Abstract
DNA methylation plays an important role in transcriptional repression. To gain insight into the dynamics of demethylation and de novo methylation, we introduced a proviral reporter, premethylated at different densities, into a defined chromosomal site in murine erythroleukemia cells and monitored the stability of the introduced methylation and reporter gene expression. A high density of methylation was faithfully propagated in vivo. In contrast, a low level of methylation was not stable, with complete demethylation and associated transcriptional activation or maintenance-coupled de novo methylation and associated silencing occurring with equal probability. Deletion of the proviral enhancer increased the probability of maintenance-coupled de novo methylation, suggesting that this enhancer functions in part to antagonize such methylation. The DNA methyltransferases (MTases) Dnmt3a and Dnmt3b are thought to be the sole de novo MTases in the mammalian genome. To determine whether these enzymes are responsible for maintenance-coupled de novo methylation, the unmethylated or premethylated proviral reporter was introduced into DNA MTase-deficient embryonic stem cells. These studies revealed the presence of a Dnmt3a/Dnmt3b-independent de novo methyltransferase activity that is stimulated by the presence of preexisting methylation.
DNA methylation is essential for mammalian development (27, 37), playing an important role in maintaining transcriptional silencing of genes on the inactive X chromosome, imprinted genes, and parasitic elements (2, 5). In mammals, DNA methylation occurs predominantly on cytosines (m5C) in the context of the 5′-CpG-3′ dinucleotide, and this epigenetic mark is propagated on both parent and nascent strands after DNA replication. Recent experiments suggest that DNA methylation may serve as a tag for the recruitment of methyl DNA binding domain proteins and the histone deacetylase complexes with which they interact (24, 35) to generate a chromatin structure that is repressive for transcription (33). However, why some genes or CpG sites are susceptible to methylation while others remain methylation free remains to be determined.
Using in vitro-methylated constructs and transient-transfection assays, a number of studies have shown that enhancers and associated transcription factor complexes play an important role in overcoming the repressive effects of methylation (25, 51). Transcription of an α-globin reporter construct methylated at low density, for example, only occurs in the presence of an enhancer (6, 7), but even in the presence of this enhancer, dense methylation prevents transcription. Similarly, the degree of repression of an episomal Rous sarcoma virus promoter construct is correlated with increasing methylation density, with a high level of methylation extinguishing expression completely (20). While these experiments reveal the antagonistic relationship between DNA methylation density and transcriptional activity, the methods used preclude analysis of the dynamic relationship between transcription and propagation of the methylation imprint. Furthermore, while Dnmt1 is regarded as the “maintenance” DNA methyltransferase (MTase), based on its preference for hemimethylated CpGs (3), and the Dnmt3a and Dnmt3b MTases show strong “de novo” activity (38), the interplay among these enzymes in vivo is not well understood (28, 42).
Using a proviral construct, we showed previously that a high density of methylation is stably propagated in vivo (33). In contrast, we show here that in the same integration site, a low density of proviral methylation is inherently unstable, with daughter cells harboring proviral cassettes that are completely demethylated and transcriptionally active or de novo methylated and transcriptionally silent. Elaboration of these distinct states occurs stochastically but with equal probability. Deletion of the Moloney murine leukemia virus 5′ long terminal repeat (LTR) enhancer decreases the probability of demethylation, suggesting that the enhancer antagonizes maintenance of the epigenetic imprint.
The observed de novo methylation is dependent upon existing methylation, since it does not occur on initially unmethylated templates. To address whether Dnmt3a and/or Dnmt3b is solely responsible for the methylation-dependent de novo methylation, unmethylated and low density methylated provirus was introduced into an embryonic stem (ES) cell line in which the catalytic domains of both of these MTases have been deleted genetically (37). Surprisingly, de novo methylation of the provirus was still detected, particularly in the presence of preexisting methylated sites, suggesting the existence of a Dnmt3a/b-independent de novo MTase activity in mammalian cells.
MATERIALS AND METHODS
Generation and in vitro methylation of L1-MFGhGFP-1L constructs.
The Moloney murine leukemia virus-based construct L1-MFGhGFP-1L which includes the humanized green fluorescent protein (hGFP)-encoding proviral genome flanked by mouse genomic sequence (11) and loxP sites, was derived from the proviral vector MFGhGFP (1, 32), as described previously (33). To generate a 5′LTR enhancer deletion (Δenh) construct, an XhoI/BglII fragment containing the 5′LTR of the MFGhGFP vector was cloned into the vector SL1180 (Pharmacia), generating SL1180-5′LTR. This construct was digested with NheI and XbaI and self-ligated, yielding a truncated 5′LTR lacking the tandem repeat enhancer and upstream region of U3 (Fig. 1). The XhoI/BglII fragment from this construct was subsequently cloned into the L1-MFGhGFP-1L vector, generating L1−ΔenhMFGhGFP-1L.
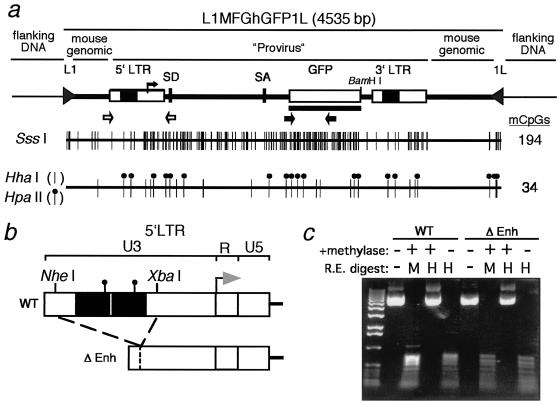
FIG. 1.
