Abstract
Lentivirus envelope glycoproteins have unusually long cytoplasmic domains compared to those of other retroviruses. To identify cellular binding partners of the simian immunodeficiency virus (SIV) envelope transmembrane protein (gp41) cytoplasmic domain (CD), we performed a yeast two-hybrid screen of a phytohemagglutinin-activated human T-cell cDNA library with the SIV gp41 CD. The majority of positive clones (50 of 54) encoded the prenylated Rab acceptor (PRA1). PRA1 is a 21-kDa protein associated with Golgi membranes that binds to prenylated Rab proteins in their GTP-bound state. While the cellular function of PRA1 is presently unknown, this protein appears to participate in intracellular vesicular trafficking, based on its cellular localization and ability to bind multiple members of the Rab protein family. Mammalian two-hybrid assays confirmed the interaction between the SIV gp41 CD and PRA1. Furthermore, gp41 sequences important for PRA1 binding were mapped to a central leucine-rich, amphipathic α-helix in the SIV gp41 cytoplasmic tail. Although the human immunodeficiency virus (HIV-1) gp41 CD failed to interact with PRA1 in the yeast two-hybrid system, its interaction with PRA1 was significantly better than that of the SIV gp41 CD in mammalian two-hybrid assays. Interestingly, PRA1 also interacted with the Env CDs of HIV-2, bovine immunodeficiency virus, equine infectious anemia virus, and feline immunodeficiency virus. Thus, PRA1 associates with envelope glycoproteins from widely divergent lentiviruses.
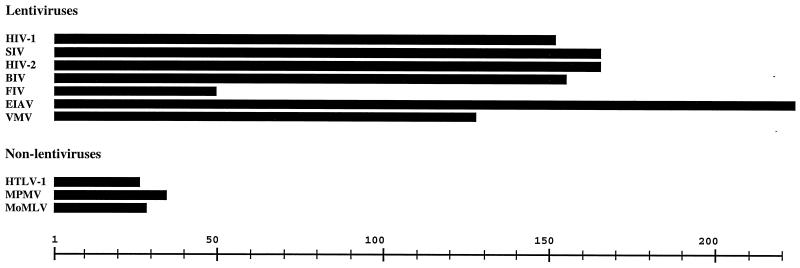
Retroviral envelope glycoproteins (Envs) are expressed as polyprotein precursors that are cleaved into surface (SU) and transmembrane (TM) subunits by cellular proteases. Env complexes of SU and TM are anchored in cellular and viral membranes by a membrane-spanning region in TM that divides TM topologically into an internal cytoplasmic domain (CD) and an external fusion domain. A distinguishing feature of lentivirus envelope glycoproteins is the unusual length of their cytoplasmic domain sequences. With the exception of feline immunodeficiency virus (FIV), the Env CDs of all lentiviruses are over 120 amino acids in length, while those of other types of retroviruses typically range between 20 and 40 amino acids (Fig. 1). The retention of unusually long Env CD sequences by lentiviruses suggests that this domain has a function specific to lentivirus replication that is, as yet, poorly understood.
FIG. 1.
Comparison of relative lengths of envelope glycoprotein cytoplasmic domain sequences from 10 different retroviruses. Scale is in numbers of amino acids. HTLV-1, human T-cell lymphotropic virus type 1; MPMV, Mason-Pfizer monkey virus; MoMLV, Moloney murine leukemia virus.
The contribution of the human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) TM (gp41) CD to virus replication appears to be species and cell type dependent. In SIV, the gp41 CD is not absolutely required for viral replication. Passage of SIV in human T-cell lines selects for premature stop codons that truncate the gp41 CD within approximately 18 residues of the membrane-spanning region (13, 19). However, viruses with these changes rapidly revert to restore the full-length gp41 CD during replication in macaque peripheral blood mononuclear cells (PBMC) or infected animals, indicating that the full-length gp41 CD confers a selective advantage to SIV replication in macaque PBMC (19). In contrast, truncations of the HIV-1 gp41 CD severely inhibit viral replication in PBMC, macrophages, and most T-cell lines (2, 6, 11, 30, 34). Yet there are a few cell lines, including MT-4 and M8166 cells, that are permissive for replication of HIV-1 mutants with shortened gp41 cytoplasmic tails (2, 30, 34). These cell-dependent differences in HIV replication appear to be related to differences in the ability to incorporate Env into virus particles and suggest a possible role for host cell factors in Env incorporation (2, 30).
Considerable genetic and biochemical evidence supports an interaction between the HIV-1 gp41 CD and the matrix (MA) domain of Pr55gag. MA mutations inhibited the incorporation of Env with a full-length CD into virus particles, and conversely, gp41 CD mutations creating Env incorporation defects were rescued by second-site mutations in MA (10, 29). A detergent-stable biochemical association between the gp41 CD and Pr55gag was also demonstrated in virus cores isolated from immature HIV-1 particles (43). Interactions between gp41 and Pr55gag thus appear to play a role in virus assembly. These interactions may also influence the intracellular site of virus release.
Expression of HIV-1 Gag protein in polarized epithelial cells resulted in particle release from both the apical and basolateral membranes, while coexpression of both Env and Gag proteins redirected virus particle release to the basolateral membrane (32). The ability of Env to influence the site of virus assembly was dependent on a tyrosine-based basolateral targeting sequence in the Env cytoplasmic domain (25).
Several host cell proteins have been identified as binding partners of the HIV and SIV gp41 CDs. Of these, the best-understood interactions are with the clathrin-associated adapter complexes AP-1 and AP-2. The μ1, μ2, and β2 subunits of AP-1 and AP-2 bind to the gp41 CDs of HIV-1 and SIV (3). Furthermore, μ1 and μ2 have been shown to bind to a highly conserved tyrosine-based endocytosis motif (YXXΦ, where Φ is hydrophobic and X can be almost any amino acid) near the gp41 membrane-spanning domain (3, 31). Binding of μ2 facilitates the recruitment of transmembrane proteins into clathrin-coated pits and appears to be a major determinant of Env endocytosis. Substitutions for tyrosine in the membrane-proximal YXXΦ motif of the HIV and SIV gp41 CDs greatly increased cell surface expression of CD8-gp41 CD chimeras with truncated CD sequences (3). However, this effect was significantly diminished in chimeras bearing full-length cytoplasmic tails, suggesting that there are additional endocytosis signals in the gp41 tail (3).
These results supported earlier findings that SIV mutants with amino acid substitutions for tyrosine in the conserved YXXL motif expressed abnormally high levels of Env on the surface of infected cells and virions and that this phenotype was more pronounced in the context of a truncated gp41 CD (22, 23, 36). In HIV-1, coexpression of the Gag protein was shown to suppress Env endocytosis, thereby facilitating the assembly and release of virus particles from the plasma membrane (7). Thus, the HIV-1 and SIV gp41 CDs interact with cellular and viral factors in a highly coordinated fashion to regulate the expression of Env on the cell surface prior to virus assembly.
Calmodulin, p115-RhoGEF, and α-catenin have also been identified as cellular binding partners of the HIV-1 gp41 CD. However, the significance of these interactions for viral replication is less clear. Calmodulin was shown to bind to peptides derived from conserved, positively charged, amphipathic α-helices of both the HIV-1 and SIV gp41 CDs (28, 37, 41). Interactions with calmodulin have been proposed to suppress T-cell activation by sequestering activated calmodulin away from downstream cellular partners and may induce apoptosis in certain cell types (17, 28, 37). p115-RhoGEF was identified as a cellular binding partner of the HIV-1 gp41 CD (45) but was found not to interact detectably with the SIV gp41 CD (L. Su, personal communication).
p115-RhoGEF regulates cytoskeletal rearrangements and cell cycle progression through activation of RhoA. Furthermore, coexpression of the HIV-1 gp41 CD with p115-RhoGEF inhibited RhoA-mediated stress fiber formation and serum response factor activation (45). Thus, it has been proposed that interactions between the HIV-1 gp41 CD and p115-RhoGEF may suppress T-cell activation and facilitate the dissemination of infected macrophages and dendritic cells in HIV-infected individuals (45). α-Catenin was identified as a cellular binding partner of the HIV-1 gp41 CD in a yeast two-hybrid screen of a HeLa cell cDNA library and was subsequently demonstrated to also interact with the SIV gp41 CD (18). α-Catenin is a component of cadherin cell adhesion complexes, and its role in viral replication is presently unclear.
