Abstract
Expression of most viral genes during productive infection by herpes simplex virus is regulated by the viral protein ICP4 (also called IE175 or Vmw175). The N-terminal portion of ICP4 contains well-defined transactivation, DNA binding, and dimerization domains that contribute to promoter regulation. The C-terminal half of ICP4 contributes to the activity of ICP4, but the functional motifs have not been well mapped. To localize functional motifs in the C-terminal half of ICP4, we have compared the relative specific activities of ICP4 variants in transient-transfection assays. Deletion of the C-terminal 56 residues reduces the specific activity more than 10-fold. Mutational analysis identified three consecutive residues (1252 to 1254) that are conserved in ICP4 orthologs and are essential for full activity, especially in the context of ICP4 variants with a deletion in the N-terminal transactivation domain. Recombinant viruses that encode variants of ICP4 with mutations in the N-terminal transactivation domain and/or the extreme C terminus were constructed. The phenotypes of these recombinant viruses support the hypothesis that efficient promoter activation by ICP4 requires motifs at both the N and C termini. The data suggest that the C terminus of ICP4 functions not as an independent transactivation domain but as an enhancer of the ICP4 N-terminal transactivation domain. The data provide further support for the hypothesis that some ICP4 motifs required for promoter activation are not required for promoter repression and suggest that ICP4 utilizes different cellular factors for activation or repression of viral promoters.
During productive infection by herpes simplex virus type 1 (HSV-1), approximately 75 genes encoded within the linear 152-kbp viral genome are transcribed by RNA polymerase II in three sequential phases designated immediate early (IE or α), early (E or β), and late (L or γ) (for a review, see reference 52). The immediate-early protein ICP4 (infected-cell polypeptide 4) is required for efficient transcription of early and late viral genes and thus is essential for productive infection. The immediate-early proteins ICP0, ICP22, and ICP27 enhance expression of early and late genes at both transcriptional and posttranscriptional levels and contribute to some functions of ICP4 (52).
ICP4 is a 1,298-amino-acid (aa) phosphoprotein that binds DNA in a sequence-specific manner as a homodimer (14, 16, 17, 34). ICP4 represses transcription from three viral genes (LAT, ICP4, and ORF-P) that have a high-affinity ICP4 binding site spanning the transcription initiation site (1, 16, 20, 21, 28, 30, 41, 42). ICP4 stimulates transcription from early and late viral promoters through interactions with viral DNA and cellular proteins that are poorly understood. Although the ICP4 DNA binding domain is essential for activation of transcription (39, 45), extensive analyses have not revealed a specific sequence that binds ICP4 with high affinity and is common to all promoters activated by ICP4 (13, 52). The observation that the minimal cis-acting elements required for stimulation of transcription by ICP4 are a TATA box and/or an initiator element (5, 11, 12, 19, 22, 24–26, 44, 46) suggests that ICP4 facilitates formation of the preinitiation complex through interactions with one or more components of the cellular transcription machinery. This model is consistent with observations that ICP4 interacts with several cellular transcription factors, including TATA binding protein (TBP), TFIIB, and TAFII250 (3, 47).
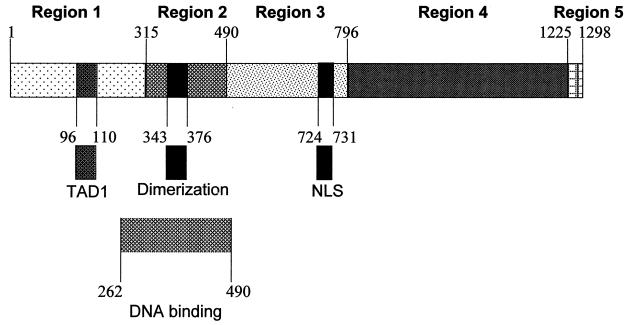
A series of genetic and biochemical analyses have been conducted to determine which of the observed interactions between ICP4, viral DNA, and cellular proteins are relevant to the function of ICP4 as a coactivator. Similar studies have been conducted with orthologs of ICP4 encoded by other members of the alphaherpesvirus family, including the pseudorabies virus IE180 protein, the varicella-zoster virus ORF62 protein, and the equine herpesvirus immediate-early protein. Based on sequence homologies, these ICP4 orthologs have been linearly divided into five conceptual regions (Fig. 1) (4, 6, 18). The orthologs are highly homologous in regions 2 and 4 but exhibit little or no homology in region 1, 3, or 5, except for a conserved serine- and acidic residue-rich motif located in either region 1 or 5. Mutational analyses of one or more of these orthologs have revealed the following: (i) residues in region 1 (ICP4 aa 1 to 315) function as a transactivation domain (32, 40, 49, 55), bind to the cellular EAP protein (EBER-associated protein) (31), contribute to the regulation of ICP4 functions by ICP27 (43), and are required for formation of a complex with DNA that contains TBP and TFIIB (47); (ii) residues in region 2 (ICP4 aa 316 to 490) are necessary for homodimerization (14, 17) and DNA binding (15, 28, 39, 53, 54); (iii) region 3 (ICP4 aa 491 to 796) contains a nuclear localization signal (38, 48); and (iv) residues in regions 4 and 5 (ICP4 aa 797 to 1298) interact with TAFII250 (3), ICP0 (56), and ICP27 (36), govern the effect of ICP0 and ICP27 on the localization of ICP4 in transfected Vero cells (35, 57), and are required for efficient transcriptional activation (2, 8, 9, 38).
FIG. 1.
Schematic diagram of ICP4. Numbers indicate amino acid residues relative to the N terminus of ICP4. Regions 1 to 5 are subdivisions based on sequence homology among ICP4 orthologs. The locations of functional domains are indicated. NLS, nuclear localization signal.
We previously observed that ICP4 residues 1 to 139 can function as a transcription activation domain when fused to the DNA binding domain of Gal4 (55) and determined that ICP4 residues 97 to 109 are essential for this activity. Deletion of these residues in the context of native ICP4 resulted in loss of approximately half of the transcriptional activation activity of ICP4 in transient-transfection assays. Observations by others that residues in the C-terminal halves of ICP4 orthologs contribute to transcriptional activation activity (8, 9, 55) led us to the hypothesis that residues in ICP4 region 4 and/or 5 either function as an independent transactivation domain or contribute to the activity of the N-terminal transactivation domain, which we designated TAD1 (55). In order to focus on portions of ICP4 regions 4 and 5 that contribute to the activity of ICP4 in the absence of other viral proteins, we initiated the present investigation using transient-transfection assays in uninfected cells. Because we anticipated that mutations in the C-terminal portion of ICP4 would reduce but not eliminate the transcriptional activity of ICP4, we defined conditions in which the response of a reporter gene is linearly dependent on the dose and activity of ICP4 variants. After we identified specific residues in region 5 that make a significant contribution to the transcriptional activity of ICP4 in the context of transient-transfection assays, we constructed a series of viral mutants that encode variants of ICP4. Quantitative analysis of the kinetics of viral gene expression in cells infected with these variants confirmed the results from the transient-transfection assays. Our results indicate that the C terminus of ICP4 does not function as an independent transactivation domain but is required for efficient activation of transcription by ICP4.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATCC CCL-81) and E5 cells (a strain of Vero cells transformed with the ICP4 gene by Neal DeLuca) (8) were grown as monolayers in Dulbecco’s modified Eagle medium supplemented with bovine calf serum (10%) and gentamicin (25 μg/ml). HSV-1 McKrae was used as the parental virus for generation of recombinant viruses.
DNA purification.
Plasmid DNA was purified from Escherichia coli by the Qiagen Qiafilter protocol and then centrifuged at 80,000 × g for 45 min at 14°C to remove insoluble material that interfered with the transient-transfection assays. Viral DNA was purified from viral capsids as described previously (10).
Reporter plasmids.
Luciferase reporter plasmids were constructed by inserting the promoters from HSV-1 genes into the multiple cloning sites located immediately upstream of the luciferase gene in either the pGL2-Basic or pGL3-Basic vector (Promega). Oligonucleotides were added where needed to facilitate cloning. To construct the reporter plasmid with the thymidine kinase (TK) gene promoter (pXK865), a PvuII-to-BglII DNA fragment extending from −207 to +57 relative to the TK mRNA cap site was inserted into pGL3-Basic. The reporter plasmid with the UL26.5 gene promoter (pXK859) was constructed by inserting a BclI-to-BstEII fragment from −464 to +42 relative to the UL26.5 mRNA cap site into pGL3-Basic along with an oligonucleotide extending from +42 to the UL26.5 translation initiation codon at +99, which also serves as the initiation codon for luciferase in pXK859. The reporter plasmid with the VP5 gene promoter (pXK877) was constructed by inserting a HincII-to-NcoI fragment from −367 to +239 relative to the VP5 mRNA cap site into pGL3-Basic. The NcoI site spans the translation initiation codon in the VP5 gene and also provides the translation initiation codon for luciferase in pXK877. The reporter plasmid with the glycoprotein D (gD) gene promoter (pXK557) was constructed by inserting a SacI-HindIII fragment spanning the HSV-1 gD gene from position −392 to + 53 relative to the gD mRNA cap site into pGL3-Basic. The reporter plasmid with the glycoprotein C (gC) gene promoter (pXK548) was constructed by inserting an SgrAI-NheI fragment spanning −82 to +110 relative to the gC mRNA cap site in the HSV-1 gC gene into pGL3-Basic. The reporter plasmid with the LAT gene promoter (pXK815) was constructed by inserting a PstI fragment spanning −142 to +67 relative to the LAT mRNA cap site into pGL2-Basic. The LAT-mut reporter plasmid (pXK816) is identical to the LAT reporter plasmid except it contains a mutation introduced by Batchelor et al. (1) that abolishes the ICP4 binding site in the LAT promoter. Construction of the reporter plasmid p5GE1b-Luc, which contains five Gal4 binding sites upstream of the E1b TATA box and the luciferase gene, has been previously described (55). The pCMV-β-gal plasmid was constructed by inserting the β-galactosidase gene from the pSV β-galactosidase control vector (Promega) into pCMV-mcs (55). The reporter plasmid with the gC gene promoter linked to the β-glucuronidase (GUS) gene (pXK1076) was constructed by inserting an NsiI-NheI fragment spanning bp −1,257 to +110 relative to the gC mRNA cap site upstream of a hybrid GUS gene that consists of the GUS-coding region followed by the simian virus 40 (SV40) small-t-antigen intron and poly(A) signal. The hybrid GUS gene was obtained as a NcoI-to-SacI fragment from pBgdTK (a kind gift from Thomas Jones [27]). The hybrid GUS gene in plasmid pXK1076 is flanked on the downstream side by sequences that span the gC gene from an XbaI site at codon 453 to a SalI site in the 3′ untranslated region (UTR) of the gC gene.
