Abstract
Utilizing a genetic screen in the yeast Saccharomyces cerevisiae, we identified a novel autoactivation region in mammalian MEK1 that is involved in binding the specific MEK inhibitor, PD 184352. The genetic screen is possible due to the homology between components of the yeast pheromone response pathway and the eukaryotic Raf-MEK-ERK signaling cascade. Using the FUS1::HIS3 reporter as a functional readout for activation of a reconstituted Raf-MEK-ERK signaling cascade, randomly mutagenized MEK variants that were insensitive to PD 184352 were obtained. Seven single-base-change mutations were identified, five of which mapped to kinase subdomains III and IV of MEK. Of the seven variants, only one, a leucine-to-proline substitution at amino acid 115 (Leu115Pro), was completely insensitive to PD 184352 in vitro (50% inhibitory concentration >10 μM). However, all seven mutants displayed strikingly high basal activity compared to wild-type MEK. Overexpression of the MEK variants in HEK293T cells resulted in an increase in mitogen-activated protein (MAP) kinase phosphorylation, a finding consistent with the elevated basal activity of these constructs. Further, treatment with PD 184352 failed to inhibit Leu115Pro-stimulated MAP kinase activation in HEK293T cells, whereas all other variants had some reduction in phospho-MAP kinase levels. By using cyclic AMP-dependent protein kinase (1CDK) as a template, an MEK homology model was generated, with five of the seven identified residues clustered together, forming a potential hydrophobic binding pocket for PD 184352. Additionally, the model allowed identification of other potential residues that would interact with the inhibitor. Directed mutation of these residues supported this region's involvement with inhibitor binding.
The mitogen-activated protein (MAP) kinase cascade, comprised of MAP kinase (ERK), MAP kinase kinase (MEK), and MAP kinase kinase kinase (Raf), is an evolutionarily conserved signaling module that regulates growth, differentiation, and movement in eukaryotic cells in response to extracellular stimulation (reviewed in reference 23). A key regulatory component of this pathway is MAP kinase kinase, or MEK, a dual-specificity kinase that phosphorylates MAP kinase (ERK) on specific threonine and tyrosine residues. This phosphorylation activates MAP kinase and induces a host of downstream cellular responses (reviewed in reference 10).
MEK itself, is subject to regulation and activation by Raf phosphorylation on two serine residues, Ser218 and Ser222 (2, 33), which lie in a regulatory loop between conserved kinase subdomains VII and VIII (16). Substitution of these serine residues with negatively charged amino acids, such as aspartate or glutamate, partially mimics the phosphorylation modification and results in a constitutively active kinase, presumably through stabilization of the regulatory loop, allowing the enzyme to retain an active conformation (2).
Another regulatory feature of MEK is a proline-rich region, located carboxy terminal to the regulatory loop, which appears to be critical for Raf association (6), and may play an important role in the ability of MEK to efficiently activate MAP kinase (8). Additionally, this polyproline region contains phosphorylation sites that are phosphorylated in vitro by a number of other kinases, including MAP kinase, which may further regulate MEK activity (14).
The region of MEK amino-proximal to the catalytic core (approximately amino acids 1 to 67) has also been shown to play an important role in regulating kinase activity. The putative MAP kinase docking site is found within the first 32 amino acids of MEK, and these residues alone are sufficient for in vitro binding of MAP kinase (15). In particular, sequences of two to four positively charged residues in the extreme amino terminus of MEK (Lys3, -4, and -5 in MEK1) are conserved in all MEK family members (MKK1 to MKK7) and are considered essential for interaction with MAP kinases (3, 31). Additionally, the amino termini of MEK1 and -2 contain a recognition sequence for anthrax lethal factor, a proteolytic component of the Bacillus anthracis virulence factor. Lethal factor cleaves the first seven amino acids from MEK1, which impairs MEK enzymatic activity both in vitro and in vivo (12). Finally, amino acids 44 to 51 of MEK have been characterized as an autoactivation domain, since deletion of these residues results in an 80-fold increase in MEK activity relative to wild-type enzyme (20). When this deletion is coupled with the activating serine substitutions (Ser218Asp/Ser222Asp), the increase in activity is 640-fold over that of wild-type enzyme.
A key role for MEK in the development of tumors has been described. We have reported that a small molecule inhibitor of MEK, PD 184352, is capable of inhibiting up to 80% of tumor growth of human and murine colon carcinomas in mice (26). The compound is selective for MEK1 and -2 and is noncompetitive for ATP and MAP kinase (26). PD 184352 can block the activation of MEK by Raf, as well as inhibiting the active (Raf phosphorylated or mutationally activated) form of the kinase. However, in the present study, we show PD 184352 does not block Raf phosphorylation of MEK either in vitro or in vivo. In order to gain insight into the molecular mechanism of PD 184352-mediated inhibition of MEK kinase activity, we devised a genetic approach by using the budding yeast Saccharomyces cerevisiae to identify amino acids involved in the interaction of PD 184352 and MEK. Here we report the identity of a structural motif that interacts with the MEK specific inhibitor and describe a novel autoactivation domain within the kinase catalytic core that regulates overall MEK activity.
MATERIALS AND METHODS
Plasmids.
Yeast expression vectors pRS314 (TRP1, CEN6, and ARSH4) and pRS425 (LEU2 2μ) (29) were modified by insertion of a 0.7-kb fragment encoding the GAL1/10 promoter from pBJ247 (American Type Culture Collection). Human Raf-1 (GenBank no. NM002880) was rendered constitutively active by deletion of regulatory amino acids 27 to 302 (5) via PCR and inserted into the BamHI/SstI sites of pRS314, 3′ of the GAL1/10 promoter, generating pRS314GALRafΔNT. Human MEK1 cDNA (GenBank no. L11284) was modified via PCR by the addition of a 5′ BamHI site (underlined), a yeast Kozak sequence (italicized), and codon changes for yeast tRNA bias (boldface) with the following oligonucleotide: 5′-GGATCCACACATAAATAAACAAAATGCCAAAGAAGAAGCCAACGCCAA-3′. In addition, a sequence corresponding to the FLAG epitope (italicized), followed by an SstI site (underlined) was added to the 3′ terminus of MEK with the following oligonucleotide: 5′-AAGCTTTTACTTGTCATCGTCGTCCTTGTAGTCGACGCCAGCAGCATGG. The BamHI/SstI PCR product encoding the modified MEK cDNA was inserted into pRS425 3′ of the GAL1/10 promoter, generating pRS425GALMEK. For expression of recombinant protein in bacteria, modified MEK cDNA was subcloned into the NdeI and HindIII sites of pET21b plasmid (Novagen/CN Biosciences). Site-specific mutagenesis (QuikChange Mutagenesis kit; Stratagene, Inc.) was used to generate various point mutations in the MEK coding sequence in pET21b. For mammalian cell transfection assays, MEK cDNA was subcloned into the BamHI/HindIII sites of pCMV4A (Stratagene) by standard techniques, with a 5′ mammalian Kozak sequence (−3ACC) added by PCR.
