Abstract
Rho family GTPases activate intracellular kinase cascades to modulate transcription of multiple genes. Previous studies have examined the roles of the ubiquitously expressed Rho GTPase, Rac1, in regulation of gene expression in cell lines and implicated NF-κB, serum response factor, and kinase signaling pathways in this regulation. To understand the role of the closely related but hematopoiesis-specific Rho GTPase, Rac2, in regulation of gene transcription, we compared the gene expression profiles between wild-type and Rac2−/− bone marrow-derived mast cells. Our data demonstrate remarkable specificity in the regulation of gene expression by Rac2 versus Rac1. Microarray analysis demonstrated that expression of 38 known genes was significantly altered in Rac2−/− mast cells after cytokine stimulation compared with those in wild-type cells. Of these, the expression of the mouse mast cell protease 7 (MMCP-7) gene in wild-type cells was highly induced at the transcriptional level after stimulation with stem cell factor (SCF). In spite of compensatorily increased expression of Rac1 in Rac2-deficient cells, SCF-induced MMCP-7 transcription did not occur. Surprisingly, the loss of MMCP-7 induction was not due to decreased activation of NF-κB, a transcription factor postulated to lie downstream of Rac1 and known to play a critical role in hematopoietic cell differentiation and proliferation. However, the activities of c-Jun N-terminal kinases (JNKs) were markedly decreased in Rac2−/− mast cells. Our results suggest that cytokine-stimulated activation of MMCP-7 gene transcription is selectively regulated by a Rac2-dependent JNK signaling pathway in primary mast cells and imply a remarkable specificity in the regulation of transcriptional activity by these two highly related Rho GTPases.
Rac proteins, members of the Rho-related small GTPase family, play important roles in multiple cellular events, including actin cytoskeletal organization, cell proliferation and survival, cell cycle progression, and gene transcription regulation (24, 45, 53, 70, 75). The Rac subfamily of Rho GTPases comprises three members, Rac1, Rac2, and Rac3, which show >80% sequence identity (14, 22). The primary difference between Rac1 and Rac2, which are over 92% homologous, is in the carboxy-terminal region, where Rac1 but not Rac2 contains a six-amino-acid polybasic domain. Unlike Rac1 and Rac3, which are widely expressed, Rac2 expression is restricted to cells of hematopoietic lineages (15, 44, 57). With respect to regulation of gene expression, Rac1 can induce the activation of transcription factors such as serum response factor and nuclear factor κB (NF-κB) to stimulate transcription of multiple genes (26, 59).
In vitro studies primarily examining Rac1 have also demonstrated activation of several intracellular signaling pathways, including c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase (MAPK) (41, 48). However, nearly all studies to date have used dominant negative or activated mutants, the effects of which may not demonstrate the functional specificities of the endogenous Rac proteins, which are normally regulated by the upstream Dbl family guanine nucleotide exchange factors and guanine nucleotide dissociation inhibitors and downstream signaling effectors (35, 40, 68, 76). In particular, expression of these mutants may lead to abnormal intracellular localization of the exogenous proteins, masking subtle but important cell-cell and cell-membrane interactions (40).
Rac2-deficient mice generated by gene-targeting methods demonstrate cellular defects in multiple hematopoietic lineages, including stem and progenitor cells, neutrophils, mast cells, T cells, and B cells (10, 37, 52, 74, 75). In addition to its essential function in actin cytoskeletal rearrangements, these studies have also demonstrated that Rac2 participates in multiple kinase signal transduction pathways. Rac2-deficient neutrophils and T cells demonstrate decreased cytokine- or chemokine-induced activation of mitogen-activated protein kinases p42/p44 and p38 (33, 52). In addition, studies in cells derived from Rac2−/− mice demonstrate that Rac2 is required for phosphatidylinositol 3-kinase (PI-3K) activation of Akt after growth factor stimulation and mediates growth factor-induced cell survival (75). Interestingly, expression of Rac1 appears to be increased during the in vitro culture of the Rac2-deficient hematopoietic cells, but this increased expression of Rac1 appears to rescue only some of the abnormal phenotypes seen in blood cells deficient in Rac2 (20, 52). These studies, and the abnormal cellular phenotypes seen in vivo in Rac2−/− mice, suggest specificity of Rac2 function in blood cells. However, to date, few studies have examined the specificity of different Rac proteins with regard to regulation of gene expression.
Mouse mast cell proteases (MMCPs) are major components of the secretory granules of mature mast cells. At least nine MMCPs have been identified in a family of serine proteases, and they are differentially expressed in tissue-specific populations of mast cells (29, 30, 38, 39, 51, 55, 56). Based on their substrate specificities, serine proteases have been designated chymases or tryptases. There is considerable evidence suggesting that MMCPs play an important role in allergic and inflammatory reactions. Mast cell tryptases are responsible for airway smooth muscle hyperresponsiveness in dogs, which is a key feature of asthma (54). Genetic linkage studies in mice suggest that two tryptases (MMCP-6 and -7) are potential candidate genes involved in airway responsiveness (12). MMCP-6 has been implicated in the regulation of neutrophil extravasation into tissues during mast cell-mediated inflammatory reactions (27). In the V3 mastocytosis mouse, fibrinogen is a physiologic substrate of MMCP-7, suggesting that this trypase may regulate fibrin-platelet clot formation and fibrinogen- and integrin-dependent cellular responses during a mast cell-mediated inflammatory event (28).
MMCPs are expressed abundantly but transiently during mouse mast cell differentiation and maturation from bone marrow precursors in in vitro cultures. The expression of some MMCP genes is regulated by cytokines. Stem cell factor (SCF), the ligand of the c-Kit receptor tyrosine kinase, plays an important role in the proliferation, survival, and differentiation of mast cells (34, 63). Bone marrow-derived mast cells (BMMCs) express the MMCP-4 gene in response to SCF (21) or express the MMCP-1 and MMCP-2 genes in the presence of interleukin-10 (IL-10) or IL-9 (16, 17). BMMCs cultured in IL-3-containing medium express high steady-state levels of the MMCP-5 and MMCP-6 mRNAs, which are referred to as “early-expressed” protease genes (39, 51).
Although the MMCP-7 and MMCP-6 genes show high nucleotide sequence homology in their coding regions, the expression patterns of these proteases are differentially regulated during mast cell development. The MMCP-7 gene is transiently expressed in immature BMMCs in the presence of IL-3. Expression of MMCP-7 is markedly diminished after 3 weeks of culture. In contrast, MMCP-6 is continuously expressed during in vitro culture (72). Attenuated MMCP-7 gene expression has been noted in the C57BL/6 mouse strain due to a point mutation in the exon 2 splice donor consensus sequence, leading to mRNA instability and rapid degradation (31). However, transcription of the MMCP-7 gene is normal compared with that in other strains. In addition, transcription of the MMCP-7 gene has been demonstrated to be attenuated in mast cells cultured in IL-3 and derived from the mutant microphthalmia (mi/mi) mouse. The basic helix-loop-helix zipper transcription factor encoded by the mi locus (MITF) regulates the transcription of the MMCP-7 gene through an interaction with c-Jun transcription factors (47).
Our laboratory has defined phenotypic abnormalities in hematopoietic cells deficient in Rac2 in spite of continued expression of the highly related Rac1 GTPase. To investigate whether Rac2 plays a unique or specific role in the regulation of gene transcription, we studied the expression profiles of wild-type and Rac2-deficient mast cells in response to SCF stimulation. RNA expression of 38 known genes was found to be significantly altered in Rac2-deficient mast cells compared with that in wild-type cells. Of particular interest, the expression of genes encoding several murine mast cell proteases was studied in detail. SCF-induced MMCP-7 gene transcription was not seen in Rac2-deficient mast cells in spite of increased activation of both Rac1 and NF-κB. In these cells, Rac2 was required for the SCF-induced activation of JNKs and the c-Jun transcriptional pathway. Our results suggest that Rac2 is a primary physiological regulator of the JNK-mediated transcriptional pathway in bone marrow-derived mast cells and that the specificity of Rac2 function is not based entirely on its restricted expression profile.
MATERIALS AND METHODS
Cell preparations, SCF stimulation, and retroviral transduction.
Rac2−/− mice were backcrossed to C57BL/6J mice (Jackson Laboratory, Bar Harbor, Maine), and the animals used were derived from the F ≥ 13 backcross generation. 129Sv, C57BL/6, or C57BL/6 × 129Sv F13, F14, or F15 Rac2-deficient mice and their normal littermates were used in the experiments. Mouse low-density bone marrow was isolated as described previously (20). BMMCs were generated by culturing low-density bone marrow in RPMI 1640 medium (Life Technologies, Rockville, Md.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), 100 U of penicillin per ml, and 100 μg of streptomycin per ml, 2 mM glutamine (Life Technologies), and 10 ng of recombinant murine interleukin-3 (PeproTech, Rocky Hill, N.J.) per ml (complete RPMI) for 4 to 6 weeks at 37°C in 5% CO2. BMMCs were then analyzed for cellularity by flow cytometry on a FACScan (Becton Dickinson, Mountain View, Calif.), and the cell morphology was analyzed by cytospin and staining with the Diff-Quik stain set (Dade Behring Inc., Newark, Del.).
To stimulate BMMCs with SCF, wild-type and Rac2−/− mast cells were starved in RPMI plus 2% bovine serum albumin (Roche Diagnostics, Indianapolis, Ind.) for 16 h at 37°C and incubated with 10 ng of recombinant rat SCF (rrSCF; Amgen, Thousand Oaks, Calif.) per ml for the indicated times in each experiment. Retroviral transduction of C57BL/6 mast cells with MIEG3-Rac1 (hemagglutinin [HA]-tagged wild-type Rac1) or MIEG3-FR2 (Flag-tagged wild-type Rac2) vectors was performed as previously described by Yang et al. (75). Green fluorescent protein-positive cells were sorted with a FACStar Plus (Becton Dickinson, Mountain View, Calif.).
DNA microarray analysis.
Total RNAs were isolated from the bone marrow-derived C57BL/6 mast cells after rrSCF treatment with the RNeasy mini kit (Qiagen, Valencia, Calif.); 10 μg of total RNA was quantitatively amplified and biotin labeled as described (6). Briefly, RNA was converted to double-stranded cDNA with an oligo(dT) primer that has a T7 RNA polymerase site at the 5′ end. The cDNA was used directly in an in vitro transcription reaction in the presence of biotinylated nucleotides to produce antisense RNA, which was hybridized overnight to Genechips (Affymetrix, San Jose, Calif.) displaying probes for 11,000 genes and expressed sequence tags (ESTs). Chips were washed, stained with phycoerythrin-streptavidin, and read with an Affymetrix Genechip scanner and accompanying gene expression software. Labeled bacterial RNAs of known concentration were spiked into each chip hybridization mix to generate an internal standard curve, allowing normalization between chips and conversion of raw hybridization intensity values to mRNA frequency (mRNA molecules per million).