Effector plasmids.
The pCMV-mcs vector has multiple cloning sites between the human cytomegalovirus (CMV) immediate-early promoter and the SV40 small-t-antigen intron and poly(A) signal (55). A control plasmid (pDAN3) that expresses glutathione S-transferase with a C-terminal nuclear localization signal (GST-nls) was constructed by inserting the glutathione S-transferase gene from the pGEX vector (Pharmacia) into pCMV-mcs along with an oligonucleotide that encodes the SV40 T-antigen nuclear localization signal (PPKKKRKVEDP). Plasmid pXK350 contains the entire ICP4-coding region and 3′ UTR from HSV-1 strain 17 (bp 147104 to 151457 in the sequence of McGeoch et al. [33]) in pCMV-mcs and has been previously described (55). Plasmid pXK643 is a derivative of pXK350 that lacks ICP4 codons 97 to 109 (ΔTAD1) and was constructed as described previously for pXK668 (55).
Plasmid pXK909, which was used as the vector for construction of effector plasmids encoding ICP4 variants with C-terminal mutations, was generated by replacing both the SV40 intron and poly(A) signal in pCMV-mcs with an oligonucleotide that spans the 27 nucleotides from the ICP4 stop codon to a 3′ BsaI site at position 151026 in the viral genome followed by a DNA fragment that extends from the BsaI site to a Sau3AI site at bp 151454 in the viral genome. The oligonucleotide introduced a HindIII site and four (underlined) nucleotides (TAAGCTT) at the 3′ end of the ICP4-coding region. Transcripts expressed from portions of the ICP4 gene inserted into pXK909 have the entire ICP4 3′ UTR and utilize the ICP4 gene poly(A) signal. Plasmid pXK912 contains the coding region and 3′ UTR for HSV-1 strain 17 wild-type ICP4 inserted downstream from the CMV promoter in pXK909. Plasmid pXK925 is a derivative of pXK912 that lacks ICP4 codons 1242 to 1298. A series of oligonucleotides with silent mutations were sequentially introduced into pXK925 to extend the ICP4 gene to codons 1252, 1258, 1270, 1279,1293, and 1298 (pXK940, pXK941, pXK942, pXK943, pXK944, and pXK945, respectively). In pXK945, the sequence from codon 1242 to 1298 is referred to as synthetic region 5 (SR5). Plasmids pXK928, pXK929, pXK931, and pXK946, encoding the Mut1, Mut2, Mut3, and Mut4 variants of ICP4, respectively, were derived from pXK945 by an oligonucleotide replacement strategy in SR5. Plasmid pXK644, which encodes an ICP4 variant with both the ΔTAD1 and Mut1 mutations in the context of the pXK909 vector, was derived from pXK928 and pXK643.
Plasmids encoding Gal4-ICP4 fusion proteins have been previously described (55). Briefly, a fragment encoding the Gal4 DNA binding domain (Gal4 aa 1 to 147) was inserted into pCMV-mcs to make pCMV-Gal4. Coding portions of the ICP4 gene were then inserted downstream of the Gal4 gene in pCMV-Gal4 using oligonucleotides to facilitate cloning and maintain correct reading frames between the Gal4 and ICP4 genes. Plasmids pXK397, pXK630, and pXK631 encode Gal4 fused to ICP4 residues 1 to 139, 731 to 1298, and 1154 to 1298, respectively. The positive control plasmid containing the VP16 transactivation domain fused to Gal4 has been previously described (55).
Transient-transfection assays.
Vero cells (5 × 105 cells in 60-mm-diameter dishes) were transfected in duplicate by the calcium phosphate precipitation procedure. A constant amount (100 ng) of the pCMV-βgal plasmid was included in each transfection to control for transfection efficiency. The total transfected DNA per dish was adjusted to 10 μg by addition of calf thymus DNA (Worthington). Transfected cells were incubated at 37°C for 42 to 48 h, harvested by being scraped into 400 μl of reporter lysis buffer (Promega), and lysed by a freeze-thaw procedure. All three enzymes were quantified by chemiluminescence assays in a luminometer. For firefly luciferase assays, a sample (20 μl) of the cell extract was mixed with 100 μl of luciferase assay substrate in luciferase assay buffer (Promega) and assayed immediately. For β-galactosidase assays, a sample (5 μl) of the cell extract was incubated with Galacton-star substrate (PE Biosystems) in 150 μl of reaction buffer for 1 h and then assayed. For GUS assays, a sample (10 μl) of the cell extract was incubated with the Glucuron chemiluminescent substrate (PE Biosystems) in 90 μl of the GUS reaction buffer for 1 h and then supplemented with 150 μl of light emission accelerator and assayed immediately. Values obtained from the luciferase and GUS assays were divided by the values obtained from the β-galactosidase assays to normalize for transfection efficiency. Fold activation for a given effector-reporter combination is defined as the normalized luciferase value obtained from cells transfected with the reporter and effector plasmids divided by the normalized luciferase value obtained from cells transfected with the reporter alone. Fold repression for a given effector-reporter combination is defined as the inverse of the ratio used to calculate fold activation. These ratios were always determined using the same amount of reporter DNA in both the test and control transfections. Control transfections were routinely performed with the reporter plasmid and a control plasmid (pDAN3) in which the CMV promoter drives expression of a nonrelevant nuclear protein.
Generation of recombinant viruses.
For insertion of genes into the ICP4 locus of the HSV-1 genome, we constructed the plasmid pXK990, which contains a multiple cloning site flanked on the upstream side by 291 bp that extend from the EcoRI site at −110 in the ICP4 promoter to the SalI site at +181 in the ICP4 gene 5′ UTR and on the downstream side by 456 bp that extend from a HindIII site inserted at the ICP4 translation stop codon through the ICP4 3′ UTR to the Sau3AI site at position 151454 in the viral genome. Plasmid pXK976 was constructed by insertion of the coding sequence for bacterial β-galactosidase into the multiple cloning site of pXK990. The β-galactosidase gene with the flanking ICP4 components was excised from pXK976 and cotransfected with the HSV-1 McKrae genome into E5 cells using Lipofectamine (Life Technologies). Gene replacement via homologous recombination in the transfected cells generated the recombinant virus HSV-976, in which the ICP4-coding sequence has been replaced with the β-galactosidase-coding sequence (50).
Plasmid pXK1072 contains the luciferase gene and SV40 poly(A) signals from pGL3-Basic flanked on the upstream side by the HSV-1 TK promoter from −1366 to +57 and on the downstream side by the 3′ portion of the TK gene corresponding to bp 47358 to 45570 in the viral genome. Strain HSV-1072 was generated by cotransfection of E5 cells with the TK-luciferase insert from pXK1072 and HSV-976 DNA. Recombinant viruses were obtained from plaques in E5 cell monolayers grown in medium supplemented with acycloguanosine (25 mM) and subjected to two rounds of plaque purification in E5 cells.
For generation of recombinant HSV expressing N-terminal enhanced green fluorescent protein (EGFP)-ICP4 fusion proteins, we constructed plasmid pXK992 by inserting the EGFP-coding region (Clontech) downstream from the ICP4 promoter in pXK990. The ICP4-coding regions from pXK912, pXK643, pXK928, and pXK644 were then inserted downstream from the EGFP gene in pXK992 to generate plasmids pXK984, pXK989, pXK988, and pXK645, which encode EGFP fused in frame with the wild-type, ΔTAD1, Mut1, and ΔTAD1 + Mut1 variants of ICP4, respectively. The ICP4 promoter and 3′ flanking sequences facilitate homologous recombination with the viral genome. The EGFP-ICP4 genes with flanking ICP4 sequences were excised from pXK984, pXK989, pXK988, and pXK645 and cotransfected with the HSV-976 genome into E5 cells using Lipofectamine. Gene replacement via homologous recombination in the transfected cells resulted in replacement of the β-galactosidase-coding sequence in HSV-976 with the variant EGFP-ICP4 genes to generate recombinant viruses encoding EGFP fused to wild-type ICP4 (HSV-984), ICP4 ΔTAD1 (HSV-989), ICP4 Mut1 (HSV-988), and ICP4 ΔTAD1 + Mut1 (HSV-990), respectively. The recombinant strains produced green fluorescent plaques on E5 cell monolayers. DNAs obtained from recombinant viruses after three to five rounds of plaque purification were analyzed by Southern hybridization with oligonucleotide probes specific for the ICP4 variants to confirm the genotype. HSV-987 contains a reversion of the ΔTAD1 mutation in HSV-990 and was obtained by cotransfection of E5 cells with HSV-990 DNA and a DNA fragment from pXK984 (EGFP-ICP4) that encodes EGFP fused to ICP4 residues 1 to 207. HSV-983 contains a reversion of the Mut1 mutation in HSV-990 and was obtained by cotransfection of E5 cells with HSV-990 DNA and a DNA fragment containing wild-type ICP4 codons 694 to 1298. Derivatives of HSV-984, -989, -988, and -990 that express luciferase from the TK gene locus were generated by transfecting cells with the corresponding viral DNA plus the luciferase gene and flanking TK sequences obtained from pXK1072. Recombinants were selected with acycloguanosine as described for HSV-1072 above.