Generation of ste11Δ ste7Δ ste5Δ mutant yeast strain.
S. cerevisiae haploid strain SY2002 (MATaHIS3::FUS1::HIS3 mfa2-Δ1::FUS1-lacZ ade1 leu2-3,112 trp1-DH1 ura3-52), a generous gift from G. F. Sprague, Jr., was modified by standard yeast genetic protocols (19) to disrupt the mating response pathway genes STE11, STE7, and STE5, generating a ste11Δ ste7Δ ste5Δ mutant strain. Unless otherwise noted, we used the genetic procedures and media described by Sherman et al. (28). Disruption of the loci was confirmed by PCR, growth on appropriate nutrient deficient media, and histidine auxotrophy conferred by disruption of the mating response pathway. Complementation of the disrupted pheromone response pathway was confirmed by transforming the ste11Δ ste7Δ ste5Δ strain with the Raf and MEK constructs and restoring the ability of the yeast to propagate on medium lacking histidine.
Random mutagenesis of MEK.
Escherichia coli XL-1 Red (Stratagene, Inc.) were used to generate random mutations in MEK cDNA due to deficiency in three primary DNA repair pathways (muts, mutD, and mutT) that results in a mutation rate ∼5,000-fold higher than the wild-type strain. Plasmid pRS425GALMEK1 was transformed into E. coli XL-1 Red by standard methods and propagated on Luria-Bertani agar medium containing 100 μg of ampicillin/ml at 37°C for 24 h, allowing mutations to accumulate in the plasmid DNA. Colonies were scraped from the agar plates and transferred to 2 liters of Luria-Bertani liquid culture containing 100 μg of ampicillin/ml and allowed to expand for 18 h at 37°C. Cells were harvested by centrifugation, and plasmid DNA prepared by using standard reagents (Qiagen, Inc.).
MEK mutant library transformation and identification of PD 184352-resistant variants.
The ste11Δ ste7Δ ste5Δ FUS1::HIS3 strain harboring pRS314GALRafΔNT was transformed with 300 μg of randomly mutagenized MEK DNA by using the lithium acetate method of Ito et al. (17) and plated onto synthetic medium lacking tryptophan and leucine (SD−Trp,Leu). Yeast cells containing functional MEK were selected by growth in yeast nitrogen base (YNB) lacking Trp, Leu, and His (YNB−Trp,Leu,His), supplemented with 2% galactose to induce expression of both RafΔNT and MEK. PD 184352-resistant clones were selected by propagating histidine prototrophs in YNB−Trp,Leu,His supplemented with 2% galactose and containing 1 μM PD 184352. From this population, 275 separate yeast colonies were obtained. These were restreaked on YNB-2% galactose−Trp,Leu,His plus 1 μM PD 184352 to confirm resistance. Mutant-MEK plasmid DNA from these colonies was isolated (Bio 101) and rescued by transformation into E. coli KC8 (Clontech Laboratories, Inc.) grown on minimal medium lacking leucine and supplemented with 100 μg of ampicillin/ml. Mutations in the MEK coding sequence were identified by sequence analysis by using an automated sequencer (Perkin-Elmer Biosystems).
Expression and purification of recombinant MEK mutants.
Plasmid pET21b (Novagen) encoding a MEK His6-tagged fusion was transformed into E. coli BL21(DE3) by standard methods. Expression of recombinant His6-tagged MEK in bacterial culture was induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 200 μM) and purified by using IMAC methods (Clontech Laboratories, Inc.). The molecular mass and protein concentration were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
In vitro kinase assays.
The kinase activities of recombinant MEK(His)6 proteins were determined by using a glutathione S-transferase (GST) fusion protein of kinase-inactive ERK1 (GSTERK1K71R) as a substrate. Recombinant MEK protein (50 ng) was incubated with various concentrations of PD 184352 in 20 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 10 μM ATP, and 5 μCi of [γ-32P]ATP in a total volume of 25 μl. Raf-1 (0.5 U; Upstate Biotechnology) was added, and the reaction initiated with the addition of GSTERK1(K71R) fusion protein (1 μg). Working within the linear range of the reaction, the samples were incubated at 27°C for 20 min and then quenched with 5 μl of 6× Laemmli sample buffer. The proteins were resolved by SDS-PAGE, and phosphoproteins were visualized by autoradiography. Radiolabel incorporated into ERK was determined by excision of the corresponding protein band and quantitated by using a standard scintillation counter (Beckman Instruments, Inc.).
3H-PD 184352 binding assay.
A scintillation proximity assay (SPA) was utilized to evaluate direct interaction of PD 184352 with the mutant MEK proteins. Recombinant MEK(His)6 proteins were prepared as described. 3H-PD 184352 was custom synthesized (Amersham) with a specific activity of 22 Ci/mmol. MEK proteins were incubated with 3H-PD 184352 in binding buffer (10 mM K2HPO4, 10 mm KH2PO4, 50 mM NaCl, 2 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 10% glycerol; pH 7.6) in 96-well flat-bottom tissue culture plates in a final volume of 100 μl. The plates were shaken for 30 min at room temperature, followed by the addition of 50 μl of YSI-Copper-His TAG SPA beads (Amersham) in binding buffer containing 1.25 mg of bovine immunoglobulin G (IgG; Sigma)/ml. Samples were shaken an additional 45 min, and the amount of 3H-PD 184352 bound to the MEK-SPA bead complex was determined in a Wallac MicroBeta plate counter.
Analysis of phospho-MEK levels in cultured cells.