In vitro methylation and targeting of L1-MFGhGFP-1L. (a) Map of the L1-MFGhGFP-1L cassette, including the proviral LTRs, splice donor (SD), splice acceptor (SA), GFP gene, loxP sites (L1 and 1L), and flanking mouse genomic DNA. Primers specific for the 5′LTR (open arows) and GFP coding sequence (solid arrows) used for bisulfite sequencing, and the GFP probe (black line) used for Southern analysis are also shown. Flanking plasmid DNA includes vector sequence flanking the L1-HyTK-1L vector at the RL5 integration site. CpG sites methylated in vitro with SssI (194 sites) or HpaII and HhaI (34 sites) methylases are also shown. (b) Map of the 5′LTRs of the wild-type and 5′ LTR enhancer deletion (Δenh) L1-MFGhGFP-1L constructs. The transcription start site (arrow), U3, R, and U5 regions are shown, along with the NheI and XbaI restriction sites used to generate the deletion. The HpaII sites within the deleted tandem enhancer (solid squares) are also shown. (c) Methylation state of the unmethylated (−) or HpaII and HhaI methylated (+) wild-type and Δenh constructs was determined by digestion with the methylation-sensitive restriction enzyme HpaII (H) and the methylation-insensitive restriction enzyme MspI (M). Digestion products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
In vitro methylation of the Δenh and wild-type L1-MFGhGFP-1L constructs with SssI methylase (NEB), which methylates all CpGs, was performed as described previously (45). In vitro “partial” methylation of these constructs with HpaII and HhaI methylases (NEB) was performed simultaneously according to the manufacturer's protocol for HhaI methylation. To determine whether the reactions were carried to completion, following organic extraction and ethanol precipitation, methylated DNA was digested with the methylation-sensitive restriction enzymes HpaII and HhaI or the methylation-insensitive enzyme MspI and visualized by electrophoresis on a 0.7% agarose gel.
Tissue culture, transfection, and selection.
MEL cells were cultured in growth medium as described previously (33). Approximately 4 × 106 cells were electroporated in the presence of 15 μg of CMV-CRE, 100 μg of sonicated salmon sperm DNA, and 25 μg of L1-MFGhGFP-1L plasmid, as previously described (45). After 3 days in nonselective medium, the cultures were supplemented with 10 μM ganciclovir and cultured for 7 days to select against cells expressing the hygromycin-thymidine kinase (HYTK) fusion protein. Ganciclovir-resistant cells were cloned by limiting dilution and screened for Cre-mediated exchange by Southern blot. For all experiments, greater than 80% of the clones analyzed contained a cassette integrated in one of the two possible orientations at the RL5 genomic site.
Dnmt1c/c−/− (c stands for disruption of the catalytic domain) (26), Dnmt2−/− (39), Dnmt3a/b−/− double-knockout (37), and wild-type J1 ES cells were cultured in the presence of leukemia inhibitory factor, as described previously (27), with the exception of the omission of feeder cells. ES cells were electroporated with 20 μg of linearized proviral vector and 5 μg of supercoiled BSKSIIβactinhis (encoding the selectable marker gene histidinol dehydrogenase [hisD]) at 250 V and 500 μF using a BTX 300 electroporator. After 48 h in culture, 3 mM l-histidinol (Sigma) was added to the electroporated cells. Histidinol-resistant clones were pooled after 10 to 12 days of selection and cultured for further analyses in the absence of histidinol.
Flow cytometry and cell sorting.
For flow cytometry analysis, MEL (32) or ES (27) cells were harvested as described previously and resuspended in staining medium (phosphate-buffered saline [PBS] supplemented with 3% calf serum) supplemented with 1 μg of propidium iodide per ml for live/dead discrimination. Data were collected on a FACSCalibur (Becton Dickinson) equipped with the standard fluorescein filter set. Data were collected on a minimum of 10,000 live cells, and fluorescence distribution was determined with FlowJo software (Treestar). MEL cells were sorted by electronic gating based on GFP fluorescence, using a Vantage cell sorter (Becton Dickinson).
Southern blot hybridization and methylation blotting.
Preparation of high-molecular-weight wild-type genomic DNA, restriction digests, membrane transfers, and preparation of the DNA probe were performed as described previously (33). The GFP probe used for Southern hybridization was generated by digestion of MFGhGFP with NcoI and BamHI, yielding a restriction fragment including the 720-bp hGFP gene. Clones harboring a single-copy integrant at the RL5 integration site were identified by digestion of genomic DNA with BglII, which cuts once in the MFGhGFP provirus, followed by Southern blot analysis with the GFP probe.
The methylation status of the provirus was determined by digestion of 5 to 10 μg of genomic DNA with BamHI alone or in combination with the methylation-sensitive enzyme HpaII. To estimate the proviral copy number in transfected ES cells, genomic DNA was digested with NcoI and XbaI (both of which cut within the proviral construct, yielding a 1.1-kb fragment, including the GFP gene) and subjected to Southern blot hybridization with the GFP probe. Subsequently, the blot was stripped and reprobed with a fragment specific for a 1.8-kb fragment of the endogenous β-globin locus.
Bisulfite analysis.
Bisulfite conversion was carried out, with minor modifications, by the protocol of Clark et al. (10), as described previously (32). Briefly, mixtures containing 5 μl of bisulfite-treated DNA (50 μl final volume) were subjected to 27 to 30 amplification cycles with a GeneAmp PCR system 9700 (Perkin Elmer), with denaturation at 94°C, annealing at 49 to 56°C, and extension at 72°C. Nested or seminested amplification was performed with 2 μl of product from the first round in a 50-μl reaction. Primers were designed to favor amplification of bisulfite-converted DNA. If the template strand included a CpG, degeneracy was incorporated in the primer at the nucleotide position corresponding to the cytosine so that no bias for amplification of methylated template was introduced. Primers used for the 5′LTR were bis+25+ (TAGGTTTGGTAAGTTAGTTTAAGTAAYGTT), bis+1080− (TAAAAAAATAATAACAAACTAACCCRAAC), bis+25+ (TTGTAAGGTATGGAAAAATATATAATTG), and bis+665− (TAAATTACTAACCAACTTACCTCCCRATAA) in the second round. Primers used for the GFP gene were GFP+1870+ (ATTATTTTCTAGATTGTTATGGTGAGTAAGGG) and GFP+2300− (CTCAAGCTTATAATTATACTCCAACTTATACCCCA) in the first round and GFP+1903+ (GAGGAGTTGTTTATYGGGGTGGTGTTT) and GFP+2300− in the second round.