In order to gain new insights into the function of lentivirus Env CDs, we performed a yeast two-hybrid screen of an activated human T-cell cDNA library using a bait construct expressing the SIV gp41 CD fused to the GAL4 DNA-binding domain (BD). To our knowledge, this represents the first screen for cellular binding partners of the SIV gp41 tail. Here we report the identification of the prenylated Rab acceptor as a cellular binding partner to the SIV gp41 CD and show that Env CDs derived from phylogenetically diverse lentiviruses, including HIV-1, HIV-2, bovine immunodeficiency virus (BIV), feline immunodeficiency virus (FIV), and equine infectious anemia virus (EIAV), also interact with PRA1.
MATERIALS AND METHODS
Yeast two-hybrid constructs. The gp41 CD coding regions of SIVmac239 (nucleotides 8746 to 9243 [35]) and HIV-1 NL4-3 (env nucleotides 2110 to 2565 [1]) were PCR amplified with primers RISENV/BHISENV (GCGAATTCGCTAAGTTAAGGCAGGGG/GCGGATCCTCACAAGAGAGTGAGCTC) and RIHENV/PIHENV (GCGAATTCAATAGAGTTAGGCAGGGA/AAACTGCAGTTATAGCAAAATCCTTTC). These sequences were then cloned into the EcoRI/BamHI and EcoRI/PstI sites of pGBKT7 in frame with the GAL4 BD to create pGBKT7-SIVEnvCD and pGBKT7-HIVEnvCD. Western blot analysis confirmed that these constructs expressed fusions of the GAL4 BD to the N terminus of the SIV and HIV gp41 CDs in yeast cells (data not shown).
Yeast two-hybrid screen.
A yeast two-hybrid screen for cellular binding partners to the SIV gp41 CD was performed using the yeast two-hybrid system 3 and a phytohemagglutinin (PHA)-activated human T-cell cDNA library (Clontech, Palo Alto, Calif.) (15). Saccharomyces cerevisiae strain AH109 was sequentially cotransformed with pGBKT7-SIVEnvCD and CsCl gradient-purified library DNA by the lithium acetate method. Transformed yeast cells were plated on selective medium lacking tryptophan and leucine (SD -W and -L) to determine the efficiency of transformation and medium also lacking histidine in the presence of 10 mM 3-amino-1,2,4 triazole (3-AT) (SD -W, -L, -H, + 3-AT) to select for two-hybrid interactions between the SIV Env CD and molecules represented in the cDNA library. In AH109, complementation of chromosomal mutations in tryptophan and leucine biosynthesis genes selects for the maintenance of bait and library plasmids on SD -W and -L medium, while GAL4-activated histidine and adenine biosynthesis genes select for yeast two-hybrid interactions between bait and library molecules.
Yeast plasmid isolation and sequence analysis.
Yeast colonies were expanded overnight at 30°C in 1.5 ml of SD -W, -L, -H with shaking. Yeast cell lysates were prepared by incubating cell pellets for 2 h at 37°C with 100 U of lyticase (Sigma, St. Louis, Mo.) and adding 10 μl of 20% sodium dodecyl sulfate (SDS). Samples were then frozen and thawed twice to ensure complete cell lysis. Plasmid DNA was recovered from yeast lysates using Chroma Spin columns (Clontech) according to the manufacturer’s instructions and used to transform Escherichia coli XL-2 Blue MRF′ cells (Stratagene, La Jolla, Calif.). Plasmid DNA minipreps representing independent yeast clones were prepared and sequenced using the pGAD10 vector-specific primers pGAD10F and pGAD10R (CGATGATGAAGATACCCCACC/TGAACTTGCGGGGTTTTTCAG).
Mammalian two-hybrid constructs.
Lentivirus env clones were obtained from the following sources. Plasmids pJSP4-27 and pFIV-PPR carrying the HIV-2 ST and FIV PPR env genes were obtained from the AIDS Research and Reference Reagent Program (McKesson Bioservices, Rockville, Md.) (20, 33). Plasmid pKF-3 carrying the HIV-2 SBL/ISY env gene was provided by Genoveffa Franchini (National Cancer Institute, Bethesda, Md.) (9, 12). Plasmid pBIV/4 carrying the BIV 127 env gene was provided by Charles Wood (University of Nebraska, Lincoln). The EIAV uk env clone was provided by Ronald Montelaro (University of Pittsburgh School of Medicine, Pittsburgh, Pa.) (5), and a visna-maedivirus (VMV) 8-5 env clone was provided by Janice Clements (Johns Hopkins University School of Medicine, Baltimore, Md.).
Env CD and PRA1 sequences were PCR amplified and cloned into pM and pVP16 (Clontech) in frame with the upstream GAL4 BD and herpes simplex virus type 1 (HSV-1) VP16 activation domain (AD) reading frames of these vectors. For most constructs, PCR primers were designed to incorporate unique restriction sites so that products could be digested and ligated directly into pM and pVP16. However, to introduce mutations into central regions of the SIV gp41 CD coding region, two rounds of PCR were performed according to the method of PCR overlap extension (14).
PCRs were performed for 30 cycles of denaturing (94°C, 40 s), annealing (50°C, 50 s), and extension (72°C, 30 s), followed by a final 5-min extension at 72°C using Pfu DNA polymerase (Stratagene). All mammalian two-hybrid constructs were sequenced to confirm that Env CD and PRA1 sequences were correct and in frame with the GAL4 BD and VP16 AD reading frames using the vector primers pMFor/pMRev (CATCATCATCGGAAGAGAGTAG/CCTCTACAAATGTGGTATGGC) and pVP16For/pVP16Rev (GCCGACTTCGAGTTTGAGC/CCTCTACAAATGTGGTATGGC).
PCR primers for amplification of lentivirus Env CD sequences included RISENV/BHISENV for SIVmac239, RIHENV/PIHENV for HIV-1 NL4-3, RIHIV-2/SIHIV-2ST (GCGAATTCAGTAGACTTAGAAAGGGC/GCGTCGACTCACAGGAGGGCGATTTC) for HIV-2 ST, RIHIV-2/SIHIV-2SBL (GCGTCGACTCACAGGAGGGCAATTTC) for HIV-2 SBL/ISY, RIBIV/HIIIBIV (GCGAATTCGCCAAGGTCAGCCAGAAT/GCAAGCTTCTACTGAGAACCTCTCAGGCC) for BIV, RIEIAV/BHIEIAV (GCGAATTCACCTCTTCGCCTAAGATC/GCGGATCCCTAAACATACTGAGGCAT) for EIAV, RIFIV/BHIFIV (GCGAATTCGATTGTATTAGAAATTGT/GCGGATCCTCATTCCTCCTCTTTTTC) for FIV, and RIVMV/BHIVMV (GCGAATTCCAGGCTTACAAGCAAGTA/GCGGATCCCTATAACTCTACATAGTC) for VMV.
C-terminal deletions of the SIV gp41 CD were created in pM using the forward primer RISENV (GCGAATTCGCTAAGTTAAGGCAGGGG) and the reverse primers BHIΔC1 (GCGGATCCTCACCAGACGGCCTGGACCGC), BHIΔC2 (GCGGATCCTCATGGTTGGAGGATCTGGTA), BHIΔC3 (GCGGATCCTCACGATAGCAAGGTTCTGCA), BHIΔC4 (GCGGATCCTCAAGGCCAGGAGCTGTTGCC), BHIΔC5 (GCGGATCCCTAGAAATAAGAGGGTGGGGA), BHIΔC6 (GCGGATCCTCATTGGCGGATCAGGAAATG), and BHIΔC7 (GCGGATCCTCAGAATAGCCAAGTCAAGAG). N-terminal deletions of the SIV gp41 CD were created in pM using the reverse primer BHISENV (GCGGATCCTCACAAGAGAGTGAGCTC) and the forward primers RIΔN1 (GCGAATTCGACGGTGGAGAAGGCGGT), RIΔN2 (GCGAATTCCTGATACGCCTCTTGACT), RIΔN3 (GCGAATTCGGGTGGAGCTATTTCCAT), and RIΔN4 (GCGAATTCTGGCAGATAGAATATATT).