KBALB cells were cultured in Dulbecco modified Eagle medium supplemented with 10% newborn calf serum, 25 mM HEPES (pH 7.5), and 1× Glutamax (Life Technologies, Inc.). Cells were serum deprived for 24 h, treated with PD 184352 at various concentrations for 0.5 to 1 h, and then stimulated with 50 ng of platelet-derived growth factor (PDGF)/ml for 5 min at 37°C. Lysates were prepared in 10 mM HEPES (pH 7.4), 50 mM β-glycerophosphate, 1% Triton X-100 (Sigma-Aldrich Co.), 70 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium vanadate, and 1 μM leupeptin (Sigma-Aldrich Co.) and then clarified by centrifugation at 30,000 × g. Protein concentrations in the lysates were determined by BCA assay (Pierce; Perbio). Extracts (15 to 20 μg) were subjected to SDS-PAGE electrophoresis, transferred to nitrocellulose and probed with anti-phospho-MEK (New England Biolabs, Inc.), anti-active MAP kinase (Promega Corp.), and anti-p44MAPK1/p42MAPK2 (Santa Cruz Biotechnology, Inc.) antibodies. Immunoreactive proteins were visualized with ECL reagents (Amersham International, plc.).
Expression of MEK mutants in mammalian cells.
HEK293T cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 25 mM HEPES (pH 7.5), and 1× Glutamax (Life Technologies, Inc.). Transient transfection of 1 to 2 μg of DNA (pCMV4A clones) into HEK293T cells was performed by using Lipofectamine (Life Technologies, Inc.). At 24 h posttransfection, cell extracts were prepared as described above, subjected to SDS-PAGE electrophoresis, and transferred to nitrocellulose. The immunoreactive proteins were visualized by Western blotting with anti-FLAG-M2 (Sigma-Aldrich Co.) and anti-active MAP kinase antibodies (Promega Corp.).
Homology modeling.
Homology models of MEK1 were constructed by using either the modeling package of Look (Molecular Applications Group) or the Homology module of InsightII (MSI). The N-terminal region prior to the kinase domain and an internal proline-rich loop region (residues 268 to 308) were not included for modeling. The crystal structure (PDB: 1CDK) of the active form of cyclic AMP-dependent protein kinase A was used as a template for homology modeling (4). Structure-based multiple sequence alignment of representative protein kinases and multiple alignment of all known MEK sequences were used to generate the final alignment. The final model was minimized by using the Discover module of InsightII and was evaluated by using the Procheck and Profile3D modules of InsightII.
RESULTS
PD 184352 does not inhibit MEK phosphorylation by Raf in vitro or in vivo.
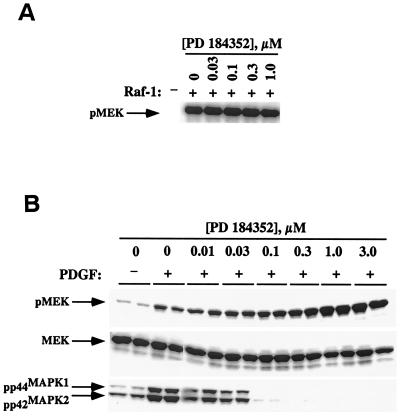
To determine whether PD 184352 inhibition of MEK occurs by blocking activation of MEK by Raf-1, we assayed the ability of Raf to phosphorylate a GST fusion protein of kinase inactive MEK (GSTMEKK97R) in the presence of PD 184352 (0 to 1 μM) in an in vitro kinase assay. This assay is a direct measurement of serine 218 and serine 222 phosphorylation in MEK by Raf-1. A similar experiment using MEK with the same residues mutated to alanine showed no detectible phosphorylation by Raf-1 (data not shown). As shown in Fig. 1A, PD 184352 was unable to block Raf-mediated MEK phosphorylation in vitro.
FIG. 1.
Effect of PD 184352 on MEK phosphorylation by Raf in vitro and in vivo. (A) Kinase-inactive MEK was incubated with Raf in the presence of increasing concentrations of PD 184352 in an in vitro kinase assay as described in Materials and Methods. PD 184352 did not inhibit Raf-mediated phosphorylation of MEK under these conditions. (B) KBALB cells were serum starved, treated for 30 min with the noted concentrations of PD 184352, and then stimulated for 5 min with PDGF. Lysates were prepared and immunoblotted for levels of phosphorylated MEK (pMEK), total MEK (MEK), and phosphorylated MAPK (pp44MAPK1 and pp42MAPK2). Samples are shown in duplicate.
To evaluate the effect of PD 184352 on MEK phosphorylation in whole cells, serum deprived (24 h) KBALB cells were treated with various concentrations of PD 184352 for 1 h at 37°C, followed by stimulation with PDGF for 5 min. Cell extracts were prepared, and the lysates were probed with antibodies recognizing phosphorylated MAP kinase and phosphorylated MEK. The latter antibody recognizes the Raf-specific MEK phosphorylation sites, Ser218 and Ser222. Unexpectedly, although treatment with PD 184352 completely abolished phospho-MAPK levels at a 0.1 μM concentration, the level of phospho-MEK actually increased with increasing inhibitor concentration. The highest dose of PD 184352 (3 μM) caused a threefold increase in phosphorylated MEK relative to PDGF-stimulated control (Fig. 1B). Increased MEK phosphorylation in the presence of PD 184352 was also observed in the absence of agonist and was found to be time dependent as well (data not shown). Additionally, the hyperphosphorylation of MEK with PD 184352 treatment was not merely an artifact of the KBALB cell line. The same effects were also observed in Colon26, H61, and NIH 3T3 fibroblast cell lines (data not shown).
A possible explanation for the elevated levels of MEK phosphorylation in the presence of PD 184352 is the existence of negative feedback loops, whereby the inhibition of MAP kinase leads to hyperactivation of Raf-1 or some other signaling component upstream of Raf (27). However, while the dose dependency of PD 184352-stimulated phospho-MEK accumulation and MEK inhibition are distinct, it is clear that this compound does not inhibit Raf kinase activity.
An S. cerevisiae genetic screen to identify the residues of MEK that are involved in PD 184352-mediated inhibition.