RT-PCR analysis.
Total RNA was isolated with the Trizol reagent (Gibco-BRL) according to the manufacturer's protocol. Superscript II reverse transcriptase was used for first-strand cDNA synthesis, and PCR was conducted according to the manufacturer's protocol (Gibco-BRL) with 10 ng of cDNA template. Reverse transcription (RT)-PCRs for the MTases (primer sequences available on request) were conducted in a 32-cycle PCR with a 59°C annealing temperature. MFGhGFP-specific primers +548LTR (TCTGAGTGATTGACTACCCGTC) and RhoGFP#2 (GCAAGCTGCCCGTGCCCT), which yield a product of 563 bp from the spliced proviral cDNA template, were used in a 28-cycle PCR. β-Actin-specific primers (Promega) G581A (TCATGAAGTGTGACGTTGACATCCGT) and G588A (CTTAGAAGCATTTGCGGTGCACGATG) were used in a 24-cycle PCR. The last two reactions were conducted with a 65°C annealing temperature.
RESULTS
Previously, we used the Cre/loxP-based recombination system recombinase-mediated cassette exchange (RMCE) (14, 15) to introduce a single-copy provirus encoding the green fluorescent protein (GFP), either unmethylated or methylated in vitro at all CpG sites, into the RL5 genomic site in MEL cells (33). While the methylated provirus remained densely methylated and transcriptionally silent, the unmethylated provirus remained methylation free and transcriptionally active with long-term passage. The faithful propagation of DNA methylation and the associated heritability of proviral silencing led us to address whether methylation density influences the stability of this epigenetic mark.
The wild-type proviral construct L1-MFGhGFP-1L (33) (Fig. 1a), was methylated in vitro at all CpG sites with SssI MTase (194 CpGs; 4.2 mCpGs/100 bp) or at low density with HpaII and HhaI MTases (34 CpGs; 0.7 mCpGs/100 bp), which methylate CCGG and GCGC sites, respectively. To establish that the methylation reactions were carried out to completion, aliquots of the in vitro-methylated DNA were digested with the m5C-sensitive restriction enzyme HpaII and the m5C-insensitive isoschizomer MspI (Fig. 1c) or with the m5C-sensitive restriction enzyme HhaI (data not shown). This in vitro-methylated DNA was introduced by RMCE (14) into the RL5 and RL6 integration sites in MEL cells (Fig. 2a), and ganciclovir-resistant cells were cultured for further analysis.
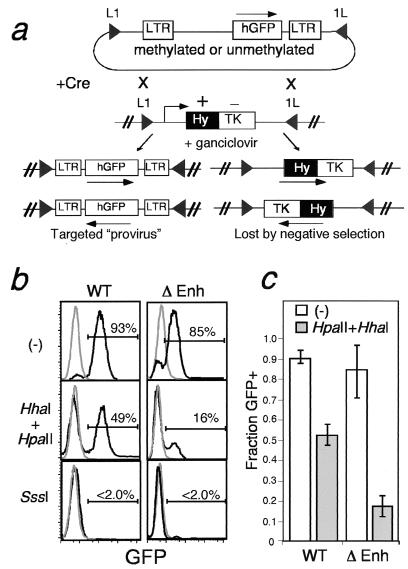
FIG. 2.
Opposing influences of the 5′LTR enhancer and CpG methylation density on the probability of proviral silencing at a defined genomic site. (a) Principle of Cre-RMCE-mediated targeting of proviral DNA. A MEL cell line containing a stably integrated HYTK fusion gene flanked by inverted loxP sites (solid triangles) is transfected with a construct (also flanked by inverted loxP sites) together with a Cre recombinase expression plasmid. Recombination between the loxP sites in the two constructs results in exchange of the cassettes and thus loss of the TK selectable marker. Alternatively, recombination between the inverted loxP sites on the same DNA molecule occurs, resulting in the inversion of the intervening DNA. Cells that have undergone the latter recombination event still express the HYTK gene and thus can be selected against with ganciclovir, allowing isolation of cells that have undergone the targeting reaction. (b) GFP expression analysis of wild-type or Δenh cassettes introduced at the RL5 integration site by RMCE. RL5 MEL cells were transfected with wild-type (WT) or Δenh constructs, either unmethylated (−), methylated at low density (with HpaII and HhaI MTases), or fully methylated (with SssI MTase). Ganciclovir-resistant cells were pooled, and GFP expression was analyzed by flow cytometry. Results of a representative experiment are shown for each of the constructs introduced. The percentage of GFP-positive cells, as determined by electronic gating, is shown for each sample. Histograms of control untransfected MEL cells are also shown (grey lines). (c) The percentage (mean ± standard deviation) of cells expressing GFP was determined for three independent transfections of the low-density-methylated (gray histograms) or unmethylated (white histograms) wild-type and Δenh constructs.
Initial methylation density influences the probability of expression and the fidelity of maintenance methylation.
To determine the influence of DNA methylation density on proviral expression from the RL5 integration site, pools of ganciclovir-resistant cells were analyzed for GFP expression by flow cytometry (Fig. 2b). While the unmethylated wild-type provirus showed expression in >90% of cells and the fully methylated cassette showed expression in <2% of cells, the cassette methylated at low density (with HpaII and HhaI) looked like a composite of the two, showing expression in ≈50% of the cells analyzed.
The influence of initial methylation density on the probability of expression was confirmed in three independent experiments (summarized in Fig. 2c). Surprisingly, GFP+ cells harboring the partially methylated wild-type provirus showed a level of GFP expression indistinguishable from that of the unmethylated wild-type provirus, with median fluorescence values of 118 and 119, respectively. These results suggest that the initial methylation density influences the probability rather than the rate of expression. Similar results were found at the RL6 integration site (data not shown).