Complementation assays.
Vero cells (60-mm-diameter dish) were transfected in duplicate with 0.1 μg of plasmid DNA plus 9.9 μg of calf thymus DNA by the calcium phosphate method, incubated at 37°C for 48 h, and then infected with HSV-976 at a multiplicity of infection of 1.0. At 24 h postinfection, cells were scraped into the medium and lysed by two freeze-thaw cycles. The titer of the lysate was determined on E5 cells. No plaques were obtained when Vero cells were infected with the same lysates.
Reporter virus assays.
Vero cells were transfected in triplicate with 0.1 μg of plasmid DNA plus 9.9 μg of calf thymus DNA by the calcium phosphate procedure, incubated at 37°C for 24 h, and then infected with HSV-1072 at a multiplicity of infection of 1.0. At 4 h postinfection, the cells were harvested and assayed for luciferase activity as described above.
RESULTS
Vectors for expression of ICP4 variants.
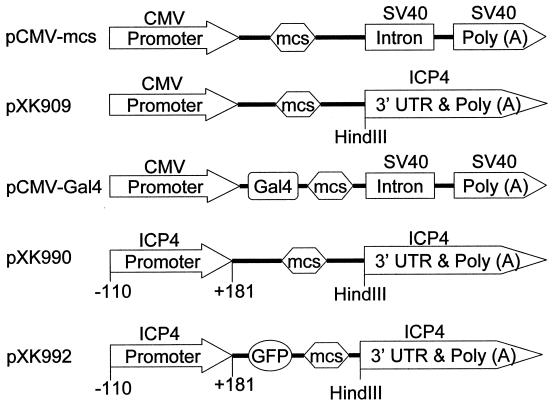
We previously used the pCMV-mcs vector to characterize the activities of ICP4 variants with N-terminal mutations in transfected cells (Fig. 1) (55). The pCMV-mcs vector contains the intron from the SV40 small-t-antigen gene downstream of the multiple cloning site (Fig. 2). Because we were concerned that this intron could lead to generation of aberrantly spliced mRNAs (23) derived from ICP4 genes with C-terminal deletions, we constructed the vector pXK909 (Fig. 2), which lacks the SV40 intron and has the ICP4 3′ UTR and poly(A) signal immediately downstream of the multiple cloning site. Plasmids with C-terminal truncations of the ICP4 gene inserted into pXK909 contain a stop codon (TAA) which overlaps the first two nucleotides of the HindIII site located immediately upstream of the ICP4 3′ UTR (Fig. 2).
FIG. 2.
Vectors used for construction of plasmids. Vectors pCMV-mcs, pXK909, and pCMV-Gal4 were used to express variants of ICP4 in transfected cells. Vectors pXK990 and pXK992 were used to construct genes for insertion into the ICP4 locus of the HSV genome by homologous recombination. Genes were inserted into the multiple cloning site (mcs). The numbers below the ICP4 promoter indicate the base position relative to the transcription initiation site.
Determination of the specific activity of ICP4.
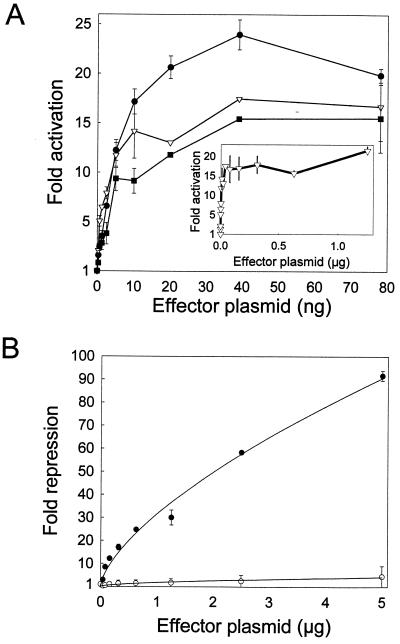
Our initial goal was to establish a quantitative assay that could measure differences in the specific activities of ICP4 variants (defined as fold activation per amount of native ICP4). For this purpose, we designed one assay (the activation assay) to measure fold activation in transfected cells and a second assay (the repression assay) to measure relative amounts of correctly folded, nucleus-localized ICP4 in transfected cells. The activation assay measures the ability of ICP4 variants to activate an HSV-1 promoter linked to a reporter gene. To define the activity of each ICP4 variant under both limiting and saturating conditions, the activation assays were conducted with 0.5 to 1,200 ng of each ICP4 effector plasmid per dish. A series of activation assays conducted with wild-type ICP4 and three different HSV-1 promoters (from the TK, VP5, and UL26.5 genes) yielded a similar pattern (Fig. 3). Increasing amounts of ICP4 effector plasmid from 0.5 to approximately 10 ng resulted in a linear increase in luciferase expression. Between 10 and 40 ng of plasmid the rate of increase in luciferase expression decreased until a maximum level of expression was reached with approximately 40 ng of plasmid (Fig. 3). We assume that the plateau in luciferase expression occurs when the amount of ICP4 is saturating. Additional experiments (data not shown) revealed that reporter plasmid is not limiting when the plateau level is reached and thus suggest that one or more cellular factors are limiting at the plateau level.
FIG. 3.
Activity of wild-type ICP4 in transient-transfection assays. (A) Vero cells (60-mm-diameter dish) were cotransfected with pCMV-βgal (100 ng/dish); a luciferase reporter plasmid (100 ng) containing either the HSV UL26.5 (circles), VP5 (triangles), or TK (squares) promoter; and increasing amounts of an effector plasmid (pXK350) that encodes wild-type ICP4. Transfected cells were harvested 48 h later and assayed for luciferase and β-galactosidase. Fold activation of the HSV promoters by ICP4 was calculated as described in Materials and Methods. (B) Vero cells were cotransfected with pCMV-βgal (100 ng), a luciferase reporter plasmid (2 μg) containing either the wild type (filled circles) or a mutant form (open circles) of the HSV-1 LAT promoter, and increasing amounts of an effector plasmid (pXK350) that encodes wild type ICP4. Fold repression of the LAT promoter by ICP4 was calculated as described in Materials and Methods. Error bars indicate standard deviations.
The repression assay measures the ability of ICP4 variants to repress basal activity of the LAT promoter linked to the luciferase gene. A repression assay conducted with 0.037 to 5 μg of effector plasmid encoding wild-type ICP4 exhibited a pattern different from that seen with the activation assay. In particular, the level of repression of the LAT promoter by ICP4 increased in an exponential fashion across the entire range of effector DNA amounts (Fig. 3B). A similar experiment using a mutant form of the LAT promoter that lacks ICP4 binding sites (1) demonstrated that repression is dependent on direct binding of ICP4 to the LAT promoter (Fig. 3B) (1). The simplest interpretation of these data is that repression of the LAT promoter is directly proportional to the amount of ICP4 that is properly folded and transported to the nuclei of transfected cells. Therefore, for a given ICP4 variant, the repression assay can provide a relative measure of the amount of functional (with respect to repression) ICP4 in transfected cells, provided that the mutation in the variant does not directly affect the ability of ICP4 to repress the LAT promoter. Published data from a variety of experimental approaches (8, 9, 21, 29, 38, 39) support the conclusion that repression of the LAT promoter is not dependent on residues in ICP4 region 4 or 5. Therefore, mutations in ICP4 region 4 or 5 should have no effect on the repression assay unless these mutations alter the folding, stability, and/or nuclear transport of the ICP4 variant. By combining the results of both the activation and repression assays, we have identified ICP4 variants with mutations in region 5 that specifically affect transcription activation.
Activities of ICP4 variants with C-terminal truncations in region 5.
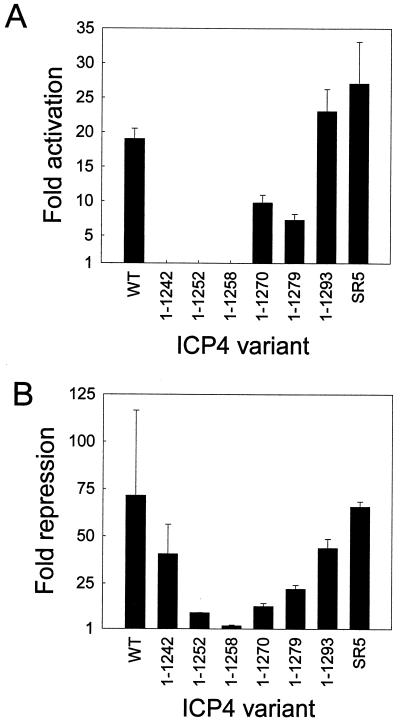
ICP4 variants that terminate at residue 1270 or 1279 exhibited a two- to threefold reduction in activation activity and a three- to fivefold reduction in repression activity (Fig. 4). Therefore, in terms of specific activity, these variants were approximately as active as wild-type ICP4 (Table 1). Variants that terminate at residue 1252 or 1258 exhibited no activation activity and a 8- to 40-fold reduction in repression activity (Table 1; Fig. 4), which implies that these variants are misfolded, unstable, and/or improperly localized. This interpretation is supported by our observation that a fusion protein consisting of GFP linked to ICP4 residues 1 to 1258 accumulates in the cytoplasm of transfected cells (data not shown). An ICP4 variant that terminates at residue 1242 exhibited no activation activity and a 2-fold reduction in the repression assay (Fig. 4) and thus has a specific activity that is at least 12-fold below that of wild-type ICP4 (Table 1). These data reveal that ICP4 residues 1270 to 1298 are not critical for activation or repression by ICP4 and suggest that residues between positions 1242 and 1270 contribute to folding, nuclear localization, and/or activation of transcription by ICP4.