Since PD 184352 is noncompetitive for ATP and exerts unexpected effects on MEK phosphorylation by Raf, we devised a screen to identify interacting residues in the protein. Homology between the mammalian MAP kinase signaling module and the budding yeast pheromone response pathway allows functional complementation of some members of this yeast signaling pathway with mammalian homologues. Genes for the yeast homologues of the mammalian MAP kinase cascade, ste11 (Raf), ste7 (MEK), and ste5 (scaffolding protein), were disrupted in a strain harboring a FUS1::HIS3 reporter, stably integrated at the FUS1 locus (SY2002). Strain SY2002 is auxotrophic for histidine, and growth on histidine-deficient medium is dependent on an intact pheromone response pathway. Disruption of any member of the pheromone response cascade renders this strain incapable of growing on a medium lacking histidine (30). However, transformation of the ste11Δ ste7Δ ste5Δ mutant strain with the mammalian counterparts of the MAP kinase pathway, specifically MEK1 and a constitutively active form of Raf (RafΔNT), restored the integrity of the signaling pathway and rendered the yeast prototrophic for histidine. Complementation of ste11 and ste7 required high-level expression of both MEK and RafΔNT, and this was accomplished by placing MEK and RafΔNT under the control of a galactose-inducible promoter. However, when yeast expressing RafΔNT and MEK1 were propagated on histidine-deficient medium containing 1 μM PD 184352, complementation was blocked, suggesting that PD 184352 was efficiently inhibiting MEK activity. PD 184352 was not merely toxic to the yeast, since the parental strain (SY2002) was able to propagate in the presence of 1 μM PD 184352. This control also demonstrates that PD 184352 did not inhibit the yeast MEK homologue ste7, since the wild-type (STE7+) strain grew normally on histidine-deficient medium containing PD 184352 (data not shown).
Isolation of PD 184352-resistant MEK mutants.
Random mutagenesis was performed on MEK cDNA by transforming pRS425GALMEK into E. coli XL-1 Red, a bacterial strain defective in three major DNA repair mechanisms (mutD, mutS, and mutT). The subsequent pool of mutagenized MEK plasmids was then transformed into the ste11Δ ste7Δ ste5Δ FUS1::HIS3 strain harboring pRS314GALRafΔNT. Mutations generating inactive forms of MEK, such as premature stop codons, were removed from the screen through the inability of the defective kinase to complement the ste7 deletion and return SY2002 to histidine prototrophy. Transformants capable of growing in the absence of histidine were then screened for their ability to grow on histidine-deficient medium containing 1 μM PD 184352.
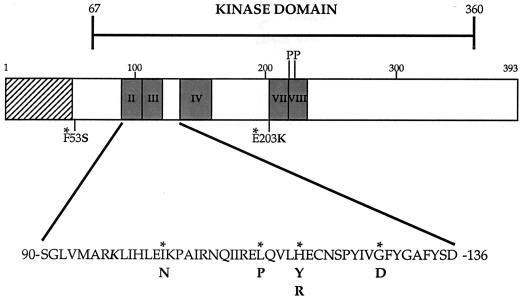
Seven different mutant alleles were identified from the screen, all of which were single-base-pair mutations in the MEK coding sequence, summarized schematically in Fig. 2. Interestingly, five of the seven mutations were clustered in the region of the enzyme spanning kinase subdomains III and IV (Fig. 2). When the isolated mutant alleles were transformed into a naive ste11Δ ste7Δ ste5Δ strain, the MEK variants allowed growth on media containing PD 184352 (data not shown), confirming that a spontaneous mutation of the yeast host had not conferred PD 184352 resistance to this strain.
FIG. 2.
Identification of PD 184352-resistant MEK mutants by using a S. cerevisiae genetic screen. An illustration of MEK gene indicating mutations found in the genetic screen is shown. Native amino acids are marked by an asterisk, with mutations indicated in boldface. Also depicted are the putative MAP kinase binding region (▨), relevant kinase subdomains (░⃞), Raf phosphorylation sites (P), and the lysine residue essential for phosphoryl transfer (K).
Biochemical properties of MEK mutants.
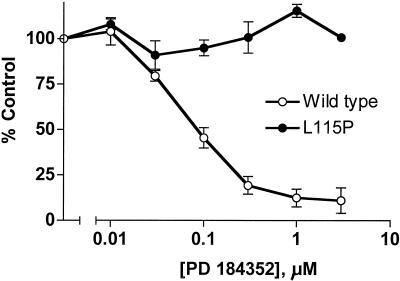
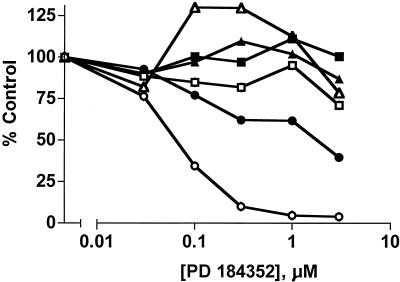
To characterize the enzymatic activities of the MEK variants identified from the screen, each was recreated by site-specific mutagenesis of a MEK(His)6 fusion construct. Recombinant MEK(His)6 proteins were expressed in bacteria, purified by immobilized metal affinity chromotography, and assayed in vitro for the ability to phosphorylate a GST fusion protein of a kinase-inactive form of mammalian ERK1 [GSTERK1(K71R)]. Surprisingly, of the seven PD 184352 resistant mutants isolated, only Leu115Pro was completely insensitive to the MEK inhibitor (Fig. 3). However, with the exception of Phe53Ser and Glu203Lys, the MEK mutants had 50% inhibitory concentration (IC50) values for PD 184352 that were up to 5.4-fold higher than that of wild-type MEK (Table 1).
FIG. 3.
PD 184352-mediated inhibition of Leu115Pro mutated MEK. Wild-type MEK and the Leu115Pro variant (50 ng) were assayed for in vitro kinase activity against a kinase-inactive form of ERK [GSTERK1(K71R)] in the presence of indicated concentrations of PD 184352. A 100% control value reflects the activity of Raf-activated, untreated samples. The results are an average of four independent experiments.
TABLE 1.
PD 184352 inhibition of MEK mutant alleles identified from the yeast genetic screen
| MEK allele | IC50 (μM)a ± range (n) |
|---|---|
| Wild type | 0.067 ± 0.02 (4) |
| Phe53Ser | 0.048 ± 0.03 (2) |
| Ile103Asn | 0.163 ± 0.07 (2) |
| Leu115Pro | >10 (4) |
| His119Arg | 0.362 ± 0.20 (2) |
| His119Tyr | 0.154 ± 0.01 (2) |
| Gly128Asp | 0.200 ± 0.07 (2) |
| Glu203Lys | 0.074 ± 0.01 (2) |
Inhibition was measured in an in vitro kinase assay as described in Materials and Methods.
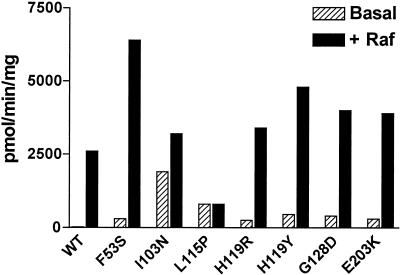
During analysis of the recombinant MEK proteins, it was noted that all seven mutants showed substantially higher basal activities against GSTERK1(K71R) compared to wild-type MEK. These basal specific activities ranged from 528.7 to 1,895.6 pmol min−1 mg−1 or were as high as 65-fold greater than wild-type enzyme (29.2 pmol min−1 mg−1) (Fig. 4). This substantially higher basal activity of the MEK mutants was further elevated by Raf activation, with total Raf-stimulated MEK kinase activity as much as 2.5 times that of Raf-activated wild-type MEK (Fig. 4). Significantly, the Leu115Pro variant did not show an increase in activity upon Raf activation, although it was still a substrate for Raf (data not shown). It is likely that the elevated basal and enhanced Raf activation of these mutants allowed them to escape the screen despite remaining sensitive to PD 184352.
