize to histone genes, or they may indicate that HIRA has functions unrelated to control of histone gene expression.
Previously we showed that HIRA is an in vivo substrate of a cyclin/cdk2 complex and that it is phosphorylated by cyclin A/cdk2 or E/cdk2 in S phase of the cell cycle (21). Moreover, De Lucia and coworkers showed that HIRA is phosphorylated during mitosis (11). The role of HIRA phosphorylation is unknown at present. It is tempting to speculate that phosphorylation regulates the subcellular localization of HIRA and/or HIRA-dependent transcriptional repression of histone genes. Consistent with the former idea, De Lucia and coworkers showed that the mitotic phosphorylation of HIRA was accompanied by release of the protein from chromatin (11). However, understanding the role of phosphorylation is complicated by the fact that HIRA contains 15 potential cyclin/cdk2 phosphorylation sites. We found that mutation of 13 of these to nonphosphorylatable residues specifically or nonspecifically destabilizes the protein (data not shown).
Regardless of the precise role of HIRA/HDAC4 foci and HIRA phosphorylation, the major conclusion of this study concerns the coordination of DNA synthesis and histone synthesis. Several lines of evidence show that HIRA-induced repression of histone gene transcription triggers S-phase arrest. First, there was a perfect correlation between the ability of HIRA mutants to repress histone gene expression and arrest in S phase. This suggests that both activities depend on the same molecular function of HIRA and is consistent with histone repression being the cause of S-phase arrest or vice versa. However, if S-phase arrest caused decreased histone mRNA abundance, then we would expect to see decreased histone mRNA stability, as shown previously (4, 25, 62). We did not observe such a decrease. Second, we failed to detect any other biochemical defect in HIRA-expressing cells that might account for S-phase arrest. Ectopic expression of HIRA did not affect other markers of normal S-phase progression, including expression and/or phosphorylation of pRB, cdk2, PCNA, E2F1, dihydrofolate reductase, and cyclin A, and cyclin A/cdk2 or E/cdk2 activity (Fig. 5 and 8). Moreover, ectopic expression of HIRA did not affect the level of expression of housekeeping genes, such as the GAPDH gene (Fig. 1). Taken together, these results strongly suggest that the effect of HIRA on histones is relatively specific and that S-phase arrest is a direct consequence of histone mRNA repression rather than an indirect consequence of gross perturbation of nuclear function. Third, histone gene repression preceded S-phase arrest. Fourth, cells arrested in S phase by HIRA contained chromatin that was markedly and reproducibly more sensitive to digestion by MNase. This is consistent with depletion of “free” histones and an inability to properly assemble chromatin being the causes of S-phase arrest. Fifth, restoration of histone mRNA abundance to normal levels by coexpression of the histone mRNA regulator, SLBP, overrode the S-phase arrest, and this required the binding of SLBP to the stem-loop of the histone mRNA 3′ UTR. Taken together, multiple lines of evidence very strongly support the conclusion that completion of DNA synthesis requires continued histone synthesis.
Conversely, it has been shown previously that inhibition of DNA synthesis with HU or AraC causes rapid destabilization of histone mRNAs (4, 25, 62). Thus sustained histone synthesis and DNA synthesis are dependent on each other during S phase. The molecular mechanism underlying the HU-induced histone mRNA destabilization has been elusive. Here we show that concerted inhibition of DNA synthesis and histone synthesis, caused by histone repression and inhibition of DNA synthesis, respectively, are related in that inhibition occurs without a prolonged inhibition of cyclin/cdk2 activity. Thus, although cyclin/cdk2 activity is rate-limiting for activation of both DNA and histone synthesis at the start of S phase, both processes can be inhibited in a cdk2-independent manner during S phase. Interestingly, and in contrast to inhibition of DNA synthesis at the end of a normal S phase, HU-induced histone mRNA destabilization was not accompanied by degradation of SLBP (71). Taken together, the evidence is consistent with a cyclin/cdk2- and SLBP-dependent pathway that regulates histone mRNA stability in a normal S phase and a distinct pathway that triggers mRNA destabilization in response to replication stress.