DNA microarray data were filtered for genes with RNA expression changes of threefold or greater at one or more SCF stimulation time points in Rac2−/− cells compared with wild-type cells, and 183 annotated mouse ESTs were found in all three experimental conditions. The experiment was then subjected to normalization so that unstimulated wild-type cells represented the baseline condition, with relative expression equal to 1.0. Gene expression altered by SCF or Rac2 genotype was thus characterized relative to that in the untreated wild-type cells. An additional restriction was then applied to eliminate genes that had poor correlation between adjacent time points or low dynamic changes (under threefold relative to the unperturbed wild-type). This screen resulted in 93 genes. With the K-means algorithm in the GeneSpring software (Silicon Genetics, Redwood City, Calif.), initial clustering approaches with hierarchical tree analysis applied to the expression ratio measurements suggested that there were 16 major classes of gene behavior. Then, we eliminated the repeated sequences (ESTs) for the same known genes and selected the final 38 genes with known functions in nine clusters.
Northern blot, nuclear run-on, and RNA stability analyses.
Northern blot analysis was performed as described previously (61) with some modifications. Briefly, total RNA samples were denatured in formaldehyde-formamide and electrophoresed in 1.3% formaldehyde-agarose gels. The separated RNAs were transferred to MagnaGraph membranes (Micron Separations, Westover, Mass.), and the resulting blots were individually hybridized with gene-specific probes for MMCP-4 (18), MMCP-7 (39), and mouse β-actin (58) labeled with [α-32P]dCTP (Dupont/NEN, Boston, Mass.) by random oligonucleotide priming with the NEBlot Kit (New England BioLabs, Beverly, Mass.). The hybridization was carried out in Hybrisol I (Intergen, Purchase, N.Y.) at 42°C for 16 h. The membranes were washed three times (15 min each) at 42°C with 2× SSC (300 mM NaCl and 30 mM sodium citrate, pH 7.0) and 0.1% sodium dodecyl sulfate (SDS), and then once at 65°C (15 min) with 0.1× SSC-0.1% SDS. Autoradiography was done on Kodak X-OMAT films (Eastman, Rochester, N.Y.) at −80°C for 1 h to 3 days.
To assess MMCP-7 mRNA stability, C57BL/6 BMMCs from 6-week-old cultures were starved and then stimulated with 10 ng of rrSCF per ml for 4 h, washed, and resuspended in complete RPMI medium with or without SCF. Actinomycin D (Sigma, St. Louis, Mo.) was then added to both cultures of mast cells at a final concentration of 1 μg/ml (73). Then 10 million cells were collected for total RNA isolation at 0, 1, 5, and 10 h after the addition of actinomycin D. Equal amounts of total RNA for the different time points were used for Northern blot analysis to detect the steady-state level of the MMCP-7 transcript.
For the nuclear run-on assay, 5 μg of DNA plasmids containing the 168-bp MMCP-7 cDNA fragment or mouse β-actin in pBluescript (Stratagene, San Diego, Calif.) were linearized with restriction enzyme EcoRI (New England BioLabs) and denatured in 0.1 M NaOH for 30 min at room temperature. After neutralization in 6× SSC, DNA samples were spotted onto a MagnaGraph membrane (Micron Separations) with a slot blot apparatus (Life Technologies). The membrane was UV cross-linked and air dried. Nuclei were isolated from both wild-type and Rac2−/− C57BL/6 mast cells, and the run-on transcription assay was performed as previously described by Pollok et al. (50). The blot was hybridized and exposed as mentioned above for Northern blot analysis.
Analysis of c-Kit receptor expression and internalization.
Mast cells were starved for 16 h and incubated with 10 ng of rrSCF (Amgen, Thousand Oaks, Calif.) per ml for 10 min or 1 or 4 h. The presence of the c-Kit receptor on the cell surface was detected with fluorescein isothiocyanate-conjugated anti-mouse CD117 (c-Kit) (PharMingen, San Diego, Calif.) incubated with the SCF-treated cells for 30 min at 4°C. Flow cytometric analysis was performed with a FACScan (Becton Dickinson). The internalization of the c-Kit receptor was determined by the decrease in mean fluorescence intensity after incubation with ligand.
Kinase inhibitor assay.
Wild-type C57BL/6 BMMCs were starved for 16 h. SCF (10 ng/ml) was added to the cells for 4 h with or without kinase inhibitors. Kinase inhibitors, including Ly294002 (20 mM) for phosphatidylinositol 3-kinase, PD98059 (50 mM) for MEK, 420116 (1 mM) for JNKs 1, 2, and 3, and SB203580 (40 mM) for p38 MAPK (all from Calbiochem, La Jolla, Calif.), were dissolved in dimethyl sulfoxide as 1,000× stocks.
Western blot, GST effector pulldown, and JNK assays.
Wild-type and Rac2−/− C57BL/6 mast cells were stimulated with 10 ng of SCF per ml for 0, 5, or 15 min or as indicated before whole-cell lysates were made in 1× cell lysis buffer (10 mM K2HPO4, 1 mM EDTA, 5 mM EGTA, 10 mM MgCl2, 1 mM Na3VO4, 50 mM sodium β-glycerophosphate, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 1 μg of pepstatin A per ml, pH 7.2; all from Sigma). Nuclear extracts were prepared as described previously (7) and kept frozen at −80°C until used.
Immunoblot analyses were carried out with rabbit antibodies for phospho-p44/p42 (Thr202/Tyr204) MAP kinase (1:2,000; New England BioLabs), phospho-p38 MAPK (1:2,000; BioLabs), phospho-JNKs (Thr183/Tyr185) (1:1,000; BioLabs), total JNKs (1:2,000; BioLabs), total p38 MAPK (1:2,000; BioLabs), NF-κB (1:1,000; Santa Cruz Biotechnology, Santa Cruz, Calif.), mouse antibody specific for Rac1 (23A8, 1:2,000; Upstate Biotechnology, Lake Placid, N.Y.), and anti-Rac monoclonal antibody (1:2,000; BD Transduction Laboratories, San Diego, Calif.) (67). Glutathione S-transferase (GST) effector pulldown assays were performed as previously described by Gu et al. (20). Lysate from 106 cells was used for each reaction.
To assay endogenous JNK activity, clear cell lysates from 107 wild-type or Rac2−/− mast cells were used for the immunocomplex kinase assay after immunoprecipitation of JNKs with antibody for total JNKs (10 μg per reaction; BioLabs) (42). JNK activity was measured by using GST-c-Jun fusion protein (5 μg per reaction; BioLabs) as a substrate in the presence of [γ-32P]ATP (Dupont/NEN). The kinase reaction samples were electrophoresed on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) gel, and the gel was dried for autoradiography.
Electrophoretic mobility shift assay.
Five micrograms of nuclear extracts from wild-type or Rac2−/− mast cells stimulated with 10 ng of SCF per ml for 1 h were analyzed for NF-κB DNA-binding activity with an electrophoretic mobility shift assay as described previously (19, 64). Briefly, the binding reaction was done for 30 min in 10 mM Tris (pH 7.5)-5 mM MgCl2-5% glycerol-1 mM EDTA-1 mM dithiothreitol-2 μg of poly(dI:dC). The NF-κB consensus sequence 5′-AGT TGA GGG GAC TTT CCC AGGC-3′ was end labeled with [α-32P]dCTP (Dupont/NEN). For the competition assay, unlabeled wild-type oligonucleotide was added. DNA-protein complexes were electrophoresed on a 7% polyacrylamide gel.
RESULTS
Rac2 deficiency affects the profile of growth factor-induced gene expression in cultured mast cells.
Rho GTPases, particularly Rac1, have previously been implicated in the activation of transcription factors and regulation of gene expression in fibroblasts (26, 32, 70). To determine whether Rac2 plays a role in the regulation of gene expression in primary cells, we studied the gene expression profiles of bone marrow-derived Rac2-deficient mast cells in response to SCF by DNA microarray analyses. Low-density bone marrow cells from Rac2−/− and wild-type C57BL/6 mice were cultured for 6 weeks in the presence of IL-3 to derive primary mast cells.
Rac2−/− and wild-type bone marrow-derived mast cells displayed similar granule content, a marker of differentiation, with side scatter on flow analysis and similar cell morphology on Wright-Giemsa-stained cytospin preparations (data not shown). Also, these cells expressed similar levels of the c-Kit receptor tyrosine kinase (CD117), another marker of mast cell development and differentiation (mean fluorescence intensity, 202 ± 14 versus 210 ± 17, wild-type versus Rac2−/−, mean intensity units ± standard deviation [SD], N = 3, P > 0.5). These data suggest that the wild-type and Rac2−/− mast cells studied were at an equivalent state of mast cell differentiation after culture in IL-3.
The mRNA expression profile of SCF-stimulated Rac2−/− mast cells was determined at 0, 1, and 4 h with Affymetrix DNA chips (Mu11k SubA and B), which simultaneously monitored the RNA levels of 11,000 murine genes and ESTs from the GenBank database. Murine Rac2 gene-specific oligonucleotides on the DNA chip indicated the absence of Rac2 mRNA in Rac2−/− mast cells, demonstrating the specificity of the microarray assay. Filtering the DNA microarray data for genes with RNA expression changes of threefold or greater at one or more SCF stimulation time points in Rac2−/− cells compared with wild-type cells yielded a list of 183 annotated mouse ESTs and full-length gene sequences.
