Abstract
Polycomb group (PcG) proteins function through cis-acting DNA elements called PcG response elements (PREs) to stably silence developmental regulators, including the homeotic genes. However, the mechanism by which they are targeted to PREs remains largely unclear. Pleiohomeotic (PHO) is a sequence-specific DNA-binding PcG protein and therefore may function to tether other PcG proteins to the DNA. Here, we show that PHO can directly bind to a Polycomb (PC)-containing complex as well as the Brahma (BRM) chromatin-remodeling complex. PHO contacts the BRM complex through its zinc finger DNA-binding domain and a short N-terminal region. A distinct domain of PHO containing a conserved motif contacts the PcG proteins PC and Polyhomeotic (PH). With mobility shift assays and DNA pulldown experiments, we demonstrated that PHO is able to link PC, which lacks sequence-specific DNA-binding activity, to the DNA. Importantly, we found that the PC-binding domain of PHO can mediate transcriptional repression in transfected Drosophila Schneider cells. Concomitant overexpression of PC resulted in stronger PHO-directed repression that was dependent on its PC-binding domain. Together, these results suggest that PHO can contribute to PRE-mediated silencing by direct recruitment of a PC complex to repress transcription.
Cellular differentiation and development of multicellular organisms is the result of the temporal and spatial orchestration of gene expression patterns. The Polycomb group (PcG) of repressors and trithorax group (trxG) of activators are required for maintenance of the determined gene expression patterns of several genes, including the homeotic genes (4, 5, 9, 22, 44, 47, 49, 61). Early in Drosophila development, the transiently expressed gap and pair-rule proteins establish the expression patterns of the homeotic genes that determine the identity of the body parts. These early regulators disappear later during development, and their function is taken over by the PcG and trxG proteins. The PcG proteins act to perpetuate silencing of the homeotic genes outside their expression domains, whereas the trxG proteins are necessary for the maintenance of transcriptional activity. Since PcG and trxG proteins conserve a state of gene expression over multiple rounds of cell division, this process is often referred to as epigenetic regulation.
PcG proteins act in concert as components of defined multiprotein complexes that are believed to silence gene transcription by inducing a higher-order chromatin structure (1, 9, 22, 24, 46, 49, 64, 66, 67, 68). Currently, two functionally distinct classes of PcG protein complexes have been identified. First, biochemical analysis uncovered a 3-MDa PRC1 complex that harbors PcG proteins Polycomb (PC), Polyhomeotic (PH), Sex combs on midleg, Posterior sex combs (PSC), and several other proteins, including components of the basal transcription factor TFIID and Zeste, a sequence-specific DNA-binding protein (22, 67, 68). Coimmunoprecipitation experiments and protein-protein interaction studies suggested that the mammalian homologues of the PcG proteins in PRC1 also form a complex (1, 66).
A second type of PcG complex contains the PcG proteins Enhancer of Zeste [E(z)], extra sex combs (Esc), the histone deacetylase Rpd3, and the histone-binding protein p55 that is also part of the chromatin assembly factor CAF1 and the chromatin remodeling factor NURF (56, 75). The association of E(z) and ESC is conserved in mammals, and repression by the mammalian ESC/E(z) complex involves histone deacetylation (66, 79). In contrast, repression by vertebrate PC homologues is resistant to inhibitors of histone deacetylation, suggesting that PC repression occurs through a distinct molecular mechanism (72, 79). Instead, PRC1 may act by inhibition of chromatin remodeling by the SWI/SNF complex in a process that does not require the histone tails (23, 68). Thus, there are at least two distinct PcG complexes, each of which represses transcription by a different mechanism. The ESC/E(z) complex appears to direct deacetylation of the histone tails, whereas PRC1 may induce a stabilized SWI/SNF-resistant chromatin structure. Recent coimmunoprecipitation experiments have indicated that there might be a transient interaction between these two distinct PcG complexes during early development (64).
An outstanding question is how PcG proteins act in a gene-specific manner. In Drosophila melanogaster, PcG-dependent silencing is mediated by large, rather poorly defined DNA sequences that are named Polycomb response elements (PREs) or cellular memory modules (5, 22, 47, 49, 61). PREs were identified by their PcG protein-dependent silencing effect on linked reporter genes in transgenic flies (12, 15, 21, 28, 41, 54, 62, 71, 82). Indeed, chromatin immunoprecipitation experiments revealed that PREs are bound by PcG proteins (57, 58, 73, 74), and immunostaining of polytene chromosomes showed that the insertion of a P-element containing a PRE creates a new PcG protein-binding site (15, 16, 82). Collectively, these results demonstrate that PcG proteins associate with PREs to mediate transcriptional silencing.
The majority of PcG proteins appear to lack sequence-specific DNA-binding activity, suggesting that protein-protein interactions play an important role in PcG complex formation on PREs. The exception so far is the PcG protein Pleiohomeotic (PHO), which contains a zinc finger DNA-binding domain (DBD), which is related to that of the mammalian transcription factor YY1 (9). There is a second region of about 25 residues that shows similarity, corresponding to a small portion of the so-called spacer region in YY1 (69).
The ability of PHO to bind DNA makes it an attractive candidate for a PcG tethering factor. Indeed, PHO has been shown to bind to PREs from the engrailed (en) gene (9), the Ultrabithorax (Ubx) gene (24), and the Abdominal-B (Abd-B) region (13, 52). Sequence inspection has revealed that most PREs contain potential PHO binding sites, suggesting that PHO might be involved in the targeting of PcG silencing (51). The PHO binding sites in the en PRE are essential for its function as a pairing-sensitive silencer of a miniwhite reporter gene, and silencing is partially impaired in pho mutants (10). Point mutations in PHO sites in the Ubx PRE abolish PcG silencing in imaginal disks, and PHO was also shown to synergize with PC to repress the Ubx gene in vivo (25, 76). Likewise, both the PHO and GAGA sites in the MCP silencer and iab-7 are required for the maintenance of repression (12, 51). However, although PHO sites are necessary in these studies, by themselves they are not sufficient to reconstitute PRE activity in vivo. Instead, the activity of additional DNA-binding proteins, such as the trxG protein GAGA, appears to be required for PcG silencing (13, 30, 35, 52, 63, 70).
Recent studies have indicated that PHO might associate with the ESC/E(z) complex (64). Moreover, the related YY1 protein has been reported to interact with the mammalian ESC/E(z) complex (65). However, these results do not exclude the possibility of a transient interaction with components of a PC-containing PRC1-related complex. Indeed, YY1 has been found to bind RYBP, a component of the vertebrate PC complex (26). Furthermore, as discussed above, PHO and PC appear to cooperate in vivo during fly development. A simple explanation for this cooperation would be a direct interaction between PHO and a PC-containing repressive complex.
Because transcription factors in general are not stably associated with their coactivators or corepressors (55), we set out to identify a putative repression domain in PHO and test whether it interacted with a PC complex. We found that PHO can directly bind a PC complex as well as the Brahma (BRM) chromatin remodeling complex in Drosophila embryo nuclear extracts. Distinct protein domains of PHO are involved in targeting either the PC or the BRM complex. PHO specifically targets PC and PH. We used mobility shift assays and DNA pulldown experiments to assess the ability of PHO to link PC to the DNA. Finally, we tested the ability of PHO to direct PC-mediated transcriptional repression in transfected Drosophila Schneider cells. Our results suggest that PHO contributes to PcG repression by connecting PC to gene-regulatory DNA elements.
MATERIALS AND METHODS
DNA constructs.
