Abstract
Cleavage of eukaryotic translation initiation factor 4GI (eIF4GI) is required for shutoff of host cell translation during poliovirus (PV) infection of HeLa cells. Reports published by several groups have led to confusion whether this cleavage is mediated by viral 2A protease (2Apro) or a putative cellular enzyme (termed eIF4Gase) which is activated by 2Apro or other aspects of viral infection. Here we have further investigated eIF4Gase activities in PV-infected cells. Column purification of eIF4GI cleavage activity separated two activities which generated N-terminal cleavage products of different lengths. Both activities were detected using either native eIF4G or radiolabeled recombinant eIF4G as the substrate. Analysis of cleavage products formed by each activity on native and mutant substrates suggests that one activity cleaves eIF4G1 at or very near the 2Apro cleavage site and the other activity cleaves approximately 40 residues upstream of the 2Apro cleavage site. When PV infections in HeLa cells were supplemented with 2 mM guanidine, which indirectly limits expression of 2Apro, two distinct C-terminal cleavage fragments of eIF4GI were detected. These C-terminal cleavage fragments of eIF4GI were purified from infected cells, and a new eIF4GI cleavage site was mapped to a unique site 43 amino acids upstream of the known 2Apro cleavage site. Further, eIF4GI cleavage in vivo could be blocked by addition of zVAD to PV-guanidine infections. zVAD is a broad-spectrum caspase inhibitor which had no effect on 2Apro cleavage activity or PV polyprotein processing. Lastly, similar types of eIF4Gase cleavage activities were also detected in uninfected cells under various conditions, including early apoptosis or during cell cycle transit. The data suggest that the same types of eIF4GI cleavage activities which are generated in PV-infected cells can also be generated in the absence of virus. Taken together, the data support a model in which multiple cellular activities process eIF4GI in PV-infected cells, in addition to 2Apro.
Poliovirus (PV), coxsackievirus B (CVB), and human rhinovirus infection of HeLa cells results in a dramatic inhibition of host translation within 1.5 to 3 h postinfection. However, only cap-dependent translation, which comprises about 95% of total cellular translation, is blocked. Most of the remaining ∼5% of cellular translation (25) as well as viral translation proceeds via cap-independent mechanisms that require a cis-acting internal ribosome entry site on the mRNA (2). Cap-dependent translation initiation requires a protein complex called eukaryotic initiation factor 4F (eIF4F). eIF4F is comprised of three subunits: eIF4E, the cap binding protein; eIF4A, an ATP-dependent helicase; and eIF4G (formerly called p220), which is a large, modular scaffolding protein. eIF4G contains binding sites for several proteins that function in translation [eIF4E, eIF3, eIF4A, mnk-1, poly(A)-binding protein (PABP)] (reviewed in reference 15). Since eIF4G can simultaneously bind eIF4E and eIF3 (situated on 40S ribosome subunits) it provides the molecular link between the 5′ cap group on the mRNA and the ribosome (15, 42).
The mechanism of translation shutoff has been extensively studied and is not yet completely understood. Early during infection, eIF4G is proteolytically cleaved to smaller products with kinetics which slightly precede translation inhibition (14). This cleavage occurs at a position that separates the eIF4E-binding domain from the eIF3-binding domain (31, 33). An attractive hypothesis for translation shutoff was derived from this observation that stated that shutoff was caused by this cleavage event which split eIF4G (and the eIF4F complex), breaking the eIF4G-mediated linkage between the capped mRNA and ribosomes. More recently it has been postulated that newly discovered cleavage of eIF4GII (a functional homolog of eIF4GI) and PABP is also required for host translation shutoff, though the functional importance of either event has not yet been established (18, 24, 29, 46).
The mechanism of eIF4GI cleavage itself has been controversial. Early reports showed that an eIF4GI cleavage activity, which could be measured in extracts from PV-infected HeLa cells, did not copurify with PV 2A protease (2Apro) or 3Cpro (35, 37). However, translation of 2Apro in vitro caused eIF4G cleavage, and antibody that inhibited 2Apro activity did not block eIF4GI cleavage activity after its induction (30). Thus, it was hypothesized that 2Apro indirectly induced eIF4G cleavage activity, which was proposed to be due to activation of a cellular protease (30, 37). Subsequent experiments using recombinant viral 2Apro from CVB, human rhinovirus, and PV demonstrated that eIF4G does serve as a direct substrate of 2Apro, which cleaves between the eIF4E and eIF3 binding domains of eIF4GI (33, 34, 45) and generates eIF4GI cleavage fragments that are identical to those produced in vivo. The direct cleavage reaction, however, was shown to be very inefficient in vitro, using purified protease and purified eIF4F (4). Recently, a very efficient cotranslational direct cleavage of eIF4G by 2Apro has been suggested, and it has been proposed that eIF4GI cleavage in vivo is mediated primarily by 2Apro directly, possibly as 2Apro is released from ribosomes (16).
In contrast to these results, the major eIF4GI cleavage activity that was detectable in biochemical assays was purified from infected cells to high levels and did not contain detectable 2Apro (5). The identity of the protease responsible for this cleavage has not been determined, although protease inhibitor profiles and molecular mass estimates suggest a cysteine protease of about 55 to 60 kDa (49). Here we present further analysis of eIF4GI-specific protease activity and provide evidence that two or more protease activities comprise what was formerly thought to be a single activity. One of these activities cleaves eIF4GI at a site distinct from the 2Apro cleavage site.
MATERIALS AND METHODS
Cells and virus infection.
HeLa S3 cells were grown in spinner culture in S-MEM (Gibco) supplemented with 9% calf serum-1% fetal calf serum and penicillin-streptomycin (Sigma) and maintained at cell densities between 3 × 105 and 8 × 105 cells/ml. K562 cells were cultured in stationary suspension culture in RPMI supplemented with 8% calf serum-2% fetal calf serum and penicillin-streptomycin. PV type 1 (Mahoney) was propagated and purified as previously described (27). Serum was added to 5% final concentration at 30 min postinfection, and cells were harvested at 5 h postinfection or the times indicated in the figures. Depending on the experiment, infections were carried out at multiplicities of infection (MOI) of 20, 100, or 200 PFU/cell at 37°C. Cell lysates were prepared for direct analysis in sodium dodecyl sulfate (SDS)-polyacrylamide gels by suspension and incubation of cell pellets in NP-40-RSB (1% NP-40, 10 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1.5 mM MgCl2), and the cytoplasmic fraction was mixed with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer for gel analysis. Lysis of large amounts of HeLa cells was accomplished by Dounce homogenization in hypotonic buffer as previously described (27).
For the induction of apoptosis, various concentrations of cisplatin (Sigma-Aldrich), dexamethasone, and butyric acid stock solutions were diluted with S-MEM and then incubated with HeLa or K562 cells at 37°C for the time indicated in each figure. For in vivo inhibition experiments cells were first preincubated with the cell-permeable caspase inhibitor Z-VAD-fmk (Enzyme Systems Products) or guanidine-HCl for 10 min at 37°C before virus was added to the cell cultures.
Purification of eIF4G cleavage activity and activity assays.
Large-scale purification of eIF4G-specific cleavage activity (eIF4Gase activity) was performed using lysates taken from infected HeLa cells at 5 h postinfection as previously described (5). Briefly, S10 lysates were prepared by Dounce homogenization, and then ribosomal salt wash (RSW) 0 to 40% ammonium sulfate precipitate fractions (RSW-Acut fractions) were prepared as described previously (6). RSW-Acut fractions were subjected to sequential chromatography over Sephacryl S300, Fast Q Sepharose, and nickel affinity columns as previously described (5). Some pooled fractions were also further purified by dialysis against buffer containing 110 mM KCl, 20 mM HEPES (pH 7.4), 5% glycerol, 10 mM MgCl2, and 1 mM dithiothreitol and then applied to an m7GTP-Sepharose column (1 ml) equilibrated in the same buffer. Unbound proteins were batch eluted and washed off with the same dialysis buffer (10 ml). Bound proteins were eluted with the same buffer supplemented with 25 mM EDTA. eIF4GI cleavage assays were conducted as previously described (5) by incubation of test fractions in 50-μl reaction mixtures containing 5 mM MgCl2, 50 mM NaCl, 20 mM Tris (pH 7.4), and 7 μl of uninfected cell RSW (URSW), which is enriched in eIF4G substrate. Reactions were incubated 2 h at 37°C, proteins were separated by SDS-PAGE using 7% gels, and eIF4GI was visualized by immunoblot analysis using eIF4GI N-terminus-specific polyclonal antisera as previously described (36). Cleavage reactions using radiolabeled eIF4G substrates contained 15 μl of test lysate or fractions combined with 15 μl of cleavage buffer and 3 μl of 35S-labeled eIF4G substrate translation reaction.