Consecutive alanine substitutions were introduced into the SIVgp41 CD in the context of SCD 1-77 using the following primer pairs; DGG-AAA For and DGG-AAA Rev (AGAGCAGCAGCAGAAGGCGGTGGCAACAGC/TGCTGCTGCTCTTTCTTTGCCTTCTCTGGT), EGG-AAA For and EGG-AAA Rev (GGAGCAGCAGCAGGCAACAGCTCCTGGCCT/TGCTGCTGCTCCACCGTCTCTTTCTTTGCC), GNS-AAA For and GNS-AAA Rev (GGTGCAGCAGCATCCTGGCCTTGGCAGATA/TGCTGCTGCACCGCCTTCTCCACCGTCTCT), SWP-AAA For and SWP-AAA Rev (AGCGCAGCAGCATGGCAGATAGAATATATT/TGCTGCTGCGCTGTTGCCACCGCCTTCTCC), WQI-AAA For and WQI-AAA Rev (CCTGCAGCAGCAGAATATATTCATTTCCTG/TGCTGCTGCAGGCCAGGAGCTGTTGCCACC), EYI-AAA For and EYI-AAA Rev (ATAGCAGCAGCACATTTCCTGATCCGCCAA/TGCTGCTGCTATCTGCCAAGGCCAGGAGCT), HFL-AAA For and HFL-AAA Rev (ATTGCAGCAGCAATCCGCCAACTGATACGC/TGCTGCTGCAATATATTCTATCTGCCAAGG), IRQ-AAA For and IRQ-AAA Rev (CTGGCAGCAGCACTGATACGCCTCTTGACT/TGCTGCTGCCAGGAAATGAATATATTCTAT), LIR-AAA For and LIR-AAA Rev (CAAGCAGCAGCACTCTTGACTTGGCTATTC/TGCTGCTGCTTGGCGGATCAGGAAATGAAT), RISENV and A779A780 (GCGGATCCTCACGATAGCAAGGTTCTGCAGTTGCTGAATAGCCAAGTTGCTGCGCGTATCAGTTGGCGGAT), RISENV and BHITWL-AAA Rev (GCGGATCCTCACGATAGCAAGGTTCTGCAGTTGCTGAATGCTGCTGCCAAGAGGCGTATCAGTTG), RISENV and BHIFSN-AAA Rev (GCGGATCCTCACGATAGCAAGGTTCTGCATGCTGCTGCTAGCCAAGTCAAGAGGCG), RISENV and BHICRT-AAA Rev (GCGGATCCTCACGATAGCAATGCTGCTGCGTTGCTGAATAGCCAAGT), and RISENV and LLS-AAA Rev (GCGGATCCTCATGCTGCTGCGGTTCTGCAGTTGCTGAA).
Nonconsecutive substitutions were introduced into the SIV gp41 CD in the context of ΔC3 using the forward primer RISENV and the reverse primers A61A68A75 (GCGGATCCTCACGATAGTGCGGTTCTGCAGTTGCTGAATGCCCAAGTCAAGAGGCGTATTGCTTGGCGGATCAGGAAATG), A61AA64–65A68 (GCGGATCCTCACGATAGCAAGGTTCTGCAGTTGCTGAATGCCCAAGTTGCTGCGCGTATTGCTTGGCGGATCAGGAAATGAATATATTCTATCTG), BHIQQSSQ (GCGGATCCTCACGATTGCGAGGTTCTGCAGTTGCTGAATAGCCAAGTCGAGAGGCGTATCTGTTGGCGGATCTGGAAATGAATATATTCTATCTG), BHIQQQ (GCGGATCCTCACGATTGCAAGGTTCTGCAGTTGCTGAATAGCCAAGTCAAGAGGCGTATCTGTTGGCGGATCTGGAAATGAATATATTCTATCTG) and BHISS (GCGGATCCTCACGATAGCGAGGTTCTGCAGTTGCTGAATAGCCAAGTCGAGAGGCGTATCAGTTGGCGGATCAGGAAATGAATATATTCTATCTG).
Glutamine and serine substitutions were introduced into the full-length SIV gp41 CD in pM using the following pairs of primers: QQSSQFor and QQSSQRev (CCAGATCCGCCAACAGATACGCCTCTCGACTTGGCTATTCAGCAACTGCAGAACCTCGCAATCGAGAGTATACCAGATCCTCCAACCAAT/CGATTGCGAGGTTCTGCAGTTGCTGAATAGCCAAGTCGAGAGGCGTATCTGTTGGCGGATCTGGAAATGAATATATTCTATCTG), QQQFor and QQQRev (CCAGATCCGCCAACAGATACGCCTCTTGACTTGGCTATTCAGCAACTGCAGAACCTTGCAATCGAGAGTATACCAGATCCTCCAACCAAT/CGATTGCAAGGTTCTGCAGTTGCTGAATAGCCAAGTCAAGAGGCGTATCTGTTGGCGGATCTGGAAATGAATATATTCTATCTG), and SSFor and SSRev (CCTGATCCGCCAACTGATACGCCTCTCGACTTGGCTATTCAGCAACTGCAGAACCTCGCTATCGAGAGTATACCAGATCCTCCAACCAAT/CGATAGCGAGGTTCTGCAGTTGCTGAATAGCCAAGTCGAGAGGCGTATCAGTTGGCGGATCAGGAAATGAATATATTCTATCTG).
PRA1 C-terminal deletions were created in pVP16 using the forward primer RIPRA1 (GCGAATTCGCAGCGCAGAAGGACCAG) and the reverse primers BHIPΔC1 (GCGGATCCTCACACAGCCTCAATCTGGTG), BHIPΔC2 (GCGGATCCTCACGCTGGGCTCACCTCTCG), BHIPΔC3 (GCGGATCCTCAGTTGCTCTGGTAGTACTC), BHIPΔC4 (GCGGATCCTCAGATGACCACCAGGGTGGC), and BHIPΔC5 (GCGGATCCTCACAGATAGAGAATGTAACA). PRA1 N-terminal deletions were created using the reverse primer BHIPRA1 (GCGGATCCTCACACGGGTTCCATCTG) and the forward primers RIPΔN1 (GCGAATTCCGCCTCGTACGCAACGTG), RIPΔN2 (GCGAATTCCGCACCTTGGAGTCCAAG), and RIPΔN3 (GCGAATTCGGCTCCCACGCTGCCTTC).
Mammalian two-hybrid assays.
Constructs expressing GAL4 BD and VP16 AD fusions with lentivirus Env CD and PRA1 sequences were cotransfected into 293 T cells (American Type Culture Collection, Rockville, Md.) with the GAL4-inducible chloramphenicol acetyltransferase (CAT) reporter construct pG5CAT (Clontech) and a constitutive secreted alkaline phosphatase (SEAP) expression construct, pSV40/SEAP (Tropix, Bedford, Mass.). Calcium phosphate precipitates (1 ml) of the GAL4 BD and VP16 AD fusion constructs (7 μg each) and the pG5CAT and pSV40/SEAP reporter constructs (1.5 μg each) were prepared and added to 293 T-cell monolayers plated at 106 cells per 100-mm dish the previous day. The medium was replaced with fresh 10% Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, l-glutamine, penicillin, and streptomycin 1 day after transfection, and cells and culture supernatants were harvested 3 days after transfection.
CAT expression was quantitated in cell lysates using a CAT enzyme-linked immunosorbent assay (ELISA) kit (Boehringer-Mannheim, Indianapolis, Ind.), and SEAP activity in culture supernatants was determined using the Phospha-light chemiluminescent detection kit (Tropix). CAT induction was normalized to SEAP activity to control for variations in transfection efficiency.