Details of cloning procedures are available upon request. The glutathione S-transferase (GST) fusion constructs were generated by a PCR-based strategy. PC-, PH-, and PHO-encoding DNA fragments were cloned in pGEX-2TKN, a derivative of pGEX-2TK (Pharmacia). The GST-BRM fusion construct has been described (18). Similarly, templates for in vitro translation were generated by cloning of the corresponding coding sequences into pTβSTOP (40). The coding sequence of full-length PHO was cloned into modified versions of the shuttle vector pVL1392 (Pharmingen) expressing either an in-frame amino-terminal Flag or GST tag, and the PHO DNA-binding domain (DBD; amino acids 355 to 520) was cloned into pVL1392-Flag. The luciferase reporter contains five Gal4 binding sites located upstream of the herpes simplex virus thymidine kinase promoter in front of luciferase (pGL3; Promega). The Gal4 DNA-binding domain chimeras were constructed by subcloning the indicated cDNAs in-frame in a modified pCDNA3. The full-length coding sequence of PC was cloned in pSuper-CATCH containing an N-terminal Flag tag. The reporter used for the long-distance repression experiment (Fig. 7C) has been described (48).
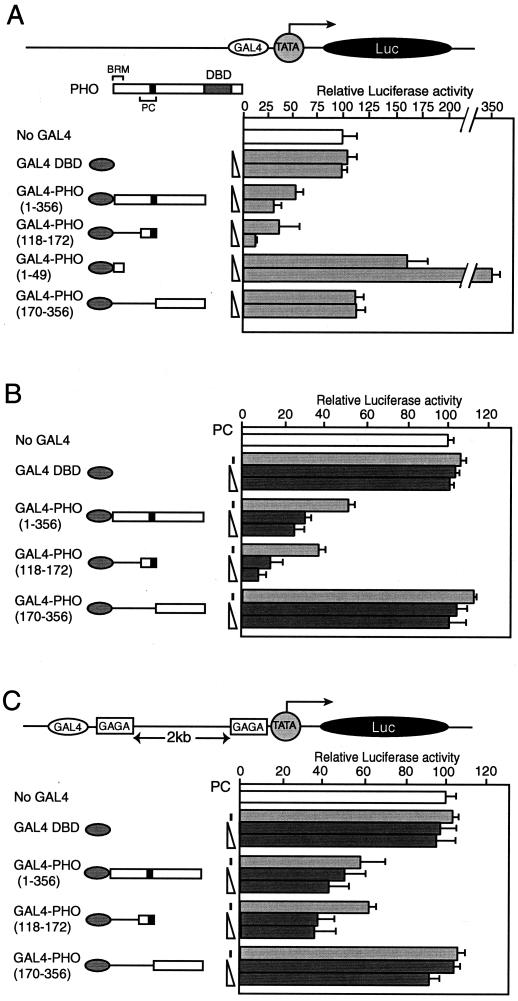
FIG. 7.
PHO represses transcription in transiently transfected Drosophila Schneider L2 cells. (A) The PC-binding domain of PHO mediates transcriptional repression. Schneider L2 cells were transfected with either 75 ng of reporter plasmid alone (no Gal4) or together with increasing amounts of the plasmid expressing the Gal4 DNA-binding domain (Gal4 DBD) alone or with various Gal4-PHO fusion constructs (50 ng and 150 ng), as indicated by the schematic representation on the left. The structure of the reporter plasmid is indicated schematically. (B) PC enhances transcriptional repression by Gal4-PHO fusions containing the PC-binding domain. L2 cells were cotransfected with 50 ng of reporter plasmid in the absence or presence of plasmids (100 ng) expressing the Gal4 DNA-binding domain (Gal4 DBD) alone or various Gal4-PHO fusion constructs. The various Gal4 constructs were cotransfected either with empty vector or together with increasing amounts of a plasmid expressing full-length PC (50 ng and 100 ng), represented by the dark grey bars. (C) The experiment described in B was repeated with a reporter harboring Gal4-binding sites flanked by GAGA sites separated by over 2 kb of intervening DNA from a promoter containing GAGA sites [pGL3-Prom(GAGA)Enh(GAGA/Gal4] (as described by Mahmoudi et al. [4848]). This reporter was cotransfected in the absence or presence of various combinations of expression vectors for Gal4 DBD, Gal4-PHO, or PC as indicated. The luciferase activities were normalized so that the reporter plasmid alone averaged at 100%. The structure of the reporter plasmid is indicated schematically.
Protein procedures.
Recombinant PHO and DBD containing an N-terminal Flag epitope was expressed in Sf9 cells in the baculovirus expression system and immunopurified with anti-Flag M2 beads (Sigma) or glutathione-Sepharose (Pharmacia) essentially as described previously (14, 40).
The Drosophila nuclear extracts and protein fractions were prepared essentially as described previously (3, 33). Briefly, all protein procedures were carried out at 4°C or on ice with HEMG buffer [25 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol (DTT), 0.2 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 1 μM pepstatin, 0.01% Nonidet P-40] containing various amounts of KCl. Nuclear extracts of dechorionated Drosophila embryos (0 to 12 h) were prepared as described previously by Kadonaga (38). The nuclear extracts were either used directly or concentrated by Poros-Heparin (Perseptive Biosystems) chromatography essentially as described previously (3, 33). The heparin-400 mM KCl fractions (H0.4) contained the vast majority of BRM, initiation switch (ISWI), and general transcription factors. The H0.4 pool was further purified by Sephacryl S-300 column chromatography guided by Western blot analysis with antibodies directed against BRM, PC, and PH. Fractions containing the bulk of the above factors were pooled and further purified on a BioScale Q10 column (Bio-Rad). It should be noted that essentially all the PC, PH, and BRM present in nuclear extracts was retained in this fraction, as judged by Western blotting analysis with the appropriate antibodies (data not shown). Most pulldown experiments were performed with crude nuclear extracts as well as with a fraction from the Q10 column with essentially similar results. The results shown were obtained with the nuclear extracts (Fig. 1) or with the partially purified Q10 fraction (Fig. 2, 3, and 6).
FIG. 1.
PHO interacts with PcG proteins and the BRM complex in Drosophila embryo nuclear extracts. (A) Coimmunoprecipitation experiments with antiserum directed against PHO with Drosophila embryo nuclear extracts. Nuclear extracts were incubated with either preimmune serum (lane 2) or antiserum directed against PHO (lane 3), followed by the addition of protein A beads. Following extensive washes with a buffer containing 250 mM NaCl and detergent, associated proteins were eluted with a buffer containing 1 M NaCl, resolved by SDS-PAGE, and analyzed by Western immunoblotting with antibodies directed against BRM (39), MOR (SN670 and SN671, pooled), OSA (74), PH (SN964), PC (SN965), PSC (49), GRO (PV1 and PV2, pooled), and the 140-kDa subunit of RNA polymerase II (Pol-II; DmRP140) (PV35). Lane 1 represents 10% of the input material used in the binding reactions. (B) The ability of a GST-tagged full-length PHO to recruit PcG proteins or the BRM complex within an embryo nuclear extract was tested by GST pulldown assays. GST alone (lane 2) or GST-PHO was immobilized on glutathione-Sepharose beads and incubated with Drosophila embryo nuclear extracts. Following a series of extensive washes with a buffer containing 150 mM NaCl, bound proteins were resolved by SDS-PAGE and analyzed by Western immunoblotting. Lane 1 represents 10% of the input material used in the binding reactions.