The presence of expressing and nonexpressing subpopulations in the pools derived from the low-density-methylated construct might be explained by the binary nature of enhancer-driven transcription, as these elements function in part by increasing the probability of expression (49). However, the role of methylation density in the formation of two distinct expression states has not been previously addressed.
In order to determine whether the underlying methylation state of the expressing cells differs from that of the nonexpressing cells, clones were derived from the pools of ganciclovir-resistant cells. Regardless of initial methylation density, greater than 80% of ganciclovir-resistant clones harbored the proviral cassette integrated at the RL5 site (Fig. 3a and data not shown), consistent with our previous experiments showing that the efficiency of the Cre-loxP-mediated recombination event is not influenced by the methylation state of the introduced cassette (33, 45).
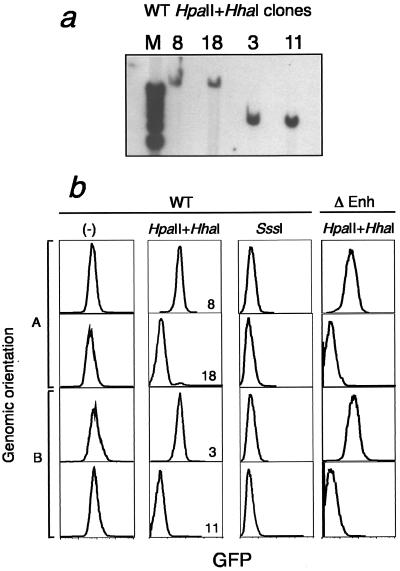
FIG.3.
Genomic orientation and expression status of RL5 L1-MFGhGFP-1L clones. (a) Clones were derived from the ganciclovir-resistant pool of cells transfected with the HpaII and HhaI methylated wild-type virus. Genomic DNA isolated from clones 3, 8, 11, and 18 was digested with BamHI and analyzed by Southern blotting with the GFP probe shown in Fig. 1. Different fragment sizes reflect the two possible genomic orientations. (b) Flow cytometry analysis of representative clones harboring the wild-type (WT) unmethylated or SssI- or HpaII- and HhaI-methylated cassette or the Δenh HpaII- and HhaI-methylated cassette. Two clones in each genomic orientation (arbitrarily labeled A and B), as determined by Southern analysis, are shown.
The methylation state of six clones from each population of transfected cells was initially evaluated with the methylation-sensitive restriction enzyme HpaII and Southern blotting (data not shown). Clones transfected with the unmethylated cassette showed no de novo methylation, while clones transfected with the fully methylated cassette revealed efficient maintenance methylation (with the exception of the proviral enhancer; see below), results consistent with our previous experiments demonstrating the stability of these methylation states at the RL5 integration site (33).
Despite the fact that the proviral cassette methylated at low density was initially uniformly methylated at all HpaII sites, HpaII digestion of genomic DNA isolated from six clones revealed two distinct digestion patterns, with some clones showing a complete lack of methylation at these initially methylated sites and others showing nearly complete maintenance methylation. Comparison of clonal methylation state and genomic orientation (Fig. 3a) with expression status (Fig. 3b) revealed that methylation state is correlated with proviral transcription rather than genomic orientation. Furthermore, the fraction of clones expressing GFP was similar to the fraction of GFP+ cells detected in the pools from which they were derived (Fig. 3b and data not shown). Similar results were found for the RL6 integration site. Taken together, these results indicate that distinct methylation states underlie the bimodal expression pattern observed.
The distinct methylation states of the provirus initially methylated uniformly at HpaII and HhaI sites were studied in greater detail by bisulfite sequencing. GFP+ and GFP− subpopulations were isolated from the ganciclovir-resistant pool by flow cytometry, and genomic DNA was extracted from the sorted cells. Analysis of the 5′LTR revealed the complete absence of methylation in cells expressing GFP (Fig. 4a). In contrast, analysis of the GFP− cells revealed that while the fidelity of maintenance methylation (at HpaII and HhaI sites) varied, depending on the CpG site, a significant number of CpGs, particularly in the promoter region and GFP gene (data not shown), became de novo methylated. Characterization of expressing (data not shown; 33) and nonexpressing (Fig. 4b) clones derived from the ganciclovir-resistant pools revealed that even with long-term culture, this binary pattern of demethylation and expression versus de novo methylation and silencing was stably maintained. Similar results were found at the RL6 integration site, suggesting that a low density of proviral methylation is generally not a stable epigenetic mark in MEL cells.
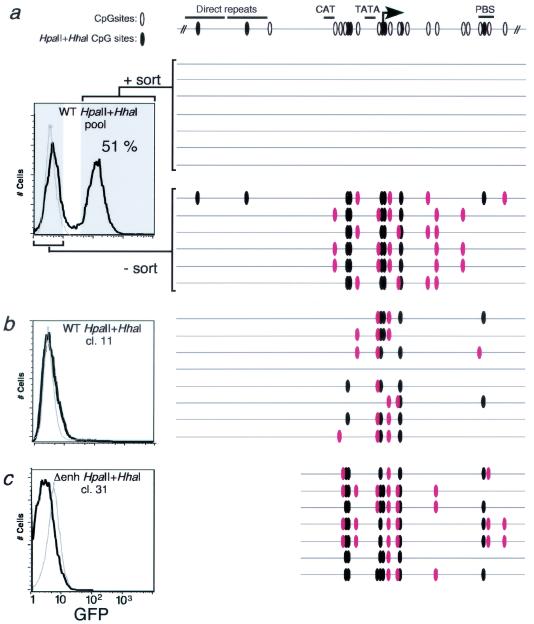
FIG. 4.