Purified proteases.
CVB3 2Apro was produced in Escherichia coli and purified as previously described (24). Human recombinant caspase 3 bearing a hexahistidine tag was also produced in E. coli and purified using metal chelate chromatography (40). PV 2Apro bearing a C-terminal histidine tag was expressed in bacteria and purified using metal chelate affinity columns as described previously (4).
Purification of eIF4GI cleavage fragments.
RSW fractions were prepared from infected cells and then further analyzed by two-dimensional (2D) gel electrophoresis (12). Migration of eIF4GI and eIF4GI cleavage products was determined by immunoblot analysis. Large-scale purification of eIF4GI-CPC (where CPC is the C-terminal cleavage product) was achieved by fractionation of RSW (20 ml) into 40 to 70% ammonium sulfate precipitates. The precipitate was resuspended in 20 mM MES (pH 6.5)-50 mM NaCl-1 mM EDTA-7 mM-2-mercaptoethanol and purified over a Sephacryl S300 column (2 by 20 cm) and then fractions containing eIF4G-CPC were subsequently fractionated over P-11 phosphocellulose (1 by 9 cm) in the same buffer. Bound proteins were eluted with a salt gradient (50 to 1,000 mM NaCl) and analyzed by SDS-PAGE and immunoblotting. Duplicate fractions containing eIF4G-CPC were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes, and proteins were visualized by staining with Coomassie brilliant blue. Protein bands representing CPC were excised and subjected to sequential Edman degradation sequencing at the University of Oklahoma Protein Chemistry Core facility.
Transcription and translation.
Transcription reactions for eIF4GI using T7 RNA polymerase (Promega) were carried out according to manufacturer’s recommendations followed by purification of the RNA using spin columns (Eppendorf-5 Prime). Plasmids used in transcription reactions were pAD-4G, pAD-4G(G486E) (32) linearized with XhoI (full-length transcripts), or SpeI (truncated transcript). A subcloned derivative of pAD-4G (pHisp120) was created by subcloning a BamHI/HindIII fragment (encoding eIF4G amino acids 661 to 1600) of pAD4G into pET-28b(+) (Novagen). The resulting pET28-4G(457-1396) plasmid was digested with BamHI, treated with Klenow, and then digested with AatII to release a 7.6-kb fragment that was purified. That DNA was ligated with a 1,145-bp fragment of pSK-HFCI (50) that was released by treatment with XmaI, followed by Klenow and final digestion with AatII. The new plasmid was termed pHisp120 and contained the fragment of eIF4GI cDNA from just upstream of the eIF4E binding site to the C terminus. For transcriptions, this plasmid was linearized with SpeI. Translation reactions were performed in rabbit reticulocyte lysate (Promega) according to the manufacturer’s recommendations in a 25-μl reaction volume and incubated for 90 min at 30°C. Samples (2 μl) were subjected to SDS-PAGE and analyzed by autoradiography or otherwise used in in vitro cleavage reactions with purified eIF4Gase fractions as described above.
RESULTS
Nature of multiple isoforms of eIF4GI.
When resolved in SDS-polyacrylamide gels, eIF4GI appears as a collection of four isoforms that differ in apparent molecular mass from each other by about 4 to 6,000 Da. The molecular explanation for these multiple isoforms has been lacking since eIF4GI was first described (14, 19). The original cDNA clone of eIF4GI produced only a single polypeptide band when expressed in vitro that migrated in SDS-polyacrylamide gels faster than all but one of the native eIF4GI isoforms (28). It has been proposed that the appearance of multiple eIF4GI isoforms was due to posttranslational modifications such as phosphorylation; however, it is possible that alternate splicing of mRNA or alternate use of initiation codons could generate the multiple isoforms (28, 50, 51). Two groups have discovered N-terminal extensions of the originally published eIF4GI cDNA sequence, thus adding another 147 amino acids to the deduced eIF4GI coding sequence (17, 26). We have recently discovered a new cDNA extension that overlaps and extends past the previous extension by an additional 40 amino acids and also contains a large, putative 5′ untranslated region. Translation of this larger eIF4GI mRNA in vitro generates a pattern of four isoforms that comigrate in SDS-polyacrylamide gels with the isoforms of native eIF4GI. Translation studies with derivative eIF4GI-EGFP fusion mRNAs bearing this upstream region reveal that alternate use of three downstream in-frame AUG codons accounts for the multiple isoforms of eIF4G (53). Thus, there is now evidence that the four isoforms of eIF4GI comprise a family of polypeptides that are 1,600, 1,560, 1,513, and 1,404 amino acids in length (Fig. 1) and are generated by alternative translation initiation. Another isoform of 1,435 amino acids may also be expressed from an additional methionine at much lower levels (53).
FIG. 1.
Schematic of eIF4GI complete amino acid sequence and isoforms derived from internal translation initiation at in-frame AUG codons. All isoforms of eIF4G are depicted cleaved at the known 2Apro cleavage site. The black box represents additional amino acids found in the new extended cDNA sequence. Hatched or gray boxes represent binding domains for the proteins PABP, eIF4E, eIF4A, eIF3, and mnk-1 as indicated. The mapped location of epitopes recognized by the polyclonal anti-eIF4GI antisera used in this study are also indicated. Numbers to the left of each isoform identify which amino acid in the open reading frame begins the isoforms, and the numbers to the right indicate total number of amino acids in the isoform, along with apparent mobility in SDS-polyacrylamide gels.
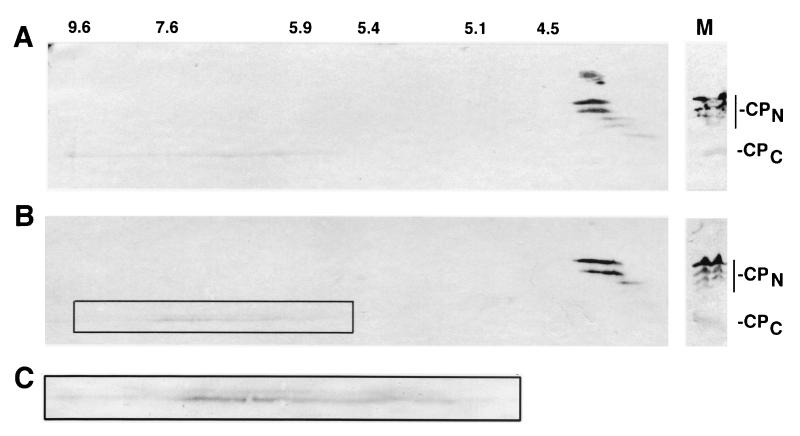
The uncertainty about the molecular nature of the eIF4GI isoforms has long hindered our analysis of eIF4GI cleavage activities since the biochemical nature of various types of cleavage products was not known. When cleaved by 2Apro, the CPN of eIF4GI also display a similar multiple banding pattern, while the CPC generally appears as a single band in SDS gels (31). In addition, migration of eIF4GI CPN in SDS-polyacrylamide gels is much slower than expected from the deduced molecular weight of the polypeptides, possibly due to a very high proline content (31, 50). To further investigate whether posttranslational modifications or alternate initiation was responsible for the multiple isoforms, we performed 2D gel analysis of eIF4GI CPN and CPC (Fig. 2). Each individual eIF4GI CPN displayed a relatively tight isoelectric focusing pattern that ranged from pH 4.3 to 4.0. Interestingly, the largest isoforms had pIs which were the least acidic. This result is inconsistent with the hypothesis that increased phosphorylation causes the appearance of isoforms with higher apparent molecular weight in SDS-polyacrylamide gels. It has recently been demonstrated that the majority of the phosphorylation of eIF4GI occurs in the C-terminal domain (43). Figure 2A shows that CPC prepared from lysates late in infection (5 h) appears as a single band which is more basic than CPN; however, it smears over a wide range of isoelectric points, ranging from 5.9 to 9.6. This pattern is consistent with a significant degree of phosphorylation of CPC, and there may be too many phosphorylation sites for discreet spots to emerge. Thus, taking these date together with our translation results (53), we now have confidence that the multiple banding pattern of eIF4GI and eIF4G CPN results from alternate use of multiple initiation codons that create polypeptides of lengths which are now defined.