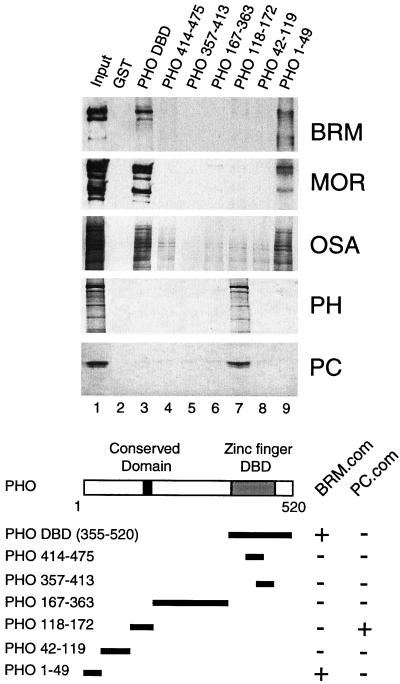
FIG. 2.
PHO interacts with PcG proteins and the BRM complex. The ability of various PHO polypeptides to recruit PC, PH, and the BRM complex from Drosophila embryo nuclear extracts was tested by GST pulldown assays. GST alone (lane 2), GST-PHO DBD (amino acids 355 to 520; lane 3), GST-PHO(414-475) (lane 4), GST-PHO(357-413) (lane 5), GST-PHO(167-363) (lane 6), GST-PHO(118-172) (lane 7), GST-PHO(42-119) (lane 8), and GST-PHO(1-49) (lane 9) were immobilized on glutathione-Sepharose beads and incubated with a partially purified column fraction containing the PC and BRM complexes. Protein complexes were washed, resolved by SDS-PAGE, and transferred to nitrocellulose. The blots were probed with antibodies directed against BRM (39), MOR (SN670 and SN671, pooled), OSA (74), PH (PV86), or PC (PV69). Lane 1 represents 5% of the input material used in the binding reactions. The domain structure of PHO, including the zinc finger DNA-binding domain (DBD), a conserved region present in YY1, and the amino acid residues present in the various derivatives are indicated. The binding of the PHO deletion constructs to either the BRM complex (BRM.com) or the PC complex (PC.com) is summarized.
Recombinant GST fusion proteins were expressed in Escherichia coli BL21 and purified by glutathione-Sepharose chromatography by standard procedures. 35S-labeled proteins were expressed with the TNT rabbit reticulocyte lysate (Promega). The GST pulldown experiments were performed as described previously (36) with the following modifications. The lysis buffer contained 25 mM HEPES-KOH (pH 7.6), 10% glycerol, 0.5 M NaCl, 0.01% NP-40, 5 mM DTT, 2.5 mM MgCl2, 50 μM ZnCl2, and protease inhibitors. Binding reactions (Fig. 2 and 4) were carried out in binding buffer [20 mM HEPES-KOH (pH 7.6), 2.5 mM MgCl2, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT] containing 70 mM KCl and 0.02% NP-40. Unbound proteins were removed with a series of washes with wash buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.2% NP-40, 100 mM NaCl]. Bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by Western blotting.
FIG. 4.
PHO interacts directly with PC, PH, and BRM. (A) 35S-labeled full-length PHO was incubated with GST alone (lane 2), GST-PH(1-595) (lane 3), GST-PH(557-855) (lane 4), GST-PH(817-1096) (lane 5), GST-PH(1077-1590) (lane 6), GST-PC(1-215) (lane 7), GST-PC(196-390) (lane 8), or GST-BRM(230-736) (lane 9) immobilized on glutathione-Sepharose beads. Protein complexes were washed and resolved by SDS-PAGE, and bound proteins were detected by autoradiography. Lane 1 represent 5% of the input material used in the binding reactions. (B) Mapping of the PC- and PH-binding domain of PHO by GST pulldown assays. GST alone (lane 2) or GST-PHO(118-172) (lane 3) was immobilized on glutathione-Sepharose beads and incubated with 35S-labeled full-length PC (residues 1 to 390), PC(1-215), PC(196-390), PH(1-595), PH(557-855), PH(817-1096), or PH(1077-1590). Protein complexes were washed and resolved by SDS-PAGE, and bound proteins were detected by autoradiography. Lane 1 represents 5% of the input material used in the binding reactions. The domain structure of PC (as described by Breiling et al. [7]) and PH (as described by Kyba and Brock [46]) and the residues present in the various GST fusion constructs are indicated.
For pulldowns with GST fused to full-length PHO, GST-PHO was purified from baculovirus-infected Sf9 cells. Binding assays (Fig. 1B) were performed as described above with the following modifications: binding was performed in HEMG containing 100 mM NaCl, and washes were performed with HEMG containing 150 mM NaCl. Far-Western analysis was carried out as described previously (39) with 35S-labeled reticulocyte-expressed protein derivatives.
Following autoradiography to detect bound proteins, blots were reprobed with antibodies against BRM, PH, and PC. All immunological procedures were performed essentially as described previously (31, 32). Rabbit antisera directed against PHO (SN842), PH (SN964), PC (SN965), Groucho (GRO) (PV1 and PV2, pooled), and Moira (MOR) (SN670 and SN671, pooled) were raised by immunization with GST fusion proteins corresponding to PHO amino acids 1 to 49, 42 to 119, and 118 to 172; PH amino acids 1 to 595, 557 to 855, 817 to 1096, and 1077 to 1590; PC amino acids 1 to 215, 196 to 390, and 1 to 390, PV1 and -2, full-length GRO, SN670 and SN671, and full-length MOR. Additional antisera (used in Fig. 2 and 3) were generated by immunization of rabbits with peptides coupled to keyhole limpet hemocyanin essentially as described previously (31, 32). The following peptides were used: PV69 anti-PC, RERDMKGDSSPVA; PV86 anti-PH, KEVPPPGEAKDPGAQ; and PV35 anti-polymerase II 140-kDa subunit (DmRP140), MSVQRIVEDSPAIELQ. The antibodies directed against BRM and ISWI (40), OSA (77) and PSC (50) have been described before.
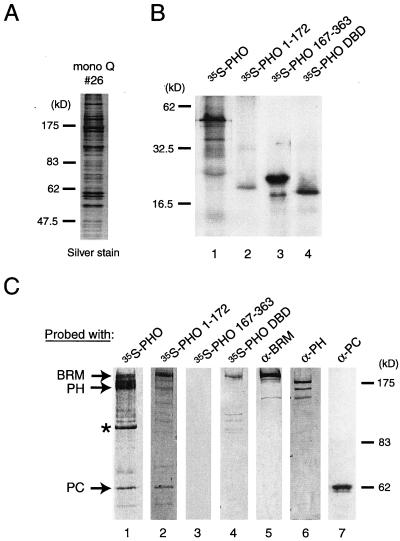
FIG. 3.