DNA methylation analysis of HpaII- and HhaI-methylated wild-type and Δenh L1MFGhGFP1L cassettes integrated at the RL5 site. (a) Ganciclovir-resistant MEL cells transfected with the HpaII- and HhaI-methylated wild-type L1MFGhGFP1L construct were sorted by electronic gating, as shown in the GFP fluorescence histogram. Control RL5 MEL parent cells (gray histogram) are also shown. Genomic DNA was isolated from the GFP+ (+sort) and GFP− (−sort) subpopulations and bisulfite sequenced with primers specific for the 5′LTR (see Fig. 1a for region amplified). CpGs (○) and CpGs in the context of HhaI or HpaII sites (•) within the region sequenced are shown at the top of the figure, along with several landmarks in the 5′LTR. Each molecule sequenced is depicted as a gray line. Those CpGs that have been de novo methylated (i.e., CpGs not in the context of HpaII or HhaI sites) are depicted as solid red ovals. Note the absence of any mCpGs in the +sort sample. (b) Bisulfite analysis of clone (cl) 11 isolated from the pool of ganciclovir-resistant cells depicted in a. (c) Bisulfite analysis of clone 31, isolated from the pool of ganciclovir-resistant cells transfected with the HpaII- and HhaI-methylated Δenh construct.
Enhancers influence the probability of propagating a repressive epigenetic mark.
While the ability of enhancers to overcome methylation-mediated repression is well documented, the role of demethylation in this process remains controversial (50). The reproducible distribution of unmethylated versus de novo-methylated wild-type proviral integrants allowed us to address whether the proviral enhancer counteracts methylation-mediated repression by disrupting propagation of the methylation mark. In order to determine the role of the Moloney murine leukemia virus 5′LTR enhancer, we generated a proviral cassette (L1−ΔenhMFGhGFP-1L; Δenh) from which this enhancer (and four CpG sites) has been deleted (Fig. 1b). The Δenh construct, methylated at different densities, was introduced into the RL5 genomic site, as described above for the wild-type construct.
Regardless of initial methylation state, the level of GFP expression in GFP+ cells was twofold lower than that detected with the wild-type construct (Fig. 2b and data not shown), revealing that the 5′LTR enhancer influences the rate of transcription. However, the Δenh construct methylated at low density also had a significantly lower probability of establishing a transcriptionally active state than the wild-type construct, ≈20% versus ≈50%, respectively (Fig. 2c). As with the wild-type construct, the probability of expression of the Δenh construct methylated at high density was reduced to <2%.
Bisulfite analysis of transcriptionally active and inactive (Fig. 3b) clones derived with the Δenh construct methylated at low density revealed that cells expressing GFP lost the methylation imprint (data not shown), while nonexpressing cells became de novo methylated (Fig. 4c). Taken together, these experiments suggest that de novo methylation of the provirus is stimulated by preexisting DNA methylation; the proviral enhancer may counteract this process by inhibiting maintenance methylation, thereby removing the epigenetic mark that triggers de novo methylation.
Dnmt3a/b-independent de novo MTase activity.
In mammalian cells, establishment and propagation of CpG methylation are thought to be carried out by different enzymes, the “de novo” MTases Dnmt3a and Dnmt3b and “maintenance” MTase Dnmt1, respectively. We wished to establish whether the apparent methylation-dependent de novo methylation activity observed in MEL cells was catalyzed by Dnmt3a and/or Dnmt3b. Determining whether de novo methylation is stimulated by preexisting methylation in MEL cells is complicated by the fact that the probability of proviral transcription at the RL5 integration site is inversely correlated with methylation density, and as described above, once an active transcription state is established, both maintenance and de novo methylation are inhibited.
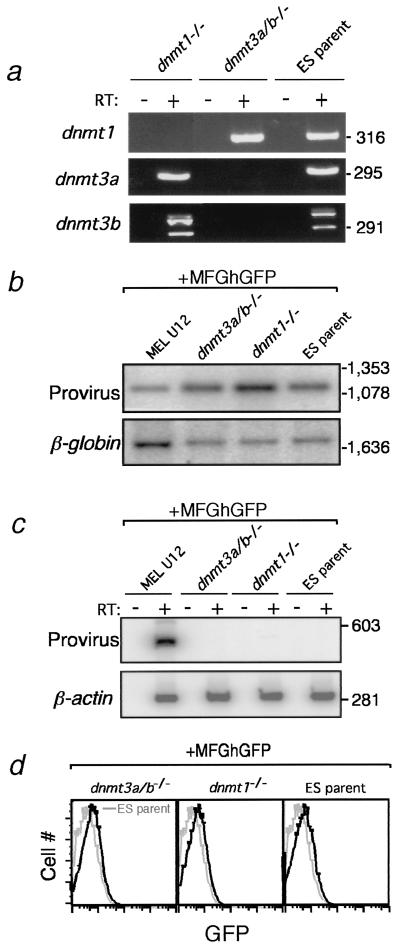
To circumvent this complication, we used murine ES cells, which do not support transcription from the wild-type Moloney murine leukemia virus LTR (17, 31) regardless of DNA methylation state (36). Furthermore, while MEL cells express each of the known MTases (M. Lorincz, unpublished observations), ES cells with genetic lesions of the MTase genes are available, permitting MTase activities to be correlated with specific MTases (26, 37). J1 ES parent, J1 Dnmt1−/− (26), and J1 Dnmt3a−/−/Dnmt3b−/− (Dnmt3a/b−/−) double-knockout cells (37) (Fig. 5a) were cotransfected with the wild-type MFGhGFP construct, either unmethylated or methylated at low density (at HpaII and HhaI sites), and the histidinol selectable marker BSKSIIβactinhis. As with the MEL RMCE system, no selection for expression from the proviral construct was applied. Southern analysis of genomic DNA isolated from histidinol-resistant ES cell pools transfected with the unmethylated MFGhGFP construct revealed a proviral copy number of at least one per cell in each of the ES pools (Fig. 5b). Nevertheless, no proviral transcription was detected (Fig. 5c,d).
FIG. 5.