FIG. 4.
Basal and Raf-stimulated in vitro kinase activity of MEK variants from the genetic screen. Recombinant MEK variants exhibited higher basal activity compared to wild-type enzyme. Basal enzyme activity is represented by hatched bars, and Raf-activated MEK activity is denoted by solid bars. The results shown are representative of two independent experiments.
We also evaluated the effect of PD 98059, a previously disclosed MEK inhibitor (11), on the various MEK mutants. Unlike PD 184352, all seven MEK mutants were resistant to inhibition by PD 98059 (data not shown). Previous work has shown that MEK activated by Raf phosphorylation (1, 13) or by mutation of S218/S222 to aspartate (data not shown) is resistant to inhibition by PD 98059. Thus, it is likely that the activating mutations found here also render these constructs resistant to PD 98059.
MEK homology model.
A homology model of the active form of MEK1 was generated based on the crystal structure of the active form of cyclic AMP-dependent protein kinase A (4) and multiple sequence alignment of several protein kinases, including the known MEK family members. All of the mutated residues, except Phe53 and Glu203, form a cluster in the model (Fig. 5), suggesting that these residues could be part of a binding pocket for the MEK1-specific inhibitor. The clustered residues are located in the interface region between the α-helix C and five-stranded β-sheet of the N-terminal lobe of the kinase domain. This pocket is distinct from the ATP binding pocket and is accessible from the rear face of the kinase domain. Analysis of the putative inhibitor binding pocket suggests it can accommodate a molecule the size of PD 184352. The conserved residues Lys97 and Glu114 form a hypothetical salt bridge in the model and are part of the wall separating the ATP binding pocket and the pocket formed by the identified mutant residues. Despite the clustering of the mutated residues in an apparent pocket for PD 184352, only the Leu115Pro substitution showed complete resistance to PD 184352, indicating that this residue may be critical for inhibitor binding. The model suggests the side chain of Leu115 lines the putative PD 184352 binding pocket; hence, altering this residue could disrupt the conformation of the pocket and prevent PD 184352 binding. To test this hypothesis, Leu115 was substituted with alanine, arginine, glutamate, or tryptophan in order to characterize the effects of different amino acid side chains at this position on inhibitor activity. The recombinant proteins were expressed in E. coli and assayed in vitro for their ability to phosphorylate GSTERK1(K71R) in the presence of increasing concentrations of PD 184352 (0 to 3 μM). As shown in Fig. 6, only the conservative leucine-to-alanine (Leu115Ala) substitution was significantly inhibited by PD 184352, and this construct was still quite resistant to inhibition (IC50 = 0.68 μM versus IC50 = 0.067 μM for wild-type). Other Leu115 substitutions exhibited IC50 values of >3 μM, indicating that the side chain of this residue is critical for PD 184352 interaction with MEK. Furthermore, similar to what was observed with the variants obtained from the genetic screen, each amino acid substitution was coupled with an increase in basal activity. Resulting basal specific activities were 197.1, 478.8, 882.3, 1,581 and 2,739 pmol min−1 mg−1 for L115R, L115P, L115W, L115E, and L115A, respectively. These ranged from 1.6- to 22.5-fold higher than wild-type MEK basal kinase activity (121.8 pmol min−1 mg−1).
FIG.5.
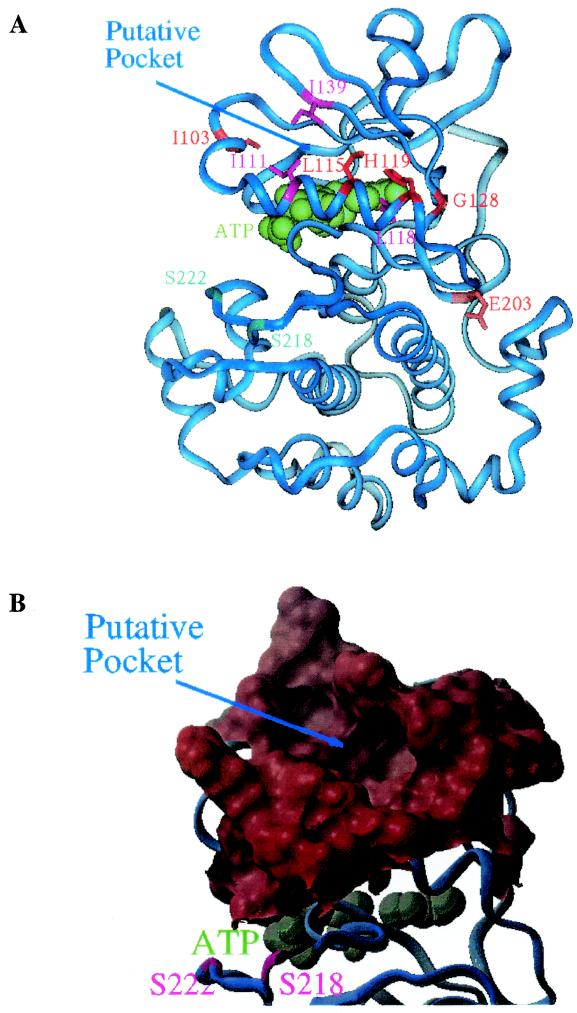
MEK homology model illustrating PD 184352 interacting mutations and putative binding pocket. (A) Ribbon diagram of a MEK1 homology model. The molecule is viewed from the rear side of the ATP binding pocket with the N-terminal lobe in the upper region and the C-terminal lobe in the lower region. Residues that were identified to perturb activity of PD 184352 are shown in sticks and colored red or magenta. Residues I103, L115, H119, G128, and E203 (red) were identified by random screening, whereas residues I111, I118, and I139 (magenta) were proposed based on the homology model. ATP is colored green and is shown in space-filling mode. Serine residues S218 and S222, which are phosphorylated when MEK1 is activated in vivo, are colored cyan. (B) Surface diagram N-terminal lobe showing the putative binding pocket. This diagram is viewed in the same orientation as for panel A. ATP is colored blue-green and is shown in space-filling mode.