RESULTS
Yeast two-hybrid interaction between SIV gp41 CD and PRA1. To identify new host cell binding partners of the SIV gp41 CD domain, we performed a yeast two-hybrid screen of a PHA-activated human T-cell cDNA library. The SIVmac239 gp41 CD coding region was PCR amplified and cloned into pGBKT7 in frame with the upstream GAL4 BD reading frame. Western blot analysis confirmed that pGBKT7-SIVEnvCD expressed a fusion of the GAL4 BD to the N terminus of the SIV gp41 CD (data not shown). Yeast strain AH109 was sequentially cotransformed with pGBKT7-SIVEnvCD and the cDNA library and plated to medium lacking tryptophan, leucine, and histidine in the presence of 10 mM 3-amino-1,2,4-triazole (SD -W, -L, -H, + 3-AT) to screen for two-hybrid interactions.
From a screen of over 9 × 106 independent cDNA clones, 60 colonies were isolated that grew on SD -W, -L, -H, + 3-AT medium. To eliminate likely false-positives, these colonies were passed to plates lacking adenine in addition to tryptophan, leucine, and histidine (SD -W, -L, -H, -A, + 3-AT). This higher-stringency selection eliminated six clones, and 29 of the remaining 54 positive clones were sequenced. Of these, 25 contained the full-length open reading frame for the human prenylated Rab acceptor (PRA1). Subsequently, the remaining 25 clones were also identified as PRA1 by PCR. Thus, 50 of 54 positive clones that we isolated encoded PRA1.
All of the PRA1 clones that were sequenced had the same seven nucleotides between the cDNA adapter and the initiating methionine of PRA1. However, we did observe heterogeneity in the size of the PRA1 cDNA inserts as a consequence of variability in the length of their poly(A) tails, indicating that multiple independent PRA1 clones were isolated. Our PRA1 clones differed by only a single nucleotide from the human PRA1 sequence in GenBank (accession number NM006423). This difference (nucleotide 416A to C) changes a glutamic acid to an alanine at position 129 in the amino acid sequence of PRA1.
To further confirm the yeast two-hybrid interaction with PRA1, 11 different PRA1 clones isolated from the library were tested for activation of the GAL4-inducible α- and β-galactosidase reporter genes in AH109, and all 11 clones were found to be positive in both of these assays (data not shown).
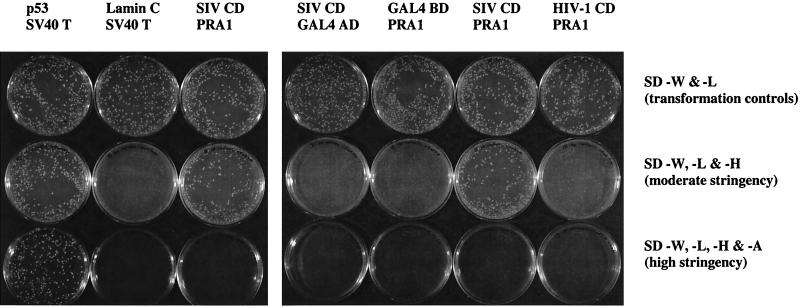
Additional yeast cotransformations were performed to test the specificity of the interaction between the SIV gp41 CD and PRA1. Yeast cells cotransformed with constructs expressing the simian virus 40 (SV40) T antigen fused to the GAL4 AD and either p53 or laminin C fused to the GAL4 BD were included as positive and negative controls, respectively. The SIV gp41 CD interaction with PRA1 supported the growth of AH109 on moderate-stringency plates (SD -W, -L, -H), but not on high-stringency plates (SD -W, -L, -H, -A) (Fig. 2). Thus, while this interaction allows growth of AH109 on medium lacking both histidine and adenine when passed from plates containing adenine, the interaction is not strong enough to rescue AH109 in a simultaneous cotransformation plated directly to medium lacking histidine and adenine (SD -W, -L, -H, -A).
FIG. 2.
Specificity of yeast two-hybrid interaction between the SIV gp41 CD and PRA1. Yeast strain AH109 was cotransformed with pairs of GAL4 BD (upper row) and GAL4 AD (lower row) expression constructs. Constructs expressing the GAL4 BD alone or fused with p53 (positive control), lamin C (negative control), or the SIV and HIV-1 gp41 CDs (SIV CD and HIV-1 CD) were cotransformed with constructs expressing GAL4 AD alone or fused with the SV40 T antigen (SV40 T) and PRA1. Yeast cells were plated to medium lacking tryptophan and leucine (SD -W and -L) as transformation controls and to medium lacking histidine and adenine to select for yeast two-hybrid interactions at moderate (SD -W, -L & -H) and high (SD -W, -L, -H & -A) stringency. Yeast plates were incubated at 30°C for 5 days.
To exclude the possibility that the SIV gp41 CD bound directly to the GAL4 AD or that the GAL4 BD bound directly to PRA1, the GAL4 BD and AD proteins without fusion partners were tested for their ability to interact with PRA1 and the SIV gp41 CD, respectively. AH109 was cotransformed with pGBKT7-SIVEnvCD and the empty GAL4 AD expression vector pGADT7 and with PRA1 clone 74 and the empty GAL4 BD expression vector pGBKT7. Importantly, the SIV gp41 CD/GAL4 AD and GAL4 BD/PRA1 transformations failed to produce colonies on either of the selective media, indicating that the SIV gp41 CD and PRA1 interact with each other and not with their GAL4 BD or AD fusion partners (Fig. 2).
The HIV-1 NL4-3 gp41 CD was also tested for yeast two-hybrid interactions with PRA1 in a cotransformation of pGBKT7-HIVEnvCD with PRA1 clone 74. Tiny colonies were visible on the HIV-1 Env CD/PRA1 plates at moderate stringency. However, these colonies failed to expand after more than a week of incubation (Fig. 2). Therefore, the gp41 CD of SIV but not HIV-1 interacted with PRA1 under the conditions of our yeast two-hybrid assay.
Mammalian two-hybrid interaction between SIV gp41 CD and PRA1.
Attempts to confirm the interaction between the SIV gp41 CD and PRA1 by coimmunoprecipitation or by glutathione S-transferase (GST) pulldown assays were unsuccessful. We therefore used mammalian two-hybrid assays (8, 40) to examine whether the SIV gp41 CD and PRA1 were capable of interacting in mammalian cells.
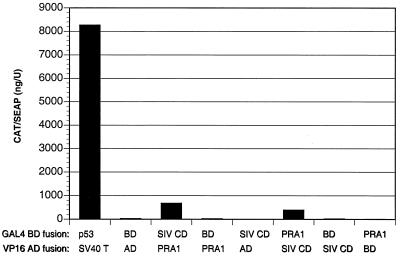
The SIV gp41 CD and PRA1 coding regions were cloned into pM and pVP16 to generate GAL4 BD and the HSV-1 VP16 AD fusion constructs. 293 T cells were cotransfected with combinations of these constructs along with the GAL4-inducible CAT reporter construct pG5CAT and the constitutive SEAP expression vector pSV40/SEAP. Three days after transfection, cells and culture supernatants were harvested, and CAT protein present in cell lysates was normalized to SEAP activity in cell culture supernatants to control for variability in the efficiency of transfection. Significant interactions between the SIV gp41 CD and PRA1 were detected when these molecules were coexpressed as either GAL4 BD or VP16 AD fusions (26- and 15-fold induction) (Fig. 3). As in the yeast two-hybrid assay, these interactions were considerably weaker than the p53/SV40 T antigen control (319-fold induction), and no interaction was detected either between the SIV gp41 CD and the VP16 AD or between PRA1 and the GAL4 BD (Fig. 3). Thus, PRA1 interacts specifically with the SIV gp41 CD in both yeast and mammalian cells.
FIG. 3.