The proviral LTR is transcriptionally inactive in ES cells, regardless of MTase activity. (a) cDNA from Dnmt1−/−, Dnmt3a/b−/−, and J1 ES parent lines was generated from total RNA with reverse transcriptase and amplified with primers specific for the catalytic domains of Dnmt1, Dnmt3a, and Dnmt3b, confirming the phenotypes of the knockout lines. (b) Dnmt1−/−, Dnmt3a/b−/−, and J1 ES cells were cotransfected with unmethylated MFGhGFP and the HisD expression vector BSKSIIβactinhis. Genomic DNA was isolated from histidinol-resistant ES cell pools derived from these transfections and from the MFGhGFP single-copy MEL clone U12 as a control. This DNA was digested and subjected to Southern blotting with the GFP probe shown in Fig. 1a. The blot was subsequently stripped and reprobed with a fragment specific for the endogenous β-globin locus. The relative intensities of the bands detected reveal that the histidinol-resistant ES cells harbor at least one copy of the proviral construct per cell. (c) cDNA from the histidinol-resistant ES cell pools and MEL clone U12 (which stably expresses the provirus) was generated and amplified with primers specific for the proviral mRNA. β-Actin primers were used as a control for the integrity of the mRNA preparation. Note the absence of proviral expression in ES cells, even in the absence of the de novo MTases Dnmt3a and Dnmt3b. (d) Analysis of GFP expression by flow cytometry from these lines and control J1 ES parent cells (gray histogram) confirms the absence of even rare GFP+ cells in these populations.
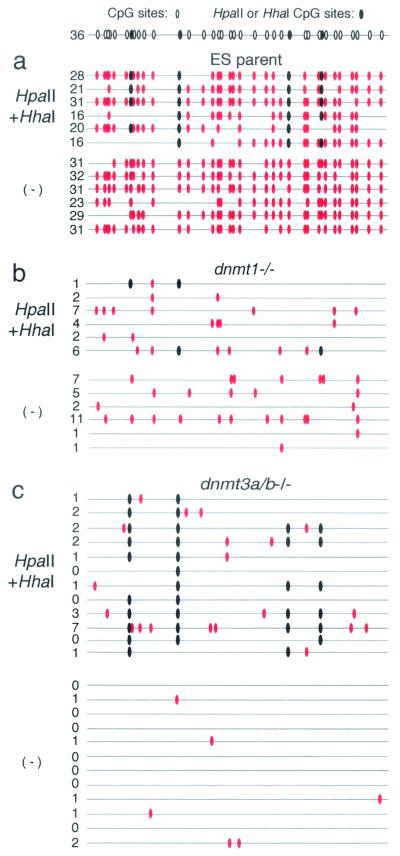
Bisulfite sequencing of genomic DNA isolated 10 days (data not shown) or 22 days after transfection was conducted on each histidinol-resistant cell pool. Analysis of J1 ES parent cells revealed that a significant number of CpG sites in the GFP gene had become de novo methylated, regardless of initial methylation state (Fig. 6a). Furthermore, methylation was maintained at most but not all of the HpaII and HhaI sites in the premethylated construct, presumably reflecting the incomplete fidelity of maintenance methylation (43). In contrast, analysis of the Dnmt1−/− cells revealed that few of the premethylated sites were still methylated 22 days postelectroporation, and significantly fewer sites were de novo methylated, regardless of initial methylation state (Fig. 6b). These results are consistent with the presumed roles of Dnmt1 and of Dnmt3a and Dnmt3b in maintenance and de novo methylation, respectively; any de novo methylation is presumably erased with each round of DNA replication in Dnmt1−/− cells, mimicking a decrease in de novo methylation efficiency.
FIG. 6.
Dnmt3a/b-independent de novo methylation activity. Genomic DNA from (a) J1 ES parent, (b) Dnmt1−/−, and (c) Dnmt3a/b−/− cell pools stably transfected with unmethylated (−) or HpaII- and HhaI-methylated MFGhGFP DNA was isolated 22 days postelectroporation. Methylation state of the GFP gene (see Fig. 1a for region amplified) was determined by bisulfite sequencing analysis. CpGs (○) and CpGs in the context of HpaII or HhaI sites (•) are shown at the top. Each molecule sequenced is depicted as a gray line. Those CpGs that are methylated as a result of de novo activity (i.e., excluding CpGs in the context of HpaII or HhaI sites in the HpaII- and HhaI-methylated samples) are shown (solid red ovals) and totaled on the left-hand side for each molecule sequenced. For the Dnmt3a/b−/− cell pools stably transfected with unmethylated or HpaII- and HhaI-methylated MFGhGFP DNA, 19 or 18 molecules were sequenced, respectively, and 12 representative molecules of each are shown.
Analysis of the Dnmt3a/b−/− cells transfected with the premethylated construct revealed that, as with the wild-type ES cells, methylation was maintained at the majority of HpaII and HhaI sites in the GFP gene (Fig. 6c). Surprisingly, a number of de novo-methylated CpGs flanking the in vitro-methylated sites were also detected, a total of 32 in the 18 clones analyzed (mean = 1.8 mCpGs/molecule, not including the premethylated CpGs). Significantly fewer de novo-methylated CpGs (10 in 19 molecules sequenced; mean = 0.53 mCpGs/molecule) were detected in the Dnmt3a/b−/− cells transfected with the unmethylated construct, with P values of 3.4 × 10−3 (one-way t test) and 5.2 × 10−4 (one-way Wilcoxon rank sum test), respectively. Fewer de novo-methylated CpGs were detected in the premethylated construct at day 10 postelectroporation (14 in 18 molecules sequenced; mean = 0.82 mCpGs/molecule; data not shown). Taken together, these results reveal the existence of a Dnmt3a/b-independent de novo MTase activity that is stimulated by preexisting mCpGs, yielding a progressive increase in methylation density with time in culture.