Identification of PHO targets within the PC and BRM complexes. (A) Polypeptide composition of a partially purified fly embryo nuclear extract containing BRM and PC (Mono Q10 fraction 26) used in the far-Western analysis. Most of the PC, PH, and BRM present in nuclear extracts is retained in this fraction, as judged by Western blotting analysis with the appropriate antibodies (data not shown). Proteins were resolved by SDS-PAGE and visualized by silver staining. The molecular masses of protein standards are indicated. (B) In vitro-translated proteins used as probes in the far-Western experiments: 35S-labeled full-length PHO (lane 1), PHO(1-172) (lane 2), PHO(167-363) (lane 3), and PHO DBD(355-520) (lane 4). (C) Far-Western blotting analysis of PHO-binding proteins. The purified BRM and PC complexes (Mono Q10 fraction 26) was resolved by SDS-PAGE and transferred to nitrocellulose. The nitrocellulose membrane was treated with 6 M guanidine-HCl, renatured, washed, and incubated with 35S-labeled reticulocyte-expressed full-length PHO(1-520) (lane 1), PHO(1-172) (lane 2), PHO(167-363) (lane 3), and PHO DBD(355-520) (lane 4). After extensive washing, the filter was exposed to film. Filters were reprobed with antibodies directed against BRM (lane 5), PH (lane 6), or PC (lane 7). The positions of BRM (39), PH (PV86), and PC (PV69) which coincide with the immunoreactive band are indicated. An unidentified protein with an estimated molecular mass of about 110 kDa that was bound by PHO is indicated with an asterisk.
Antibodies were affinity purified as described by Hancock and Evan (31). When appropriate, all critical immunoprecipitations and Western immunoblot experiments were repeated with different antisera. For coimmunoprecipitation experiments (Fig. 1A), 300 μl of Drosophila embryo nuclear extract was incubated overnight at 4°C on a spinning wheel with 30 μl of antiserum directed against PHO. Next, 50 μl of protein A-Sepharose beads (Pharmacia) was added and incubated for another hour, and following a series of extensive washes with HEMG buffer containing 250 mM NaCl, bound proteins were eluted with HEMG-1 M NaCl, resolved by SDS-PAGE, and analyzed by Western immunoblotting.
DNA-binding assays.
The DNA band shift assays were essentially performed as described previously (14). Double-stranded oligonucleotides harboring a PHO site (5′-AATTCCGGCGCAGCCATTATGGTGG-3′) (51) were end labeled with T4 polynucleotide kinase. Binding reactions were carried out in a reaction volume of 20 μl of 0.5× HEMG buffer containing 70 mM NaCl, 50 μg of bovine serum albumin per ml, 0.05% NP-40, 1 mM DTT, ≈60 fmol of double-stranded labeled probe, 1 μg of poly(dGdC)-poly(dGdC), and the indicated polypeptide. All binding reactions were carried out on ice for 90 min and were analyzed on 5% polyacrylamide gels run in 0.5× Tris-glycine-0.01% NP-40 buffer at room temperature. For supershift experiments, recombinant GST-PC or GST alone was added to the binding reaction. In the antibody supershift experiments, either affinity-purified anti-PC antiserum or preimmune antiserum was added to the reaction directly before the addition of the labeled probe.
For recruitment assays, PC or BRM complexes were immunopurified from Mono Q fraction 26 with affinity-purified antibodies directed against each of these proteins that were cross-linked to protein A beads with dimethylpimelimidate as described previously (32). Affinity resins were incubated with protein fractions for 2 h at 4°C in HEMG containing 75 mM KCl, followed by extensive washes with excess HEMG containing 150 mM KCl. Next, these beads were incubated in either the presence or absence of recombinant PHO or PHO DBD, and radiolabeled double-stranded oligonucleotides harboring five PHO sites present in natural PREs (5′-CTAGACGGCGCAGCCATTATGGTGCAGTCGGCCATGAGTGATAAAGGCAGCCATTTTCCTGTGCTGCCGCCATATTATTTTGCGGCAGCCATGTTGGATG-3′) (51) as well as an unrelated control DNA fragment that lacks PHO sites. The binding reaction was carried out in binding buffer [25 mM HEPES (pH 7.6), 12.5 mM MgCl2, 10% glycerol, 0.1 mg of bovine serum albumin per ml, 100 ng of poly(dGdC)-poly(dGdC) per μl, and 70 mM KCl] at room temperature for 30 min. After several washes with binding buffer containing 100 mM KCl, bound DNA was resolved on a 1.75% agarose gel and visualized by autoradiography.
Transient-transfection assays.
Plasmids for transfection studies in Drosophila Schneider L2 cells were isolated with Qiagen columns according to the manufacturer's instructions. SL2 cells were propagated in Ultimate Insect serum-free medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum, penicillin (100 μg/ml), streptomycin (100 μg/ml), and fungizone (amphotericin B; 250 μg/ml; Gibco-BRL). All transfections were performed with Fugene (Roche), according to the manufacturer's instructions. Empty vector was added for each transfection to a total amount of 250 ng or 1 μg of DNA (for 24- and 6-well plates, respectively).
For repression assays, SL2 cells were plated at 60 to 80% confluency in 24- or 6-well plates, fresh medium was added the following day, and the plasmids described above were transfected. The next day, the medium was replaced, and 48 h after transfection, the cells were harvested, washed in phosphate-buffered saline, and resuspended in 100 μl or 500 μl (for 24- and 6-well plates, respectively) of lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100). Luciferase activity was determined according to the manufacturer's protocol (Promega).
RESULTS
PHO binds the BRM complex and a PC complex.
To assess whether PHO might be involved in the recruitment of other PcG proteins, we tested its ability to bind endogenous PC, PH, and PSC present in Drosophila embryo extracts. We also investigated whether PHO could bind the multisubunit chromatin-remodeling BRM complex, containing at least three subunits encoded by trxG genes: BRM itself, Osa (OSA), and Moira (MOR) (17, 18, 40). First, we used either preimmune serum or antiserum directed against PHO for immunoprecipitation experiments with Drosophila embryo nuclear extracts (Fig. 1A). Following extensive washes with a buffer containing 250 mM NaCl and detergent, PHO-associated proteins were eluted with a buffer containing 1 M NaCl. Western immunoblot analysis revealed the presence of PcG proteins PC, PH, and PSC (all components of PRC1) as well as three subunits of the BRM complex, BRM, MOR, and OSA, suggesting that a PC complex as well as the BRM complex can associate with PHO in embryo extracts. In contrast, the corepressor Groucho (GRO) and the RNA polymerase II complex detected by an antibody directed against the 140-kDa subunit did not associate with PHO. As expected, none of these proteins were immunoprecipitated with beads coupled to preimmune serum.
Because we suspected a dynamic rather than a stable association of PHO with the PC or BRM complex, we next tested whether full-length recombinant PHO was able to bind either of these complexes present in embryo nuclear extracts. For these experiments, we purified GST-tagged PHO from extracts of insect Sf9 cells infected with recombinant baculoviruses. An affinity resin was generated by immobilization of GST-PHO on glutathione-Sepharose beads and incubated with Drosophila nuclear embryo extracts. Following a series of extensive washes, bound proteins were resolved by SDS-PAGE and analyzed by Western immunoblotting (Fig. 1B). In agreement with the immunoprecipitation experiments, recombinant PHO was found to efficiently bind the BRM complex as well as a PC complex. Neither GRO nor RNA polymerase II was bound by PHO, and none of the PHO-binding proteins were retained on GST beads.
To map the PHO domains involved in binding the BRM or PC complex, we expressed and purified distinct PHO deletions as GST fusion proteins and immobilized these polypeptides on glutathione-Sepharose beads. The various PHO affinity resins were incubated with a partially purified fly embryo nuclear extract (see Materials and Methods). PHO-associated proteins were resolved by SDS-PAGE and analyzed by Western immunoblotting (Fig. 2). The zinc finger DBD (Fig. 2, lane 3) and the first 49 residues of PHO (Fig. 2, lane 9) efficiently bound the BRM complex, as revealed by the presence of its BRM, MOR, and OSA subunits. Neither GST alone (Fig. 2, lane 2) nor other regions of PHO (Fig. 2, lanes 4 to 8) were able to bind the BRM complex. Conversely, the two BRM-binding domains of PHO did not bind PC or PH. However, a distinct region, comprising amino acids 118 to 172, efficiently retained both PC and PH but not the BRM complex (Fig. 2, lane 7). This domain harbors a stretch of residues conserved between PHO and YY1 (indicated with a black box in Fig. 2). None of the remaining regions of PHO or GST alone interacted with either PC or PH, indicating that the protein-protein interactions are selective.