FIG. 6.
In vitro kinase activity of Leu115 substitution mutants. Leu115 substitution variants were assayed in vitro with increasing concentrations of PD 184352 as described. Only the alanine substitution was inhibited (IC50 = 0.68 μM) at a 10-fold higher concentration than that required for wild-type MEK (IC50 = 0.067 μM). The constructs assayed included the wild-type (○), Leu115Ala (•), Leu115Glu (□), Leu115Pro (▪), Leu115Arg (▵), and Leu115Trp (▴) constructs. These data represent two independent experiments. The 100% control value reflects the Raf-activated, untreated enzyme activity.
The homology model further suggested additional residues might be interacting with PD 184352. In particular, isoleucine111 (Ile111), leucine118 (Leu118), and isoleucine139 (Ile139) appeared to comprise part of the putative binding pocket (Fig. 5). These residues were subsequently mutated to various other residues, and the recombinant proteins assayed in vitro as described previously. Several of the variants were resistant to PD 184352-mediated inhibition, while others showed reduced sensitivity to the compound, compared to wild-type MEK (Table 2). Combined with the Leu115 substitution data, these results further support the putative PD 184352 binding pocket in the MEK homology model. Interestingly, the effect of these mutations on MEK basal activity was somewhat different than previously observed with the mutations from the genetic screen and the Leu115 variants. Whereas the Ile111 variants all exhibited elevated basal activities compared to wild-type enzyme, the Ile139 substitutions did not have any effect on kinase activity, and the Leu118 variants showed a decrease in basal kinase activity (Table 2). All of the substitutions could be activated by Raf, however, and with the Leu118 variants the Raf activation exceeded that seen for wild-type MEK (data not shown).
TABLE 2.
PD 184352 inhibition of directed MEK mutants as suggested by residues lining the putative binding pocket
| MEK variant | IC50 (μM)a ± range (n) | Relative activity |
|---|---|---|
| Wild type | 0.067 ± 0.02 (2) | 1 |
| Ile111Ala | 0.151 ± 0.01 (2) | 10 |
| Ile111Asp | >3 (2) | 4 |
| Ile111Pro | >1 (2) | 9 |
| Ile111Arg | >1 (2) | 4 |
| Leu118Asp | >3 (2) | 0.3 |
| Leu118Pro | >3 (2) | 0.13 |
| Leu118Gly | 0.366 ± 0.05 (2) | 0.3 |
| Leu118Gln | >3 (2) | 0.5 |
| Ile139Gly | >3 (2) | 1 |
| Ile139Arg | 0.143 (1) | 1 |
| Ile139Asn | 0.247 (1) | 1 |
Inhibition and the activities of MEK variants were measured against GSTERK(K71R) with various concentrations of PD 184352 in an in vitro kinase assay as described in Materials and Methods. Relative activities were normalized to the average basal activity of wild-type MEK from each experiment.
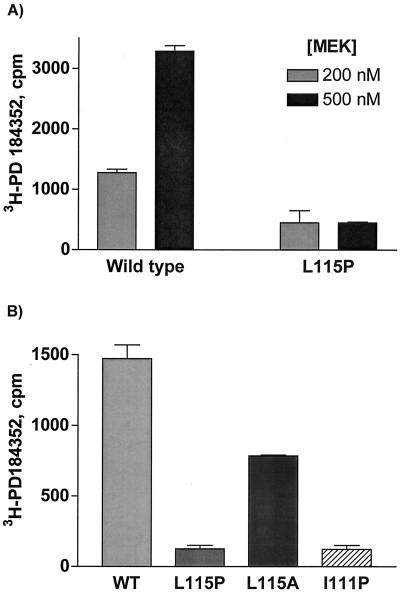
It is apparent that the region identified on MEK1 substantially affects enzyme activity. Although we suggest this is also the region where PD 184352 interacts, it is possible that the inhibitor binds to a different region and that the activity effects seen with the mutants are reflective of perturbations on basal activity. To further examine direct effects of the inhibitor, we devised a scintillation proximity binding assay utilizing 3H-PD 184352. Figure 7A demonstrates binding of the labeled compound to wild-type enzyme that correlates with increasing protein concentration. Further, no concentration-dependent binding of 3H-PD 184352 to the Leu115Pro mutant was detected. Additionally, binding to Ile111Pro was also abrogated (Fig. 7B), These two mutants were resistant to PD 184352-mediated inhibition in an in vitro kinase assay (Tables 1 and 2). In contrast, binding to Leu115Ala was decreased roughly 50% under these conditions. This particular mutant showed partial sensitivity to inhibition by the inhibitor (Fig. 6). Together, these data support the argument that these residues are part of the binding pocket for PD 184352.
FIG. 7.
Binding of 3H-PD 184352 to MEK variants. A scintillation proximity assay was used to assess binding of 3H-PD 184352 to different MEK mutants. (A) A 1 μM concentration of 3H-PD 184352 was incubated with 200 or 500 nM wild-type MEK or L115P MEK, and the amount of bound 3H was determined as described elsewhere. (B) A 500 nM concentration of MEK proteins was incubated with 300 nM 3H-PD 184352, and the amount of associated 3H was determined. Nonspecific values were determined in the absence of MEK protein and subtracted from the values shown. Values are averages of four replicate samples are presented. Error bars reflect the standard error of the mean.
Effect of MEK mutations on endogenous MAP kinase.