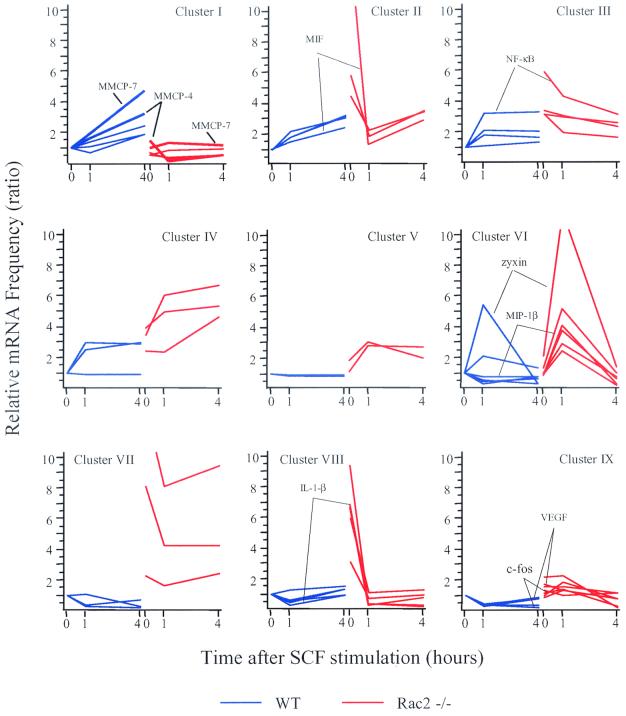
To understand the role of Rac2 in the regulation of gene expression, cluster analysis was performed based on the relative expression patterns in wild-type versus Rac2−/− cells in response to SCF with the K-means algorithm in GeneSpring software (details in Materials and Methods). Thirty-eight genes with known functions were clustered into nine groups, as shown in Fig. 1. These genes showed significantly altered expression patterns in response to SCF in Rac2-deficient cells compared with wild-type cells. The data are presented as a ratio relative to expression in wild-type mast cells at the 0-h time point. The genes in each of the nine clusters are listed in Table 1. In Rac2−/− mast cells, some genes appeared to have similar or slightly decreased baseline expression (red lines, at 0 h), and significantly decreased expression was found up to 4 h after SCF stimulation compared with that in wild-type cells (blue lines, see cluster I). Table 1 indicates that cluster I contained MMCP-4 and -7 (mouse mast cell proteases), Lipocortin I (calcium and phospholipid binding protein) and two other cytoplasmic proteins.
FIG. 1.
Gene expression clusters of Rac2-deficient and wild-type mast cells. Six-week-old BMMCs derived from wild-type (WT) and Rac2−/− C57BL/6 mice were stimulated with 10 ng of rrSCF per ml for 1 or 4 h and used to harvest total cellular RNA. Then 10 μg of total RNA per sample was used to generate biotinylated antisense RNA, which was hybridized overnight to the Affymetrix Mu11k SubA and B DNA chips. Microarray data were collected as hybridization intensity and then converted into mRNA frequency values by comparison with an internal standard curve. Microarray data were filtered for genes and ESTs with RNA expression changes of threefold or greater at one or more SCF stimulation time points in Rac2−/− cells (red lines) compared with wild-type cells (blue lines), and 38 genes with known functions were selected. These genes were clustered into nine groups (clusters I to IX) on the basis of expression patterns over the three time points with the K-means algorithm in the GeneSpring software. Gene expression levels are shown on the y axis as mRNA frequency ratios, relative to the mRNA frequency of the gene seen at 0 h in wild-type cells. VEGF, vascular endothelial growth factor.
TABLE 1.
Genes identified in nine clustersa
| Cluster | Nameb | GenBank accession no. | Function group | Ratio, Rac2−/−/wild type, at:
|
||
|---|---|---|---|---|---|---|
| 0 h | 1 h | 4 h | ||||
| I | MMCP-7 | L00654 | Mast cell-specific proteases | 0.9 | 0.7 | 0.2 |
| CAP | L12367 | Cytoplasmic proteins | 0.5 | 0.4 | 0.3 | |
| M17 | U13263 | Cytoplasmic proteins | 0.7 | 0.3 | 0.3 | |
| Lipocortin 1 | X07486 | Calcium and phospholipid binding proteins | 0.5 | 0.5 | 0.3 | |
| MMCP-4 | X68804 | Mast cell-specific proteases | 1.4 | 0.1 | 0.2 | |
| II | LDH-A | Y00309 | Metabolic enzymes | 4.5 | 1.3 | 1.1 |
| Stat4 | U06923 | Transcription factor- and DNA-binding proteins | 4.5 | 1.2 | 1.1 | |
| MIF | L39357 | Growth factors, cytokines, and chemokines | 15 | 0.9 | 1.2 | |
| III | NF-κB | L28117 | Transcription factor and DNA-binding proteins | 5.9 | 1.3 | 0.9 |
| G protein beta 2 | U34960 | G protein related | 3.3 | 1.6 | 1.3 | |
| Aldolase 1 | Y00516 | Metabolic enzymes | 3.1 | 1.4 | 1.3 | |
| M2 pyruvate kinase | X97047 | Intracellular kinases | 3.1 | 1.8 | 1.2 | |
| IV | N-cadherin | M31131 | Adhesion molecules | 2.4 | 2.6 | 5.1 |
| Calcyclin | M37761 | Calcium-binding proteins | 3.4 | 2.1 | 2.3 | |
| LAF1 transketolase | U05809 | Metabolic enzymes | 3.9 | 2.0 | 1.8 | |
| V | 2B4 | L19057 | Cell surface receptors | 1.1 | 3.5 | 3.4 |
| MMTV | M11024 | Virus | 1.9 | 3.5 | 2.3 | |
| VI | P600 | M23504 | Inflammatory proteins | 1.0 | 10 | 1.0 |
| MIP-1β | M35590 | Inflammatory proteins | 1.3 | 7.2 | 1.4 | |
| Nocturnin | U70139 | Transcription factor- and DNA-binding proteins | 1.0 | 3.7 | 1.2 | |
| Krox-20 | X06746 | Transcription factor- and DNA-binding proteins | 0.9 | 4.7 | 0.5 | |
| N10 | X16995 | Nuclear hormone-binding receptors | 0.8 | 2.0 | 0.2 | |
| Zyxin | Y07711 | Adhesion molecules | 2.1 | 2.2 | 5.8 | |
| VII | CCR5 | D83648 | Cell surface receptors | 2.3 | 4.3 | 3.8 |
| Heat-stable antigen | X53825 | Antigens | 8.1 | 15 | 20 | |
| TSC-22 | X62940 | Transcription factor- and DNA-binding proteins | 13 | 7.6 | 35 | |
| VIII | IL-1β | M15131 | Growth factors, cytokines, and chemokines | 6.8 | 1.3 | 0.9 |
| TCA3 | M17957 | Growth factors, cytokines, and chemokines | 6.6 | 0.5 | 0.6 | |
| li | X00496 | Membrane proteins | 9.3 | 0.8 | 0.2 | |
| Secretory leukocyte | ||||||
| Protease inhibitor | U73004 | Protease inhibitors | 3.0 | 1.3 | 1.0 | |
| Calcitonin | X97991 | Cytoplasmic proteins | 5.9 | 0.9 | 0.8 | |
| IX | PEBP2a1 | D14636 | Transcription factor- and DNA-binding proteins | 1.3 | 3.5 | 1.6 |
| HTK ligand | L38847 | Transmembrane proteins | 1.0 | 4.3 | 1.0 | |
| gas3 | M32240 | Transmembrane proteins | 1.6 | 4.7 | 0.9 | |
| VEGF | M95200 | Growth factors, cytokines, and chemokines | 1.7 | 3.0 | 1.0 | |
| mSLIM3 | U77040 | LIM proteins | 0.8 | 4.3 | 1.4 | |
| c-fos | V00727 | Oncogenes | 1.2 | 4.5 | 1.5 | |
| MIP | X12531 | Inflammatory proteins | 2.2 | 6.8 | 0.7 | |
According to their GenBank accession numbers obtained from DNA microarray analysis, the names and putative function of the 38 gene members in the nine clusters shown in Fig. 1 are listed here. Ratios indicate how much the particular gene is upregulated or downregulated in Rac2−/− mast cells at three different SCF stimulation time points compared with those in wild-type cells.
Abbreviations: CAP, adenylyl cyclase-associated protein; LDH-A, lactate dehydrogenase-A; MIF, migration inhibitory factor; 2B4, non-MHC-restricted associated molecule; MMTV, mouse endogenous mammary tumor virus; TCA3, T-cell activation gene 3; li, Ia-associated invariant chain; PEBP2a, polyomavirus enhancer binding protein alpha subunit type 1; HTK, hepatoma transmembrane kinase; VEGF, vascular endothelial growth factor; mSLIM3, mouse LIM protein 3; LIM, lin-11, ISl-1, and mec-3 proteins.
In contrast to cluster I, clusters II to IX contained mostly genes that had increased baseline expression levels at 0 h in Rac2−/− cells compared with those in wild-type cells (Fig. 1). When stimulated with SCF, the expression of some of these genes also appeared to be increased early (at 1 h) in Rac2−/− cells (see clusters VI and IX). The magnitude of increased gene expression at 1 h was higher in cluster VI compared with cluster IX. Cluster VI included genes such as macrophage inflammatory protein MIP-1β and zyxin (an adhesion molecule), and cluster IX contained genes such as that for vascular endothelial growth factor and c-fos (an oncogene) (Table 1).
The expression of other genes was increased both early and late (at 1 h and 4 h) in Rac2−/− cells (see clusters III, IV, V, and VII). Cluster III contained four genes that had their peak expression at 0 h and their lowest expression at 4 h after SCF stimulation in Rac2−/− cells. This cluster included the genes for NF-κB (a transcription factor; see below), whereas cluster VII contained three genes that had their lowest expression at 1 h after SCF stimulation in Rac2−/− cells. Cluster IV contained three genes that had their peak expression at 4 h after SCF stimulation in Rac2−/− cells. Cluster V represented genes with a peak expression at 1 h that declined slightly at 4 h in Rac2−/− cells.
In spite of their markedly increased baseline expression in Rac2−/− cells, the expression of some genes subsequently declined to levels similar to those in wild-type cells after SCF stimulation (see clusters II and VIII), including genes encoding migration inhibitory factor (a growth factor) and IL-1β (a cytokine). Cluster II differed from cluster VIII in a secondary induction of expression at 4 h after SCF stimulation. Taken together, these data suggest that Rac2 function may be important, either directly or indirectly, in the regulation of expression of multiple genes in mast cells after SCF stimulation and that this function is not subserved by Rac1.
Rac2 regulates SCF-induced expression of the MMCP-7 gene at a transcriptional level.
In order to understand how Rac2 regulates a specific transcriptional signaling pathway, we focused on the genes demonstrating the most defective SCF-induced expression in Rac2−/− cells compared with wild-type cells. For example, the expression levels of genes shown in cluster I were induced to a peak level at 4 h by SCF stimulation in wild-type cells, but showed no response to SCF in Rac2-deficient cells. In particular, expression of the MMCP-4 and -7 genes increased three- and fivefold, respectively (bold blue lines in cluster I, Fig. 1) in wild-type cells, but the mRNA levels of these two MMCP genes did not increase after SCF stimulation in Rac2−/− cells (bold red lines in cluster I, Fig. 1).