FIG. 2.
D gel analysis of eIF4GI cleavage fragments. (A) RSW from PV-infected HeLa cells (harvested at 4 h postinfection) was subjected to isoelectric focusing in the first dimension and SDS-PAGE in the second dimension, followed by immunoblot analysis with a mixture of antibodies specific for N-terminal or C-terminal domains of eIF4GI. Positions of isoelectric points (indicated above the panel) were determined by migration of known isoelectric focusing protein standards. The right panel shows migration of eIF4G cleavage products in the one-dimensional SDS-PAGE lane of the same gels. (B) Migration of CPCs from guanidine-PV infection. The right panel shows migration of eIF4G cleavage products in the one-dimensional SDS-PAGE lane of the same gels. (C) Appearance of eIF4GI-CPC doublet in guanidine infections. Shown is an enlarged view of the boxed region in panel B.
Multiple eIF4GI cleavage activities in PV-infected cells.
In our previous studies of eIF4GI cleavage activity present in PV-infected cells, we have shown that 2Apro can be separated by column chromatography from the predominant eIF4G cleavage activity measured in in vitro cleavage assays (5). In contrast, infected cell fractions containing large amounts of native 2Apro exhibited little or no cleavage activity in vitro on native eIF4GI substrate contained in RSW preparations (5). In the present study, only purified eIF4Gase fractions that did not contain detectable 2Apro were selected for further analysis (5). We have often observed different types of eIF4GI cleavage products that were produced in assays using purified eIF4Gase fractions yet were unable to explain the biochemical mechanism for these differences since there was such uncertainty about the nature of the eIF4GI isoforms. Figure 3 shows examples where two types of cleavage products were obtained in assays after chromatography of such eIF4Gase preparations. The PV-RSW contains cleavage products of eIF4GI that comigrate with those produced by 2Apro (37, 52). Figure 3A shows chromatography of purified eIF4Gase fractions over m7GTP-Sepharose yielded an activity in fractions 4 and 5 that produced CPN that comigrated with CPN produced in infected cells (PvRSW) and that also comigrated exactly with 2Apro cleavage products (4). In contrast, fraction 8 contained an activity that generated CPN that migrated slightly faster in SDS-polyacrylamide gels. Two of the bands in fraction 5 (bands 2 and 3) and 8 (bands 1 and 2) also appear to overlap. To alleviate confusion, we have named these two activities eIF4Gase-α and eIF4Gase-β, which generate the larger and smaller set of N-terminal cleavage fragments, respectively.
FIG. 3.
Chromatographic separation of different eIF4G cleavage activities. (A) Separation on m7GTP-Sepharose. Pooled partly purified material was applied to m7GTP-Sepharose, and fractions were eluted and assayed for eIF4G cleavage activity as described in Materials and Methods using antisera specific for N-terminal domain of eIF4GI. Immunoblot controls are URSW and PvRSW incubated alone for the period of the assay. The flowthrough fraction is in lane F, and eluted fractions are in lanes labeled with numbers. Arrows indicate CPN produced by eIF4Gase-α or 2Apro activity, and arrowheads indicate CPN cleavage products produced by eIF4Gase-β. (B) Pooled material from S300 Sepharose was applied to a nickel-affinity column, and then fractions were eluted with either pH 6.0 buffer (fraction 1) or 25 mM EDTA (fraction 2). Fractions were tested for eIF4G cleavage activity as described in Materials and Methods, and cleavage products were compared with 2Apro cleavage products by immunoblotting using antiserum specific for eIF4GI CPN (left panel) or eIF4GI CPC (right panel).
Figure 3B shows eIF4Gase-β activity can also be purified by nickel-affinity chromatography. In this procedure, the eIF4Gase-α activity does not efficiently bind the column and is recovered in flowthrough fractions (data not shown). Immunoblots of cleavage assays probed with antibodies specific for the N-terminal domain of eIF4GI revealed the same faster-migrating eIF4Gase-β cleavage fragments shown in Fig. 3A which migrate faster than 2Apro cleavage fragments (fraction 1 and fraction 2). However, probing the same cleavage assay immunoblots with antibodies specific for the C-terminal domain of eIF4GI revealed that a larger, slower-migrating CPC is produced by eIF4Gase-β that does not comigrate with the CPC produced by 2Apro. These data are consistent with eIF4Gase-β activity cleaving native eIF4GI at a novel position upstream of the mapped 2Apro cleavage site. Further, since the two largest isoforms of eIF4GI differ in length by 40 amino acids, the shift in band migration of CPNs is consistent with an eIF4Gase-β activity cleaving eIF4GI approximately 40 amino acids upstream of the known 2Apro cleavage site. Since PV 2Apro cleaves very inefficiently in in vitro cleavage reactions (4, 16), and since starting fractions used in these final chromatographic separations did not contain detectable 2Apro, it is unlikely that the cleavage activity measured here that produces either the slower-migrating CPN or faster-migrating CPN is actually 2Apro. Thus, using conventional chromatographic procedures we can separate two distinct types of eIF4GI cleavage activities present in PV-infected cell extracts that cleave eIF4GI in distinct locations.
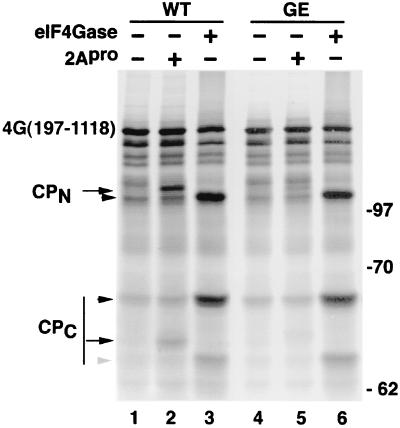
To further investigate eIF4Gase activities, we sought to use defined radiolabeled eIF4GI substrates generated by in vitro translation of truncated eIF4GI mRNA. The use of truncated forms of eIF4GI further enhanced the separation in SDS gels of cleavage products resulting from proteolysis at closely spaced cleavage sites. Figure 4A shows that translation of 4G(197-1118)-WT RNA generated a polypeptide with an Mr of 147, as expected. Several smaller translation products arise due to aberrant internal translation initiation with this construct. When this substrate was incubated with purified recombinant PV His-2Apro, partial cleavage resulted in generation of two new cleavage products, representing C- and N-terminal products. The migration of each product was consistent with expected results if 2Apro cleaved at the well-defined and mapped cleavage site between amino acids 681 and 682. Only a single CPN and CPC was produced, showing 2Apro cleaves at a single site on this substrate. We also expressed a similar eIF4G substrate containing a point mutation which changes the required P1 glycine in the 2Apro cleavage site sequence to glutamic acid [4G(197-1118)-GE]. This mutation causes a 10-fold reduction in 2Apro cleavage activity (32). Cleavage by 2Apro was inhibited by this substrate, confirming that recombinant 2Apro cleaved the truncated eIF4G substrate at the normal site. Note that the cleavage activity of recombinant PV 2Apro, which is prone to denaturation, is usually much lower than that of CVB3 2Apro; hence, only partial cleavage of eIF4GI substrate was observed in this experiment. Subsequent experiments in this study were conducted with the more stable CVB3 2Apro, which cleaves eIF4GI at the exact same site (31, 33), typically with greater efficiency.
FIG. 4.