This is, to our knowledge, the first demonstration that completion of DNA synthesis requires histone synthesis. Moreover, it could not have been easily predicted that inhibition of histone synthesis would block DNA synthesis for two reasons. First, in vitro replication of plasmid DNA in mammalian cell extracts or Drosophila melanogaster embryo extracts does not require assembly of the newly replicated DNA into chromatin (7, 63). In both of these systems presynthesized histones are already present in cell extracts or added exogenously, and the replication of a plasmid in an extract is obviously significantly different from replication of a whole genome in an intact cell. However, despite these differences, if DNA synthesis is independent of chromatin assembly, there is no reason to believe it is dependent on synthesis of histones, the essential precursors to chromatin assembly. Second, yeast expressing histones from conditional promoters can seemingly replicate their entire genomes even when new histone synthesis is repressed (22, 31). In these studies, yeast lacking either histone H2B or histone H4 appeared to arrest in G2 phase after DNA replication, rather than in S phase itself. Although it is possible that the arrest was in late S phase rather than G2, this was not apparent from the data, and the authors concluded that a full round of replication occurred after repression of histone synthesis. Similarly, Drosophila embryos with hypomorphic alleles of the dSLBP gene and dramatically reduced levels of histone mRNA have no obvious defect in DNA synthesis but display failures in chromosome condensation that block normal anaphase (66). Thus we have shown for the first time that completion of DNA synthesis in intact mammalian cells requires ongoing histone synthesis and/or chromatin assembly. Taken together, our data and the published data are consistent with the idea that in intact mammalian somatic cells tight controls operate to coordinate DNA synthesis and histone synthesis such that these processes are dependent on each other. This may not be the case in lower eukaryotes, mammalian-cell extracts, or developing embryos with simpler cell cycles. Consistent with this idea, the eggs, oocytes, and embryos of a variety of species build up large pools of free histones and to do so must undergo a large amount of histone synthesis in the absence of DNA synthesis, indicating that, in contrast to what is found for mammalian somatic cells, histone synthesis is not dependent on DNA synthesis (68).
Since both histone gene expression and DNA synthesis are, like most S-phase events, thought to be triggered directly or indirectly by cyclin/cdk2 activity (17, 39, 74), it is interesting that concerted regulation of these two processes during S phase should be independent of cyclin/cdk2 activity. We speculate that the reason for this stems from the fundamental role played by cyclin/cdk2 activity in cell cycle control. In eukaryotes S-phase cyclin/cdk activity not only activates DNA synthesis but also inhibits relicensing of replication origins and, therefore, rereplication within S phase (5). If a block to histone synthesis or chromatin assembly blocked DNA synthesis through inhibition of cyclin/cdk2 activity, this would be expected to promote the relicensing of replication origins and, potentially, rereplication of portions of the genome when replication resumed. This would contribute to genome instability.
What is the cdk2-independent mechanism that underlies concerted control and mutual dependence of DNA and histone synthesis? As has been proposed previously, the cell might monitor the size of the pool of free histones (8). According to this model, when DNA replication is inhibited, the pool of free histones rises and represses histone gene expression through an autoregulatory mechanism. Conversely, when histone synthesis is inhibited, the pool of free histones falls and DNA replication is inhibited. Consistent with this idea, expression of histones in yeast is subject to autoregulation. That is, elevated levels of histones H2A and H2B trigger a negative feedback that reduces histone transcription of the HTA1-HTB1 locus that codes for these two histones. Interestingly, this autoregulation requires the yeast homologues of human HIRA, Hir1p and Hir2p (48, 51, 55). Alternatively, stalled replication forks induced by HU might trigger repression of histone synthesis through the same cell cycle checkpoint pathways that are known to mediate viable cell cycle arrest in HU (1, 75). Conversely, when DNA replication occurs in the absence of histones, accumulation of unusually long stretches of newly replicated DNA that are not fully incorporated into nucleosomes may inhibit further DNA synthesis through a putative “chromatin assembly checkpoint.” This too might depend on known cell cycle checkpoint pathways. Current efforts are directed toward addressing these various possibilities.
Whatever the monitoring mechanism, this concerted regulation and mutual dependence of DNA and histone synthesis may ensure that free histones and DNA are both synthesized at the same rate. Since newly synthesized DNA is quickly incorporated into nucleosomes, this would account for the observed small pool of free histones present in somatic mammalian cells (68). The ability to coordinate the relative rates of DNA synthesis and histone synthesis in S phase facilitates the ordered assembly of chromatin, the physiological substrate for most nuclear processes.