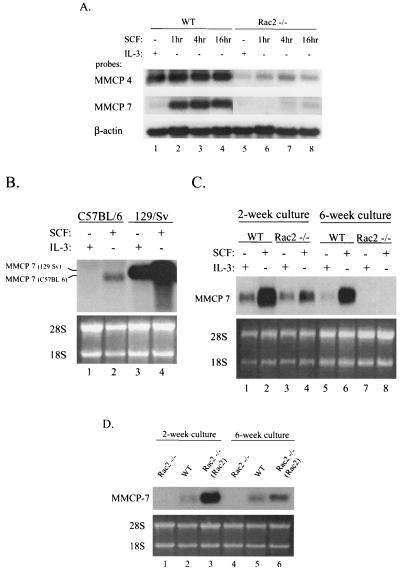
To confirm that the expression of MMCP genes is modulated via Rac2, as suggested by DNA microarray analysis, we examined the change in mRNA levels by Northern blot analysis in both Rac2−/− and wild-type mast cells after SCF stimulation with MMCP-4 and -7 gene-specific cDNA probes. As shown in Fig. 2A, the transcripts of MMCP-4 and -7 genes were induced by SCF stimulation in wild-type C57BL/6 BMMCs. In Rac2−/− cells, expression of MMCP-4 was markedly diminished and expression of MMCP-7 was only detectable at 16 h after SCF stimulation and at significantly reduced levels compared with those in wild-type cells (lanes 5 to 8 versus lanes 1 to 4). In addition, this stimulation of expression of MMCP-7 was specific for SCF and was not seen after IL-3 stimulation (Fig. 2A, MMCP-7, lanes 2 to 4 versus lane 1). In contrast, MMCP-4 mRNA expression was less differentially stimulated by SCF compared with IL-3 (Fig. 2A, MMCP-4, lanes 2 to 4 versus lane 1), although the expression of MMCP-4 was also clearly affected by Rac2.
FIG. 2.
Defective SCF-induced protease expression in Rac2-deficient mast cells. (A) Steady-state levels of MMCP-4 and MMCP-7 mRNAs in response to SCF and IL-3. Wild-type (WT) and Rac2−/− C57BL/6 BMMCs maintained in IL-3-containing medium were stimulated with 10 ng of SCF per ml, and 10 μg of each total RNA sample from 6-week-old C57BL/6 BMMCs was used per lane. Levels of mouse β-actin mRNA were used as a loading control. (B) MMCP-7 mRNA levels in wild-type C57BL/6 and 129/Sv BMMCs. Cells cultured in the presence of IL-3 for 2 weeks were stimulated with SCF for 4 h. We used 10 μg of each total RNA sample per lane, and ethidium bromide-stained 28S and 18S RNAs were used as a loading control. (C) MMCP-7 mRNA levels in wild-type (WT) and Rac2−/− 129/Sv BMMCs. 129/Sv BMMCs from 2- or 6-week-old cultures were stimulated with 10 ng of SCF per ml for 4 h. We used 5 μg of each total RNA sample in lanes 1 to 4 and 10 μg of each total RNA sample in lanes 5 to 8. (D) Restoration of SCF-induced MMCP-7 mRNA levels in Rac2−/− C57BL/6 BMMCs. Rac2−/− BMMCs were transduced with a retroviral vector, MIEG3-FR2, expressing wild-type Rac2 [designated Rac2−/− (Rac2)]. Cells from 2- or 6-week-old cultures were stimulated with 10 ng of SCF per ml for 4 h. We used 5 μg of total RNA sample per lane. All results shown are representative of three experiments.
Next, in order to further assess the role of Rac2 in regulating SCF-induced gene expression, we carried out more detailed studies focusing on MMCP-7 expression. The mRNA level of the MMCP-7 gene has been shown to be reduced in the C57BL/6 mouse strain due to a point mutation at the splice donor sequences of exon 2 (31). This mutation leads to its rapid mRNA degradation and an approximately 100-bp-smaller transcript than that in 129/Sv mice, but has no effect on its transcription rate. Consistent with this previous study, mRNA levels of the MMCP-7 gene were not detectable in IL-3-cultured BMMCs from the C57BL/6 mouse by Northern blot analysis, as shown in Fig. 2A (MMCP-7, lane 1) and Fig. 2B (MMCP-7, lane 1). However, after SCF stimulation, wild-type C57BL/6 BMMCs expressed low but detectable levels of an MMCP-7 mRNA which was also smaller in size (Fig. 2B, lane 2 versus 3).
To determine whether MMCP-7 expression is also differentially regulated by SCF and IL-3 in a different mouse strain, we performed Northern blot analysis on RNA isolated from 129/Sv mouse BMMCs after treatment with SCF or IL-3 following culture for 2 and 6 weeks in vitro. After 2 weeks of culture in the presence of IL-3, wild-type 129/Sv BMMCs contained much higher levels of MMCP-7 mRNA than the C57BL/6 BMMCs, and the transcript was larger, as expected (Fig. 2B, lane 1 versus lane 3 and lane 3 versus lane 2). SCF increased the levels of the MMCP-7 transcript in both C57BL/6 and 129/Sv BMMCs (Fig. 2B, lanes 2 and 4; Fig. 2C, lanes 2 and 6), suggesting that expression of the MMCP-7 gene can be stimulated by SCF in both early (immature) and late (mature) primary BMMCs maintained in IL-3-containing medium.
Confirming the data from C57BL/6 mice, the deficiency Rac2 in BMMCs derived from 129/Sv mice was associated with significantly decreased levels of MMCP-7 mRNA compared with wild-type cells, particularly after stimulation with SCF (Fig. 2C, MMCP-7, lane 4 versus lane 2). The difference in SCF stimulatory effect on MMCP-7 expression between wild-type and Rac2−/− mast cells was more clearly seen in 6-week-old culture (Fig. 2C, MMCP-7, lane 8 versus lane 6). These data further suggest that MMCP-7 gene expression is differentially affected by SCF versus IL-3 cytokines and that SCF-induced expression is largely regulated by Rac2 activity.
Since the regulation of MMCP-7 expression by SCF via Rac2 was consistent in the two mouse strains tested, C57BL/6 and 129/Sv, we focused on the C57BL/6 BMMCs in subsequent studies. To further confirm that the defective MMCP-7 expression in Rac2-deficient mast cells was a direct result of Rac2 deficiency, we expressed the murine Rac2 cDNA in the Rac2−/− BMMCs with the retroviral vector MIEG3-FR2, expressing wild-type Rac2 and enhanced green fluorescent protein from a bicistronic message (71). Compared with wild-type cells, the transduced Rac2−/− cells expressed Rac2 protein at levels similar to or slightly higher than those in the wild type, as described previously (20, 75). As shown in Fig. 2D, the transduced Rac2−/− BMMCs expressing the Rac2 cDNA demonstrated a restoration of MMCP-7 mRNA levels when analyzed 4 h after SCF stimulation in both 2- and 6-week-old cultures (lanes 3 and 6 versus lanes 1 and 4).
Next, to determine whether Rac2-mediated SCF signaling increases the expression of MMCP-7 at a transcriptional level, we performed nuclear run-on assays with wild-type and Rac2−/− BMMCs. Similar results were observed in the nuclear run-on assays with the cells of 2- and 6-week-old cultures. In wild-type BMMCs, MMCP-7 transcription was increased fourfold in the presence of SCF (Fig. 3A, panel 1 versus panel 2). In contrast, MMCP-7 transcription in Rac2−/− BMMCs did not increase after SCF stimulation for 1 h and was less than 30% of the wild-type transcriptional level (Fig. 3A, panels 3 and 4). The reduced MMCP-7 gene expression noted after SCF stimulation in Rac2−/− BMMCs was not due to differences in c-Kit receptor cell surface expression (75) (see above). In addition, both wild-type and Rac2−/− BMMCs demonstrated similar c-Kit receptor internalization in response to SCF binding (data not shown). These data suggested that a deficiency of Rac2 does not affect ligand binding or receptor activation.
FIG. 3.
SCF and Rac2 regulate expression of MMCP-7 gene at the transcriptional level. (A) Nuclear run-on analyses. 32P-labeled nuclear run-on transcripts from C57BL/6 BMMCs (6-week-old cultures) were used to hybridize to 5 μg of gene-specific DNA probes for MMCP-7, β-actin, and vector only on the membrane. The hybridization intensity is represented in arbitrary units. Wild-type (WT) and Rac2−/− cells were stimulated with 10 ng of SCF per ml for 1 h. (B) RNA stability analysis. C57BL/6 BMMCs from 6-week-old cultures were stimulated with 10 ng of SCF per ml for 4 h. The cells were then treated with actinomycin D at a final concentration of 1 μg/ml and further cultured with or without SCF. Total RNA was isolated at 0, 1, 5 and 10 h after the addition of actinomycin D. We used 10 μg of total RNA sample for Northern blot analysis examining MMCP-7 transcript levels in the upper panel. The ethidium bromide-stained 28S and 18S RNAs were used as a loading control in the middle panel. The remaining levels of the MMCP-7 transcript are shown in the bottom panel as a percentage of the starting level of the transcript. The result shown is representative of three experiments.
We also examined the effect of SCF on the stability of the MMCP-7 transcript. After 4 h of SCF stimulation, wild-type BMMCs were exposed to actinomycin D, an inhibitor of transcription, and the cells were further cultured in medium with or without SCF. As shown in Fig. 3B, in the presence of actinomycin D, SCF had no significant effect on MMCP-7 mRNA half-life. Thus, SCF regulates MMCP-7 expression at least in part via Rac2, and this regulation is mainly transcriptional in cultured BMMCs. These data do not completely rule out the possibility of an additional posttranscriptional mechanism, as previously reported for the expression of MMCP-4 in BMMCs (73).
Rac2 is not associated with activation of NF-κB in BMMCs.
Rac1 GTPase activity has been shown to activate the transcription factor NF-κB, which mediates the regulation of expression of multiple genes (3, 32). In hematopoietic cells, NF-κB-dependent transcription plays essential roles in cellular proliferation and differentiation (4). Cytoplasmic NF-κB forms an inactive complex with an inhibitory protein of the IκB family. Activation of NF-κB leads to nuclear translocation and subsequent transcriptional activities.