The eIF4Gase-β cleavage site is distinct from the 2Apro cleavage site. Defined radiolabeled eIF4G substrates were translated in vitro and then incubated with recombinant PV His-2Apro (0.5 μg), CVB3 2Apro, or eIF4Gase-β-enriched fractions (metal chelate pool) as indicated. Multiple smaller bands in starting substrate preparation result from aberrant internal initiation of translation on transcript RNA. Cleavage products produced by 2Apro are indicated by arrows, and cleavage products from eIF4Gase-β are indicated by arrowheads. The gray arrowhead indicates a minor eIF4Gase CPC not detected with native protein. WT indicates the wild-type 2Apro cleavage site in eIF4G substrate, whereas GE indicates the form of eIF4G containing a point mutation of Gly682 to Glu, which inhibits 2Apro cleavage.
When one of the highly purified, eIF4Gase-β-enriched fractions was incubated with radiolabeled eIF4GI substrate, two new major cleavage products appeared that did not comigrate with 2Apro cleavage products. Further, this cleavage activity was not impaired by the GE mutation in the substrate (Fig. 4A, lane 6). The migration of the new cleavage products was consistent with cleavage occurring approximately 40 amino acids upstream of the mapped 2Apro cleavage site, similar to the results seen in Fig. 3. This provides evidence that eIF4Gase-β proteolytic activity is distinct from 2Apro. We also analyzed cleavage of the recombinant form of the smallest of the naturally occurring eIF4GI isoforms [eIF4GI(197-1600)] and obtained similar results (data not shown).
When a third smaller eIF4Gase fragment [4G(580-1118)] was incubated with eIF4Gase fractions, the upstream cleavage site was not utilized; rather, an alternate cleavage site downstream of the 2Apro cleavage site was cleaved preferentially. With this truncated substrate, cleavage with 2Apro still produced the two expected fragments (data not shown). This indicated that a conformation change on this extremely truncated substrate masked or altered the upstream eIF4Gase cleavage site and/or exposed a new secondary cleavage site. This alternate type of CPC cleavage product (also seen in Fig. 4A, lanes 3 and 6) may be aberrant since it has not been observed with native eIF4GI.
eIF4GI is cleaved at two sites in vivo.
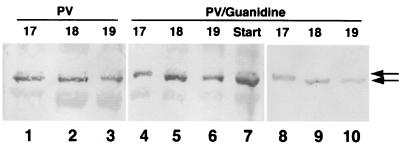
We next wanted to determine the proteolytic site where eIF4GI is cleaved in vivo during virus infection. The site of in vitro eIF4GI cleavage by 2Apro has been well documented in the past (31, 32, 33), but the native protein in infected cells has not been examined. Thus, we used a CPC-specific antiserum to purify eIF4GI CPC from PV-infected cells that was then sequenced by Edman degradation procedures. During the course of this work, we noticed that the first fraction eluted from phosphocellulose columns contained a faint slower-migrating CPC isoform (Fig. 5, lane 1) whose significance and identity were unknown; however, the majority of CPC appeared as a slightly faster-migrating isoform. The CPC bands from these fractions were pooled and sequenced. The polypeptides contained an N terminus beginning with a Gly that corresponded with the previously mapped 2Apro cleavage site downstream sequence (GPPR) (Fig. 6). This suggested most CPC was generated by cleavage with 2Apro or eIF4Gase-α or both. Consistent with this, CPC from PV-infected cells harvested late in infection shows only one band in 2D gel analysis (Fig. 2A).
FIG. 5.
Elution of eIF4G CPC from phosphocellulose. PV infections with or without guanidine were used as source cells for purification of eIF4GI-CPC as detailed in Materials and Methods. Pooled fractions chromatographed over P-11 phosphocellulose were analyzed by immunoblot analysis for eIF4GI CPC. Sequential chromatography fractions (numbered 17 to 19) from normal PV infections are shown (lanes 1 to 3) as well as equivalent fractions from two separate purifications from guanidine-supplemented infections (lanes 4 to 10). The left two panels are also counterstained with amido black to identify all protein in the sample.
FIG. 6.
Sequence analysis of eIF4GI CPC. (A) Purified eIF4GI-CPC was subjected to Edman degradation sequence analysis. Protein was purified from cells infected for 2.5 or 5 h before harvesting or cells from 4-h infections supplemented with guanidine-HCl. In addition, protein from PV-guanidine infections separated by SDS-PAGE into fast (lower band)- and slower (upper band)-migrating forms was analyzed individually. p.i., postinfection. (B) Comparison of known 2Apro cleavage sites on viral polyproteins and cellular proteins and a consensus recognition sequence.
We also purified and sequenced CPC from infected cells that were harvested early in infection (2.5 h). Interestingly, analysis of this polypeptide revealed a new sequence beginning with a Val that did not correspond to the sequence beginning at the 2Apro cleavage site (Fig. 6). We then purified eIF4GI CPC from HeLa cells infected in the presence of 2 mM guanidine-HCl. This drug inhibits viral RNA replication and serves to reduce expression of 2Apro 20- to 50-fold; however, eIF4G cleavage still occurs (5). This would be expected to lower production of CPC beginning with the GPPR sequence by 20- to 50-fold if all of this product was gained from 2Apro cleavage and might increase the relative concentration of any alternate larger CPC isoform arising from eIF4Gase-β activity. Analysis of the CPC in cell extracts by 2D gel analysis did reveal a doublet that smears across the entire isoelectric focusing range of CPC (Fig. 2B and C). This is consistent with production of the new CPC from alternate cleavage rather than phosphorylation; thus, there was an increase in quantity of the larger band in guanidine infections that was only faintly visible in normal late infection extracts. When CPC from guanidine cell extracts was purified for sequencing, the first fraction eluting from phosphocellulose was comprised of mostly the slow-migrating, larger isoform (Fig. 5, lanes 4 and 8). The smaller isoform still comprised the majority of CPC in the fraction, suggesting that approximately 70% of eIF4G was still cleaved at or near the 2Apro cleavage site. However, the relative increase in the larger isoform was not the 20- to 50-fold or more expected from depletion of 2Apro, but rather a modest 4- to 5-fold. We separately harvested large and small CPC isoforms and subjected them to sequence analysis (Fig. 6). The data revealed an apparent mixture of polypeptides beginning at the 2Apro cleavage site and identified a new sequence, VVLDKA, that is located 43 amino acids upstream of the 2Apro cleavage site. The upper band showed a lesser degree of contamination with the isoform beginning with GPPR. Apparently, our methods failed to completely isolate one isoform from another, possibly due to factors such as phosphorylation or close migration of the isoforms in gels. However, these data do identify a novel N terminus of eIF4GI CPC that appears early in PV infection and is emphasized in infections carried out in the presence of guanidine. The position of this putative cleavage site is consistent with eIF4Gase-β activity and, based on the mapping results presented above, appears to cleave approximately 40 amino acids upstream of the 2Apro cleavage site.
Figure 6B shows the complete sequence at this site compared to known 2Apro cleavage sites on viral and cellular polypeptides. Sequence requirements for 2Apro recognition and cleavage include a hydrophobic L or M at the P4 position, a hydroxyl-containing T or S at the P2 position and a crucial G at the P1′ position. Few of these sequence requirements are contained in the new putative cleavage site, that contains a charged His at P4 and Val at the P1′ position. Alternatively, caspases are known to require Asp at the P1 position, this residue is conserved in the eIF4G cleavage site. However, the HISD sequence in eIF4GI does not match sequence requirements any of the caspases whose cleavage specificities have been well characterized to date (Fig. 6).