Mammalian two-hybrid interaction between the SIV gp41 CD and PRA1. 293 T cells were cotransfected with pM and pVP16 constructs expressing GAL4 BD and VP16 AD fusions with the SIV gp41 CD (SIV CD) and PRA1. Additional transfections with constructs expressing p53 and SV40 T antigen fusions and the empty pM and pVP16 vectors expressing the GAL4 BD and VP16 AD proteins without fusion partners (BD and AD) were included as positive and negative controls, respectively. The GAL4-responsive CAT reporter pG5CAT and the constitutive SEAP reporter pSV40/SEAP were included in each transfection. Three days posttransfection, cells were harvested, and CAT expression in cell lysates was normalized to SEAP activity in cell culture supernatants.
PRA1 interacts with envelope glycoprotein cytoplasmic domains of widely divergent lentiviruses.
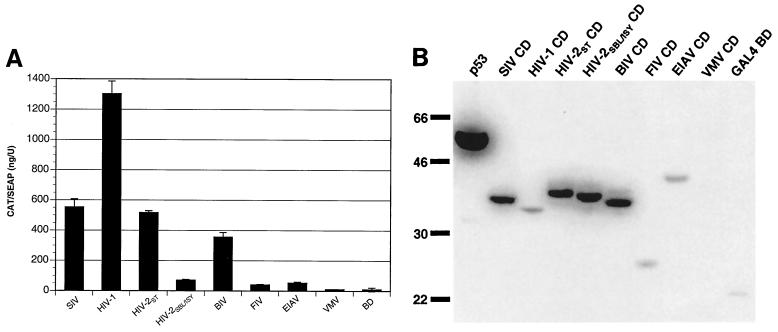
Since extended envelope glycoprotein cytoplasmic domains are common to all lentiviruses, we tested the ability of Env CDs of other lentiviruses to interact with human PRA1. In addition to the SIV gp41 CD, the Env CD coding sequences of seven different lentiviruses were cloned into pM and tested for interactions with PRA1 in mammalian two-hybrid assays. These included Env CDs from HIV-1 NL4-3, the HIV-2 ST and SBL/ISY isolates, BIV 127, FIV PPR, EIAV uk, and VMV 8.5. Remarkably, PRA1 interacted not only with the gp41 CDs of HIV-1 and HIV-2 but also with the Env CD of BIV (Fig. 4A). Weaker responses were also detected with the EIAV and FIV Env tails (Fig. 4A). Only a single Env CD from VMV failed to interact with PRA1.
FIG. 4.
Envelope glycoprotein CDs from diverse lentiviruses interact with PRA1. (A) The Env CDs of SIVmac239, HIV-1 NL4-3, HIV-2 ST, HIV-2 SBL/ISY, BIV 127, FIV PPR, EIAV uk, and VMV 8.5 were expressed as fusions with the GAL4 BD and tested for their ability to interact with PRA1 in a mammalian two-hybrid assay. An additional transfection with vectors expressing the GAL4 BD and VP16 AD without fusion partners was included as a negative control for background levels of CAT production (BD). Error bars indicate the standard deviations of duplicate transfections. (B) Expression of each of the lentivirus envelope cytoplasmic domains fused to the GAL4 BD. The sizes (shown in kilodaltons) of each of the expression products are consistent with the expected sizes of fusions of the different lentivirus envelope cytoplasmic domains with the GAL4 BD. Lysates from transfected 293 T cells were separated on a 10% polyacrylamide gel, transferred to a nylon membrane, and detected with a GAL4 BD-specific monoclonal antibody (Clontech).
The lack of an interaction with PRA1 does not appear to be the result of an error in the VMV Env CD construct, since sequence analysis indicated that the VMV Env CD sequence was correct and in frame with the upstream GAL4 BD coding region of pM. However, in contrast to the other lentivirus Env CD expression constructs, we were unable to detect expression of the GAL4 BD/VMV Env CD fusion protein by Western blot analysis of transfected 293 T cells (Fig. 4B). While the reason for this is unclear, the absence of detectable protein expression may account for the inability of the VMV Env CD to interact with PRA1. Thus, mammalian two-hybrid interactions with PRA1 were detected for all of the lentivirus Env CDs that were well expressed.
Of the two HIV-2 gp41 CDs tested, we noticed a considerable difference in the strength of their interactions with PRA1. The response with the gp41 CD derived from the HIV-2 ST isolate was similar to that of the SIV gp41 CD, while the HIV-2 SBL/ISY gp41 CD response was substantially lower (Fig. 4A). Alignment of the predicted amino acid sequences for the gp41 CDs of these two isolates revealed a number of differences, the most notable being an 11-amino-acid deletion in the gp41 tail of HIV-2 SBL/ISY (21).
To further examine the influence of these 11 residues on the interaction with PRA1, a deletion mutation eliminating the corresponding residues of the SIV gp41 CD (SCD Δ86–96) was created and tested. We also tested an additional 4-amino-acid deletion of the SIV gp41 CD in the same region that corresponds to another naturally occurring variant of the HIV-2 gp41 cytoplasmic tail (SCD Δ91–94) (21). Mammalian two-hybrid responses were reduced by the 11-amino-acid deletion in the SIV gp41 CD similar to the HIV-2 SBL/ISY gp41 CD, while the smaller 4-amino-acid deletion did not dramatically affect interaction with PRA1 (Fig. 5A, SCD Δ86–96 and SCD Δ91–94). These results indicate that the residues missing from the gp41 CD of HIV-2 SBL/ISY probably account for its weaker interaction with PRA1 and that there are polymorphic differences in the ability of gp41 CDs from different HIV-2 isolates to associate with PRA1.
FIG. 5.
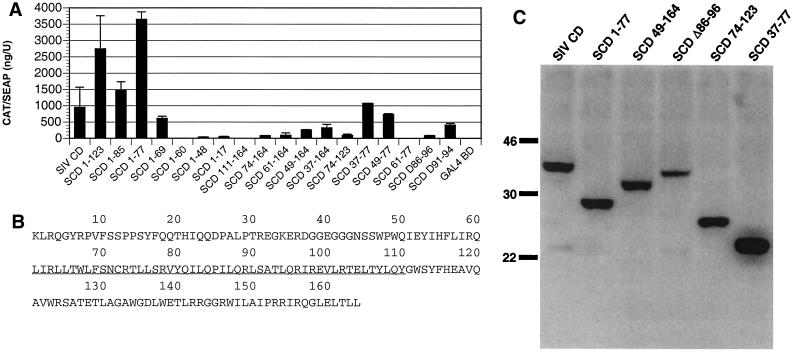
Deletion mapping of PRA1 binding region in SIV gp41 CD. (A) A panel of deletion mutants of the SIV gp41 CD (SCD) were expressed as GAL4 BD fusions and tested for their ability to interact with PRA1 in a mammalian two-hybrid assay. The residues retained by each deletion construct are indicated as subscripts and correspond to the SIV gp41 CD sequence shown in panel B. Error bars indicate the standard deviations of duplicate transfections. (B) The predicted amino acid sequence of the SIV gp41 CD is numbered beginning with the first residue of the cytoplasmic domain. The underlined sequence indicates the region predicted to interact with PRA1. (C) Western blot showing the relative expression levels of selected deletions of the SIV gp41 CD. The sizes (shown in kilodaltons) of all expression products are consistent with the predicted sizes for each segment of the SIV gp41 CD fused to the GAL4 BD. Lysates from transfected 293 T cells were separated on a 14% polyacrylamide gel, transferred to a nylon membrane, and detected with a GAL4 BD-specific monoclonal antibody (Clontech).
Deletion mapping of PRA1 binding region in SIV gp41 cytoplasmic domain.
To define regions of the SIV gp41 CD that interact with PRA1, a panel of SIV gp41 CD deletion mutants was created and tested for mammalian two-hybrid interactions with PRA1. Interestingly, two of the C-terminal deletions, SCD 1-123 and SCD 1-77, interacted with PRA1 even better than the full-length SIV gp41 CD, suggesting that sequences at the C-terminal end of gp41 are not required for, and may even interfere with, PRA1 binding (Fig. 5A). A further deletion of all but the first 69 residues of the SIV gp41 tail (SCD 1-69) reduced but did not eliminate interactions with PRA1. However, removal of an additional 9 amino acids (SCD 1-60) eliminated mammalian two-hybrid responses, indicating that residues 61 to 69 of the SIV gp41 CD are important for PRA1 association (Fig. 5A).