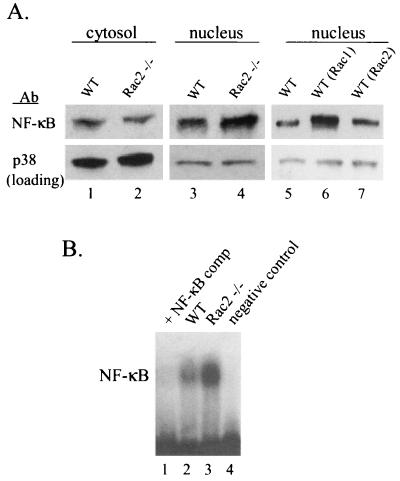
To determine whether the regulation of MMCP-7 expression by Rac2 involves the activation of NF-κB, we next examined the distribution of cytoplasmic and nuclear NF-κB protein after SCF stimulation in both wild-type and Rac2−/− BMMCs by immunoblot analysis. There was no significant difference in the level of NF-κB protein in the cytosol between wild-type and Rac2−/− cells stimulated with SCF for an hour (Fig. 4A, lanes 1 and 2). However, unexpectedly, the amount of nuclear NF-κB protein was slightly and consistently increased in Rac2−/− mast cells after stimulation (Fig. 4A, lane 4 versus lane 3). To confirm this surprising result, electrophoretic mobility shift assays were performed. Nuclear extracts from Rac2−/− cells consistently demonstrated increased NF-κB binding to a consensus DNA sequence compared with nuclear extract from wild-type cells after SCF stimulation (Fig. 4B, lane 3 versus lane 2).
FIG. 4.
NF-κB activity in Rac2-deficient mast cells. Wild-type (WT) and Rac2−/− C57BL/6 BMMCs were stimulated with 10 ng of SCF per ml for 1 h and then harvested for nuclear extracts and cytosol lysates. (A) Western blot analysis with an anti-NF-κB antibody (Ab). Levels of total p38 MAPK proteins were used as a loading control. Wild-type BMMCs were transduced with retroviral vectors expressing wild-type Rac2 or wild-type Rac1 [designated WT (Rac2) and WT (Rac1)]. (B) Electrophoretic mobility shift assay. We used 5 μg of nuclear extracts for binding to the NF-κB consensus sequence labeled with [32P]dCTP, and DNA-protein complexes were examined on a 7% polyacrylamide gel. A 50-fold excess of unlabeled oligonucleotide (NF-κB comp) was added to the competition assay with wild-type (WT) nuclear extracts. No nuclear extract protein was used in the negative control lane. The results shown are representative of three experiments.
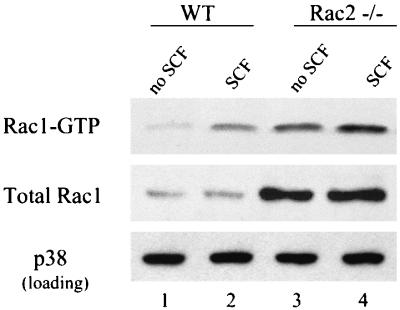
Since we have previously demonstrated that Rac2−/− hematopoietic myeloid progenitor cells cultured in vitro displayed increased expression and activation of Rac1 (20), we hypothesized that the increased NF-κB transcriptional activity in Rac2−/− BMMCs could be due to increased Rac1 expression. To test this hypothesis, we examined Rac1 protein expression and activation in Rac2−/− BMMCs before and after SCF stimulation. As shown in Fig. 5, Rac2−/− mast cells displayed an increased level of total Rac1 protein with or without SCF stimulation by immunoblot. In addition, active GTP-bound Rac1, as measured with a GST-p21-activated kinase (PAK) effector pulldown assay (5), was increased more than threefold compared with cultured wild-type mast cells both at baseline and after stimulation with SCF (Fig. 5, lanes 2 and 4 versus lanes 1 and 3). These data further suggest that Rac1 but not Rac2 is responsible for NF-κB activation in mast cells after SCF stimulation.
FIG. 5.
Rac1 expression and activity are upregulated in Rac2-deficient mast cells. BMMCs from wild-type (WT) and Rac2−/− C57BL/6 mice were stimulated with 10 ng of SCF per ml for 5 min. Mast cell lysates from 4 × 106 cells were used in the GST-PAK effector pulldown assay. Levels of GTP-bound Rac1 protein were examined by immunoblot with antibody specific for Rac1 (upper panel). Immunoblot analysis of total Rac1 protein and total p38 MAPK (as a loading control) in wild-type and Rac2−/− C57BL/6 BMMCs is shown in the middle and lower panels. Similar results were seen in three experiments.
This hypothesis is further supported by the fact that a significant increase in the level of nuclear NF-κB was also found in transduced wild-type mast cells overexpressing wild-type Rac1 but not wild-type Rac2 after SCF stimulation for 1 h (Fig. 4A, lane 6 versus lane 7). Also, inhibition of NF-κB activity by its specific inhibitory peptide did not significantly block MMCP-7 expression in response to SCF stimulation in wild-type mast cells (Fig. 7, lane 7 versus lane 1). Thus, our data suggest that unlike Rac1, Rac2 does not lead to activation of the NF-κB transcription pathway in BMMCs, and the decrease in MMCP-7 expression does not appear to be related to defective activation of NF-κB in these cells. Expression of the MMCP-7 gene is specifically not responsive to increased Rac1 activity.
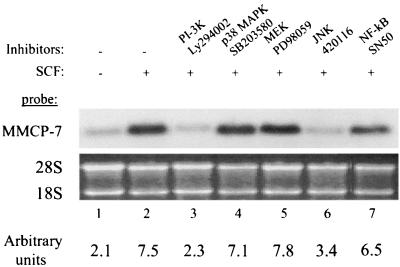
FIG. 7.
Phosphatidylinositol 3-kinase and JNK activities are required for SCF-induced MMCP-7 expression in mast cells. Wild-type C57BL/6 BMMCs were stimulated with 10 ng of SCF per ml with or without the kinase inhibitors Ly294002 (20 μM) for phosphatidylinositol 3-kinase, PD98059 (50 μM) for MEK, 420116 (1 μM) for JNKs, SN50 (20 μM) for NF-κB, and SB203580 (40 μM) for p38 MAPK, for 4 h. We used 10 μg of total RNA for Northern blot analysis with a gene-specific probe for MMCP-7. The ethidium bromide-stained 28S and 18S RNAs were used as a loading control. The hybridization intensity is represented in arbitrary units in the bottom panel. Similar results were seen in two other experiments.
Activation of JNK, a known upstream regulator of gene expression, is defective in Rac2-deficient mast cells after SCF stimulation.
A number of studies have demonstrated that Rac protein (primarily Rac1) also regulates the transcriptional activity of multiple genes in response to different growth factors via kinase pathways (8, 41, 65). To determine if Rac2 mediates the regulation of MMCP-7 transcription after SCF stimulation via a known kinase pathway, we examined the activation of JNKs, and p38, p42, and p44 MAPKs in Rac2−/− BMMCs.
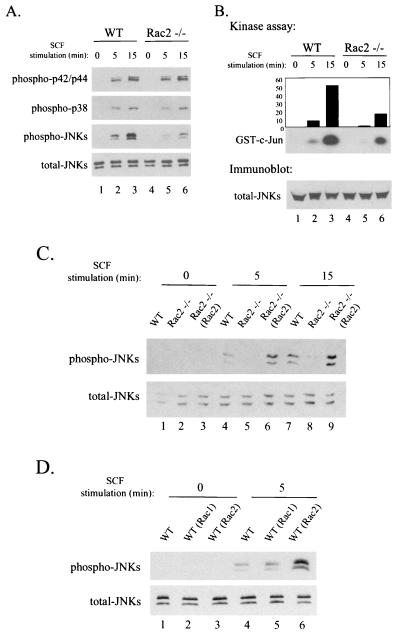
As shown in Fig. 6A, Rac2−/− mast cells displayed markedly decreased phosphorylation and activation of JNKs in response to SCF compared with wild-type cells (lanes 4 to 6 versus lanes 1 to 3) for up to 15 min of stimulation. In contrast to the defective signaling reported previously in Rac2-deficient neutrophils after stimulation with chemoattractants (52), phosphorylation of p38, p42, and p44 MAPKs appeared normal in Rac2−/− mast cells following SCF stimulation.
FIG. 6.
Decreased JNK activity in Rac2-deficient mast cells. (A) Immunoblot analyses with phosphorylation-specific anti-p42/p44, anti-p38 MAPK, or anti-JNK antibodies. Wild-type (WT) and Rac2−/− C57BL/6 BMMCs were stimulated with 10 ng of SCF per ml for 0, 5, and 15 min. (B) JNK activity measured by the JNK kinase assay. JNK activity was measured by the immunocomplex kinase assay with cell lysates from 107 wild-type or Rac2−/− mast cells. GST-c-Jun fusion protein was used as a substrate in the presence of [γ-32P]ATP. (C) Restoration of JNK activity in Rac2−/− C57BL/6 BMMCs. Rac2−/− BMMCs were transduced with a retroviral vector expressing wild-type (WT) Rac2 [designated Rac2−/− (Rac2)]. Cells from 2-week-old cultures were stimulated with 10 ng of SCF per ml for 0, 5, and 15 min. We used 20 μg of cell lysates per lane for immunoblot analysis with the phosphorylation-specific anti-JNK antibodies. (D) JNK activity in response to overexpression of Rac1 and Rac2 in primary wild-type mast cells. Wild-type (WT) C57BL/6 BMMCs were transduced with retroviral vectors expressing wild-type Rac2 or wild-type Rac1 [designated WT (Rac2) and WT (Rac1)]. Levels of total JNKs were used as loading controls. The results shown are representatives of three experiments.
JNK has been identified as a critical kinase for phosphorylating the transactivating domain of the c-Jun transcription factor, which has potent AP-1 activity and can regulate the expression of a number of genes (2, 13, 36). Recent studies have demonstrated that binding of the c-Jun transcription factor on the AP-1 site at the promoter of the MMCP-7 gene activated its transcription (47). To further evaluate this pathway in Rac2−/− mast cells, JNK activity was assayed with c-Jun as a substrate. The kinase assay showed that the deficiency of Rac2 in BMMCs was associated with a 70% decrease in endogenous JNK activity compared with wild-type cells after SCF stimulation (Fig. 6B, lanes 5 and 6 versus lanes 2 and 3). The restored activation of JNKs found in Rac2−/− cells transduced with and expressing wild-type Rac2 confirmed that the decreased JNK activity in response to SCF was specifically due to Rac2 deficiency (Fig. 6C, lanes 6 and 9). In contrast, unlike Rac2, wild-type Rac1 expressed in the transduced wild-type mast cells did not significantly induce the activation of endogenous JNKs after SCF stimulation (Fig. 6D, lane 2 versus lane 3).