FIG. 4.
Activity of C-terminal deletion variants of ICP4. (A) Vero cells were transfected with pCMV-βgal (100 ng), the VP5 promoter-luciferase reporter plasmid (100 ng), and 7.8 ng of an effector plasmid encoding the portion of ICP4 indicated below the x axis. Wild-type (WT), full-length ICP4; SR5, full-length ICP4 with multiple silent mutations in region 5. Fold activation of the VP5 promoter by ICP4 was calculated as described in Materials and Methods. (B) Vero cells were transfected with pCMV-βgal (100 ng), the LAT promoter-luciferase reporter plasmid (2 μg), and 1.25 μg of an effector plasmid encoding the portion of ICP4 indicated below the x axis. Fold repression of the LAT promoter by ICP4 was calculated as described in Materials and Methods. Error bars indicate standard deviations.
TABLE 1.
Specific activities of ICP4 variants with the VP5 promotera
| Expt | ICP4 variant | Fold activationb | Fold repressionb | Relative activationc | Relative repressiond | Sp acte |
|---|---|---|---|---|---|---|
| 1 | 1–1298 wild type | 21 | 72 | 1.0 | 1.0 | 1.0 |
| 1–1242 | 0.84 | 41 | <0.048 | 0.56 | <0.085 | |
| 1–1252 | 1.0 | 9.5 | <0.048 | 0.12 | <0.40 | |
| 1–1258 | 0.86 | 2.6 | <0.048 | 0.023 | <2.07 | |
| 1–1270 | 12 | 13 | 0.55 | 0.17 | 3.2 | |
| 1–1279 | 11 | 22 | 0.5 | 0.3 | 1.7 | |
| 1–1293 | 24 | 44 | 1.2 | 0.61 | 2.0 | |
| 1–1298 (SR5) | 18 | 66 | 0.85 | 0.92 | 0.92 | |
| 2 | 1–1298 wild type | 19 | 18 | 1.0 | 1.0 | 1.0 |
| 1–1298 Mut 1 | 1.6 | 11 | 0.033 | 0.65 | 0.051 | |
| 1–1298 Mut 2 | 8.4 | 16 | 0.41 | 0.88 | 0.47 | |
| 1–1298 Mut 3 | 1.7 | 9.5 | 0.39 | 0.50 | 0.078 | |
| 1–1298 Mut 4 | 16 | 13 | 0.83 | 0.71 | 1.2 | |
| 3 | 1–1298 wild type | 30 | 50 | 1.0 | 1.0 | 1.0 |
| 1–1298 ΔTAD1 | 15 | 54 | 0.48 | 1.1 | 0.44 | |
| 1–1298 Mut1 | 3.7 | 32 | 0.093 | 0.63 | 0.15 | |
| 1–1298 ΔTAD1 + Mut1 | 1.0 | 44 | <0.033 | 0.88 | <0.038 |
Vero cells were transfected with the VP5-luciferase reporter plasmid (100 ng) or the LAT-luciferase reporter (2 μg) and effector plasmids (32.5 ng for activation assays or 1.25 μg for repression assays) encoding the indicated variants of ICP4.
Relative to reporter alone.
Relative activation is defined as fold activation by variant minus one divided by fold activation by wild type minus one.
Relative repression is defined as fold repression by variant minus one divided by fold repression by wild type minus one.
Specific activity is defined as relative activation divided by relative repression.
Identification of a functional motif in region 5.
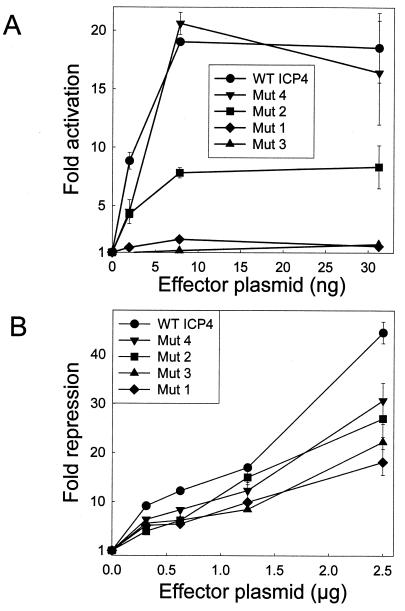
Comparison of the extreme C-terminal sequences of ICP4 orthologs encoded by five members of the alphaherpesvirus family revealed some limited areas of homology, including a TVL motif at ICP4 residues 1252 to 1254, an AAAG motif at residues 1255 to 1258, and a VEV motif at residues 1260 to 1262 (Fig. 5). To determine the functional relevance of some of these conserved residues, variants of ICP4 in which the TVL motif was altered to NGA (Mut1), the VEV motif was changed to TKT (Mut2), or glutamates at 1261 and 1266 were changed to glutamines (Mut4) were constructed. The ICP4 variant designated Mut3 has both the Mut1 and Mut2 alterations. Our choices for the specific amino acid substitutions were based on a table, compiled by Dayhoff et al. (7), showing the frequency of amino acid substitutions at similar positions in protein orthologs. We chose substitutions that occurred with a frequency of 5 to 10% and did not replace a nonpolar residue with a charged residue or vice versa.
FIG. 5.
Comparison of amino acid sequences within region 5 of ICP4 orthologs. (A) The amino acid sequence of HSV-1 ICP4 was individually aligned with those of the ICP4 orthologs from HSV-2, pseudorabies virus (PRV), bovine herpesvirus (BHV), and feline herpesvirus (FHV). The amino acid position relative to the N terminus is indicated for each ICP4 ortholog. Positions that are identical or conservative in at least two ICP4 orthologs are capitalized. Residues that are identical or conservative in at least four ICP4 orthologs are capitalized and in boldface. (B) Amino acid substitutions were introduced into ICP4 region 5 as indicated to create full-length ICP4 variants designated Mut1, Mut2, Mut3, and Mut4.
When tested in transient-transfection assays, the Mut1 and Mut3 variants were severely impaired in the activation assay but only moderately less active in the repression assay (Fig. 6). The Mut2 variant exhibited twofold less activity in the activation assay and nearly wild-type activity in the repression assay. The Mut4 variant exhibited nearly wild-type activity in both assays (Fig. 6). The specific activities of the Mut1 and Mut3 variants were 13- to 20-fold lower than that of wild-type ICP4 (Table 1). These data suggest that of the conserved residues which were tested, the TVL motif at residues 1252 to 1254 makes the most significant contribution to activation and the valines at positions 1260 and 1262 provide a slight contribution to activation. Replacement of the glutamates at positions 1261 and 1266 with glutamines has no effect on ICP4 activity.
FIG. 6.
Activities of region 5 variants of ICP4. (A) Vero cells were transfected with pCMV-βgal (100 ng), the VP5 promoter-luciferase reporter plasmid (100 ng), and the indicated amounts of an effector plasmid encoding the indicated ICP4 variant. Fold activation of the VP5 promoter by ICP4 was calculated as described in Materials and Methods. (B) Vero cells were transfected with pCMV-βgal (100 ng), the LAT promoter-luciferase reporter plasmid (2 μg), and the indicated amounts of an effector plasmid encoding the indicated ICP4 variant. Fold repression of the LAT promoter by ICP4 was calculated as described in Materials and Methods. WT, wild type. Error bars indicate standard deviations.
Relative role of TAD1 and region 5 in activation by ICP4.
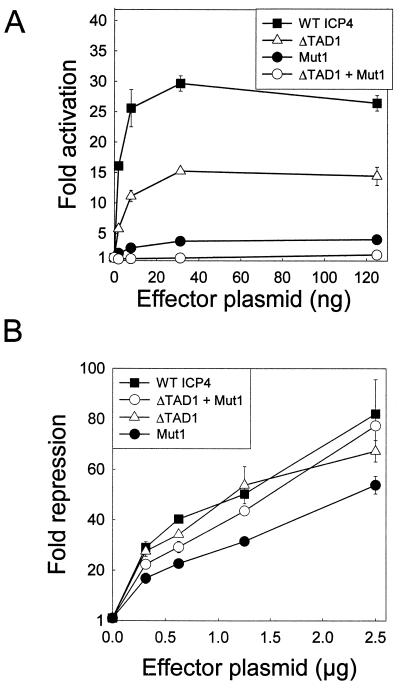
Although severely impaired, the Mut1 variant retained detectable activity in the activation assay. To determine whether this residual activity was provided by TAD1 (the N-terminal transactivation domain in ICP4), we constructed the double mutant ICP4 ΔTAD1 + Mut1, which is a derivative of Mut1 that lacks ICP4 TAD1 (residues 97 to 109). When tested in the activation assay, the single mutants ICP4 ΔTAD1 and ICP4 Mut1 were approximately 50 and 12% as active as wild-type ICP4, respectively (Fig. 7). The double mutant ICP4 ΔTAD1 + Mut1 did not detectably activate the VP5 promoter in this assay (Fig. 7 and Table 1) but was nearly as active as wild-type ICP4 in the repression assay (Fig. 7 and Table 1).