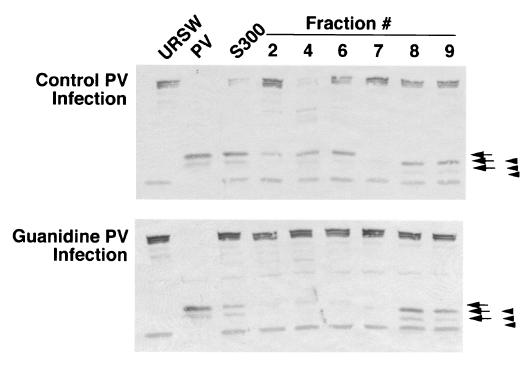
To further characterize the effect of guanidine on the production or activation of eIF4Gase we examined cells early in infection when levels of 2Apro are very low and compared the types of eIF4Gase in normal infected cells with cells infected in the presence of guanidine. Cell lysates were harvested after only 2.5 h infection, processed through several rounds of column chromatography to separate all detectable 2Apro from the samples (5) and finally purified by metal chelate chromatography to resolve eIF4Gase-α from eIF4Gase-β activity. Comparison of partly purified S300 pools revealed eIF4Gase activity was stronger in normal infection lysate than in guanidine lysates. As seen earlier, two types of cleavage activities eluted from the column and the eIF4Gase-α activity was significantly diminished in the guanidine lysate material, whereas the eIF4Gase-β activity was not diminished (Fig. 7). This suggests that eIF4Gase-α and eIF4Gase-β activity may not be induced by the same mechanism in infected cells and that eIF4Gase-α is more dependent on the level of viral protein expression or RNA replication. Further, the diminution of eIF4Gase-α type activity in guanidine infections is consistent with the cleavage site sequence data discussed above.
FIG. 7.
Comparison of early eIF4Gase activities in normal and guanidine-inhibited PV infections. PV infections with (bottom panel) or without (top panel) guanidine were performed for 2.5 h, and eIF4Gase activity was sequentially purified on Sepharose S300, Fast Q Sepharose, and metal chelate columns. Immunoblot analysis (CPN specific) of eIF4Gase activity in fractions from the latter is shown. Arrows indicate cleavage products produced by 2Apro/eIF4Gase-α, and arrowheads indicate cleavage products produced by eIF4Gase-β. PV, RSW fraction from PV infected cells (4.5 h infection).
Caspase inhibitor can block production of eIF4Gase in vivo.
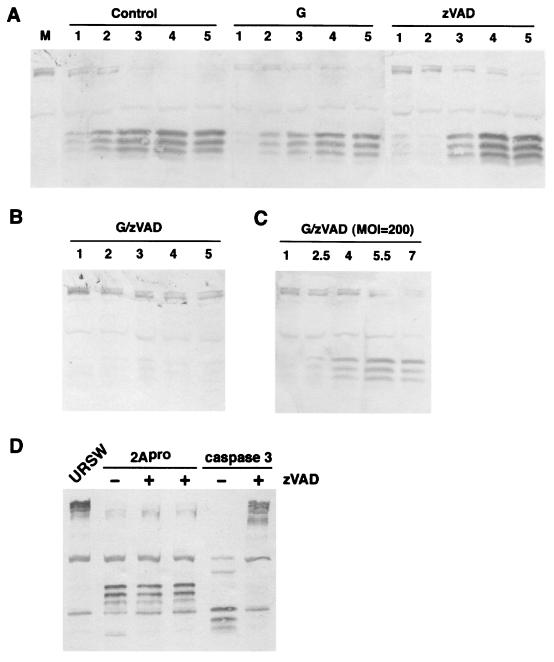
The new putative eIF4Gase cleavage site containing an Asp residue in the P1 position caused us to consider a role for caspases in eIF4G cleavage in PV infection. We have previously shown that induction of apoptosis in uninfected HeLa cells results in cleavage of eIF4GI with kinetics that correlate with translation shutoff and that caspase 3 is the dominant protease that cleaves eIF4GI in vivo. The caspase 3 cleavage of eIF4GI has been mapped to two new sites (7) and generates unique N-terminal cleavage fragments not ordinarily observed in PV-infected cells (Fig. 8D). Several reports have indicated that PV infection induces some apoptotic markers early in infection, then further development of apoptosis is blocked until late in infection (1, 47, 48). It is possible that an early apoptosis activation cascade results in activation of eIF4Gase(s) in PV-infected cells without development of caspase 3 activation, which is delayed until late in infection (reference 8 and data not shown). Thus, we decided to determine if the broad-specificity caspase inhibitor zVAD-fmk could block or delay eIF4G cleavage in PV-infected cells. Figure 8A shows kinetics of eIF4GI cleavage in three parallel infections. Addition of guanidine to infected cells has the effect of reducing 2Apro concentrations 20- to 50-fold and only slightly delays eIF4G cleavage as reported previously (5). When zVAD was added to infected cells, eIF4G cleavage was not blocked, but the kinetics of cleavage were slightly delayed in some experiments. Pulse-label analysis with [35S]methionine demonstrated that complete shutoff of host translation still occurred in zVAD-treated cells; however, the kinetics were delayed somewhat (data not shown). Investigation of the proteolytic processing of PV proteins revealed no obvious alterations compared to control infections; all PV polypeptides were produced as normal, suggesting that zVAD did not inhibit 3Cpro- or 2Apro-mediated processing of the viral polyprotein (data not shown). However, when both guanidine and zVAD were used together, eIF4Gase cleavage was inhibited significantly (Fig. 8B). If the multiplicity of infection was raised to very high levels, this blockage of eIF4G cleavage by combined inhibitor treatment could be partly overcome or reversed (Fig. 8C), although longer infections were required to demonstrate greater than 80% cleavage. We tested whether zVAD could inhibit 2Apro-mediated cleavage of eIF4G in vitro, which it failed to do; however, effective inhibition of caspase 3-mediated cleavage of eIF4GI was observed as expected (Fig. 8D). Thus, zVAD had no measurable effect on 2Apro-mediated eIF4G cleavage yet was able to block eIF4Gase activity in vivo. However, this result does not suggest that eIF4Gase must be a caspase but rather only implies that caspases may be involved in activation of eIF4Gase activity. Taken together, the data provide direct evidence for cellular eIF4G cleavage activity in vivo that is not attributable to 2Apro yet also provide evidence that in highly productive infections in HeLa cells both 2Apro and eIF4Gase(s) likely contribute significantly to eIF4G cleavage.
FIG. 8.
Combined treatment with guanidine and zVAD blocks development of eIF4Gase cleavage activity in PV-infected cells. (A) PV-infected cells (MOI = 20) were collected at the indicated time points postinfection or mock-infected cells were collected at 5 h (lane M) and analyzed for eIF4G cleavage by immunoblotting. Infections were unsupplemented (control) or supplemented with 2 mM guanidine-HCl (G) or 75 μM zVAD-fmk (zVAD) 10 min prior to infection. (B) Same as panel A, except cells were supplemented with both inhibitors. (C) Infection and analysis is conducted the same as for panel B, except the MOI was increased to 200. (D) Purified recombinant caspase 3 and 2Apro (0.5 μg each) were incubated with 5 μg of URSW for 90 min at 37°C before immunoblot analysis. Indicated reactions were supplemented with 200 μM zVAD.
eIF4Gase activity in apoptotic cells.
To further investigate relationships between eIF4Gase and apoptosis we conducted several experiments with uninfected HeLa cells treated with apoptosis inducers. We have previously noted transient production of eIF4Gase-like cleavage products in HeLa cells induced to undergo apoptosis with 100 μM cisplatin (40); however, all eIF4GI CPNs quickly converted to faster-migrating forms produced by caspase 3, and in many experiments only caspase 3 cleavage products were observed. Figure 9A shows a similar experiment using lower levels of cisplatin in order to delay caspase 3 induction. In this case, at 3 to 8 h after induction faint eIF4Gase-α cleavage products were detected in cell lysates that were replaced with caspase 3 products at late time points. When those cell lysates were incubated with URSW in eIF4Gase assays, a modest eIF4Gase-α activity was detected by renewed production of the eIF4Gase-α cleavage products. The eIF4Gase-α activity measured by this assay peaked by 3 h postinduction. This provides evidence that eIF4G can be cleaved by a cellular enzyme(s) in the total absence of 2Apro and those reactions produce cleavage products that comigrate with those produced by 2Apro. We also examined the effects of apoptosis inducers on K562 erythroblastoid cells, which in our hands have proven to be more resistant to apoptosis inducers than HeLa cells. Figure 9B shows that eIF4Gase-α cleavage products and probably eIF4Gase-β cleavage products can be detected along with caspase 3 cleavage products in K562 cells that were treated with a number of apoptosis inducers. Attempts to inhibit eIF4Gase-α activity induced in apoptotic lysates with DEVD-CHO have not proven successful (Fig. 9C), suggesting that this enzyme(s) is not closely related to caspase 3. This is not surprising since there are no aspartic acid residues in the vicinity of the 2Apro/eIF4Gase cleavage site(s) on eIF4GI.