Rac proteins have also been linked to signaling through the PI-3K pathways, and PI-3K has been implicated in SCF/c-Kit signaling in BMMCs (5, 46, 69, 75). In primary Rac2-deficient mast cells derived from C57BL/6 mice, activation of PI-3K in response to SCF has been shown to be equivalent or slightly increased compared with that in wild-type cells (75), while Akt activation by PI-3K is defective. We therefore studied the PI-3K signaling cascade and MAPK pathways in order to ascertain their involvement in SCF-dependent activation of MMCP-7 transcription in BMMCs.
We treated wild-type BMMCs with inhibitors specific to p38 MAPK, MEK (MAP or ERK kinase), JNKs, NF-κB, and PI-3K before SCF stimulation. Total RNAs were isolated from these treated cells for Northern blot analysis with an MMCP-7-specific probe. As shown in Fig. 7, addition of Ly294002, an inhibitor of PI-3K, and 420116, an inhibitor of JNKs, significantly blocked the SCF-induced increase in steady-state MMCP-7 mRNA (lanes 5 to 6 versus lanes 2 and 1), while inhibitors of p38 MAPK, MEK, and NF-κB had no significant effect (lanes 3, 4, and 7).
Taken together, these data demonstrate that Rac2 is required for full activation of JNKs following SCF stimulation in BMMCs and suggest that Rac2-dependent activation of JNKs subsequently regulates transcription of the MMCP-7 gene via the c-Jun pathway. The defect in MMCP-7 gene activation lies downstream of the previously described defect in PI-3K signaling in Rac2-deficient mast cells after c-Kit activation (75). Therefore, our results suggest that in spite of the high degree of sequence homology between the Rac1 and Rac2 proteins, Rac1 and Rac2 differentially regulate the activation of gene transcription signaling pathways.
DISCUSSION
The studies reported here, with a genetic approach, suggest that Rac2 specifically activates the JNK signaling cascade, which is associated with SCF-induced expression of several mast cell protease genes, in primary mast cells. Our data demonstrate a remarkable specificity of activation of downstream pathways between Rac subfamily members that was not previously appreciated. Thus, in primary mast cells deficient in Rac2, JNK activation is defective in spite of increased expression and activation of the highly related Rac1. Rac1 has been shown in some but not all studies in cell lines to effectively activate JNKs with the activated or dominant negative forms of Rac1 (reviewed in reference 66).
Previous studies have implicated localization of GTPases to discrete subcellular pools that may thus be differentially regulated as a possible mechanism of the specificity of GTPase function (60). For instance, stimulation of neutrophils with N-formyl-methionyl-leucyl phenylalanine leads to recruitment of Rac to the membrane, which is associated with GTP-GDP exchange. Rac molecules that are unable to translocate are defective in activation responses (11). It has also recently been shown that the constitutively active or dominant negative mutant Rac1 blocks binding to Rho guanine nucleotide dissociation inhibitors and redirects their subcellular distribution in living cells (40). In addition, guanine nucleotide exchange factors such as Vav, a known guanine nucleotide exchange factor for Rac, have been shown to translocate to the membrane after activation via upstream growth factor receptors (49). Overexpressed mutant Rac1 proteins could be mislocated in the cell, leading to a wide variety of responses which do not reflect normal physiological outcomes. Taken together, these data suggest that Rac2 is the physiologic regulator of JNK activation in mast cells.
To understand the roles of Rac2 in the regulation of transcription, we studied gene expression patterns in primary mast cells deficient of Rac2 protein in response to SCF stimulation. Based on the data from DNA microarray analysis, we identified 38 genes with known functions whose expression patterns were markedly altered in Rac2-deficient mast cells compared with wild-type cells. As summarized in Fig. 1 and Table 1, most of these genes displayed upregulated expression in Rac2-deficient cells in both the absence and presence of SCF stimulation, including several genes encoding transcription factors such as NF-κB, Stat4, and TSC-22. The upregulation of gene expression by loss of Rac2 activity could suggest that Rac2 had an inhibitory effect on the transcription of these genes. However, there is no previous evidence to support this hypothesis.
A second possibility is that the expression of these genes is secondarily affected by levels of other Rac proteins in these mast cells. Supporting this possibility, our results show that the protein level and activity of another highly related Rho GTPase, Rac1, is significantly increased in Rac2-deficient cells with or without SCF stimulation (Fig. 5) (20, 75), and the increased expression of Rac1 but not Rac2 appears to be directly related to increased NF-κB activity. Interestingly, the expression of four genes, including the gene encoding MMCP-7 (listed in cluster I) was markedly decreased at baseline and did not respond to SCF stimulation in Rac2−/− cells in contrast to wild-type cells. In particular, the defective expression of MMCP-7 in response to SCF was confirmed by Northern blot analyses and was completely rescued by the expression of wild-type Rac2 protein in Rac2−/− cells (Fig. 2). These data suggest that Rac2 but not Rac1 protein may be important in the regulation of expression of a subset of genes in mast cells.
In bone marrow-derived mast cells, as previously described, the MMCP-7 gene is transiently expressed during the first 3 weeks of in vitro culture in the presence of IL-3 (72). Our studies demonstrated that MMCP-7 expression is also induced by SCF at a transcriptional level, and this induction requires the activity of Rac2, a hematopoiesis-specific Rho GTPase. Rac1, although highly related, apparently cannot subserve this function at physiologic levels. It has been shown previously that SCF binding to the c-Kit receptor tyrosine kinase on mast cells activates receptor autophosphorylation and downstream signaling through the PI-3K and Src pathways (62). These pathways are summarized in Fig. 8. In agreement with these studies, we showed that PI-3K inhibitors completely blocked the ability of SCF to induce MMCP-7 expression (Fig. 7). Our results showed that loss of Rac2 is associated with markedly decreased JNK activation and subsequently reduced expression of MMCP-7.
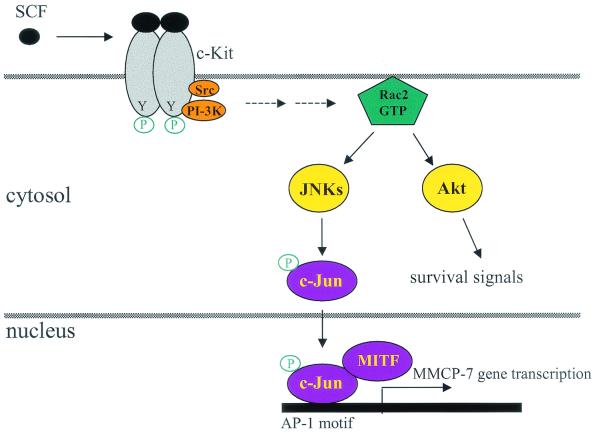
FIG. 8.
Proposed model for Rac2-mediated regulation of gene expression in mast cells. Studies by us and others suggest that in mast cells, SCF binding to c-Kit activates receptor dimerization, intrinsic tyrosine kinase activity, and autophosphorylation. Phosphatidylinositol 3-kinase (PI-3K) and Src kinase, binding to the activated c-Kit receptor, activate cytoplasmic signaling cascades through the Rac2 GTPase, JNKs, and Akt (62, 75). While Akt regulates cell survival signals, activated JNKs phosphorylate and activate downstream transcription factors, such as c-Jun, which subsequently translocate to the nucleus to regulate gene transcription (47).
Consistent with this, it has recently been reported that transcription of the MMCP-7 gene is controlled by an AP-1 binding motif in its promoter region, and binding of c-Jun transcription factor to the AP-1 motif significantly increases its transcription. The expression of MMCP-7 can also be synergistically transactivated by another transcription factor encoded by the mi locus (MITF) by forming a complex with c-Jun (47) (see summary in Fig. 8). However, our studies here with an antibody for phospho-specific JNKs and an inhibitor of JNKs could not distinguish the specific roles of the three JNK family members JNK 1, JNK 2, and JNK 3 on Rac2-dependent expression of MMCP-7 in response to SCF. This can be addressed in the future with JNK gene-targeted knockout animals.
Previously, we also reported that while activation of PI-3K after SCF stimulation appears normal in Rac2-deficient mast cells, loss of Rac2 expression is associated with decreased survival due to the absence of Akt activation, a downstream effector of PI-3K (75) (Fig. 8). The mechanism by which the PI-3K and Src kinase signaling pathways activate Rac2 after SCF stimulation in mast cells is still not clear. In human hematopoietic cells, SCF/c-Kit receptor activation results in phosphorylation and activation of the Vav GDP/GTP exchange factor (1). PI-3K has also been demonstrated to regulate the activation of Vav (25). Similar to Rac2, Vav1 expression is restricted to hematopoietic cells, and Vav is a known guanine nucleotide exchange factor for Rac (9, 43). Whether Vav acts as the major activator of Rac2 following SCF stimulation in primary mast cells is currently under study with Vav1−/− mast cells.
Rac1 and Rac2 show a high degree of homology in amino acid sequence and are both expressed in hematopoietic cells (14, 44). The area of greatest sequence divergence between all mammalian Rac proteins is the carboxy terminus. Haataja et al. recently demonstrated specific interaction of Rac3 but not Rac1 or Rac2 with the calcium- and integrin-binding protein (CIB) and subsequent translocation to the Triton-insoluble fraction of the cell via the carboxy-terminal sequence (23). Our previous studies with Rac2-deficient mice generated by gene targeting have demonstrated that Rac2 has some agonist-specific nonoverlapping functions in NADPH oxidase activation in phagocytes and cell adhesion, migration, and survival in multiple hematopoietic cell lineages (52, 74, 75). Here, we show that Rac2 also functions with specificity in the regulation of transcription of some genes in bone marrow-derived mast cells. Surprisingly, defective induction of MMCP-7 expression in response to SCF is not due to decreased NF-κB activation in these cells. On the contrary, NF-κB activation is increased in response to the increased Rac1 activity.
Taken together, our results with the primary Rac2-deficient mast cells suggest that these two closely related GTPases regulate gene expression via different transcriptional signaling pathways.