FIG. 7.
Activities of ΔTAD1 and Mut1 variants of ICP4. (A) Vero cells were transfected with pCMV-βgal (100 ng), the VP5 promoter-luciferase reporter plasmid (100 ng), and the indicated amounts of an effector plasmid encoding the indicated ICP4 variant. Fold activation of the VP5 promoter by ICP4 was calculated as described in Materials and Methods. (B) Vero cells were transfected with pCMV-βgal (100 ng), the LAT promoter-luciferase reporter plasmid (2 μg), and the indicated amounts of an effector plasmid encoding the indicated ICP4 variant. Fold repression of the LAT promoter by ICP4 was calculated as described in Materials and Methods. WT, wild type. Error bars indicate standard deviations.
To determine whether the contributions of TAD1 and region 5 to transactivation by ICP4 were promoter specific, we conducted activation assays with four additional viral promoters (UL26.5, TK, gD, and gC). As previously reported (55), we observed that ICP4 ΔTAD1 exhibited approximately 50% of wild-type activation activity when tested with the VP5, UL26.5, TK, and gC promoters but was nearly inactive when tested with the gD promoter (Table 2). ICP4 Mut1 exhibited 25 to 50% of wild-type activity when tested with the TK and UL26.5 promoters, approximately 15% of wild-type activity with the VP5 and gC promoters, and no activity with the gD promoter (Table 2). The double mutant ICP4 ΔTAD1 + Mut1 exhibited 8 to 12% of wild-type activity with the TK and UL26.5 promoters and less than 4% of wild-type activity with the VP5 and gC promoters, and it repressed the gD promoter approximately fivefold (Table 2). These data suggest that the relative contributions of TAD1 and region 5 to activation of HSV promoters by ICP4 varies among the promoters tested and thus imply that these two motifs may function somewhat independently.
TABLE 2.
Activation of HSV-1 promoters by ICP4 variantsa
| ICP4 | Fold activation of the following promoterb:
|
|||||
|---|---|---|---|---|---|---|
| VP5 | UL26.5 | TK | gD | gC + luciferasec | gC + GUSd | |
| Wild type | 30 | 36 | 17 | 6.3 | 4.7 | 71 |
| ΔTAD1 | 15 | 14 | 8.6 | 1.3 | 3.0 | 38 |
| Mut1 | 3.7 | 15 | 4.1 | 0.46 | 1.1 | 12 |
| ΔTAD1 + Mut1 | 1.0 | 4.5 | 1.5 | 0.17 | 0.43 | 2.5 |
Vero cells were transfected with a reporter plasmid (100 ng) and effector plasmids (32.5 ng) encoding the indicated variants of ICP4.
Fold activation relative to reporter alone.
The promoter was linked to the luciferase gene.
The promoter was linked to the GUS gene.
Contributions of ICP4 TAD1 and region 5 to viral replication.
We used several different approaches to determine whether the phenotypes of the TAD1 and region 5 mutants observed in transient-transfection assays reflect the role of these motifs in productive HSV infection. For the first approach, Vero cells were transfected with plasmids encoding ICP4 variants and subsequently infected with HSV-1072 (a strain of HSV-1 which lacks the ICP4 gene and has the luciferase gene driven by the HSV TK promoter). HSV-1072 does not replicate in Vero cells and expresses a very low level of luciferase due to the absence of ICP4. Cells transfected with a plasmid encoding wild-type ICP4 and subsequently infected with HSV-1072 exhibited a 76-fold increase in luciferase expression at 4 h postinfection compared to cells transfected with a control plasmid (Table 3). Cells transfected with a plasmid encoding the ΔTAD1 or Mut1 variant of ICP4 and subsequently infected with HSV-1072 exhibited a 59- or 20-fold increase, respectively, in luciferase expression at 4 h postinfection (Table 3). The ΔTAD1 + Mut1 double mutant was approximately 5% as active as wild-type ICP4 in this assay (Table 3). These results are quite similar to the pattern observed in transient transfections (Table 2) and thus argue that the effects of the ΔTAD1 and Mut1 mutations on the activity of ICP4 are similar in transfected and HSV-infected cells.
TABLE 3.
Activities of ICP4 variants in HSV-1-infected cellsa
| Transfected ICP4 | Cells infected with:
|
|||
|---|---|---|---|---|
| HSV-1072
|
HSV-976
|
|||
| Fold activationc | % of wild type activation | PFU/dish | Fold complementationd | |
| Noneb | 1.0 | 0.0 | 5.5 × 103 | 1.0 |
| Wild type | 77 | 100 | 5.5 × 105 | 100 |
| ΔTAD1 | 59 | 76 | 2.2 × 105 | 40 |
| Mut1 | 20 | 24 | 9.6 × 104 | 18 |
| ΔTAD1 + Mut1 | 5.2 | 5.5 | 2.4 × 104 | 4.4 |
Vero cells were transfected with a plasmid (100 ng) encoding the indicated ICP4 variant and infected with the indicated virus 24 h posttransfection.
Cells were transfected with a control plasmid.
Fold activation of the TK-luciferase gene in HSV-1072 4 h postinfection.
Ratio of virus yields from cells transfected with the indicated ICP4-encoding plasmid versus a control plasmid.
For the second approach, Vero cells were transfected with plasmids encoding ICP4 variants and subsequently infected with HSV-976 (a strain of HSV-1 that lacks the ICP4 gene and thus does not replicate in Vero cells). The relative yield of infectious virus at 24 h postinfection (determined by plaque assays on E5 cells) provides a measure of the ability of ICP4 variants to complement the ICP4-null virus. The ΔTAD1 and Mut1 variants were 40 and 18%, respectively, as effective as wild-type ICP4 in this complementation assay (Table 3). The ΔTAD1 + Mut1 double mutant was approximately 4% as effective as wild-type ICP4 (Table 3). These values agree remarkably well with the values obtained in the transient-transfection assays and suggest that there is a direct correlation between the relative contributions of these two motifs to the ability of ICP4 to activate gene expression and the ability of HSV to productively infect cells.
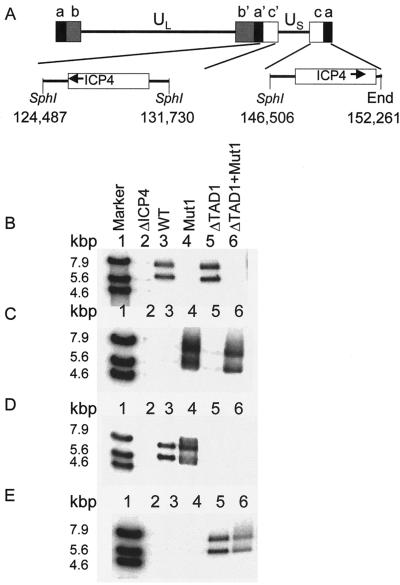
For the third approach, we constructed derivatives of HSV-1 McKrae in which both copies of the ICP4-coding region were replaced with sequences encoding EGFP fused to each of the ICP4 variants used for the assays shown in Fig. 7 and Tables 2 and 3. It should be noted that we have not detected any effect of the N-terminal EGFP protein on the activation activity of any variant of ICP4 in transfected or infected cells (data not shown). The genome of HSV-976 contains the codons for bacterial β-galactosidase inserted in place of the codons for ICP4 (50); this ΔICP4 virus was used as the parent virus for isolation of recombinants encoding EGFP-ICP4 fusion proteins. Insertion of the appropriate EGFP-ICP4 variant into both ICP4 loci was confirmed by Southern analysis (Fig. 8). SphI digestion of HSV-1 McKrae DNA yielded a 7.2-kbp fragment containing the ICP4 gene from the internal repeat and a 5.8-kbp fragment from the terminal repeat (Fig. 8A). SphI digestion of DNA from the EGFP-ICP4 recombinants yielded fragments of approximately 7.9 and 6.4 kbp due to the presence of the additional codons for EGFP (Fig. 8B). A radiolabeled oligonucleotide probe corresponding to wild-type ICP4 codons 1250 to 1258 hybridized to SphI fragments of 7.9 and 6.4 kbp obtained from the genomes of recombinant viruses expressing wild-type EGFP-ICP4 and EGFP-ICP4ΔTAD1 but did not hybridize to DNAs obtained from recombinant viruses expressing EGFP-ICP4Mut1 or EGFP-ICP4ΔTAD1+Mut1 (Fig. 8B). Conversely, a radiolabeled oligonucleotide probe spanning codons 1250 to 1258 with the alterations that correspond to the Mut1 mutation (Fig. 5) hybridized to SphI fragments obtained from the genomes of recombinant viruses expressing EGFP-ICP4Mut1 and EGFP-ICP4ΔTAD1+Mut1 but did not hybridize to DNAs from recombinant viruses expressing EGFP-ICP4 or EGFP-ICP4ΔTAD1 (Fig. 8C). These results are consistent with the expected genotype of the Mut1 variants. A radiolabeled probe spanning ICP4 codons 95 to 102 hybridized to SphI fragments obtained from the genomes of recombinant viruses expressing EGFP-ICP4 and EGFP-ICP4Mut1 but did not hybridize to DNAs from recombinant viruses expressing EGFP-ICP4ΔTAD1 or EGFP-ICP4ΔTAD1+Mut1 (Fig. 8D). Conversely, a radiolabeled oligonucleotide probe corresponding to codons 92 to 96 plus 110 to 114 (i.e., spanning the ΔTAD1 deletion) hybridized to SphI fragments obtained from the genomes of recombinant viruses expressing EGFP-ICP4ΔTAD1 and EGFP-ICP4ΔTAD1+Mut1 but did not hybridize to DNAs from recombinant viruses expressing EGFP-ICP4 or EGFP-ICP4Mut1 (Fig. 8E). These results are consistent with the expected genotypes of the ΔTAD1 variants.