FIG. 9.
Generation of eIF4Gase activities during early apoptosis. (A) HeLa cells treated with 25 mM cisplatin for the indicated time points were analyzed by immunoblotting for eIF4GI cleavage (left panel) or lysates were incubated with URSW for 6 h prior to immunoblot analysis to measure eIF4Gase activities in vitro. (B) eIF4Gase activation by various apoptosis inducers in K562 cells. K562 cells were mock treated or incubated with dexamethasone (1 μM), cisplatin (10 μM), or butyric acid (2.5 mM) for 16 h, and then lysates were analyzed by immunoblotting. (C) Extracts taken from HeLa cells treated with cisplatin (10 μM) for 16 h were incubated with URSW for 2 h at 37°C before immunoblot analysis. One assay also contained 10 μM DEVD-CHO. Controls included incubated and nonincubated URSW and extract from PV-infected cells. Arrows indicate eIF4Gase-α-type cleavage products, filled arrowheads indicate eIF4Gase-β- or -γ-type cleavage products, and open arrowheads indicate caspase 3 cleavage products.
eIF4Gase activity in uninfected cells.
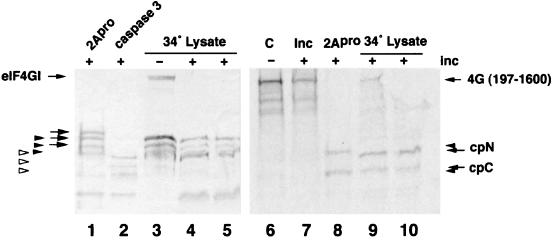
eIF4GI is well known to be degraded during preparation from uninfected cells; however, consistently, two to three types of cleavage products are observed, which we now recognize to be from caspase 3 and the eIF4Gases described above. Interestingly, the appearance of degradation products are cell batch specific. Over time we have noticed that eIF4GI in some cell extracts and the subcellular fractions derived from them is inherently unstable and produces more and more cleavage products upon handling and incubation whereas other cell lysates and derived fractions are extremely stable under the same conditions. This suggests that cell growth conditions at the time of cell harvest may lead to limited activation of eIF4Gase activities in some, but not all, batches of cell extract. To explore this, we incubated a panel of S10 cell extracts that had been produced without inclusion of the normal panel of protease inhibitors. These were then incubated at 34°C for 5 h and examined for appearance of eIF4Gase cleavage products. The data showed that the eIF4G in about 60% of S10 lysates was very stable during this incubation; however, eIF4G was partly degraded to various extents in about 40% of the lysates. Mixtures of faint eIF4G cleavage products were observed in several lysates (data not shown); however, some produced strong eIF4Gase cleavage products that we initially termed eIF4Gase-γ since activation occurred in normal uninfected cell lysates. Examination of these cleavage fragments revealed they were indistinguishable from eIF4Gase-β cleavage products produced in PV-infected lysates (Fig. 10). A few lysates also produced detectable caspase 3 fragments; however, there was no positive correlation with eIF4Gase activity (data not shown). To determine if a spontaneous eIF4Gase cleavage activity could be assayed in vitro, one lysate was incubated together with radiolabeled 4G(197-1600) substrate for an additional period. The data show that during this additional incubation, the remainder of uncleaved native eIF4GI in the lysate was cleaved to eIF4Gase-β cleavage products (Fig. 10, lanes 4 and 5). Furthermore, the radiolabeled substrate was also cleaved, again producing cleavage products consistent with eIF4Gase-β type activity (Fig. 10, lanes 9 and 10).
FIG. 10.
IF4Gase activity in uninfected cell lysates. Uninfected HeLa cell S10 lysate was incubated at 34°C for 5 h to generate spontaneous eIF4Gase cleavage products (lane 3). URSW samples were also incubated with recombinant 2Apro or caspase 3 to generate specific cleavage products (lanes 1 and 2). Lysate in lane 3 (15 μg) was incubated an additional 8 h with 35S-radiolabeled 4G(197-1600) substrate (lanes 4, 5, 9, and 10) as indicated in Materials and Methods. The same 4G(197-1600) substrate was also held on ice, incubated in buffer alone, or incubated with 2Apro (lanes 6, 7, and 8, respectively).The left panel shows an eIF4GI-specific immunoblot. The right panel shows an autoradiograph of duplicate lanes on the same gel. The migration of cleavage products is indicated with arrows, keyed as explained in the legend to Fig. 9.
To determine if cell cycle transit had any influence on appearance of eIF4Gase activity, we synchronized HeLa cells with a double thymidine block and, after release, collected S10 lysates from cells as they transited the cell cycle. Examination of the lysates revealed that caspase 3 cleavage products and eIF4Gase cleavage products appeared during discrete times of the cell cycle, being more concentrated in S phase and mitosis (data not shown). eIF4G cleavage was never complete and never approached even 50%. This result was reproducible, but it should be noted that not every thymidine block experiment we attempted resulted in production of significant discernible eIF4G cleavage products, suggesting additional factors or conditions other than simple cell cycle transit may be involved in eIF4Gase activation. However, taken together, the data show that both eIF4Gase-α and eIF4Gase-β activities can be generated in uninfected HeLa cells under a variety of conditions.
DISCUSSION
The nature of the activities that process eIF4GI in PV-infected cells has been controversial. Significant evidence exists to support processing by both 2Apro and a cellular activity termed eIF4Gase. Purified recombinant 2Apro has been shown to directly process purified eIF4G in vitro both in eIF4F complexes and eIF4F-eIF3 complexes; however, the cleavage was inefficient and required high levels of protease (4, 20). More recent data suggest 2Apro cleavage efficiency towards eIF4GI may be significantly enhanced in the context of ribosomes and active translation (16). Other data suggest that cellular eIF4Gase activities also process eIF4GI in vivo and in vitro. These data include the separation of in vitro eIF4G cleavage activity from all detectable 2Apro and poor correlation between levels of 2Apro expression and levels of eIF4G cleavage in guanidine-PV-infected cells. Previously, only one type of eIF4Gase activity was described that produced cleavage products similar to those detected in PV-infected cells. We now propose that eIF4GI is processed in HeLa cells by two cellular proteases in addition to 2Apro. The data in this manuscript provide the first evidence that eIF4GI is cleaved at two sites during PV infection.
We have not yet examined eIF4GII, the newly discovered homolog of eIF4GI, in regards to cleavage by 2Apro or eIF4Gase activities described here. eIF4GII represents a minor constituent of the total eIF4G pool in HeLa cells—we estimate 10% or less of the total since it is not easily detectable in silver-stained purified eIF4F preparations or 2Apro cleavage products derived thereof (reference 4 and data not shown). Delayed cleavage of eIF4GII has been correlated with a lack of complete translation inhibition in guanidine infections (18); however, a lack of cleavage of PABP under the same conditions could also prevent complete translation shutoff (24). More investigation is required to determine the relative significance of eIF4GII and PABP cleavage on translation inhibition during infection; however, cleavage of eIF4GI seems to be required.