Acknowledgments
We thank Eva Meunier and Kerry Holleran for excellent manuscript preparation. We thank Richard L. Stevens, Yi Zheng, David Skalnik, and Michael Jansen for helpful discussion. Also, we thank Gary M. Bokoch for providing the anti-Rac2 antibody.
This work was supported by the Howard Hughes Medical Institute (Y.G. and D.A.W.) and NIH grant R01 DK48605.
REFERENCES
- 1.Alai, M., A. L.-F. Mui, R. L. Cutler, X. R. Bustolo, and M. Barbacid. 1992. Steel factor stimulates the tyrosine phosphorylation of the proto-oncogene product, p95vav, in human hematopoietic cells. J. Biol. Chem. 267:18021-18025. [PubMed] [Google Scholar]
- 2.Angel, P., E. A. Allegretto, S. T. Okino, K. Hattori, W. J. Boyle, T. Hunter, and M. Karin. 1988. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature 332:166-171. [DOI] [PubMed] [Google Scholar]
- 3.Arbibe, L., J. P. Mira, N. Teusch, L. Kline, M. Guha, N. Mackman, P. J. Godowski, R. J. Ulevitch, and U. G. Knaus. 2000. Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat. Immunol. 1:533-540. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, A. S., Jr. 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 5.Benard, V., B. P. Bohl, and G. M. Bokoch. 1999. Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils with a novel assay for active GTPases. J. Biol. Chem. 274:13198-13204. [DOI] [PubMed] [Google Scholar]
- 6.Byrne, M. C., M. Z. Whitley, and M. T. Follettie. 2000. Preparation of mRNA for expression monitoring, 4th ed., p. 22.2.1-22.2.13. In F. M. Ausubel (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 7.Camoretti-Mercado, B., H. W. Liu, A. J. Halayko, S. M. Forsythe, J. W. Kyle, B. Li, Y. Fu, J. McConville, P. Kogut, J. E. Vieira, N. M. Patel, M. B. Hershenson, E. Fuchs, S. Sinha, J. M. Miano, M. S. Parmacek, J. K. Burkhardt, and J. Solway. 2000. Physiological control of smooth muscle-specific gene expression through regulated nuclear translocation of serum response factor. J. Biol. Chem. 275:30387-30393. [DOI] [PubMed] [Google Scholar]
- 8.Coso, O. A., M. Chiariello, J. C. Yu, H. Teramoto, P. Crespo, N. Xu, T. Miki, and J. S. Gutkind. 1995. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81:1137-1146. [DOI] [PubMed] [Google Scholar]
- 9.Crespo, P., X. R. Bustelo, D. S. Aaronson, O. A. Coso, M. Lopez-Barahona, M. Barbacid, and J. S. Gutkind. 1996. Rac-1 dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene 13:455-460. [PubMed] [Google Scholar]
- 10.Croker, B. A., D. M. Tarlinton, L. A. Cluse, A. J. Tuxen, A. Light, F. C. Yang, D. A. Williams, and A. W. Roberts. 2002. The Rac2 guanosine triphosphatase regulates B lymphocyte antigen receptor responses and chemotaxis and is required for establishment of B-1a and marginal zone B lymphocytes. J. Immunol. 168:3376-3386. [DOI] [PubMed] [Google Scholar]
- 11.del Pozo, M. A., L. S. Price, N. B. Alderson, X. D. Ren, and M. A. Schwartz. 2000. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19:2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Sanctis, G. T., M. Merchant, D. R. Beier, R. D. Dredge, J. K. Grobholz, T. R. Martin, E. S. Lander, and J. M. Drazen. 1995. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nat. Genet. 11:150-154. [DOI] [PubMed] [Google Scholar]
- 13.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 14.Didsbury, J., R. F. Weber, G. M. Bokosh, T. Evans, and R. Synderman. 1989. Rac, a novel ras-related family of proteins that are botulinum toxin substrates. J. Biol. Chem. 264:16378-16382. [PubMed] [Google Scholar]
- 15.Diekmann, D., C. D. Nobes, P. D. Burbelo, A. Abo, and A. Hall. 1995. Rac GTPase interacts with GAPs and target proteins through multiple effector sites. EMBO J. 14:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eklund, K. K., N. Ghildyal, K. F. Austen, and R. L. Stevens. 1993. Induction by IL-9 and suppression by IL-3 and IL-4 of the levels of chromosome 14-derived transcripts that encode late-expressed mouse mast cell proteases. J. Immunol. 151:4266-4273. [PubMed] [Google Scholar]
- 17.Ghildyal, N., H. P. McNeil, M. F. Gurish, K. F. Austen, and R. L. Stevens. 1992. Transcriptional regulation of the mucosal mast cell-specific protease gene, MMCP-2, by interleukin 10 and interleukin 3. J. Biol. Chem. 267:8473-8477. [PubMed] [Google Scholar]
- 18.Ghildyal, N., H. P. McNeil, S. Stechschulte, K. F. Austen, D. Silberstein, M. F. Gurish, L. L. Somerville, and R. L. Stevens. 1992. IL-10 induces transcription of the gene for mouse mast cell protease-1, a serine protease preferentially expressed in mucosal mast cells of Trichinella spiralis-infected mice. J. Immunol. 149:2123-2129. [PubMed] [Google Scholar]
- 19.Ghislain, J. J., and E. N. Fish. 1996. Application of genomic DNA affinity chromatography identifies multiple interferon-alpha-regulated Stat2 complexes. J. Biol. Chem. 271:12408-12413. [DOI] [PubMed] [Google Scholar]
- 20.Gu, Y., B. Jia, F. C. Yang, M. D'Souza, C. E. Harris, C. W. Derrow, Y. Zheng, and D. A. Williams. 2001. Biochemical and biological characterization of a human Rac2 GTPase mutant associated with phagocytic immunodeficiency. J. Biol. Chem. 276:15929-15938. [DOI] [PubMed] [Google Scholar]
- 21.Gurish, M. F., N. Ghildyal, H. P. McNeil, K. F. Austen, S. Gillis, and R. L. Stevens. 1992. Differential expression of secretory granule proteases in mouse mast cells exposed to interleukin 3 and c-kit ligand. J. Exp Med. 175:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haataja, L., J. Groffen, and N. Heisterkamp. 1997. Characterization of RAC3, a novel member of the Rho family. J. Biol. Chem. 272:20384-20388. [DOI] [PubMed] [Google Scholar]
- 23.Haataja, L., V. Kaartinen, J. Groffen, and N. Heisterkamp. 2002. The small GTPase Rac3 interacts with the integrin-binding protein CIB and promotes integrin alpha(IIb)beta(3)-mediated adhesion and spreading. J. Biol. Chem. 277:8321-8328. [DOI] [PubMed] [Google Scholar]
- 24.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 25.Han, J., K. Luby-Phelps, B. Das, X. Shu, Y. Xia, R. D. Mosteller, U. M. Krishna, J. R. Falck, M. A. White, and D. Broek. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279:558-560. [DOI] [PubMed] [Google Scholar]
- 26.Hill, C. S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159-1170. [DOI] [PubMed] [Google Scholar]
- 27.Huang, C., W. Y. Ma, M. R. Young, N. Colburn, and Z. Dong. 1998. Shortage of mitogen-activated protein kinase is responsible for resistance to AP-1 transactivation and transformation in mouse JB6 cells. Proc. Natl. Acad. Sci. USA 95:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, C., G. W. Wong, N. Ghildyal, M. F. Gurish, A. Sali, R. Matsumoto, W. T. Qiu, and R. L. Stevens. 1997. The tryptase, mouse mast cell protease 7, exhibits anticoagulant activity in vivo and in vitro due to its ability to degrade fibrinogen in the presence of the diverse array of protease inhibitors in plasma. J. Biol. Chem. 272:31885-31893. [DOI] [PubMed] [Google Scholar]
- 29.Huang, R. Y., T. Blom, and L. Hellman. 1991. Cloning and structural analysis of MMCP-1, MMCP-4 and MMCP-5, three mouse mast cell-specific serine proteases. Eur. J. Immunol. 21:1611-1621. [DOI] [PubMed] [Google Scholar]
- 30.Hunt, J. E., D. S. Friend, M. F. Gurish, E. Feyfant, A. Sali, C. Huang, N. Ghildyal, S. Stechschulte, K. F. Austen, and R. L. Stevens. 1997. Mouse mast cell protease 9, a novel member of the chromosome 14 family of serine proteases that is selectively expressed in uterine mast cells. J. Biol. Chem. 272:29158-29166. [DOI] [PubMed] [Google Scholar]
- 31.Hunt, J. E., R. L. Stevens, K. F. Austen, J. Zhang, Z. Xia, and N. Ghildyal. 1996. Natural disruption of the mouse mast cell protease 7 gene in the C57BL/6 mouse. J. Biol. Chem. 271:2851-2855. [DOI] [PubMed] [Google Scholar]
- 32.Kheradmand, F., E. Werner, P. Tremble, M. Symons, and Z. Werb. 1998. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science 280:898-902. [DOI] [PubMed] [Google Scholar]
- 33.Kim, C., and M. C. Dinauer. 2001. Rac2 is an essential regulator of neutrophil nicotinamide adenine dinucleotide phosphate oxidase activation in response to specific signaling pathways. J. Immunol. 166:1223-1232. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura, Y. 1989. Heterogeneity of mast cells and phenotypic change between subpopulations. Annu. Rev. Immunol. 7:59-76. [DOI] [PubMed] [Google Scholar]
- 35.Knaus, U. G., Y. Wang, A. M. Reilly, D. Warnock, and J. H. Jackson. 1998. Structural requirements for PAK activation by Rac GTPases. J. Biol. Chem. 273:21512-21518. [DOI] [PubMed] [Google Scholar]
- 36.Kyriakis, J. M., P. Banerjee, E. Nikolakaki, T. Dal, E. A. Rubie, M. F. Ahmad, J. Avruch, and J. R. Woodett. 1994. The stress-activated protein kinase subfamily of c-jun kinases. Nature 369:156-160. [DOI] [PubMed] [Google Scholar]
- 37.Li, B., H. Yu, W. Zheng, R. Voll, S. Na, A. W. Roberts, D. A. Williams, R. J. Davis, S. Ghosh, and R. A. Flavell. 2000. Role of the guanosine triphosphatase Rac2 in T helper 1 cell differentiation. Science 288:2219-2222. [DOI] [PubMed] [Google Scholar]
- 38.Lutzelschwab, C., M. R. Huang, M. C. Kullberg, M. Aveskogh, and L. Hellman. 1998. Characterization of mouse mast cell protease-8, the first member of a novel subfamily of mouse mast cell serine proteases, distinct from both the classical chymases and tryptases. Eur. J. Immunol. 28:1022-1033. [DOI] [PubMed] [Google Scholar]