FIG. 8.
Southern blot analysis of viral genomes. (A) Schematic diagram of the HSV-1 genome indicating the locations of the two copies of the ICP4 gene relative to flanking SphI cleavage sites and the terminus. Numbers indicate base pair positions in the sequence of HSV-1 strain 17 (33). (B to E) DNA isolated from the indicated recombinant virus was digested with SphI, subjected to electrophoresis in an agarose gel (lanes 2 to 6), transferred to nylon, and then hybridized to a 32P-end-labeled oligonucleotide probe specific for wild-type (WT) ICP4 codons 1250 to 1258 (B), codons 1250 to 1258 with the Mut1 mutation (C), wild-type codons 95 to 102 (D), or codons 92 to 96 and 110 to 114 (E). Bound probes were detected with a phosphorimager. Marker DNA (lanes 1) consisted of a mixture of plasmids pXK984 (wild-type ICP4 gene) and pXK645 (ICP4 ΔTAD1 + Mut1) digested with either XhoI alone, XhoI and NdeI, or XbaI and NdeI. This mixture generates bands at 7.9, 5.6, and 4.6 kbp that react with all four radiolabeled oligonucleotide probes.
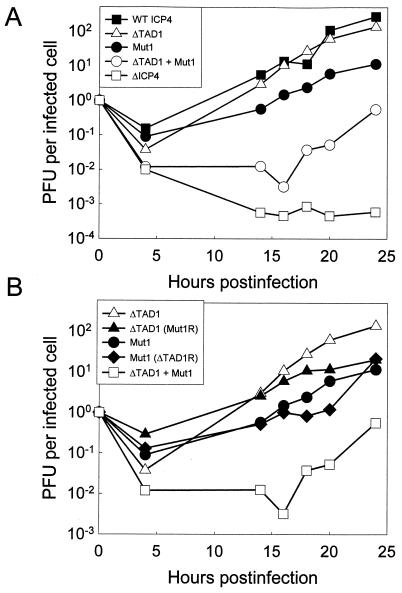
Each of these isolates was initially isolated by propagation in E5 cells. The rates of productive infection for the viral mutants were then compared in Vero cells infected at a low multiplicity (Fig. 9). All variant EGFP-ICP4 proteins were expressed to similar levels (based on immunoblot analysis with anti-EGFP [data not shown]) and localized to the nucleus (based on the distribution of EGFP-ICP4 in infected cells [data not shown]). Compared to the virus encoding wild-type EGFP-ICP4, the ΔTAD1 mutant exhibited less than a 2-fold reduction in virus yield over the course of the experiment, whereas the Mut1 variant exhibited approximately a 20-fold reduction in virus yield at 20 h postinfection (Fig. 9A). Of particular interest was the observation that the ΔTAD1 + Mut1 mutant exhibited a 2,000-fold reduction in virus yield at 20 h postinfection (Fig. 9A). However, in contrast to the ΔICP4 virus, which produced no infectious virus in Vero cells even at 48 h postinfection, the ΔTAD1 + Mut1 virus was capable of productive infection in Vero cells (Fig. 9A) and produced small plaques visible to the unaided eye in approximately 7 days. In Vero cell monolayers infected with EGFP-ICP4 variants, the second round of infection (as defined by the appearance of EGFP-ICP4 in neighboring cells) begins at around 12 h with the wild-type variant and at around 24 h with the ΔTAD1+ Mut1 virus. To determine whether the greatly reduced virulence of the ΔTAD1 + Mut1 variant was a consequence of sporadic mutations in another portion of the viral genome, we generated revertants of the ΔTAD1 + Mut1 mutant in which either the Mut1 or ΔTAD1 mutation was restored to the wild-type sequence. These revertants exhibited a growth phenotype that was similar to that of the corresponding single mutant (Fig. 9B).
FIG. 9.
Growth curves for mutant viruses. Vero cells (5 × 105 cells in 60-mm-diameter dishes) were infected with an HSV strain encoding the indicated EGFP-ICP4 variant. The infected cells were incubated at 37°C in Dulbecco’s modified Eagle medium supplemented with 10% bovine calf serum for the indicated times, harvested by being scraped into the medium, and lysed by two freeze-thaw cycles. Titers of input virus (4 × 102 to 7 × 104 PFU, depending on the virus) and viral yields were determined on E5 cell monolayers. The results are plotted as PFU per initially infected cell. WT, wild type.
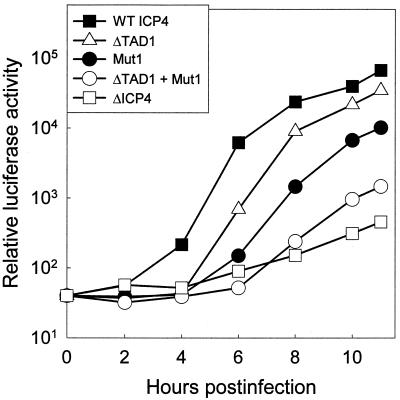
To specifically examine the kinetics of early viral gene expression in cells infected with the HSV mutants encoding the EGFP-ICP4 variants described above, we replaced the TK gene in each of the mutants with a recombinant gene in which expression of firefly luciferase is under the control of the HSV-1 TK promoter. As a negative control, we used HSV-1072, which lacks the entire ICP4-coding region and has the luciferase gene under the control of the TK promoter. Cells infected with the viral mutants were harvested at 2-h intervals postinfection and assayed for luciferase. Compared to the virus encoding wild-type EGFP-ICP4, the ΔTAD1 mutant exhibited less than a twofold reduction in luciferase activity at 10 h postinfection, whereas the Mut1 variant exhibited a sixfold reduction in luciferase activity (Fig. 10). The ΔTAD1 + Mut1 mutant exhibited a 43-fold reduction in luciferase activity at 10 h postinfection compared to the wild-type virus. Although the ΔICP4 virus (HSV-1072) produced 150-fold less luciferase than the wild-type virus, the luciferase activity in cells infected with HSV-1072 was above the background level. This result demonstrates that the HSV-1 TK promoter functions at a low level in HSV-infected cells in the absence of ICP4. Taken together, these results consistently demonstrate that residues near both termini of ICP4 contribute to the ability of ICP4 to activate expression of viral promoters.
FIG. 10.
Kinetics of early gene promoter activity in cells infected with ICP4 mutants. Vero cells (5 × 105 cells in 60-mm-diameter dishes) were infected with 5 × 103 PFU of HSV strains encoding the indicated ICP4 variants as an EGFP fusion protein. Each of these strains also contains a luciferase gene under the control of the TK promoter. Lysates prepared from cells harvested at the indicated times were assayed for luciferase as described in Materials and Methods. WT, wild type.
ICP4 region 5 does not function as an independent transactivation domain.
We previously postulated that the C-terminal portion of ICP4 might contain an independent transactivation domain that we provisionally designated TAD2 (55). Based on the results shown above, we suspected that we had mapped TAD2 to region 5. To determine whether the C-terminal portion of ICP4 can function as an independent transactivation domain, we conducted a series of transient-transfection assays with Gal4-ICP4 fusion proteins and the 5GE1b-luc reporter plasmid as described previously (55). The Gal4-ICP4 (residues 1 to 139) fusion protein contains TAD1 and activated the reporter 270-fold (Table 4). In contrast, Gal4 fusion proteins containing ICP4 residues 731 to 1298 or 1154 to 1298 failed to activate the reporter above the levels observed for Gal4 alone (Table 4). A Western immunoblot of proteins from cells transfected with these plasmids revealed that each of the Gal4-ICP4 fusion proteins was expressed (data not shown). Therefore, at least as defined by this experimental approach, the C-terminal portion of ICP4 does not function as an independent transactivation domain. Alternative models for the function of residues in region 5 are presented in Discussion.
TABLE 4.
Activation of reporter gene by Gal4-ICP4 fusion proteinsa
| Effector | Fold activationb |
|---|---|
| None | 1.0 |
| Gal4 | 5.0 |
| Gal4-VP16 | 3,700.0 |
| Gal4-ICP4(1–139) | 270.0 |
| Gal4-ICP4(731–1298) | 2.1 |
| Gal4-ICP4(1154–1298) | 4.1 |
Vero cells were cotransfected with the reporter plasmid p5GE1b-Luc (100 ng) and effector plasmids (10 ng) encoding the indicated Gal4-ICP4 fusion proteins.
Fold activation relative to reporter alone.