The eIF4Gase preparations used in this study were all sufficiently purified to remove detectable 2Apro. The eIF4Gase-α activity described above generates cleavage products that are indistinguishable from 2Apro cleavage products, and its activity was inhibited by the GE mutation in eIF4GI that blocks 2Apro cleavage (data not shown). If two proteases shared the same cleavage site specificity, this result would be expected. However, this also leaves open the possibility that despite our efforts to remove all contaminating 2Apro from fractions tested here, eIF4Gase-α may represent a biochemically altered form of 2Apro with enzymatic activity much greater than the bulk of 2Apro present in cell lysates. In contrast, evidence also exists for a cellular eIF4Gase-α activity since it is induced in apoptotic cells in the total absence of 2Apro. Also, 2Apro-mediated cleavage of eIF4GI was not inhibited by zVAD, yet the eIF4Gase-α type activity present in low-MOI guanidine infections was blocked. Three feasible explanations are that (i) little to none of the eIF4G cleavage observed in low-MOI guanidine infections (which restricts expression of 2Apro) was due to 2Apro itself; (ii) the ability of 2Apro to efficiently cleave eIF4GI is caspase dependent, possibly from caspase-mediated modification of 2Apro; and (iii) an eIF4Gase activity may be activated by 2Apro. No further processing of 2Apro, potentially carried out by a caspase, has been observed previously in PV-infections and no effect of zVAD treatment on PV polyprotein processing was observed in this study. Likewise, an examination of caspase zymogen amino acid sequences has not revealed any obvious 2Apro cleavage sites near known sites of caspase cleavage that result in caspase zymogen activation.
The new eIF4Gase-β activity described here in infected cell lysates produces shorter N-terminal cleavage fragments than 2Apro and may be the same as the eIF4Gase-γ activity detected in uninfected cell extracts. Using defined eIF4G substrates, we could find no evidence for 2Apro cleavage at the upstream site recognized by eIF4Gase-β activity. The new cleavage site that was mapped in this study does not contain any of the known sequence requirements for PV 2Apro recognition; thus, cleavage is not expected at this site. Evidence that this site is utilized during PV infections in vivo comes from detection of two closely migrating forms of eIF4GI CPC in 2D gels, direct sequencing of eIF4G fragments purified from infected cells, and purification of eIF4Gase-β activity from infected cells which produced alternate CPC and CPC.
These data also provide the first evidence that more than one eIF4Gase activity is present in PV-infected cells and uninfected cells. The multiple activities were not recognized previously since the product bands of N-terminal cleavage overlap in SDS immunoblots. In many assays of eIF4Gase cleavage activity and other immunoblots of eIF4G cleavage products in infected cells, cleavage products similar to those produced by eIF4Gase-β activity were noted or what appeared to be lessening of the intensity of the upper or slowest migrating eIF4GI CPN in relationship to the staining of other CPN bands was observed. However, no explanation was available for alternate isoforms of eIF4GI; thus, a biochemical explanation for what was apparently alternate forms of cleavage products was not available. Now with knowledge that multiple isoforms result from use of alternate initiation codons, the generation of different forms of CPN or CPC can be ascribed to use of different cleavage sites. The fact that eIF4Gase activity can be separated into at least two types of cleavage activities also helps explain some of the difficulties encountered with purification and identification of the small amount of enzyme(s) responsible.
Recently, other investigators used zVAD-fmk in a similar attempt to block eIF4G cleavage in PV-infected cells but saw no inhibition of eIF4GI cleavage in the presence of zVAD and concluded that caspases were not involved in the processing of eIF4GI (44). We also observed little effect on eIF4GI cleavage in our experiments carried out at normal to high MOI and using zVAD alone. However, to minimize the impact of 2Apro in the system, and expecting 2Apro not to be inhibited by caspase inhibitors, we also carried out infections which included guanidine to limit the production of 2Apro in the cells. Under these conditions, eIF4Gase cleavage was severely inhibited by zVAD, providing evidence that much of the processing of eIF4Gase in guanidine-supplemented infections was due to cellular enzymes and not 2Apro. Interestingly, addition of DEVD and zVAD itself did not result in inhibition of the eIF4Gase activity after it is induced (Fig. 9 and data not shown), suggesting that eIF4Gase cleavage activity may reside in not a caspase itself, but rather a distinct protease that may be dependent upon a caspase for activation.
Both types of eIF4Gase activities (α and β) can be detected in uninfected cell lysates as well. In this case the overall level of cleavage activity measured was weaker than the corresponding activity level measured in virus-infected cell lysates. Certainly, any protease activity that leads to cleavage of eIF4GI and down-regulates cap-dependent translation would be tightly regulated to avoid promiscuous eIF4GI cleavage. It is not surprising that eIF4Gase cleavage activity may be linked to apoptosis, since down-regulation of cellular translation is always observed in apoptotic cells. Investigation of apoptotic cells has begun to reveal a complex scheme of translation regulation in that caspase 3 modification of eIF4GI, eIF4GII, eIF2a, eIF3, and eIF4B alters the function of many of these factors but does not necessarily inactivate all of them (9, 38, 39, 40). Caspase 3 modification of eIF4GI results in release of the central core fragment of eIF4G that has been shown to be able to support cap-dependent and cap-independent translation (7, 11, 41). Thus, eIF4GI cleavage by caspase 3 may not destroy its basal function in translation initiation. However, activation of eIF4Gase-α at early times in apoptosis would serve to down-regulate cap-dependent translation only. The mRNAs of two key apoptosis regulatory proteins, XIAP and APAF-1, have been shown to be translated via internal ribosome entry site elements that would not be down-regulated by this alteration in eIF4G or may be enhanced in their expression (10, 22, 23). Thus, normal cellular functions of eIF4Gase may include partial down-regulation of translation in apoptosis that may favor translation of certain polypeptides over others in the early regulation of apoptosis. Activation of caspase 3 and the further proteolytic modification of eIF4GI, eIF4GII, eIF2, etc. are largely late events in apoptosis that occur after commitment to complete translation shutoff and death occurs. Another possible normal cellular function of eIF4Gase may also be to cause down-regulation of translation during certain phases of the cell cycle or times of cell stress. Since 2Apro largely duplicates this eIF4G cleavage activity, it is possible that eIF4Gase is not easily activated in all cell types that PV infects in humans; thus, 2Apro provides a backup or enables complete eIF4GI cleavage in those cells. Even so, infection of erythroleukemic K562 cells with PV results in only partial eIF4G cleavage despite efficient production of 2Apro (3). Thus, it is possible that enteroviruses have evolved 2Apro in order to grow efficiently and enhance virus replication in cell types that may not express these enzymes. The ability to cleave eIF4GI may aid viral pathogenesis and virus spread throughout the body. Alternatively, the apparently duplicate function of eIF4Gase and 2Apro may be an illusion. It is possible that each enzyme preferentially cleaves different subcellular pools of eIF4GI. Since eIF4GI is a scaffolding protein with multiple binding partners, it likely exists in distinct types of multiprotein complexes that could easily influence protease cleavage. 2Apro preferentially cleaves eIF4GI complexed to eIF4E, not free eIF4GI (20). However, eIF4G may exist in molar excess over eIF4E (13, 21), providing a pool of unassociated eIF4GI that may be resistant to 2Apro cleavage. This specific effect, which may be conformational, may be restricted to eIF4E, which binds very close to the 2Apro cleavage site. Binding of other proteins to eIF4GI, such as eIF3, has not been shown to have any effect on 2Apro-mediated cleavage (4).
Lastly, it has long been thought that activation of eIF4Gase in PV-infected cells was due to production of 2Apro itself (30). Although this may be true, it is also possible that eIF4Gase induction may be stimulated by other events that occur during the viral replication cycle. With the growing sense that apoptosis may serve the function of a virus defense system to limit viral replication in host animals similar to the interferon system, it is possible that eIF4Gase activation represents a portion of the apoptotic response that is activated early yet not further controlled by expression of the viral program and that otherwise limits full development of apoptosis (1). Since several apoptosis activation cascades proceed via activation of death domains on the cytoplasmic domain of receptors upon ligand binding, it is possible that interaction of PV with the PV receptor or coreceptors during binding and uptake may serve to activate apoptosis or eIF4Gase. More experiments are required to test this hypothesis.
Acknowledgments
We are grateful to R. E. Rhoads for furnishing the plasmid constructs pAD4G, pAD4G(GE), and pSK-HFCI. L. Carasco is acknowledged for providing anti-eIF4G antiserum. Thanks are also provided to E. Ehrenfeld for critical review of the manuscript.
This work was supported by NIH grants AI27914 and GM 59803.