- 39.McNeil, H. P., D. S. Reynolds, V. Schiller, N. Ghildyal, D. S. Gurley, K. F. Austen, and R. L. Stevens. 1992. Isolation, characterization, and transcription of the gene encoding mouse mast cell protease 7. Proc. Natl. Acad. Sci. USA 89:11174-11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush, and M. R. Philips. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minden, A., A. Lin, F. X. Claret, A. Abo, and M. Karin. 1995. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81:1147-1157. [DOI] [PubMed] [Google Scholar]
- 42.Minden, A., A. Lin, M. McMahon, C. Lange-Carter, B. Derijard, R. J. Davis, G. L. Johnson, and M. Karin. 1994. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science 266:1719-1723. [DOI] [PubMed] [Google Scholar]
- 43.Miranti, C. K., L. Leng, P. Maschberger, J. S. Brugge, and S. J. Shattil. 1998. Identification of a novel integrin signaling pathway involving the kinase Syk and the guanine nucleotide exchange factor Vav1. Curr. Biol. 8:1289-1299. [DOI] [PubMed] [Google Scholar]
- 44.Moll, J., G. Sansig, E. Fattori, and H. van der Putten. 1991. The murine rac1 gene: cDNA cloning, tissue distribution and regulated expression of rac1 mRNA by disassembly of actin microfilaments. Oncogene. 6:863-866. [PubMed] [Google Scholar]
- 45.Nobes, C. D., and A. Hall. 1995. Rho, Rac and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 46.Nobes, C. D., P. Hawkins, L. Stephens, and A. Hall. 1995. Activation of the small GTP-binding proteins rho and rac by growth factor receptors. J. Cell Sci. 108:225-233. [DOI] [PubMed] [Google Scholar]
- 47.Ogihara, H., E. Morii, D. K. Kim, K. Oboki, and Y. Kitamura. 2001. Inhibitory effect of the transcription factor encoded by the mutant mi microphthalmia allele on transactivation of mouse mast cell protease 7 gene. Blood 97:645-651. [DOI] [PubMed] [Google Scholar]
- 48.Olson, M. F., A. Ashworth, and A. Hall. 1995. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 269:1270-1272. [DOI] [PubMed] [Google Scholar]
- 49.Pande, A., J. Pande, N. Asherie, A. Lomakin, O. Ogun, J. A. King, N. H. Lubsen, D. Walton, and G. B. Benedek. 2000. Molecular basis of a progressive juvenile-onset hereditary cataract. Proc. Natl. Acad. Sci. USA 97:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollok, K., J. van der Loo, R. Cooper, L. Kennedy, and D. Williams. 1999. Costimulation of transduced T lymphocytes via T-cell receptor/CD3 complex and CD28 leads to increased transcription of integrated retrovirus. Hum. Gene Ther. 10:2221-2236. [DOI] [PubMed] [Google Scholar]
- 51.Reynolds, D. S., D. S. Gurley, K. F. Austen, and W. E. Serafin. 1991. Cloning of the cDNA and gene of mouse mast cell protease-6. Transcription by progenitor mast cells and mast cells of the connective tissue subclass. J. Biol. Chem. 266:3847-3853. [PubMed] [Google Scholar]
- 52.Roberts, A. W., C. Kim, L. Zhen, J. B. Lowe, R. Kapur, B. Petryniak, A. Spaetti, J. D. Pollock, J. B. Borneo, G. B. Bradford, S. J. Atkinson, M. C. Dinauer, and D. A. Williams. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 10:183-196. [DOI] [PubMed] [Google Scholar]
- 53.Sanders, L., F. Matsumura, G. Bokoch, and P. de Lanerolle. 1999. Inhibition of myosin light chain kinase by p21-activated kinase. Science 283:2083-2085. [DOI] [PubMed] [Google Scholar]
- 54.Sekizawa, K., G. H. Caughey, S. C. Lazarus, W. M. Gold, and J. A. Nadel. 1989. Mast cell tryptase causes airway smooth muscle hyperresponsiveness in dogs. J. Clin. Investig. 83:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serafin, W. E., D. S. Reynolds, S. Rogelj, W. S. Lane, G. A. Conder, S. S. Johnson, K. F. Austen, and R. L. Stevens. 1990. Identification and molecular cloning of a novel mouse mucosal mast cell serine protease. J. Biol. Chem. 265:423-429. [PubMed] [Google Scholar]
- 56.Serafin, W. E., T. P. Sullivan, G. A. Conder, A. Ebrahimi, P. Marcham, S. S. Johnson, K. F. Austen, and D. S. Reynolds. 1991. Cloning of the cDNA and gene for mouse mast cell protease 4. Demonstration of its late transcription in mast cell subclasses and analysis of its homology to subclass-specific neutral proteases of the mouse and rat. J. Biol. Chem. 266:1934-1941. [PubMed] [Google Scholar]
- 57.Shirsat, N. V., R. J. Pignolo, B. L. Kreider, and G. Rovera. 1990. A member of the ras gene superfamily is expressed specifically in T, B and myeloid hemopoietic cells. Oncogene 5:769-772. [PubMed] [Google Scholar]
- 58.Spiegelman, B. M., M. Frank, and H. Green. 1983. Molecular cloning of mRNA from 3T3 adipocytes. Regulation of mRNA content for glycerophosphate dehydrogenase and other differentiation-dependent proteins during adipocyte development. J. Biol. Chem. 258:10083-10089. [PubMed] [Google Scholar]
- 59.Sulciner, D. J., K. Irani, Z. X. Yu, V. J. Ferrans, P. Goldschmidt-Clermont, and T. Finkel. 1996. rac1 regulates a cytokine-stimulated, redox-dependent pathway necessary for NF-κB activation Mol. Cell. Biol. 16:7115-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Symons, M., and J. Settleman. 2000. Rho family GTPases: more than simple switches. Trends Cell Biol. 10:415-419. [DOI] [PubMed] [Google Scholar]
- 61.Thomas, P. S. 1980. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. USA 77:5201-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timokhina, I., H. Kissel, G. Stella, and P. Besmer. 1998. Kit signaling through PI 3-kinase and Src kinase pathways: an essential role for Rac1 and JNK activation in mast cell proliferation. EMBO J. 17:6250-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsujimura, T., U. Koshimizu, H. Katoh, K. Isozaki, Y. Kanakura, T. Tono, S. Adachi, T. Kasugai, H. Tei, Y. Nishimune, et al. 1993. Mast cell number in the skin of heterozygotes reflects the molecular nature of c-kit mutation. Blood 81:2530-2538. [PubMed] [Google Scholar]
- 64.Uddin, S., E. N. Fish, D. Sher, C. Gardziola, O. R. Colamonici, M. Kellum, P. M. Pitha, M. F. White, and L. C. Platanias. 1997. The IRS-pathway operates distinctively from the Stat-pathway in hematopoietic cells and transduces common and distinct signals during engagement of the insulin or interferon-alpha receptors. Blood 90:2574-2582. [PubMed] [Google Scholar]
- 65.Uddin, S., F. Lekmine, N. Sharma, B. Majchrzak, I. Mayer, P. R. Young, G. M. Bokoch, E. N. Fish, and L. C. Platanias. 2000. The Rac1/p38 mitogen-activated protein kinase pathway is required for interferon alpha-dependent transcriptional activation but not serine phosphorylation of Stat proteins. J. Biol. Chem. 275:27634-27640. [DOI] [PubMed] [Google Scholar]
- 66.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 67.van Oers, N. S., W. Tao, J. D. Watts, P. Johnson, R. Aebersold, and H. S. Teh. 1993. Constitutive tyrosine phosphorylation of the T-cell receptor (TCR) zeta subunit: regulation of TCR-associated protein tyrosine kinase activity by TCR zeta. Mol. Cell. Biol. 13:5771-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vojtek, A. B., and J. A. Cooper. 1995. Rho family members: activators of MAP kinase cascades. Cell 82:527-529. [DOI] [PubMed] [Google Scholar]
- 69.Vosseller, K., G. Stella, N. S. Yee, and P. Besmer. 1997. c-Kit receptor signaling through its phosphatidylinositide-3′-kinase-binding site and protein kinase C: role in mast cell enhancement of degranulation, adhesion, and membrane ruffling. Mol. Biol. Cell 8:909-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol. Cell Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams, D. A., W. Tao, F. C. Yang, C. Kim, Y. Gu, P. Mansfield, J. E. Levine, B. Petryniak, C. W. Derrrow, C. Harris, B. Jia, Y. Zheng, D. R. Ambruso, J. B. Lowe, S. J. Atkinson, M. C. Dinauer, and L. Boxer. 2000. A dominant negative mutation of the hematopoiesis-specific RhoGTPase, Rac 2, is associated with a human phagocyte immunodeficiency. Blood 96:1646-1654. [PubMed] [Google Scholar]
- 72.Wong, G. W., Y. Tang, E. Feyfant, A. Sali, L. Li, Y. Li, C. Huang, D. S. Friend, S. A. Krilis, and R. L. Stevens. 1999. Identification of a new member of the tryptase family of mouse and human mast cell proteases which possesses a novel COOH-terminal hydrophobic extension. J. Biol. Chem. 274:30784-30793. [DOI] [PubMed] [Google Scholar]
- 73.Xia, Z., N. Ghildyal, K. F. Austen, and R. L. Stevens. 1996. Posttranscriptional regulation of chymase expression in mast cells. A cytokine-dependent mechanism for controlling the expression of granule neutral proteases of hematopoietic cells. J. Biol. Chem. 271:8747-8753. [DOI] [PubMed] [Google Scholar]
- 74.Yang, F. C., S. J. Atkinson, Y. Gu, J. B. Borneo, A. W. Roberts, Y. Zheng, J. Pennington, and D. A. Williams. 2001. Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc. Natl. Acad. Sci. USA 98:5614-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, F. C., R. Kapur, A. J. King, W. Tao, C. Kim, J. Borneo, R. Breese, M. Marshall, M. C. Dinauer, and D. A. Williams. 2000. Rac 2 stimulates Akt activation affecting BAD/Bcl-XL expression while mediating survival and actin-based cell functions in primary mast cells. Immunity 12:557-568. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, Y. 2001. Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26:724-732. [DOI] [PubMed] [Google Scholar]