Acknowledgments
This work was funded by NIH grant RO1-GM62281. J.W.H. was supported by Public Health Service grant GM54137. T.J.Y. was supported by NIH grants CA75138 and GM44762.
We thank William Marzluff for pSP64T β-globinSLBP and anti-SLBP antibody and Ken Zaret, Randy Strich, Erica Golemis, Maureen Murphy, Paul Kaufman, and colleagues at Fox Chase Cancer Center for advice during the course of this work.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albig, W., P. Kioschis, A. Poustka, K. Meergans, and D. Doenecke. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314-322. [DOI] [PubMed] [Google Scholar]
- 4.Baumbach, L. L., G. S. Stein, and J. L. Stein. 1987. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry 26:6178-6187. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 6.Bond, U., and T. A. Yario. 1994. The steady state levels and structure of the U7 snRNP are constant during the human cell cycle: lack of cell cycle regulation of histone mRNA 3′ end formation. Cell. Mol. Biol. Res. 40:27-34. [PubMed] [Google Scholar]
- 7.Bulger, M., T. Ito, R. T. Kamakaka, and J. T. Kadonaga. 1995. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc. Natl. Acad. Sci. USA 92:11726-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, W. B., and G. C. Mueller. 1973. Control of histone synthesis in HeLa cells. Biochim. Biophys. Acta 294:481-496. [DOI] [PubMed] [Google Scholar]
- 9.Connell-Crowley, L., S. J. Elledge, and J. W. Harper. 1998. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr. Biol. 8:65-68. [DOI] [PubMed] [Google Scholar]
- 10.DeLisle, A. J., R. A. Graves, W. F. Marzluff, and L. F. Johnson. 1983. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol. Cell. Biol. 3:1920-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Lucia, F., S. Lorain, C. Scamps, F. Galisson, J. MacHold, and M. Lipinski. 2001. Subnuclear localization and mitotic phosphorylation of HIRA, the human homologue of Saccharomyces cerevisiae transcriptional regulators Hir1p/Hir2p. Biochem. J. 358:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 13.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 21:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominski, Z., J. A. Erkmann, X. Yang, R. Sanchez, and W. F. Marzluff. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 16:58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 16.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewen, M. E. 2000. Where the cell cycle and histones meet. Genes Dev. 14:2265-2270. [DOI] [PubMed] [Google Scholar]
- 18.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 19.Gick, O., A. Kramer, A. Vasserot, and M. L. Birnstiel. 1987. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. USA 84:8937-8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard, F., U. Strausfeld, A. Fernandez, and N. J. Lamb. 1991. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell 67:1169-1179. [DOI] [PubMed] [Google Scholar]
- 21.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, M., M. Chang, U. J. Kim, and M. Grunstein. 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48:589-597. [DOI] [PubMed] [Google Scholar]
- 23.Harper, J. W., and P. D. Adams. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511-2526. [DOI] [PubMed] [Google Scholar]
- 24.Harris, M. E., R. Bohni, M. H. Schneiderman, L. Ramamurthy, D. Schumperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintz, N., H. L. Sive, and R. G. Roeder. 1983. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 3:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann, I., and M. L. Birnstiel. 1990. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature 346:665-668. [DOI] [PubMed] [Google Scholar]
- 27.Hunter, T., and J. Pines. 1994. Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell 79:573-582. [DOI] [PubMed] [Google Scholar]
- 28.Kao, G. D., W. G. McKenna, and T. J. Yen. 2001. Detection of repair activity during the DNA damage-induced G2 delay in human cancer cells. Oncogene 20:3486-3496. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 31.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirov, N., A. Shtilbans, and C. Rushlow. 1998. Isolation and characterization of a new gene encoding a member of the HIRA family of proteins from Drosophila melanogaster. Gene 212:323-332. [DOI] [PubMed] [Google Scholar]
- 33.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krek, W., and J. A. DeCaprio. 1995. Cell synchronization. Methods Enzymol. 254:114-124. [DOI] [PubMed] [Google Scholar]
- 35.Lorain, S., S. Demczuk, V. Lamour, S. Toth, A. Aurias, B. A. Roe, and M. Lipinski. 1996. Structural organization of the WD repeat protein-encoding gene HIRA in the DiGeorge syndrome critical region of human chromosome 22. Genome Res. 6:43-50. [DOI] [PubMed] [Google Scholar]