DISCUSSION
We introduced a proviral construct, methylated in vitro at different densities, into the RL5 genomic site in MEL cells. Analysis of the stability of these predetermined methylation patterns revealed that propagation of this epigenetic mark is dependent on the initial methylation density. Unmethylated or densely methylated epigenetic states were stably maintained, coincident with transcriptional activity or silencing, respectively. Although methylation of specific CpGs was not necessarily maintained in the latter case, the methylation status of the proviral cassette was faithfully propagated. In contrast, a low-density methylation imprint was inherently unstable, with stochastic demethylation or de novo methylation occurring with equal probability. In the former case, demethylation coincided with transcriptional activation, while in the latter case, de novo methylation was apparently stimulated by preexisting methylation, effectively consolidating the methylation imprint.
In order to determine if cis-acting regulatory elements influence the probability of methylation-mediated de novo methylation, we introduced a proviral cassette lacking the 5′LTR enhancer and methylated at low density into the same integration site. Compared with the wild-type construct, the probability of maintenance and de novo methylation and concomitant transcriptional silencing increased significantly, results consistent with the hypothesis that transcriptionally silent chromatin is targeted for de novo methylation (5). We propose that the probability of expression from a given locus is influenced by local DNA methylation density in addition to the dynamic interplay between sequence-specific transcriptional activators and repressors.
For some genes, a low methylation density may be a metastable state, with allelic demethylation or de novo methylation occurring during differentiation. A predicted consequence would be stochastic inactivation of expression from one or both alleles with a defined probability; such “allelic bias” has been observed for the interleukin-4 (IL-4) gene (22). In contrast, sites particularly prone to de novo methylation (16, 34), such as repetitive elements, may be methylated early in development at a density that ensures transcriptional silencing regardless of enhancer strength.
The proviral enhancer studied here was preferentially demethylated and rarely de novo methylated regardless of initial methylation density, suggesting that this enhancer, like other DNA-regulatory domains (18, 45), is somehow protected from de novo methylation. The presence of a DNase I-hypersensitive site in the proviral enhancer, irrespective of proviral expression state (33), reveals that factors constitutively bound to the Moloney murine leukemia virus LTR enhancer may belong to a putative group of transcription factors, like Sp1 (8, 46), that not only bind their cognate binding site regardless of methylation state (19), but also target bound sites for demethylation (30) and protect them from subsequent de novo methylation (21). Localized demethylation may be a prerequisite for transcriptional activation, as has been shown for other regulatory elements (29, 40, 45).
The use of wild-type and DNA MTase-deficient ES cell lines allowed us to determine which of these enzymes are responsible for de novo methylation of an unmethylated template and maintenance and de novo methylation of a premethylated template in vivo. While a higher density of methylation was detected in the wild-type ES cells than either the Dnmt1−/− or Dnmt3a/b−/− cell lines, a significant level of de novo methylation was detected in the Dnmt1−/− cells. These results are consistent with a recent study of repetitive elements with the same MTase knockout cells used here (28), in which the authors proposed that ongoing de novo methylation of these elements by Dnmt3a and/or Dnmt3b compensates for the inefficient maintenance methylation mediated by Dnmt1 (28).
Here, we reveal a de novo MTase activity that is stimulated by preexisting methylation in cells lacking both of the known “de novo” MTases. Presumably, Dnmt3a/b-independent de novo methylation was not detected in the original study of the Dnmt3a/b−/− cells because Southern blotting rather than the more sensitive bisulfite sequencing method was used to analyze DNA methylation state (37). Furthermore, the preferred substrate of the Dnmt3a/b−/− independent de novo MTase activity, namely, DNA already containing mCpGs, was not surveyed in this study (37).
Dnmt1 and/or Dnmt2 may be responsible for the Dnmt3a/b-independent de novo methylation described here. However, while Dnmt2 shows no CpG MTase activity in vitro (39), Dnmt1 has de novo activity in vitro (41, 52), and this activity is stimulated by the presence of preexisting methylation (9, 13, 47). Furthermore, overexpression of Dnmt1 in ES cells (4) or in human fibroblasts (48) results in de novo methylation of a subset of normally unmethylated genes or CpG islands, respectively. Taken together, these results suggest that Dnmt1 is responsible for the observed Dnmt3a/b-independent de novo MTase activity.
Intriguingly, partially methylated CpG islands are particularly susceptible to de novo methylation in breast cancer cells (23). Although overexpression of Dnmt1 does not appear to be responsible for tumor-associated hypermethylation (12, 44), it is possible that such de novo methylation is mediated by aberrant Dnmt1 activity.
Acknowledgments
This work was supported by NIH fellowship GM 19767/01to M.L., a fellowship from the Rett Syndrome Research Foundation to D.S., and DK44746, HL57620, and CA54337 to M.G.
We thank E. Li for the ES cell lines and helpful discussions, Eric Bouhassira for the RL5 and RL6 MEL cell lines, and the members of the Groudine laboratory for suggestions; Wendy Paulsene and Urszula Maliszewski for technical assistance; and Tomoyuki Sawado, Mike Teitell, and Fred van Leeuwen for comments on the manuscript.