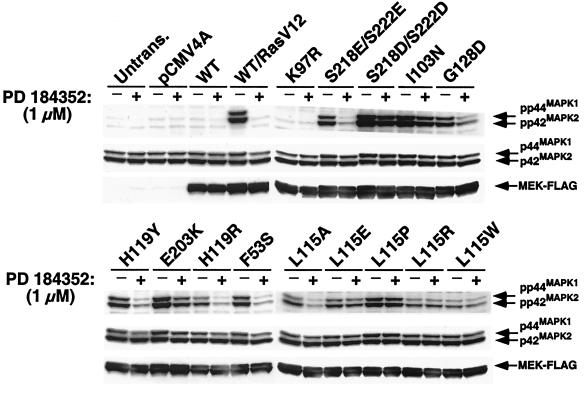
Since the in vitro kinase activity data indicated that the MEK variants possessed elevated basal activity, the effect of these mutations on endogenous MAP kinase was characterized in a whole-cell system. MEK alleles were cloned into a mammalian expression vector containing a carboxyl-terminal FLAG epitope and then transiently transfected into HEK293T cells. For comparison purposes, the constitutively active forms of MEK, Ser218Glu/Ser222Glu (2E), and Ser218Asp/Ser222Asp (2D) were also used. Lysates were prepared and activated MAP kinase was detected by Western blotting with antibodies specific for the phosphorylated forms of p44MAPK1 and p42MAPK2. As shown in Fig. 8, overexpression of the MEK variants caused activation of MAP kinase in HEK293T cells under serum-starved conditions, with levels comparable to, or exceeding those seen with wild-type MEK cotransfected with a constitutively active form of Ras (RasV12). Several variants displayed activity that was even higher than the Ser218Glu/Ser222Glu (2E) mutant, but only Ile103Asn and Glu203Lys appeared to have levels of activity comparable to Ser218Asp/Ser222Asp (2D). Consistent with the in vitro kinase data, treatment with 1 μM PD 184352 reduced phosphorylated MAP kinase levels in cells transfected with all of the variants except Leu115Pro. Of the directed Leu115 substitutions, only the Leu115Ala variant was significantly inhibited by PD 184352, in agreement with the in vitro observations (Fig. 6). These data further support the hypothesis that this region is not only essential for PD 184352 inhibition of MEK but also plays a key role in regulating MEK activity.
FIG. 8.
Effects of MEK variants in HEK293T cells. HEK293T cells were transiently transfected with MEK variants, serum deprived, and then harvested in the absence of additional stimulation. Extracts were probed with anti-phospho p44MAPK1 and -p42MAPK2 antibodies as described in Materials and Methods. Cells were treated with 1 μM PD 184352 for 1 h prior to harvesting. FLAG-tagged MEK expression and total p44MAPK1/p42MAPK2 levels were assessed by immunoblotting with anti-FLAG and anti-p44 MAPK1/p42MAPK2 antibodies, respectively. After treatment with 1 μM PD 184352, the Leu115Pro variant retained greater than 95% activity, as determined by densitometry quantitation (Kodak Imaging Systems) of a phospho-p44MAPK1/p42MAPK2 Western blot. The results are representative of three independent experiments.
DISCUSSION
To gain insight into the molecular mechanism of inhibition by PD 184352, we utilized a S. cerevisiae genetic screen to identify the region of MEK involved in inhibitor interaction. The screen revealed several amino acids that when mutated either hindered or completely abolished PD 184352-mediated inhibition of MEK kinase activity. Since PD 184352 does not compete for ATP (or MAP kinase) binding to MEK, it was interesting that five of the seven mutations clustered within kinase subdomains III and IV (Fig. 2). This region of the kinase harbors essential residues involved in nucleotide binding, including the catalytic lysine residue (Lys97) involved in phosphoryl-transfer from ATP to substrate, and the conserved glutamate (Glu114), which is thought to form a stabilizing salt bridge with Lys97 (32). However, since these mutations did not compromise kinase activity, it is apparent that they did not interfere with the ability of MEK to bind ATP. In fact, all of the MEK variants isolated in the screen exhibited a substantial increase in basal kinase activity.
Interestingly, the screen also identified Phe53, which falls outside of the usual kinase domain. The only significant homology between MEK and cyclic AMP-dependent protein kinase is within the kinase domain; thus, the MEK homology model can only be reasonably constructed for this domain. Phe53 is roughly 15 residues N-terminal to this domain and is not included in the modeling effort. However, Phe53 is very close to an N-terminal region comprising residues 32 to 51 that was identified by Ahn's group as acting to possible stabilize an inactive state of MEK (20, 21). Deletion of the region from residues 32 to 51 markedly activates MEK, as does the Phe53 mutation. Clearly, the N-terminal region of MEK also plays an important regulatory role in addition to substrate recognition (15).
Of the mutants identified in the yeast screen, the Leu115Pro mutation exerted the greatest effect on inhibitor activity. In the homology model of MEK1, Leu115 is part of α-helix C and its side chain is located inside the putative PD 184352 binding pocket. Proline is not favored in α-helices due to the lack of an amide hydrogen bond donor and to the steric constraints imposed upon the α-helical structure by the proline side chain. Thus, the Leu115Pro mutation could disrupt a region of the α-helix C and change the conformation of the pocket to prevent the binding of PD 184352 to MEK. Additionally, the site-specific mutagenesis and biochemical analysis of residues Ile111, Leu118, and Ile139, which were suggested by the model to be part of the inhibitor binding pocket, lend further support that this region is where PD 184352 interacts with MEK.
A possible explanation for the increased basal activity of the mutants is that a more active conformation is obtained due to changes in localized subdomains within the kinase core. X-ray crystal structures of several protein kinases have revealed catalytic activity of these enzymes involves two primary conformational changes (7). The first occurs due to phosphorylation of residues that lie within the activation lip of the kinase, which helps to stabilize the kinase in an active form. Second, the two lobes that comprise the kinase core domain rotate with respect to each other to close the catalytic cleft, thereby bringing the essential residues into the correct conformation to complete catalysis (7). However, other studies have indicated that subtle dynamic motion involving various secondary structures may also be important for catalysis. Specifically, Ahn's group have shown by deuterium exchange that significant dynamic movement and flexibility in the N-terminal lobe of the kinase core was observed in the mutationally activated form of MEK (24). In particular, the region of the kinase containing five of the seven mutated residues identified in the screen was shown to undergo the most movement when the enzyme was activated, with the exception of the activation segment.
We believe that the apparent dynamic nature of this region has profound implications for regulation of this enzyme. Thus, perturbations introduced by the mutations revealed by the screen could result in a loss of ordering in this region resulting in an increase in flexibility and hence catalytic activity. This could possibly also lead to elevated autophosphorylation at the activation loop, which would increase catalytic activity. Indeed, preliminary experiments suggest that Ile103Asn has a substantially higher level of autophosphorylation than with wild-type or the other mutants (data not shown). Conversely, any imposition on flexibility could have a significant impact on the catalytic activity of MEK. Binding of PD 184352 may lock the enzyme into an inactive conformation, preventing catalysis but still allowing phosphorylation of Ser218 and Ser222 in the activation segment by Raf. Additionally, this might interfere with recognition by MEK-specific phosphatases (27), possibly explaining the MEK hyperphosphorylation observed in KBALB cells treated with PD 184352.