It is well established that PC and PH are part of a large multiprotein complex (24, 65, 66). Indeed, coimmunoprecipitation and size exclusion chromatography experiments confirmed that PC and PH were stably associated in our extracts (data not shown). This PcG protein complex is likely to be similar or related to the previously described PRC1 (65, 66). However, since we have not characterized it further, we will refer to it as the PC complex. In summary, these experiments established that distinct regions of PHO can mediate binding to either the BRM or PC complex. The PHO N-terminal domain (amino acids 1 to 49) and its DBD can bind independently to the BRM complex. A separate domain of 55 residues (amino acids 118 to 172) mediates PC binding.
Identification of targets of PHO within the PC and BRM complexes.
To identify the molecular weights of potential PHO targets, we performed a far-Western experiment with a partially purified column fraction containing both the BRM and PC complexes (Mono Q fraction 26; Fig. 3A). Proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Following denaturation and renaturation, the membrane was probed with radiolabeled full-length PHO or various deletion constructs (Fig. 3B and C). Autoradiography of the membrane suggested direct binding of PHO to proteins that precisely comigrated with BRM, PH, or PC (Fig. 3C, lane 1).
The presence and position of BRM, PH, and PC were established by reprobing the far-Western blots with antibodies directed against these proteins (Fig. 3C, lanes 5 to 6). In addition, we observed binding to a protein of around 110 kDa. None of the other proteins present in the protein fraction used (see Fig. 3A) were significantly bound by PHO, indicating that the interactions detected in the far-Western analysis were selective. PHO(1-172) (Fig. 3C, lane 2), which contains the putative PC-binding domain and the N-terminal BRM complex-binding region, efficiently bound to both BRM and PC, while weak binding to PH was observed. Although PHO(167-363) was efficiently expressed and labeled, it failed to recognize any protein present on the membrane (Fig. 3C, lane 3). The far-Western analysis suggested that the PHO DBD can bind directly to BRM (lane 4) but not to PC or PH. Thus, in agreement with the pulldown assays with embryo extracts (Fig. 1), these experiments suggest that separate PHO domains mediate association with the BRM and PC complexes.
The far-Western analysis indicates that BRM, PC, and PH are the most likely targets contacted by PHO. To obtain direct evidence for binding to PHO, we expressed and purified various polypeptides corresponding to PC, PH, and BRM as GST fusion proteins. These fusion proteins were immobilized on glutathione-Sepharose beads and tested for their ability to bind radiolabeled full-length PHO. As shown in Fig. 4A, PHO bound efficiently to the N-terminal half of PC but not to its C-terminal half. Moreover, PHO associated with a central portion (amino acids 230 to 736) of BRM and the N-terminal and C-terminal domains of PH but not with the central regions of PH or with GST alone. Thus, these experiments with recombinant polypeptides provide further evidence for the notion that PHO interacts specifically with BRM, PC, and PH.
Since our experiments with embryo extracts (Fig. 2) suggested that PHO residues 118 to 172 could recruit an endogenous PC complex, we tested whether this domain could directly recognize recombinant PC or PH in a pulldown assay (Fig. 4B). Indeed, the polypeptide PHO(118-172) efficiently retained full-length PC or its N-terminal half but not its C-terminal half. Moreover, this region of PHO bound the N-terminal portion of PH(1-595) but not to the remainder of the protein. We conclude that PHO(118-172) constitutes a PC-binding domain that associates with both PC and PH.
PHO can link PC to the DNA.
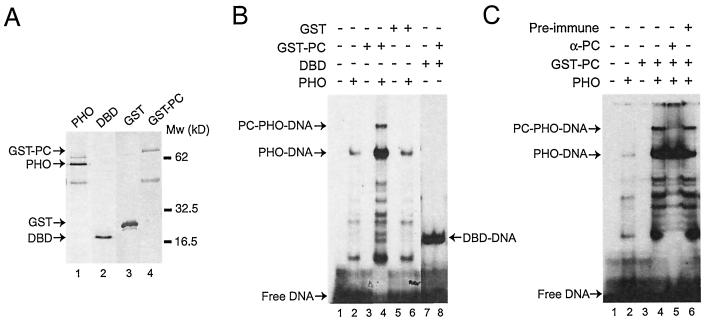
Since PHO but not PC possesses sequence-specific DNA-binding ability, we wondered whether PHO could tether PC to a PHO recognition DNA sequence. Full-length PHO and the C-terminal portion harboring the DBD were expressed in Sf9 insect cells infected with recombinant baculoviruses. PHO polypeptides were immunopurified from Sf9 cell extracts to near homogeneity with their N-terminal Flag epitopes (Fig. 5A). PC was expressed as a GST fusion protein and purified from E. coli extracts (Fig. 5, lane 4). Next, we performed electrophoretic mobility shift assays with the PHO polypeptides either alone or in the presence of GST-PC (Fig. 5B). As expected, full-length PHO (Fig. 5B, lane 2) as well as the DBD (Fig. 5B, lane 7) could bind efficiently a DNA fragment bearing a PHO-binding site. Addition of GST-PC to the binding reaction containing PHO led to the appearance of a novel slower-migrating species, which we interpret as a PC-PHO-DNA complex. Indeed, GST-PC by itself failed to bind DNA (Fig. 5B, lane 3), and GST alone did not induce a PHO supershift (lane 6). Thus, the appearance of the supershifted species depends on the presence of both PHO and PC. Moreover, PHO DBD, which lacks the PC-binding domain, was not supershifted by GST-PC (Fig. 5B, lane 8).
FIG. 5.
PHO can link PC to the DNA. (A) Recombinant Flag-tagged PHO and PHO DBD (residues 355 to 520) were immunopurified from extracts of baculovirus-infected Sf9 cells with an anti-Flag column and eluted under native conditions with a peptide corresponding to the Flag epitope (lanes 1 and 2). Recombinant GST and GST-PC were expressed in E. coli BL21, purified by glutathione-Sepharose chromatography, and eluted with reduced glutathione (lanes 3 and 4). Proteins were resolved by SDS-PAGE and visualized by silver staining. (B) The DNA-binding activity of recombinant PHO and PHO DBD was tested in the absence and presence of GST-PC or GST by electrophoretic mobility shift assays with a radiolabeled double-stranded oligonucleotide containing a single PHO site. Binding reactions were done either in the absence of protein (lane 1) or in the presence of recombinant PHO alone (lane 2), GST-PC (lane 3), both PHO and GST-PC (lane 4), GST (lane 5), both GST and PHO (lane 6), DBD alone (lane 7), or DBD and PC (lane 8). (C) Mobility shift experiment similar to that in B. Binding reactions were done either in the absence of protein (lane 1) or in the presence of recombinant PHO (lane 2), GST-PC (lane 3), or PHO and GST-PC (lanes 4 to 6). Incubations were done either in the absence of antibodies (lane 4) or in the presence of affinity-purified anti-PC (PV69) (lane 5) or preimmune serum (lane 6). The positions of free DNA, PHO-DNA, DBD-DNA, and the ternary PC-PHO-DNA complex are indicated.