None of the N-terminal deletions interacted as well as the full-length SIV gp41 CD. Nevertheless, N-terminal deletions of up to 73 amino acids were still positive. However, the PRA1 interaction was lost with SCD 111-164, which eliminated 110 residues from the N terminus of the SIV gp41 CD (Fig. 5A). Thus, SCD 1-60 and SCD 111-164 define a 50-amino-acid region between residues 61 and 110 of the SIV gp41 CD that is critical for interactions with PRA1 (Fig. 5B).
Four additional combinations of N- and C-terminal deletions were tested along with the internal deletions corresponding to variants of the HIV-2 gp41 CD. The overlapping segments SCD 37-77 and SCD 49-77 resulted in two-hybrid responses that were comparable to that with the full-length SIVgp41 CD (Fig. 5A). Thus, a nearly complete response was observed with a fragment including only 29 amino acids. A weaker response was also observed with residues 74 to 123 (Fig. 5A). Since SCD 74-123 and SCD 49-77 overlap, it is possible that the 4 amino acids shared by these two segments could account for the weaker interaction with SCD 74-123. However, the reduction in the mammalian two-hybrid response observed with SCD Δ86-96 suggests that additional residues may also participate.
Western blot analysis demonstrated that the relative level of protein expression from selected SIV gp41 CD deletion constructs was comparable and does not represent a significant determinant of the strength of mammalian two-hybrid responses (Fig. 5C). Therefore, we conclude that two regions of the SIV gp41 tail including residues 49 to 77 and 74 to 123 contribute to interactions with PRA1. However, residues 49 to 77 appear to be sufficient for most of the mammalian two-hybrid activity.
Analysis of amino acid substitutions in PRA1 binding region of SIV gp41 CD.
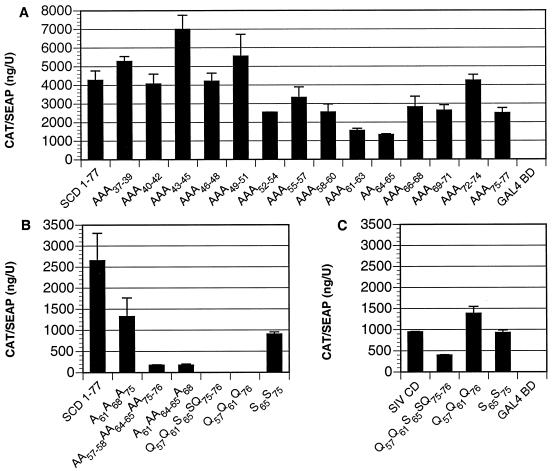
To identify residues of the SIV gp41 tail that contribute to interactions with PRA1, consecutive double or triple alanine substitutions were introduced into the SIV gp41 CD. These replacements were made in the context of SCD 1-77, since this construct consistently resulted in higher mammalian two-hybrid responses with PRA1. Furthermore, these alanine substitutions were made in the region defined by SCD 37-77, since these sequences were sufficient for responses comparable to the full-length SIV gp41 tail. Substitutions for residues 52 through 77 resulted in an approximately 50% decrease in CAT induction, supporting a role for this region in associating with PRA1 (Fig. 6A). However, none of the consecutive alanine replacements were able to eliminate or even substantially reduce interactions with PRA1 (Fig. 6A).
FIG. 6.
Identification of SIV gp41 residues that participate in interactions with PRA1. Amino acid substitutions were introduced into the SIV gp41 CD in the context of the C-terminal deletion SCD 1-77 and the full-length gp41 cytoplasmic tail (SIV CD) and tested for the ability to associate with PRA1 in mammalian two-hybrid assays. These mutations included a series of consecutive double and triple alanine substitutions (A) and combinations of nonconsecutive alanine, glutamine, and serine substitutions in SCD 1-77 (B) and the full-length SIV gp41 CD (C). Amino acid replacements are indicated as subscripts and correspond to the amino acid sequence shown in Fig. 4B. Error bars indicate the standard deviations of duplicate transfections.
Since the central region of the SIV gp41 CD is rich in dileucine (LL) and leucine repeat (LX6L) elements that might contribute to protein-protein interactions, we introduced combinations of nonconsecutive alanine substitutions at each of these positions in a further attempt to disrupt the interaction with PRA1. Alanine replacements for three leucines in an LX6L element (A61A68A75) resulted in a modest reduction in the ability to associate with PRA1 (Fig. 6B). However, replacement of an isoleucine-leucine pair and both dileucine elements (AA57–58AA64–65AA75–76) reduced mammalian two-hybrid responses with PRA1 to nearly background levels (Fig. 6B). Similar reductions were also observed when one of the dileucine elements was replaced together with the leucine repeats (A61AA64–65A68, Fig. 6B). Overall, these results are consistent with the participation of multiple leucine residues in the central region of the SIV gp41 CD in molecular interactions with PRA1.
We next tested an additional series of glutamine and serine substitutions to identify changes that would inhibit the ability of the SIV gp41 CD to associate with PRA1 and could be introduced into the SIV genome without affecting the overlapping rev reading frame. Two mutants, Q57Q61S65SQ75–76 and Q57Q61Q76, greatly reduced mammalian two-hybrid responses in the context of SCD 1-77 (Fig. 6B). However, these changes did not have a significant impact on interactions with PRA1 in the context of the full-length SIV gp41 CD (Fig. 6C). Thus, interactions between the SIV gp41 CD and PRA1 appear to be complex and probably involve multiple nonconsecutive gp41 residues.
Analysis of PRA1 domains important for gp41 association.
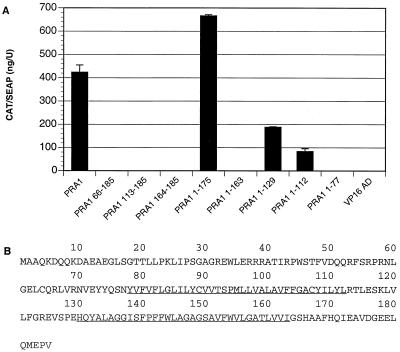
PRA1 contains two internal hydrophobic regions that represent potential membrane-spanning domains and that are predicted to divide the molecule into five distinct domains. To examine which of these domains may be important for interactions with the SIV gp41 CD, PRA1 deletion mutants were constructed in pVP16 and tested for the ability to interact with the full-length SIV gp41 CD.
None of the three N-terminal deletions were able to associate with the SIV gp41 CD, suggesting that sequences at the N terminus of PRA1 are important for interactions with the SIV gp41 CD (Fig. 7). Truncation of the C-terminal hydrophilic domain (PRA1 1-175) did not inhibit interactions with the SIV gp41 CD. However, removal of the second hydrophobic domain in both PRA1 1-129 and PRA1 1-112 significantly reduced mammalian two-hybrid responses (Fig. 7). The failure of PRA1 1-163 to interact with the SIV gp41 CD probably reflects changes in the cellular distribution of PRA1, since a similar deletion of the entire C-terminal hydrophilic domain was shown to cause PRA1 to behave as an integral membrane protein (16). An additional C-terminal deletion eliminating both internal hydrophobic domains and leaving only the N-terminal hydrophilic domain (PRA1 1-77) also abolished mammalian two-hybrid responses with the SIV gp41 tail (Fig. 7). Overall, N-terminal sequences as well as both internal hydrophobic domains of PRA1 appear to contribute to the association with the SIV gp41 CD.
FIG. 7.
Deletion mapping of SIV gp41 CD binding region in PRA1. (A) PRA1 deletion mutants were expressed as fusions with the VP16 AD and tested for mammalian two-hybrid interactions with the SIV gp41 CD. The residues retained by each deletion construct are indicated and correspond to the PRA1 residues shown in panel B. (B) The predicted amino acid sequence of PRA1 is shown, with the underlined residues indicating the two internal hydrophobic regions.