REFERENCES
- 1.Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, L. I. Romanova, L. V. Sladkova, and E. A. Tolskaya. 2000. Competing death programs in poliovirus-infected cells: commitment switch in the middle of the infectious cycle. J. Virol. 74:5534–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G., and R. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869–900. In N. Sonenberg, J. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Benton, P. A., J. W. Murphy, and R. E. Lloyd. 1995. K562 cell strains differ in their response to poliovirus infection. Virology 213:7–18. [DOI] [PubMed] [Google Scholar]
- 4.Bovee, M. L., B. Lamphear, R. E. Rhoads, and R. E. Lloyd. 1998. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology 245:241–249. [DOI] [PubMed] [Google Scholar]
- 5.Bovee, M. L., W. E. Marissen, M. Zamora, and R. E. Lloyd. 1998. The predominant eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology 245:229–240. [DOI] [PubMed] [Google Scholar]
- 6.Brown, B. A., and E. Ehrenfeld. 1980. Initiation factor preparations from poliovirus-infected cells restrict translation in reticulocyte lystates. Virology 103:327–339. [DOI] [PubMed] [Google Scholar]
- 7.Bushell, M., D. Poncet, W. E. Marissen, H. Flotow, R. E. Lloyd, M. J. Clemens, and S. J. Morley. 2000. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 7:628–636. [DOI] [PubMed] [Google Scholar]
- 8.Carthy, C. M., D. J. Granville, K. A. Watson, D. R. Anderson, J. E. Wilson, D. C. Yang, D. W. C. Hunt, and B. M. McManus. 1998. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. J. Virol. 72:7669–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, M. J., M. Bushell, I. W. Jeffrey, V. M. Pain, and S. J. Morley. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7:603–615. [DOI] [PubMed] [Google Scholar]
- 10.Coldwell, M., S. Mitchell, M. Stoneley, M. MacFarlane, and A. Willis. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19:899–905. [DOI] [PubMed] [Google Scholar]
- 11.De Gregorio, E., T. Preiss, and M. W. Hentze. 1999. Translation driven by an eIF4G core domain in vivo. EMBO J. 18:4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, B. S., H. Kimura, and T. M. Timmons. 1990. Protein analysis using high resolution two-dimensional polyacrylamide gel electrophoresis. Methods Enzymol. 182:441–459. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, R., S. C. Milburn, and J. W. B. Hershey. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 262:380–388. [PubMed] [Google Scholar]
- 14.Etchison, D., S. C. Milbur, I. Edery, N. Sonenberg, and J. W. B. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806–14810. [PubMed] [Google Scholar]
- 15.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, W., and T. Skern. 2000. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Lett. 480:151–155. [DOI] [PubMed] [Google Scholar]
- 17.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifo, J. A., S. A. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258:5804–5810. [PubMed] [Google Scholar]
- 20.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiremath, L. S., N. R. Webb, and R. E. Rhoads. 1985. Immunological detection of the messenger RNA cap-binding protein. J. Biol. Chem. 260:7843–7849. [PubMed] [Google Scholar]
- 22.Holcik, M., and R. G. Korneluk. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holcik, M., C. Lefebvre, C. Yeh, T. Chow, and R. Korneluk. 1999. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1:190–192. [DOI] [PubMed] [Google Scholar]
- 24.Joachims, M., P. C. van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannes, G., M. Carter, M. Eisen, P. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNA that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP and eIF4G conferred by internal ribosome entry sites. RNA 4:1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, C., and E. Ehrenfeld. 1983. The effect of poliovirus infection on the translation in vitro of VSV messenger ribonucleoprotein particles. Virology 129:415–430. [DOI] [PubMed] [Google Scholar]
- 28.Joshi, B., R. Q. Yan, and R. E. Rhoads. 1994. In vitro synthesis of human protein synthesis initiation factor-4 gamma and its localization on 43-S and 48-S initiation complexes. J. Biol. Chem. 269:2048–2055. [PubMed] [Google Scholar]
- 29.Kerekatte, V., B. D. Keiper, C. Bradorff, A. Cai, K. U. Knowlton, and R. E. Rhoads. 1999. Cleavage of poly(A)-binding protein by Coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kräusslich, H. G., M. J. H. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eukaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases: implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975–21983. [DOI] [PubMed] [Google Scholar]
- 32.Lamphear, B. J., and R. E. Rhoads. 1996. A single amino acid change in protein synthesis initiation factor 4G renders cap-dependent translation resistant to picornaviral 2A proteases. Biochemistry 35:15726–15733. [DOI] [PubMed] [Google Scholar]
- 33.Lamphear, B. J., R. Q. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor-eIF-4g of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200–19203. [PubMed] [Google Scholar]
- 34.Liebig, H. D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, R. Rhoads, E. Kuechler, and T. Skern. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry 32:7581–7588. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd, R. E., D. Etchison, and E. Ehrenfeld. 1985. Poliovirus protease does not mediate cleavage of the 220,000-Da component of the cap binding protein complex. Proc. Natl. Acad. Sci. USA 82:2723–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd, R. E., H. G. Jense, and E. Ehrenfeld. 1987. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. J. Virol. 61:2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloyd, R. E., H. Toyoda, D. Etchison, E. Wimmer, and E. Ehrenfeld. 1986. Cleavage of the cap binding protein complex polypeptide p220 is not effected by the second poliovirus protease 2A. Virology 150:229–303. [DOI] [PubMed] [Google Scholar]
- 38.Marissen, W., Y. Guo, A. Thomas, R. Matts, and R. Lloyd. 2000. Identification of caspase 3-mediated cleavage and functional alteration of eukaryotic initiation factor 2α in apoptosis. J. Biol. Chem. 275:9314–9323. [DOI] [PubMed] [Google Scholar]
- 39.Marissen, W. E., A. Gradi, N. Sonenberg, and R. E. Lloyd. 2000. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ. 7:1234–1243. [DOI] [PubMed] [Google Scholar]
- 40.Marissen, W. E., and R. E. Lloyd. 1998. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18:7565–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal on-third functions as modulatory region. Mol. Cell. Biol. 20:468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morley, S. J., P. S. Curtis, and V. M. Pain. 1997. eIF4G: translation’s mystery factor begins to yield its secrets. RNA 3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 43.Raught, B., A.-C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19:434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, L., A. Boxall, L. Lewis, G. Belsham, and G. Kass. 2000. Caspases are not involved in the cleavage of translation initiation factor eIF4GI during picornavirus infection. J. Gen. Virol. 81:1703–1707. [DOI] [PubMed] [Google Scholar]
- 45.Sommergruber, W., H. Ahorn, H. Klump, J. Seipelt, A. Zoephel, F. Fessl, E. Krystek, D. Blaas, E. Kuechler, H. D. Liebig, and T. Skern. 1994. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4 γ via a common recognition motif. Virology 198:741–745. [DOI] [PubMed] [Google Scholar]
- 46.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, T. A. Ivannikova, and V. I. Agol. 1996. Final checkpoint in the drug-promoted and poliovirus-promoted apoptosis is under post-translational control by growth factors. J. Cell. Biochem. 63:422–431. [DOI] [PubMed] [Google Scholar]
- 48.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, T. A. Ivannikova, E. A. Smirnova, N. T. Raikhlin, and V. I. Agol. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 69:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyckoff, E. E., R. E. Lloyd, and E. Ehrenfeld. 1992. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. J. Virol. 66:2943–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, R., W. Rychlik, D. Etchison, and R. E. Rhoads. 1992. Amino acid sequence of the human protein synthesis initiation factor eIF-4 γ. J. Biol. Chem. 267:23226–23231. [PubMed] [Google Scholar]
- 51.Yan, R. Q., and R. E. Rhoads. 1995. Human protein synthesis initiation factor eIF-4 gamma is encoded by a single gene (EIF4G) that maps to chromosome 3q27-qter. Genomics 26:394–398. [DOI] [PubMed] [Google Scholar]
- 52.Yu, S. F., and R. E. Lloyd. 1991. Identification of essential amino acid residues in the functional activity of poliovirus 2A-protease. Virology 182:615–625. [DOI] [PubMed] [Google Scholar]
- 53.Zamora, M., M. Byrd, and R. E. Lloyd. 2000. Generation of multiple isoforms of eIF4GI by use of alternate translation initiation codons. Submitted. [DOI] [PMC free article] [PubMed]