Following incubation with affinity-purified antibodies directed against PC, the PC-PHO-DNA complex but not the PHO-DNA complex disappeared (Fig. 5C, lane 5). The appearance of label just below the well may indicate the presence of an antibody-PC-PHO-DNA complex that has difficulty entering the gel. As expected, formation of the ternary PC-PHO-DNA complex was not blocked by the addition of preimmune serum (Fig. 5C, lane 6).
To test whether PHO could link an endogenous PC complex to the DNA, we used beads coated with affinity-purified antibodies directed against either PC or BRM to purify the complexes from embryo nuclear extracts. Next, we assessed the ability of the immobilized PC and BRM complexes to associate with specific DNA sequences in either the presence or absence of PHO. The affinity resins were incubated with a radiolabeled DNA fragment containing five PHO-binding sites and an unrelated control fragment in the presence of an excess of poly(dGdC)-poly(dGdC) competitor DNA. After a series of washes, bound DNA was recovered and analyzed by agarose gel electrophoresis, followed by autoradiography (Fig. 6).
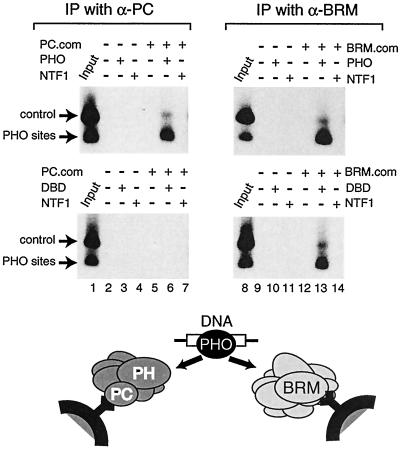
FIG. 6.
PHO can tether an endogenous PC complex to a PRE. PHO links PC and BRM complexes to DNA. The PC or BRM complex was purified with beads coated with affinity-purified antibodies directed against either PC or BRM. The immobilized PC complex (left-hand panels) or BRM complex (right-hand panels) was incubated with a radiolabeled DNA fragment containing PHO-binding sites and an unrelated control fragment in the presence of an excess of poly(dGdC)-poly(dGdC) competitor DNA. Binding reactions were done, as indicated, in the presence of either no additional protein, PHO, or the unrelated transcription factor NTF-1. Following a series of washes, bound DNA was recovered and analyzed by agarose gel electrophoresis, followed by autoradiography. Lanes 1 and 8 represent 5% of the input material used in the binding reactions.
Although the PC complex alone was unable to bind DNA (Fig. 6, lane 5, upper and lower panels), selective association with the PHO sites but not with the control DNA was observed in the presence of PHO (Fig. 6, upper panel, lane 6). An unrelated transcription factor (NTF-1) was unable to link the PC complex to the DNA (Fig. 6, lane 7). Furthermore, PHO was able to tether the BRM complex to the PHO elements but not to the control DNA (Fig. 6, upper panel, lane 13). The PHO DBD, lacking the PC-binding domain, failed to connect the PC complex to the DNA, whereas it still efficiently recruited the BRM complex (Fig. 6, compare lanes 6 and 13, bottom panel). We conclude that PHO can act as a tethering factor that uses distinct domains to link the BRM complex as well as the PC complex to a regulatory DNA element.
PC binding domain of PHO mediates transcriptional repression.
So far, our results have shown that PHO is able to recruit a PC complex to the DNA. Next, we wished to investigate the functional consequences of the PHO-PC interaction in Drosophila cells. Previously, it was demonstrated that PcG proteins tethered to the DNA by fusion to the Gal4 DBD act as transcriptional repressors in transiently transfected cells (11). We took a similar approach and replaced the PHO DBD with that of Gal4 and tested the effect of the fusion protein on gene expression in transfected Drosophila Schneider L2 cells. As a reporter, we used a plasmid containing five Gal4-binding sites located upstream of a strong basal promoter (herpes simplex virus thymidine kinase) driving expression of the luciferase gene.
Cotransfection of the reporter plasmid with a vector expressing Gal4-PHO(1-356) resulted in a clear dose-dependent transcriptional repression (Fig. 7A). As expected, expression of the Gal4-DBD alone did not significantly influence reporter activity. Importantly, the PC-binding domain [Gal4-PHO(118-172)] was sufficient to mediate repression. While the PHO polypeptide containing both the PC- and BRM-binding regions [Gal4-PHO(1-356)] repressed transcription, the isolated BRM-binding domain of PHO (residues 1 to 49) functioned as an activation domain. Thus, the repressive function mediated by the PC-binding domain was dominant in the longer PHO polypeptide. Finally, Gal4-PHO(170-356), which binds neither PC nor BRM, did not affect transcription.
These results show that the PC-binding domain of PHO can repress transcription, possibly through recruitment of an endogenous PC complex. In order to obtain additional support for this notion, we tested whether the overexpression of PC could enhance PHO-directed repression. Indeed, transcriptional repression by Gal4 fusions to PHO polypeptides that could bind PC [PHO(1-356) and PHO(118-172)] was markedly enhanced by overexpression of PC (Fig. 7B). In contrast, neither the Gal4 DBD nor Gal4-PHO(170-356) was able to mediate PC repression.
Next, we tested whether Gal4-PHO was able to mediate repression from a distal position. For these experiments, we used a reporter containing Gal4 binding sites flanked by GAGA sites separated by over 2 kb of intervening DNA from a promoter containing GAGA sites (48). This reporter was cotransfected in the absence or presence of various combinations of expression vectors for the Gal4 DBD, Gal4-PHO, or PC (Fig. 7C). In this setting, Gal4-PHO was again able to mediate transcriptional repression, which was dependent on the presence of the PC-binding domain. Moreover, concomitant expression of PC led to stronger repression. From these results, we conclude that the ability of PHO polypeptides to bind PC in vitro correlates well with their capacity to mediate PC repression in Drosophila cells.
DISCUSSION
PcG-mediated gene silencing is accomplished via the cis-acting PREs, which are the DNA targets for the PcG proteins. However, it is not yet understood how PC and other PcG proteins that lack any apparent sequence-specific DNA-binding ability are directed towards PREs. In this study, we show that PHO can link PC and the BRM chromatin-remodeling complex to the DNA. A small domain in PHO mediates the recruitment of the PC complex by binding to PC and PH. This PC-binding domain directs transcriptional repression in transfected cells, which is enhanced by concomitant overexpression of PC. These results support the notion that PHO contributes to PRE-mediated silencing by tethering a PC complex to repress transcription.
Consistent with a direct role for PHO in PcG silencing, the phenotypes of pho mutants show similarity to those of mutants with changes in other PcG genes (6, 27, 29). Due to a large maternal contribution, animals homozygous for pho null alleles survive up to the pupal stage but display homeotic transformations. In the absence of maternal pho mRNA, embryos die early during development and exhibit segmentation defects as well as severe homeotic transformations. Previous in vivo studies have shown that mutations in PHO DNA-binding sites or in the PHO protein itself compromise PcG silencing (10, 13, 25, 29, 52, 70), and PHO silencing has been observed to be PC dependent in in vivo genetic experiments (25, 76). These observations suggest that PHO DNA-binding elements are important components of at least a subclass of PREs.