DISCUSSION
We performed a yeast two-hybrid screen of a PHA-activated human T-cell cDNA library to identify cellular factors that associate with the SIV gp41 CD. The overwhelming majority of positive clones that we recovered encoded the prenylated Rab acceptor. Controls in both the yeast and mammalian two-hybrid systems confirmed the specificity of the interaction between the SIV gp41 CD and PRA1. The specificity of the interaction was further demonstrated by the identification of individual deletion and substitution mutations in the SIV gp41 CD and PRA1 that eliminated the ability of these molecules to interact with each other. Additionally, we mapped a region of the SIV gp41 cytoplasmic tail capable of interacting with PRA1 to a 50-amino-acid region in the middle of the SIV gp41 CD. Thus, our data identify PRA1 as a cellular factor that associates with the SIV gp41 CD.
Attempts to demonstrate a biochemical association between the gp41 CD and epitope-tagged PRA1 by coprecipitation with specific antibodies or as GST fusions with glutathione-conjugated beads have so far been unsuccessful. This could possibly reflect a low binding affinity for the gp41 CD/PRA1 interaction, which would be consistent with the relatively weak yeast and mammalian two-hybrid interactions observed for gp41 and PRA1 relative to the p53 and SV40 T antigen controls. Since we cannot exclude the participation of a third-party bridging molecule in the formation of gp41 complexes with PRA1, it is also possible that the binding of PRA1 to the SIV gp41 CD may be indirect, involving one or more intermediate factors.
PRA1 is a 21-kDa protein that has been found in all tissues examined so far and is well conserved, with more than 95% amino acid identity between the human and rat homologs (4, 26). Both the human and rat PRA1 clones were originally obtained from yeast two-hybrid screens for Rab binding partners (4, 26). The Rab proteins represent the largest family of small Ras-like GTPases, and they appear to regulate intracellular vesicular trafficking. So far, over 30 different Rab isoforms have been identified, and each isoform associates with the cytoplasmic surface of different membrane-bound cellular compartments (42). Rab proteins cycle between active (GTP bound) and inactive (GDP bound) states, and membrane association is facilitated through prenylation of a C-terminal cysteine residue. Since PRA1 preferentially binds to prenylated Rab proteins in their GTP-bound form, PRA1 is predicted to bind specifically to activated membrane-associated Rab proteins (4, 26). PRA1 also binds to VAMP2, which is a component of the SNARE vesicular fusion complexes, and to GDI1, which is responsible for sequestering GDP-bound Rab proteins to the cytosol (16, 26).
Cellular fractionation studies revealed that PRA1 is present in both membrane and cytoplasmic fractions, indicating that PRA1 can exist in both membrane-associated and soluble states (16). Furthermore, the membrane-associated fraction could be extracted in high-salt buffers, indicating that PRA1 exists as an extrinsic membrane protein despite having two internal hydrophobic domains typical of type III integral membrane proteins (16). Additionally, confocal microscopy of cells transfected with epitope-tagged PRA1 constructs revealed that the membrane-associated fraction of PRA1 localizes primarily to the Golgi (16). Thus, while the precise cellular function of PRA1 is presently unknown, it is believed to participate in vesicular trafficking, based on its interactions with Rab proteins and cellular distribution.
A region of SIV gp41 that participates in interactions with PRA1 was mapped to a 50-amino-acid sequence that spans residues 61 to 110 in the middle of the cytoplasmic domain. However, a smaller region spanning residues 49 to 77 was sufficient for a strong mammalian two-hybrid response with PRA1. Deletion of up to 87 residues from the C terminus of the SIV gp41 CD actually improved interactions with PRA1 relative to the full-length SIV gp41 tail, indicating that C-terminal gp41 sequences may partially interfere with PRA1 binding.
Central regions of the gp41 CDs of both SIV and HIV-1 are predicted to adopt an amphipathic α-helical conformation (37, 38). These sequences include dileucine (LL) and leucine repeat (LX6L) elements reminiscent of leucine zipper motifs that could contribute to protein-protein interactions with PRA1. Alanine substitutions for different combinations of these leucines partially reduced the ability of PRA1 to interact with a truncated gp41 CD. However, they had little affect on PRA1 interactions in the context of the full-length SIV gp41 tail. Thus, while these leucine elements may contribute to gp41 interactions with PRA1, other residues are probably also involved.
Overall, interactions between the SIV gp41 CD and PRA1 appear to be complex and involve multiple residues present in the central region of the SIV gp41 CD. The corresponding region of HIV-1 gp41 has been implicated in interactions with calmodulin, p115-RhoGEF, α-catenin, and the matrix domain of Gag (18, 29, 37, 45). Therefore, residues within or near the central amphipathic α-helix of the gp41 CD appear to be capable of binding multiple host cell factors.
In addition to the SIV gp41 CD, PRA1 also interacted with the gp41 CDs of HIV-1 and HIV-2. The HIV-1 gp41 CD resulted in the strongest mammalian two-hybrid response with human PRA1 despite showing negligible activity in yeast two-hybrid assays. This observation is noteworthy because it demonstrates that certain interactions between viral or mammalian proteins may be missed in the yeast two-hybrid system. In fact, PRA1 would not have been identified as a cellular partner of gp41 if we had used the HIV-1 gp41 CD as bait in our yeast two-hybrid screen. These observations may explain why PRA1 was not identified as a cellular binding partner of the HIV-1 gp41 CD in previous yeast two-hybrid screens.
The HIV-2 gp41 CD interaction with PRA1 is not surprising given the amino acid sequence similarity between the HIV-2 and SIV gp41 CDs. However, we did observe a significant difference in the strength of PRA1 interactions with gp41 CDs derived from two different HIV-2 isolates. While the HIV-2 ST gp41 CD interacted comparably to the SIV gp41 CD, use of gp41 CD sequences from HIV-2 SBL/ISY resulted in a substantial reduction in the two-hybrid interaction with PRA1. Comparison of the gp41 sequences of these two isolates revealed an 11-amino-acid deletion in the predicted PRA1 binding region of the HIV-2 SBL/ISY isolate. Interestingly, deletions in this region are common among HIV-2 isolates (21) and may represent polymorphic differences in the ability to associate with PRA1. However, we do not know to what extent passage in cell culture may have contributed to this variability observed among HIV-2 gp41 CD sequences.
CD sequences derived from the envelope glycoproteins of nonprimate lentiviruses also interacted with human PRA1. Although these responses were generally lower, significant mammalian two-hybrid interactions were detected between PRA1 and the Env CDs of BIV, EIAV, and FIV. Of these, the BIV Env CD interaction was the strongest, while interactions with the EIAV and FIV Env CDs were considerably weaker by comparison. The only lentivirus Env CD that did not interact with PRA1 was derived from VMV. However, while the sequence of the VMV Env CD construct was confirmed to be correct, it was not expressed detectably in transfected 293 T cells; this may thus explain its inability to interact with PRA1. Furthermore, since human PRA1 was used exclusively in these assays, the lack of a detectable interaction with the VMV Env tail, as well as the generally weaker interactions observed with the other nonprimate lentivirus Env CDs, may reflect host species-specific differences in PRA1.
The ability of Env CD sequences derived from nonprimate lentiviruses to interact with PRA1 was surprising given the absence of discernible sequence conservation among the Env CDs of these viruses, particularly FIV and EIAV, which encode Env cytoplasmic tails of 47 and 223 residues, respectively. However, there are several possible explanations for this observation. PRA1 may interact with conformational features that are shared among the Env CDs of these viruses that are not recognizable at the primary amino acid sequence level. Alternatively, binding of PRA1 to the Env CD may not be direct and could involve different third-party intermediates or different regions of the same cellular factor for these viruses. Yet another possibility is that while these viruses may have conserved interactions with PRA1, they may have evolved different sequence elements to achieve this interaction. As precedent for this, the domains used by the SIV Nef protein to interact with cellular factors in the downregulation of major histocompatibility complex class I molecules differ from those used by HIV-1 Nef (39).