- 36.Lorain, S., J. P. Quivy, F. Monier-Gavelle, C. Scamps, Y. Lecluse, G. Almouzni, and M. Lipinski. 1998. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 18:5546-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luscher, B., and D. Schumperli. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luscher, B., C. Stauber, R. Schindler, and D. Schumperli. 1985. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc. Natl. Acad. Sci. USA 82:4389-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, T., B. A. V. Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnaghi, P., C. Roberts, S. Lorain, M. Lipinski, and P. J. Scambler. 1998. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 20:74-77. [DOI] [PubMed] [Google Scholar]
- 41.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 42.Marzluff, W. F., and N. B. Pandey. 1988. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem. Sci. 13:49-52. [DOI] [PubMed] [Google Scholar]
- 43.Mathews, M. B., R. M. Bernstein, B. R. Franza, Jr., and J. I. Garrels. 1984. Identity of the proliferating cell nuclear antigen and cyclin. Nature 309:374-376. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto, Y., K. Hayashi, and E. Nishida. 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 9:429-432. [DOI] [PubMed] [Google Scholar]
- 45.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyake, S., W. R. Sellers, M. Safran, X. Li, W. Zhao, S. R. Grossman, J. Gan, J. A. DeCaprio, P. D. Adams, and W. G. Kaelin, Jr. 2000. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 20:8889-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molinari, M., C. Mercurio, J. Dominguez, F. Goubin, and G. F. Draetta. 2000. Human Cdc25 A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran, L., D. Norris, and M. A. Osley. 1990. A yeast H2A-H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 4:752-763. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 51.Osley, M. A., and L. M. Hereford. 1981. Yeast histone genes show dosage compensation. Cell 24:377-384. [DOI] [PubMed] [Google Scholar]
- 52.Pagano, M., R. Pepperkok, F. Verde, W. Ansorge, and G. Draetta. 1992. Cyclin A is required at two points in the human cell cycle. EMBO J. 11:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray-Gallet, D., J.-P. Quivy, C. Scamps, E. M.-D. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 55.Recht, J., B. Dunn, A. Raff, and M. A. Osley. 1996. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resnitzky, D., M. Gossen, H. Bujard, and S. I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resnitzky, D., and S. I. Reed. 1995. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol. 15:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridgway, P., and G. Almouzni. 2001. Chromatin assembly and organization. J. Cell Sci. 114:2711-2712. [DOI] [PubMed] [Google Scholar]
- 59.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 60.Sharp, J. A., A. A. Franco, M. A. Osley, P. D. Kaufman, D. C. Krawitz, T. Kama, E. T. Fouts, and J. L. Cohen. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherwood, P. W., S. V. Tsang, and M. A. Osley. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sittman, D. B., R. A. Graves, and W. F. Marzluff. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 80:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, S., and B. Stillman. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15-25. [DOI] [PubMed] [Google Scholar]
- 64.Spector, M. S., A. Raff, H. DeSilva, K. Lee, and M. A. Osley. 1997. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stauber, C., and D. Schumperli. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050-2054. [DOI] [PubMed] [Google Scholar]
- 68.van Holde, K. E. 1988. Chromatin. Springer-Verlag KG, Berlin, Germany.
- 69.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Z. F., M. L. Whitfield, T. C. Ingledue III, Z. Dominski, and W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 71.Whitfield, M. L., L. X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wimmel, A., F. C. Lucibello, A. Sewing, S. Adolph, and R. Muller. 1994. Inducible acceleration of G1 progression through tetracycline-regulated expression of human cyclin E. Oncogene 9:995-997. [PubMed] [Google Scholar]
- 73.Zaret, K. 1999. Micrococcal nuclease analysis of chromatin structure, p. 21.1.1-21.1.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. John Wiley & Sons Inc., New York, N.Y. [DOI] [PubMed]
- 74.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 76.Zindy, F., E. Lamas, X. Chenivesse, J. Sobczak, J. Wang, D. Fesquet, B. Henglein, and C. Brechot. 1992. Cyclin A is required in S phase in normal epithelial cells. Biochem. Biophys. Res. Commun. 182:1144-1154. [DOI] [PubMed] [Google Scholar]