In the work presented here, we provide a biochemical and functional link between PHO and PC. We found that PHO can bind both PC and PH through a small 55-amino-acid domain. PHO contacts the N-terminal portion of PC but not its C-terminal repression domain that interacts with nucleosomes and, possibly, other PcG proteins (7, 46). In agreement with the inhibitory function of PC, the PC-binding domain but not other portions of PHO mediates transcriptional repression in transfected Drosophila cells. Significantly, concomitant overexpression of PC leads to stronger repression, supporting the notion that PHO acts through recruitment of PC.
These experiments established a correlation between the ability of PHO polypeptides to bind PC in vitro and their capacity to mediate PC-dependent repression in cells. It should be noted that, because we have not used a purified defined PC complex in our experiments, additional factors might be involved. Furthermore, we have not addressed the role of chromatin in PcG silencing, and it is not clear whether the repression we detected in our transfection assay involves any modulation of chromatin structure. Alternatively, PC may directly block the functioning of the general transcription machinery. This possibility is of interest in light of the recently described interaction between PcG proteins and components of the basal machinery (8, 67). The various potential mechanisms of PcG repression (reviewed in reference 21) are not mutually exclusive, and stable silencing might be the result of multiple blocks to transcription, each acting at a different level.
In addition to PC recruitment, we found that PHO interacts with the BRM complex. PHO contains two BRM complex-binding domains, its N-terminal 49 amino acids and the zinc finger DBD. Interestingly, similar to the zinc finger DBDs of Sp1, EKLF, and GATA-1 (2, 37), the PHO DBD recruits the BRM complex via binding to BRM itself. Although the structural determinants are not yet clear, it appears that a class of zinc finger DBDs has evolved that can simultaneously bind DNA and target a chromatin-remodeling complex (37). Previously, we found that the trxG protein Zeste selectively recruits the BRM complex to activate transcription on chromatin templates (40). In contrast to PHO, Zeste does not contact BRM itself but rather interacts with other BRM-associated proteins, including the trxG proteins Moira and Osa. Thus, different regulators target the BRM complex by binding to distinct subunits. Although we did not directly address the role of BRM in transcriptional repression by PHO, there is evidence to indicate that ATP-dependent remodelers are involved in repression as well as activation (34, 45, 53, 80, 81). The role of the BRM complex may simply be to remodel chromatin and facilitate PHO-DNA binding. Alternatively, BRM may play a more direct role in silencing and cooperate with PcG proteins in the formation of repressive higher-order chromatin structures.
In summary, PHO has been implicated in binding the ESC/E(z) complex (64), the PC complex, and the BRM complex (this paper). Likewise, YY1 has been reported to bind the mammalian homologues of the ESC/E(z) complex (65) and the PC complex (26). Moreover, in light of the high conservation of the Kruppel-like zinc finger DBD of YY1, it seems probable that it will also interact with BRM. Most of these associations appear to be relatively weak and of a transient nature, which is typical of a transcription factor-coregulator interaction (55). Thus, the association of PHO with multiple distinct complexes does not necessarily have to occur simultaneously. For instance, one might speculate that the BRM complex helps PHO to gain access to a chromatinized PRE. Subsequent recruitment of the ESC/E(z) complex may lead to histone deacetylation, followed by recruitment of a PC-containing PRC1-related complex.
Although it has been well established that PHO contributes to PRE function, the presence of a series of PHO sites by itself does not suffice to reconstitute a PRE (10). This indicates that PHO may act in a combinatorial fashion with other tethering factors such as GAGA and Zeste. It has become clear that PREs are composed of a multitude of distinct binding elements which, depending on their context, can be redundant with, cooperate with, or antagonize each other (5, 47, 49, 61). For example, the trxG protein GAGA, generally thought of as an activator that induces chromatin remodeling (19, 20, 42, 78), has been implicated in PcG repression (13, 30, 35, 52).
Interestingly, we recently found that GAGA is required for PHO binding to a chromatinized PRE, suggesting that PHO and GAGA elements together may form a functional module (T. Mahmoudi, L. M. P. Zuijderduijn, A. Mohd-Sarip, and C. P. Verrijzer, submitted for publication). Moreover, evidence has been presented indicating that GAGA may be more directly involved in PC recruitment. In coimmunoprecipitation experiments, GAGA has been found associated with a complex containing PC, PH, PHO, E(z), ESC, and RPD3 (but not PSC) in early embryonic extracts (64). In extracts from older embryos, GAGA was found to be associated with a complex including PC, PH, PSC, and RPD3, whereas PHO coimmunoprecipitates with ESC, E(z), and RPD3. However, in other studies, PHO was not found in the purified ESC/E(z) complex (75), and GAGA was absent from the purified PRC1 complex containing PC, PSC, PH, dRING1, and many of the TATA-binding protein-associated factor components of the general transcription factor TFIID (67, 68).
Interestingly, the sequence-specific DNA-binding protein Zeste was identified as an approximately stoichiometric component of PRC1, raising the possibility that Zeste may contribute to DNA targeting (67). Although Zeste can activate transcription in a BRM-dependent manner (40), it also displays genetic interactions with PcG repressors (59, 60). Thus, similar to our results with PHO, it appears that Zeste can interact with the BRM complex as well as with a PC complex. Since neither LexA-PHO nor LexA-GAGA suffices to mediate stable PcG silencing (63) and since by themselves the binding elements for Zeste, PHO, and GAGA do not constitute a PRE, it seems clear that PRE silencing is not achieved by a single recruiter.
Finally, it should be noted that in addition to setting up the expression pattern of the homeotic genes, the Gap proteins may very well play a role in the initial recruitment of PcG complexes (5, 43). For instance, the early repressor HB binds the dMi2 chromatin remodeling and histone deacetylase complex that genetically participates in PcG repression (43). Although dMi-2 might interact directly with PcG proteins, an alternative scenario would be that the deacetylation by dMi-2 creates a chromatin structure conducive to the subsequent assembly of a silencing PcG complex.
In conclusion, current evidence suggests that PcG-mediated silencing is not achieved by a one-step mechanism. While the underlying mechanisms remain enigmatic, it has become clear that PRE function involves a highly elaborate interplay of protein-DNA and protein-protein interactions that direct the formation of a specialized higher-order chromatin structure. At least three distinct steps appear to be distinguishable: targeting to a specific gene, transcriptional repression, and heritable maintenance of the silenced state (4). In this study, we have investigated the role of one of the PRE-binding proteins, PHO, in the recruitment of a PC complex. Our results demonstrate a direct biochemical and functional link between PHO and PC-mediated transcriptional repression.
Acknowledgments
We are very grateful to Renato Paro, Hugh Brock, Eric Kalkhoven, and Rob Hoeben for the gift of plasmids, Renato Paro for the gift of anti-Polycomb antibodies, and Jesper Svejstrup, Lee Fradkin, Eric Kalkhoven, Natalie Little, and members of our laboratory for critical reading of the manuscript.