DISCUSSION
For this investigation we initially defined conditions for transient-transfection assays in which the expression of a reporter gene linked to a viral promoter would be directly proportional to the specific activity of a given ICP4 variant. We particularly wanted to avoid circumstances in which an excess of a mutant protein might compensate for the lower specific activity of that protein. We have established conditions in which the response of a reporter gene to ICP4 is linearly proportional to the amount of transfected plasmid that encodes ICP4. In past studies, we and others routinely used approximately 2 μg of effector plasmid in transfections with 106 cells, so we were surprised to find that 20 ng of effector plasmid provides a saturating amount of ICP4 for promoters that are activated by ICP4. We were also surprised to find that the amount of ICP4 plasmid required to saturate the activation assay was similar for all promoters tested (Fig. 3A). Through a variety of controls, we have established that (i) 20 ng of plasmid does not represent the maximum amount of DNA that can be transfected into cells (Fig. 3B), (ii) the amount of ICP4 that can be produced in transfected cells is much greater than the amount produced in cells transfected with 20 ng of plasmid (Fig. 3B), (iii) the amount of luciferase that can be produced in transfected cells is much greater than the amount produced under the conditions of the activation assay, and (iv) at the levels used for our assays, the promoters in the effector, reporter, and control plasmids do not compete for a limited pool of cellular transcription factors (data not shown). We have also established that the reporter plasmid is not the limiting factor in these assays. For example, the plateau in the activation assay is reached with 10 ng of ICP4 plasmid when we use either 100 or 1,000 ng of reporter plasmid per dish, and the fold activation with 10 ng of ICP4 is constant from 1 to 1,000 ng of reporter plasmid (data not shown). Therefore, we propose that the plateau in activation (Fig. 3A) occurs when ICP4 is saturating relative to one or more cellular proteins. Cellular proteins that are known to contribute to activation by ICP4 include TBP, TFIIB, TAFII250, and HMG1 (3, 47, 51). However, when we cotransfected Vero cells with a plasmid encoding ICP4 and a plasmid encoding either TBP, TFIIB, or HMG1, we did not detect any change in the fold activation or saturation point for ICP4 (data not shown).
In contrast to the activation data, the data for the repression assay (Fig. 3B) imply that cellular factors which contribute to repression by ICP4 are not limiting under the conditions of the repression assay. Our interpretation of the data presented in Fig. 3B is that repression of the LAT promoter by ICP4 is a stoichiometric process in which the rate-limiting step is formation of a stable complex between ICP4 and a high-affinity ICP4 binding site that spans the LAT promoter (1, 41). This interpretation is consistent with the finding that a cellular protein (HMG1) which augments binding of ICP4 to DNA also enhances repression by ICP4 in transient-transfection assays (37).
The kinetic data from the activation and repression assays (Fig. 3) demonstrate that the mechanism by which ICP4 activates transcription is fundamentally different from the mechanism by which ICP4 represses transcription. This conclusion is consistent with the observation that there are ICP4 mutants which are impaired for activation but are not impaired for repression (38, 45). The relatively small amount of ICP4 plasmid required to reach saturation in the activation assay is particularly intriguing because promoters which are activated by ICP4 lack well-defined, high-affinity binding sites for ICP4, whereas promoters which are repressed by ICP4 have high-affinity ICP4 binding sites. Therefore, if the rate-limiting step for activation by ICP4 were direct binding of ICP4 to low-affinity sites in viral promoters, then the amount of ICP4 required to saturate the activation assay would be much greater than the amount required to saturate the repression assay. Because this is not the case, we propose that activation by ICP4 is primarily dependent on a high-affinity interaction between ICP4 and one or more cellular proteins rather than DNA. Evidence from previous studies indicates that the N-terminal portion of ICP4 interacts in a DNA-dependent manner with TBP and TFIIB (47), whereas the C-terminal portion of ICP4 interacts in a DNA-independent manner with TAFII250 (3). The results presented in Fig. 3 do not reveal whether the proposed high-affinity interaction between ICP4 and a cellular protein is DNA dependent or DNA independent and do not distinguish between bimolecular and multimolecular interactions. These issues cannot be fully addressed until the protein interaction motifs in ICP4 and the interacting cellular proteins have been identified.
We previously reported that ICP4 residues 97 to 109 are a component of a transactivation domain referred to as TAD1 at the N terminus of ICP4. The identification of these residues as a transactivation domain was based on transient-transfection assays with Gal4 hybrid proteins (55). In the context of full-length ICP4, residues 97 to 109 contribute to, but are not essential for, transactivation of viral gene promoters (Table 2). The slightly impaired phenotype of the ΔTAD1 mutant clearly reveals that ICP4 residues 97 to 109 are not essential in the context of infected cells. Thus, either these residues do not function as a transactivation domain or there is an additional motif in ICP4 that can function as a transactivation domain.
A search for the putative TAD2 led to the identification of a motif in the C-terminal portion of ICP4 that contributes to activation but is not required for repression of viral gene promoters. In particular, we found that a combination of three missense mutations at codons 1252 to 1254 reduces the ability of ICP4 to activate promoters approximately 30-fold but has less than a 2-fold effect on the ability of ICP4 to repress the LAT promoter in the context of transient transfections. The repression assay provides strong evidence that these three missense mutations do not significantly alter the expression, stability, nuclear localization, DNA binding ability, or global conformation of ICP4. Observations of the synthesis and nuclear localization of GFP-tagged ICP4 variants are consistent with this interpretation. These same mutations, in the context of a recombinant virus, result in lower expression of the TK promoter, smaller plaques, and reduced burst size. These data are consistent with the results of the transient-transfection assays. We thus conclude that a motif that spans ICP4 residues 1252 to 1254 plays a critical role in activation of transcription by ICP4.
Previous results have provided a somewhat inconsistent view of the role for the ICP4 C terminus in activation of gene expression. Paterson and Everett (38) reported that deletion of ICP4 residues 1232 to 1269 resulted in a 10- to 15-fold reduction in the activity of ICP4 in transient-transfection assays with the gD promoter. In contrast, DeLuca and Schaffer (8) reported that deletion of ICP4 residues 775 to 1298 reduced the activity of ICP4 only twofold in transient-transfection assays with 1,000 ng of ICP4 plasmid and the TK promoter. Under the conditions of our transient-transfection assays (1 to 50 ng of ICP4 plasmid), variants of ICP4 containing residues 1 to 1298 (wild type), 1 to 1242, or 1 to 775 activated the UL26.5 promoter 21-, 3.5-, or 1.4-fold, respectively (data not shown). In transient transfections in U2OS cells, where ICP4 is a much stronger activator, variants of ICP4 containing residues 1 to 1298 (wild type), 1 to 1298 (Mut1), 1 to 1242, or 1 to 775 activated the UL26.5 promoter 153-, 19.6-, 10-, or 21.2-fold, respectively (data not shown). Thus, in transient-transfection assays under conditions where ICP4 is not in a 100-fold excess, the activity of the ICP4 variant containing residues 1 to 775 is as low as that of ICP4 variants with mutations or deletions in region 5. One interpretation of this observation is that ICP4 region 5 (aa 1225 to 1298) contains a functionally important motif that contributes to, but is not absolutely required for, activation of viral promoters by ICP4. The data with the ΔTAD1 + Mut1 double mutant suggest that the residual transactivation activity in variants that lack region 5 is provided by the transactivation domain in region 1.
To determine whether TAD1 is responsible for the residual activity exhibited by ICP4 variants with mutations in region 5, we tested the activities of ICP4 variants with deletions in TAD1 and region 5. The results from transient-transfection assays and from infections with recombinant viruses revealed that ICP4 variants with mutations in region 5 are dependent on TAD1 for activity. However, we noticed that an ICP4 variant with mutations in both TAD1 and region 5 exhibited weak but detectable activity (Fig. 10; Table 2). One explanation for this observation is that the mutations we made in both the N and C termini of ICP4 impair but do not absolutely abrogate the functions of either motif.
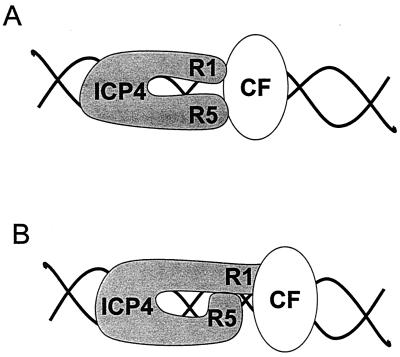
Our data are not sufficient to distinguish among several different models for the role of region 5 in activation of transcription by ICP4 (Fig. 11). Although the data with the Gal4-region 5 hybrid assays indicate that region 5 does not function as an independent transactivation domain (Table 4), these results do not exclude direct interactions between region 5 and cellular proteins that may alter the conformation of ICP4 and/or contribute to the stability of transcriptional complexes that contain ICP4 (Fig. 11, model A). Alternatively, region 5 may contribute to the activity of TAD1 either through direct interactions with TAD1 or indirectly through interactions with other portions of ICP4 (Fig. 11, model B). Because the only known ICP4 dimerization domain is in region 2 (17), model B illustrates the region 1-region 5 interaction as intramolecular. However, the experiments to define the dimerization domain in region 2 were performed exclusively with region 2 and thus would not have detected dimerization domains in other portions of ICP4. Therefore, it is possible that region 5 directly contributes to formation of ICP4 homodimers, perhaps through interactions with region 1 in homodimers that are oriented in a head-to-tail fashion. Experiments to distinguish between models A and B are in progress.
FIG. 11.
Models for the roles of ICP4 regions 1 and 5 in promoter activation. (A) ICP4 regions 1 (R1) and 5 (R5) bind independently to the same (or to a different) cellular factor (CF). (B) ICP4 region 5 interacts with another portion of ICP4 and thus enhances the interaction of the transactivation domain in region 1 with a cellular factor.
An alternative interpretation is that region 5 is simply an accessory for a critical motif in region 4. The high degree of homology in region 4 among all ICP4 orthologs and the impaired phenotypes of region 4 mutants provide compelling evidence that an essential function is provided by ICP4 region 4 (2, 8, 9, 38). Our data are consistent with a model in which the structural and functional integrity of region 4 requires the presence of region 5. If this model is correct, it will be necessary to understand the multiple functions provided by motifs in region 4 before we can elucidate the role of region 5 in transactivation by ICP4.