The reported MEK inhibitors, PD 98059, PD 184352, and U0126, have proven to be exceptionally specific. Interestingly, they do not inhibit related MKK family members MKK3, MKK4, MKK6, or MKK7, although they do inhibit the closely related MEK2 (9, 26). Recently, these compounds have been reported to also show activity, albeit weaker, against MKK5 (18, 22, 23). Of the MKK family members, MKK5 exhibits the closest homology to MEK1 and MEK2, with ca. 46% amino acid identity in the catalytic domain (22). Alignments of the MKK family reveal that all of the mutants found here, as well as the targeted residues chosen from the homology model, are identical in MEK1, MEK2, and MKK5 and diverge in the other members. It is possible that a similar binding pocket exists in other MKK members and the MKK5 pocket is close enough to be recognized by the MEK1 inhibitors.
It is interesting that basal MEK kinase activity seen in vitro with some of the variants did not correlate with the activity of these mutants when overexpressed in HEK293T cells. Combined with the hyperphosphorylated state of endogenous MEK observed in cells treated with PD 184352, it is tempting to speculate that there are additional cellular components involved in regulation of MEK activity, as well as PD 184352-mediated inhibition of MEK. Further investigation to identify these unknown components, as well as the effects of other mutations in the proposed binding pocket on PD 184352 inhibition of MEK will be required to more fully understand the molecular mechanism of MEK kinase inhibition by PD 184352.
REFERENCES
- 1.Alessi, D. R., A. Cuenda, P. Cohen, D. T. Dudley, and A. R. Saltiel. 1995. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270:27489-27494. [DOI] [PubMed] [Google Scholar]
- 2.Alessi, D. R., Y. Saito, D. G. Campbell, P. Cohen, G. Sithanandam, U. Rapp, A. Ashworth, C. J. Marshall, and S. Cowley. 1994. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 13:1610-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L., and J. Thorner. 1996. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem. Sci. 21:373-374. [PubMed] [Google Scholar]
- 4.Bossemeyer, D., R. A. Engh, V. Kinzel, H. Ponstingl, and R. Huber. 1993. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 Å structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24). EMBO J. 12:849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruder, J. T., G. Heidecker, and U. R. Rapp. 1992. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 6:545-556. [DOI] [PubMed] [Google Scholar]
- 6.Catling, A. D., H. J. Schaeffer, C. W. Reuter, G. R. Reddy, and M. J. Weber. 1995. A proline-rich sequence unique to MEK1 and MEK2 is required for Raf binding and regulates MEK function. Mol. Cell. Biol. 15:5214-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, S., E. Radzio-Andzelm, and S. S. Taylor. 1994. Domain movements in protein kinases. Curr. Opin. Struct. Biol. 4:893-901. [DOI] [PubMed] [Google Scholar]
- 8.Dang, A., J. A. Frost, and M. H. Cobb. 1998. The MEK1 proline-rich insert is required for efficient activation of the mitogen-activated protein kinases ERK1 and ERK2 in mammalian cells. J. Biol. Chem. 273:19909-19913. [DOI] [PubMed] [Google Scholar]
- 9.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhanasekaran, N., and E. Premkumar Reddy. 1998. Signaling by dual specificity kinases. Oncogene 17:1447-1455. [DOI] [PubMed] [Google Scholar]
- 11.Dudley, D. T., L. Pang, S. J. Decker, A. J. Bridges, and A. R. Saltiel. 1995. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA 92:7686-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duesbery, N. S., C. P. Webb, S. H. Leppla, V. M. Gordon, K. R. Klimpel, T. D. Copeland, N. G. Ahn, M. K. Oskarsson, K. Fukasawa, K. D. Paull, and G. F. Vande Woude. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734-737. [DOI] [PubMed] [Google Scholar]
- 13.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van-Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1999. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 14.Frost, J. A., H. Steen, P. Shapiro, T. Lewis, N. Ahn, P. E. Shaw, and M. H. Cobb. 1997. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 16:6426-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda, M., Y. Gotoh, and E. Nishida. 1997. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 16:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanks, S. K., and A. M. Quinn. 1991. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200:38-62. [DOI] [PubMed] [Google Scholar]
- 17.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamakura, S., T. Moriguchi, and E. Nishida. 1999. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274:26563-26571. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber, and J. Heitman. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158:113-117. [DOI] [PubMed] [Google Scholar]
- 20.Mansour, S. J., J. M. Candia, J. E. Matsuura, M. C. Manning, and N. G. Ahn. 1996. Interdependent domains controlling the enzymatic activity of mitogen-activated protein kinase kinase 1. Biochemistry 35:15529-15536. [DOI] [PubMed] [Google Scholar]
- 21.Mansour, S. J., W. T. Matten, A. S. Hermann, J. M. Candia, S. Rong, K. Fukasawa, G. F. Vande Woude, and N. G. Ahn. 1994. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265:966-970. [DOI] [PubMed] [Google Scholar]
- 22.Mody, N., J. Leitch, C. Armstrong, J. Dixon, and P. Cohen. 2001. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502:21-24. [DOI] [PubMed] [Google Scholar]
- 23.Pearson, G., F. Robinson, T. Beers Gibson, B. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocrine Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 24.Resing, K. A., and N. G. Ahn. 1998. Deuterium exchange mass spectrometry as a probe of protein kinase activation: analysis of wild-type and constitutively active mutants of MAP kinase kinase-1. Biochemistry 37:463-475. [DOI] [PubMed] [Google Scholar]
- 25.Schaeffer, H. J., A. D. Catling, S. T. Eblen, L. S. Collier, A. Krauss, and M. J. Weber. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281:1668-1671. [DOI] [PubMed] [Google Scholar]
- 26.Sebolt-Leopold, J. S., D. T. Dudley, R. Herrera, K. Van Becelaere, A. Wiland, R. C. Gowan, H. Tecle, S. D. Barrett, A. Bridges, S. Przybranowski, W. R. Leopold, and A. R. Saltiel. 1999. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 5:810-816. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro, P. S., and N. G. Ahn. 1998. Feedback regulation of Raf-1 and mitogen-activated protein kinase (MAP) kinase kinases 1 and 2 by MAP kinase phosphatase-1 (MKP-1). J. Biol. Chem. 273:1788-1793. [DOI] [PubMed] [Google Scholar]
- 28.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson, B. J., N. Rhodes, B. Errede, and G. F. Sprague, Jr. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 6:1293-1304. [DOI] [PubMed] [Google Scholar]
- 31.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell. Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, S. S., and E. Radzio-Andzelm. 1994. Three protein kinase structures define a common motif. Structure 2:345-355. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, C. F., and K. L. Guan. 1993. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J. Biol. Chem. 268:23933-23939. [PubMed] [Google Scholar]