REFERENCES
- 1.Alkema, M. J., M. Bronk, E. Verhoeven, A. Otte, L. J. van't Veer, A. Berns, and M. van Lohuizen. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 11:226-240. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Austin, R. J., and M. D. Biggin. 1996. Purification of the Drosophila RNA polymerase II general transcription factors. Proc. Natl. Acad. Sci. USA 93:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuchle, D., G. Struhl, and J. Muller. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993-1004. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, M., and J. Muller. 1995. Transcriptional silencing of homeotic genes in Drosophila. Bioessays 17:775-784. [DOI] [PubMed] [Google Scholar]
- 6.Breen, T. R., and I. M. Duncan. 1986. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 118:442-456. [DOI] [PubMed] [Google Scholar]
- 7.Breiling, A., E. Bonte, S. Ferrari, P. B. Becker, and R. Paro. 1999. The Drosophila Polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol. Cell. Biol. 19:8451-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. [DOI] [PubMed] [Google Scholar]
- 9.Brock, H. W., and M. van Lohuizen. 2001. The Polycomb group—no longer an exclusive club? Curr. Opin. Genet. Dev. 11:175-181. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 11.Bunker, C. A., and R. E. Kingston. 1994. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 14:1721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busturia, A., and M. Bienz. 1993. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 12:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busturia, A., A. Lloyd, F. Bejarano, M. Zavortink, H. Xin, and S. Sakonju. 2001. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128:2163-2173. [DOI] [PubMed] [Google Scholar]
- 14.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, C. S., L. Rastelli, and V. Pirrotta. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13:2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang, A., M. B. O'Connor, R. Paro, J. Simon, and W. Bender. 1995. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development 121:1681-1689. [DOI] [PubMed] [Google Scholar]
- 17.Collins, R. T., T. Furukawa, N. Tanese, and J. E. Treisman. 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18:7029-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby, M. A., C. Miller, T. Alon, K. L. Watson, C. P. Verrijzer, R. Goldman-Levi, and N. B. Zak. 1999. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croston, G. E., L. A. Kerrigan, L. M. Lira, D. R. Marshak, and J. T. Kadonaga. 1991. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science 251:643-649. [DOI] [PubMed] [Google Scholar]
- 20.Espinas, M. L., E. Jimenez-Garcia, A. Vaquero, S. Canudas, J. Bernues, and F. Azorin. 1999. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 274:16461-16469. [DOI] [PubMed] [Google Scholar]
- 21.Fauvarque, M. O., and J. M. Dura. 1993. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 7:1508-1520. [DOI] [PubMed] [Google Scholar]
- 22.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell. Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 23.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 24.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fritsch, C., J. L. Brown, J. A. Kassis, and J. Muller. 1999. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126:3905-3913. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, E., C. Marcos-Gutierrez, L. M. del Mar, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehring, W. J. 1970. A recessive lethal (l(4)29) with a homeotic effect in D. melanogaster. Drosophila Information Service 45:103. [Google Scholar]
- 28.Gindhart, J. G., Jr., and T. C. Kaufman. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girton, J. R., and S. H. Jeon. 1994. Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev. Biol. 161:393-407. [DOI] [PubMed] [Google Scholar]
- 30.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hancock, D. C., and G. I. Evan. 1998. Production and characterization of antibodies against synthetic peptides. Methods Mol. Biol. 80:15-22. [DOI] [PubMed] [Google Scholar]
- 32.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Heberlein, U., and R. Tjian. 1988. Temporal pattern of alcohol dehydrogenase gene transcription reproduced by Drosophila stage-specific embryonic extracts. Nature 33:410-415. [DOI] [PubMed] [Google Scholar]
- 34.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 35.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jimenez, G., C. P. Verrijzer, and D. Ish-Horowicz. 1999. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol. Cell. Biol. 19:2080-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadonaga, J. T. 1990. Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J. Biol. Chem. 265:2624-2631. [PubMed] [Google Scholar]
- 39.Kaelin, W. G. Jr., W. Krek, W. R. Sellers, J. A. DeCaprio, F. Ajchenbaum, C. S. Fuchs, T. Chittenden, Y. Li, P. J. Farnham, and M. A. Blanar. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70:351-364. [DOI] [PubMed] [Google Scholar]
- 40.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 41.Kassis, J. A. 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136:1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 44.Kennison, J. A. 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29:289-303. [DOI] [PubMed] [Google Scholar]
- 45.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 46.Kyba, M., and H. W. Brock. 1998. The Drosophila Polycomb group protein Psc contacts ph and Pc through specific conserved domains. Mol. Cell. Biol. 18:2712-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyko, F., and R. Paro. 1999. Chromosomal elements conferring epigenetic inheritance. Bioessays 21:824-832. [DOI] [PubMed] [Google Scholar]
- 48.Mahmoudi, T., K. R. Katsani, and C. P. Verrijzer. 2002. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahmoudi, T., and C. P. Verrijzer. 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20:3055-3066. [DOI] [PubMed] [Google Scholar]
- 50.Martin, E. C., and P. N. Adler. 1993. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development 117:641-655. [DOI] [PubMed] [Google Scholar]
- 51.Mihaly, J., R. K. Mishra, and F. Karch. 1998. A conserved sequence motif in Polycomb-response elements. Mol. Cell 1:1065-1066. [DOI] [PubMed] [Google Scholar]
- 52.Mishra, R. K., J. Mihaly, S. Barges, A. Spierer, F. Karch, K. Hagstrom, S. E. Schweinsberg, and P. Schedl. 2001. The iab-7 Polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreira, J. M., and S. Holmberg. 1999. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 18:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller, J., and M. Bienz. 1991. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 10:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 56.Ng, J., C. M. Hart, K. Morgan, and J. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlando, V., E. P. Jane, V. Chinwalla, P. J. Harte, and R. Paro. 1998. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 17:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orlando, V., and R. Paro. 1993. Mapping Polycomb-repressed domains in the bithorax complex with in vivo formaldehyde cross-linked chromatin. Cell 75:1187-1198. [DOI] [PubMed] [Google Scholar]
- 59.Pelegri, F., and R. Lehmann. 1994. A role of polycomb group genes in the regulation of gap gene expression in Drosophila. Genetics 136:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips, M. D., and A. Shearn. 1990. Mutations in polycombeotic, a Drosophila polycomb-group gene, cause a wide range of maternal and zygotic phenotypes. Genetics 125:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pirrotta, V. 1998. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93:333-336. [DOI] [PubMed] [Google Scholar]
- 62.Pirrotta, V., and L. Rastelli. 1994. White gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. Bioessays 16:549-556. [DOI] [PubMed] [Google Scholar]
- 63.Poux, S., D. McCabe, and V. Pirrotta. 2001. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development 128:75-85. [DOI] [PubMed] [Google Scholar]
- 64.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Satijn, D. P., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satijn, D. P., and A. P. Otte. 1999. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim. Biophys. Acta 1447:1-16. [DOI] [PubMed] [Google Scholar]
- 67.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 68.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 69.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 70.Shimell, M. J., A. J. Peterson, J. Burr, J. A. Simon, and M. B. O'Connor. 2000. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 218:38-52. [DOI] [PubMed] [Google Scholar]
- 71.Simon, J., A. Chiang, W. Bender, M. J. Shimell, and M. O'Connor. 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158:131-144. [DOI] [PubMed] [Google Scholar]
- 72.Strouboulis, J., S. Damjanovski, D. Vermaak, F. Meric, and A. P. Wolffe. 1999. Transcriptional repression by XPc1, a new Polycomb homolog in Xenopus laevis embryos, is independent of histone deacetylase. Mol. Cell. Biol. 19:3958-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strutt, H., G. Cavalli, and R. Paro. 1997. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J 16:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strutt, H., and R. Paro. 1997. The Polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol. Cell. Biol. 17:6773-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 76.Tillib, S., S. Petruk, Y. Sedkov, A. Kuzin, M. Fujioka, T. Goto, and A. Mazo. 1999. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol. 19:5189-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Treisman, J. E., A. Luk, G. M. Rubin, and U. Heberlein. 1997. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 11:1949-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsukiyama, T., P. B. Becker, and C. Wu. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367:525-532. [DOI] [PubMed] [Google Scholar]
- 79.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 80.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 82.Zink, D., and R. Paro. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14:5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]