Acknowledgments
We thank You-gang Lin for excellent technical assistance in all phases of this investigation and Sam Holtzman for purification of many of the plasmids used for transfections. Bill Burns and Sandra Weller provided helpful advice on isolation of recombinant viruses.
This research was supported by NIH grant AI17246.
REFERENCES
- 1.Batchelor, A. H., K. W. Wilcox, and P. O’Hare. 1994. Binding and repression of the latency-associated promoter of herpes simplex virus by the immediate early 175K protein. J. Gen. Virol. 75:753–767. [DOI] [PubMed] [Google Scholar]
- 2.Baudoux, L., P. Defechereux, S. Schoonbroodt, M.-P. Merville, B. Rentier, and J. Piette. 1995. Mutational analysis of varicella-zoster virus major immediate-early protein IE62. Nucleic Acids Res. 23:1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. J. Virol. 16:3085–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, W. J., S. M. Lin, N. A. DeLuca, and D. M. Coen. 1995. Initiator elements and regulated expression of the herpes simplex virus thymidine kinase gene. J. Virol. 69:7291–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1769–1816. [DOI] [PubMed] [Google Scholar]
- 7.Dayhoff, M., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, p. 345–352. In M. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- 8.DeLuca, N. A., and P. A. Schaffer. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 15:4491–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denniston, K. J., M. J. Madden, L. W. Enquist, and G. VandeWoude. 1981. Characterization of coliphage lambda hybrids carrying DNA fragments from herpes simplex virus type 1 defective interfering particles. Gene 15:365–378. [DOI] [PubMed] [Google Scholar]
- 11.DiDonato, J. A., and M. T. Muller. 1989. DNA binding and gene regulation by the herpes simplex virus type 1 protein ICP4 and involvement of the TATA element. J. Virol. 63:3737–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D. 1984. A detailed analysis of an HSV-1 early promoter: sequences involved in trans-activation by viral immediate-early gene products are not early-gene specific. Nucleic Acids Res. 12:3037–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D. 1987. The regulation of transcription of viral and cellular genes by herpesvirus immediate early gene products. Anticancer Res. 7:589–604. [PubMed] [Google Scholar]
- 14.Everett, R. D., M. Elliott, G. Hope, and A. Orr. 1991. Purification of the DNA binding domain of herpes simplex virus type 1 immediate-early protein Vmw175 as a homodimer and extensive mutagenesis of its DNA recognition site. Nucleic Acids Res. 19:4901–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., T. Paterson, and M. Elliott. 1990. The major transcriptional regulatory protein of herpes simplex virus type 1 includes a protease resistant DNA binding domain. Nucleic Acids Res. 18:4579–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber, S. W., and K. W. Wilcox. 1988. Association of herpes simplex virus regulatory protein ICP4 with sequences spanning the ICP4 gene transcription initiation site. Nucleic Acids Res. 16:555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallinari, P., K. Wiebauer, M. C. Nardi, and J. Jiricny. 1994. Localization of a 34-amino-acid segment implicated in dimerization of the herpes simplex virus type 1 ICP4 polypeptide by a dimerization trap. J. Virol. 68:3809–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy, F. J., R. P. Baumann, and D. J. O’Callaghan. 1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172:223–236. [DOI] [PubMed] [Google Scholar]
- 19.Gu, B., and N. DeLuca. 1994. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J. Virol. 68:7953–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, B., R. Rivera-Gonzalez, C. A. Smith, and N. A. DeLuca. 1993. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc. Natl. Acad. Sci. USA 90:9528–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu, B. H., R. Kuddus, and N. A. DeLuca. 1995. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol. Cell. Biol. 15:3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homa, F. L., J. C. Glorioso, and M. Levine. 1988. A specific 15-bp TATA box promoter element is required for expression of a herpes simplex virus type 1 late gene. Genes Dev. 2:40–53. [DOI] [PubMed] [Google Scholar]
- 23.Huang, M. T., and C. M. Gorman. 1990. The simian virus small-t intron, present in many common expression vectors, leads to aberrant splicing. Mol. Cell. Biol. 10:1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imbalzano, A. N., and N. A. DeLuca. 1992. Substitution of a TATA box from a herpes simplex virus late gene in the viral thymidine kinase promoter alters ICP4 inducibility but not temporal expression. J. Virol. 66:5453–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbalzano, A. N., A. A. Shepard, and N. A. DeLuca. 1990. Functional relevance of specific interactions between herpes simplex virus type 1 ICP4 and sequences from the promoter-regulatory domain of the viral thymidine kinase gene. J. Virol. 64:2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, P. A., and R. D. Everett. 1986. The control of herpes simplex virus type-1 late gene transcription: a ‘TATA-box’/cap site region is sufficient for fully efficient regulated activity. Nucleic Acids Res. 21:8247–8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, T. R. 1991. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 65:5860–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. K., R. H. Smith, and D. J. O’Callaghan. 1995. Characterization of DNA binding properties of the immediate-early gene product of equine herpesvirus type 1. Virology 213:46–56. [DOI] [PubMed] [Google Scholar]
- 29.Kuddus, R., B. Gu, and N. A. DeLuca. 1995. Relationship between TATA-binding protein and herpes simplex virus type 1 DNA-binding sites in complex formation and repression of transcription. J. Virol. 69:5568–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leopardi, R., N. Michael, and B. Roizman. 1995. Repression of the herpes simplex virus 1 α 4 gene by its gene product (ICP4) within the context of the viral genome is conditioned by the distance and stereoaxial alignment of the ICP4 DNA binding site relative to the TATA box. J. Virol. 69:3042–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leopardi, R., and B. Roizman. 1996. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc. Natl. Acad. Sci. USA 93:4572–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, K. J., J. W. Lillie, and M. R. Green. 1990. Transcriptional activation by the pseudorabies virus immediate early protein. Genes Dev. 4:2376–2382. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531–1574. [DOI] [PubMed] [Google Scholar]
- 34.Metzler, D. W., and K. W. Wilcox. 1985. Isolation of herpes simplex virus regulatory protein ICP4 as a homodimeric complex. J. Virol. 55:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullen, M.-A., S. Gerstberger, D. M. Ciufo, J. D. Mosca, and G. S. Hayward. 1995. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J. Virol. 69:476–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panagiotidis, C. A., E. K. Lium, and S. J. Silverstein. 1997. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J. Virol. 71:1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagiotidis, C. A., and S. J. Silverstein. 1999. The host-cell architectural protein HMG1(Y) modulates binding of herpes simplex virus type 1 ICP4 to its cognate promoter. Virology 256:64–74. [DOI] [PubMed] [Google Scholar]
- 38.Paterson, T., and R. D. Everett. 1988. Mutational dissection of the HSV-1 immediate-early protein Vmw175 involved in transcriptional transactivation and repression. Virology 166:186–196. [DOI] [PubMed] [Google Scholar]
- 39.Paterson, T., and R. D. Everett. 1988. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 16:11005–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 67:4474–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivera-Gonzalez, R., A. N. Imbalzano, B. Gu, and N. A. DeLuca. 1994. The role of ICP4 repressor activity in temporal expression of the IE-3 and latency-associated transcript promoters during HSV-1 infection. Virology 202:550–564. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, M. S., A. Boundy, P. O’Hare, M. C. Pizzorno, D. M. Ciufo, and G. S. Hayward. 1988. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol. 62:4307–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapira, M., F. L. Homa, J. C. Glorioso, and M. Levine. 1987. Regulation of the herpes simplex virus type 1 late (gamma 2) glycoprotein C gene: sequences between base pairs −34 to +29 control transient expression and responsiveness to transactivation by the products of the immediate early (alpha) 4 and 0 genes. Nucleic Acids Res. 15:3097–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shepard, A. A., A. N. Imbalzano, and N. A. DeLuca. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 63:3714–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smiley, J. R., D. C. Johnson, L. I. Pizer, and R. D. Everett. 1992. The ICP4 binding sites in the herpes simplex virus type 1 glycoprotein D (gD) promoter are not essential for efficient gD transcription during virus infection. J. Virol. 66:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, C., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, R. H., V. R. Holden, and D. J. O’Callaghan. 1995. Nuclear localization and transcriptional activation activities of truncated versions of the immediate-early gene product of equine herpesvirus 1. J. Virol. 69:3857–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, R. H., Y. Zhao, and D. J. O’Callaghan. 1994. The equine herpesvirus type 1 immediate-early gene product contains an acidic transcriptional activation domain. Virology 202:760–770. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, J. L., D. Unverrich, W. J. O’Brien, and K. W. Wilcox. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805–815. [DOI] [PubMed] [Google Scholar]
- 51.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-jun degradation in vivo is mediated by the δ domain. Cell. 78:787–798. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection, vol. 51. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 53.Wu, C.-L., and K. W. Wilcox. 1990. Codons 262 to 490 from the herpes simplex virus ICP4 gene are sufficient to encode a sequence-specific DNA binding protein. Nucleic Acids Res. 18:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, C.-L., and K. W. Wilcox. 1991. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J. Virol. 65:1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao, W., L. I. Pizer, and K. W. Wilcox. 1997. Identification of a promoter-specific transactivation domain in the herpes simplex virus regulatory protein ICP4. J. Virol. 71:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, Z., and P. A. Schaffer. 1995. Intracellular localization of the herpes simplex virus type 1 major transcriptional regulatory protein ICP4, is affected by ICP27. J. Virol. 69:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]