REFERENCES
- 1.Anderson, M. T., I. M. Tjioe, M. C. Lorincz, D. R. Parks, L. A. Herzenberg, G. P. Nolan, and L. A. Herzenberg. 1996. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell with engineered green fluorescent proteins. Proc. Natl. Acad. Sci. USA 93:8508-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 3.Bestor, T. H., and V. M. Ingram. 1983. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. USA 80:5559-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biniszkiewicz, D., J. Gribnau, B. Ramsahoye, F. Gaudet, K. Eggan, D. Humpherys, M. A. Mastrangelo, Z. Jun, J. Walter, and R. Jaenisch. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 22:2124-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 6.Boyes, J., and A. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 7.Boyes, J., and A. Bird. 1992. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 9.Carotti, D., S. Funiciello, F. Palitti, and R. Strom. 1998. Influence of preexisting methylation on the de novo activity of eukaryotic DNA methyltransferase. Biochemistry 37:1101-1108. [DOI] [PubMed] [Google Scholar]
- 10.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dranoff, G., E. Jaffee, A. Lazenby, P. Golumbek, H. Levitsky, K. Brose, V. Jackson, H. Hamada, D. Pardoll, and R. C. Mulligan. 1993. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting antitumor immunity. Proc. Natl. Acad. Sci. USA 90:3539-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eads, C. A., K. D. Danenberg, K. Kawakami, L. B. Saltz, P. V. Danenberg, and P. W. Laird. 1999. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 59:2302-2306. [PubMed] [Google Scholar]
- 13.Fatemi, M., A. Hermann, S. Pradhan, and A. Jeltsch. 2001. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 309:1189-1199. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Y. Q., M. C. Lorincz, S. Fiering, J. M. Greally, and E. E. Bouhassira. 2001. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol. Cell. Biol. 21:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, Y. Q., J. Seibler, R. Alami, A. Eisen, K. A. Westerman, P. Leboulch, S. Fiering, and E. E. Bouhassira. 1999. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 292:779-785. [DOI] [PubMed] [Google Scholar]
- 16.Graff, J. R., J. G. Herman, S. Myohanen, S. B. Baylin, and P. M. Vertino. 1997. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 272:22322-22329. [DOI] [PubMed] [Google Scholar]
- 17.Grez, M., E. Akgun, F. Hilberg, and W. Ostertag. 1990. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 87:9202-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groudine, M., and K. F. Conklin. 1985. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science 228:1061-1068. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, M. A., P. A. Jones, M. Imagawa, and M. Karin. 1988. Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. USA 85:2066-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh, C. L. 1999. Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol. 19:46-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu-Li, J., C. Pannetier, L. Guo, M. Lohning, H. Gu, C. Watson, M. Assenmacher, A. Radbruch, and W. E. Paul. 2001. Regulation of expression of IL-4 alleles: analysis with a chimeric GFP/IL-4 gene. Immunity 14:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Huang, T. H., M. R. Perry, and D. E. Laux. 1999. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 8:459-470. [DOI] [PubMed] [Google Scholar]
- 24.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 25.Knebel-Morsdorf, D., S. Achten, K. D. Langner, R. Ruger, B. Fleckenstein, and W. Doerfler. 1988. Reactivation of the methylation-inhibited late E2A promoter of adenovirus type 2 by a strong enhancer of human cytomegalovirus. Virology 166:166-174. [DOI] [PubMed] [Google Scholar]
- 26.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 27.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 28.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtenstein, M., G. Keini, H. Cedar, and Y. Bergman. 1994. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell 76:913-923. [DOI] [PubMed] [Google Scholar]
- 30.Lin, I. G., T. J. Tomzynski, Q. Ou, and C. L. Hsieh. 2000. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loh, T. P., L. L. Sievert, and R. W. Scott. 1990. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell. Biol. 10:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorincz, M. C., D. Schubeler, S. C. Goeke, M. Walters, M. Groudine, and D. I. Martin. 2000. Dynamic analysis of proviral induction and De Novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 20:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorincz, M. C., D. Schubeler, and M. Groudine. 2001. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 21:7913-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mummaneni, P., K. A. Walker, P. L. Bishop, and M. S. Turker. 1995. Epigenetic gene inactivation induced by a cis-acting methylation center. J. Biol. Chem. 270:788-792. [DOI] [PubMed] [Google Scholar]
- 35.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 36.Niwa, O., Y. Yokota, H. Ishida, and T. Sugahara. 1983. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell 32:1105-1113. [DOI] [PubMed] [Google Scholar]
- 37.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 38.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 39.Okano, M., S. Xie, and E. Li. 1998. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 26:2536-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paroush, Z., I. Keshet, J. Yisraeli, and H. Cedar. 1990. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell 63:1229-1237. [DOI] [PubMed] [Google Scholar]
- 41.Pradhan, S., A. Bacolla, R. D. Wells, and R. J. Roberts. 1999. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274:33002-33010. [DOI] [PubMed] [Google Scholar]
- 42.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 43.Riggs, A. D., Z. Xiong, L. Wang, and J. M. LeBon. 1998. Methylation dynamics, epigenetic fidelity and X chromosome structure. Novartis Found. Symp. 214:214-225. [DOI] [PubMed] [Google Scholar]
- 44.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27:2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubeler, D., M. C. Lorincz, D. M. Cimbora, A. Telling, Y. Q. Feng, E. E. Bouhassira, and M. Groudine. 2000. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20:9103-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silke, J., K. I. Rother, O. Georgiev, W. Schaffner, and K. Matsuo. 1995. Complex demethylation patterns at Sp1 binding sites in F9 embryonal carcinoma cells. FEBS Lett. 370:170-174. [DOI] [PubMed] [Google Scholar]
- 47.Tollefsbol, T. O., and C. A. Hutchison 3rd. 1997. Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J. Mol. Biol. 269:494-504. [DOI] [PubMed] [Google Scholar]
- 48.Vertino, P. M., R. W. Yen, J. Gao, and S. B. Baylin. 1996. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 16:4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters, M. C., S. Fiering, J. Eidemiller, W. Magis, M. Groudine, and D. I. Martin. 1995. Enhancers increase the probability but not the level of gene expression. Proc. Natl. Acad. Sci. USA 92:7125-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolffe, A. P., P. L. Jones, and P. A. Wade. 1999. DNA demethylation. Proc. Natl. Acad. Sci. USA 96:5894-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yisraeli, J., R. S. Adelstein, D. Melloul, U. Nudel, D. Yaffe, and H. Cedar. 1986. Muscle-specific activation of a methylated chimeric actin gene. Cell 46:409-416. [DOI] [PubMed] [Google Scholar]
- 52.Yoder, J. A., N. S. Soman, G. L. Verdine, and T. H. Bestor. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270:385-395. [DOI] [PubMed] [Google Scholar]