The finding that PRA1 interacts with the Env CDs of highly divergent lentiviruses suggests that PRA1 has a function specific to lentivirus replication. However, since the cellular function of PRA1 is not clear, we can only speculate about its role in virus replication. Since PRA1 localizes to Golgi membranes and appears to participate in vesicular trafficking, it is reasonable to speculate that PRA1 may direct newly synthesized Env proteins to the appropriate Golgi compartments for posttranslational modification.
Lentivirus Env proteins are known to undergo extensive posttranslational modification. In the case of the HIV and SIV Env proteins, this includes N- and O-linked glycosylation, cleavage into gp120 and gp41, and cysteine palmitoylation of the cytoplasmic domain (24, 44). PRA1 may facilitate one or more of these processing steps by influencing the intracellular trafficking of nascent Env proteins. Alternatively, PRA1 may participate in the regulation of Env expression at the cell surface. Cell surface expression of the HIV and SIV Env proteins is tightly regulated, in part, through a conserved YXXΦ endocytosis motif in the cytoplasmic tail of gp41 (7, 22, 23, 36). While amino acid substitutions that disrupt this motif increase the level of Env expression on the cell surface, this phenotype is more dramatic in the context of a truncated cytoplasmic domain, suggesting that there are additional C-terminal endocytosis signals (23). Thus, gp41 interactions with PRA1 could also contribute to Env endocytosis.
In support of this possibility, PRA1 binds particularly well to Rab5, which has been implicated in vesicular trafficking from the plasma membrane to early endosomes (4, 27). Yet another possibility is that PRA1 may contribute to virus assembly, either by facilitating Env incorporation into virions or by directing Env proteins to intracellular sites of virus particle assembly and release. This possibility is supported by observations of a cell type-dependent defect in Env incorporation into virus particles for certain HIV-1 gp41 CD truncation mutants (2, 30).
Acknowledgments
We thank Kathryn Penny for making some of the PRA1 deletion constructs used in this investigation.
This work was supported by Public Health Service grants AI10464-02, AI25328, and RR00168 and the Center for AIDS Research of the University of Massachusetts Medical School.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Wiley, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., T. Fukumori, and A. Adachi. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74:4891–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucci, C., M. Chiariello, D. Lattero, M. Maiorano, and C. B. Bruni. 1999. Interaction cloning and characterization of the cDNA encoding the human prenylated Rab acceptor (PRA1). Biochem. Biophys. Res. Commun. 258:657–662. [DOI] [PubMed] [Google Scholar]
- 5.Cook, R. F., C. Leroux, S. J. Cook, S. L. Berger, D. L. Lichtenstein, N. N. Ghabrial, R. C. Montelaro, and C. J. Issel. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 72:1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan, M. A., L. M. Carruth, J. F. Rowell, X. Yu, and R. F. Siliciano. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70:6547–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fearson, E. R., T. Finkel, M. L. Gillison, S. P. Kennedy, J. F. Casella, G. F. Tomaselli, J. S. Morrow, and C. V. Dang. 1992. Karyoplasmic interaction selection strategy: a general strategy to detect protein-protein interactions in mammalian cells. Proc. Natl. Acad. Sci. USA 89:7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franchini, G., K. A. Fargnoli, F. Giombini, L. Jagodzinski, A. D. Rossi, M. Bosch, G. Biberfeld, E. M. Fenyo, J. Albert, R. C. Gallo, and F. Wong-Staal. 1989. Molecular and biological characterization of a replication competent human immunodeficiency type 2 (HIV-2) proviral clone. Proc. Natl. Acad. Sci. USA 86:2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattori, N., F. Michaels, K. Fargnoli, L. Marcon, R. C. Gallo, and G. Franchini. 1990. The human immunodeficiency virus type 2 vpr gene is essential for productive infection of human macrophages. Proc. Natl. Acad. Sci. USA 87:8080–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch, V., N. Riedel, and J. I. Mullins. 1987. The genome organization of STLV-3 is similar to that of the AIDS virus except for a truncated transmembrane protein. Cell 49:307–319. [DOI] [PubMed] [Google Scholar]
- 14.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. [DOI] [PubMed] [Google Scholar]
- 15.Howe, A. Y. M., J. U. Jung, and R. C. Desrosiers. 1998. Zeta chain of the T-cell receptor interacts with Nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 72:9827–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutt, D. M., L. F. DaSilva, L. H. Chang, D. C. Prosser, and J. K. Ngsee. 2000. PRA1 inhibits the extraction of membrane-bound Rab GTPases by GDI1. J. Biol. Chem. 275:18511–18519. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, H., M. Sasaki, S. Noda, and Y. Koga. 1998. Apoptosis induction by the binding of the carboxy terminus of human immunodeficiency virus type 1 to calmodulin. J. Virol. 72:6574–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, E. M., K. H. Lee, and J. W. Kim. 1999. The cytoplasmic domain of HIV-1 gp41 interacts with the carboxy-terminal region of α-catenin. Mol. Cells 9:281–285. [PubMed] [Google Scholar]
- 19.Kodama, T., D. P. Wooley, Y. M. Naidu, H. W. D. Kestler, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 63:4709–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, L. I., S.-W. Lee, J. C. Kappes, J. S. Parkin, D. Decker, J. A. Hoxie, B. H. Hahn, and G. M. Shaw. 1988. West African HIV-2-related human retrovirus with attenuated cytopathicity. Science 240:1525–1529. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.). 1999. A compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 22.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, and J. A. Hoxie. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68:5509–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373–10382. [PubMed] [Google Scholar]
- 25.Lodge, R., J.-P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martincic, I., M. E. Peralta, and J. K. Ngsee. 1997. Isolation and characterization of a dual prenylated Rab and VAMP2 receptor. J. Biol. Chem. 272:26991–26998. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, O., and B. Goud. 1998. Rab proteins. Biochim. Biophys. Acta 1404:101–112. [DOI] [PubMed] [Google Scholar]
- 28.Miller, M. A., T. A. Mietzner, M. W. Cloyd, W. G. Robey, and R. C. Montelaro. 1993. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retrovir. 9:1057–1066. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305–315. [DOI] [PubMed] [Google Scholar]
- 32.Owens, R. J., J. W. Dubay, E. Hunter, and R. W. Compans. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 88:3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, T. R., R. L. Talbott, C. Lamont, S. Muir, K. Lovelace, and J. H. Elder. 1990. Comparison of two host cell range variants of feline immunodeficiency virus. J. Virol. 64:4605–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piller, S. C., J. W. Dubay, C. A. Derdeyn, and E. Hunter. 2000. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 human immunodeficiency virus type 1: effects on glycoprotein incorporation and infectivity. J. Virol. 74:11717–11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221–1231. [DOI] [PubMed] [Google Scholar]
- 36.Sauter, M. M., A. Pelchen-Matthews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. F. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J. Cell Biol. 132:795–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivas, S. K., R. V. Srinivas, G. M. Anantharamaiah, R. W. Compans, and J. P. Segrest. 1993. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 268:22895–22899. [PubMed] [Google Scholar]
- 38.Srinivas, S. K., R. V. Srinivas, G. M. Anantharamaiah, J. P. Segrest, and R. W. Compans. 1992. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J. Biol. Chem. 267:7121–7127. [PubMed] [Google Scholar]
- 39.Swigut, T., A. J. Iafrate, J. Muench, F. Kirchhoff, and J. Skowronski. 2000. Simian and human immunodeficiency virus nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol. 74:5691–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takacs, A. M., T. Das, and A. K. Banerjee. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. USA 90:10375–10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tencza, S. B., T. A. Mietzner, and R. C. Montelaro. 1997. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res. Hum. Retrovir. 13:263–269. [DOI] [PubMed] [Google Scholar]
- 42.Watson, E. L. 1999. GTP-binding proteins and regulated exocytosis. Crit. Rev. Oral Biol. Med. 10:284–306. [DOI] [PubMed] [Google Scholar]
- 43.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, H., L. Wang, S. Kao, I. P. Whitehead, M. J. Hart, B. Liu, K. Duus, K. Burridge, C. J. Der, and L. Su. 1999. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr. Biol. 9:1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]