Abstract
We sequenced the rhesus lymphocryptovirus (LCV) genome in order to determine its genetic similarity to Epstein-Barr virus (EBV). The rhesus LCV encodes a repertoire identical to that of EBV, with 80 open reading frames, including cellular interleukin-10, bcl-2, and colony-stimulating factor 1 receptor homologues and an equivalent set of viral glycoproteins. The highly conserved rhesus LCV gene repertoire provides a unique animal model for the study of EBV pathogenesis.
Epstein-Barr virus (EBV)-related herpesviruses in the same gamma-1, or lymphocryptovirus (LCV), genera are known to naturally infect both Old and New World nonhuman primates, and the biology of these nonhuman LCVs appears indistinguishable from that of EBV (reviewed in reference 35). The potential utility of using Old World LCV as an animal model system was demonstrated by the ability to experimentally infect naive rhesus macaques with rhesus LCVs and reproduce many aspects of acute and persistent EBV infection in humans (20).
Previous studies revealed that Old World LCV genomes are organized in a colinear fashion with EBV and that EBV DNA cross-reacts with viral DNA from simian LCVs (11, 12). Rhesus LCV homologues for most of the EBV latent infection genes have been described (reviewed in reference 35). In virtually every aspect, these rhesus LCV latent infection genes are functionally interchangeable with the EBV genes despite modest degrees of homology (27 to 50% amino acid homology). However, the gene repertoire from the rhesus LCV, or any gamma-1 herpesvirus besides EBV, has not been completely characterized, particularly those genes encoding cellular homologues and viral glycoproteins that are highly relevant for studies in an animal model system. The development of a rhesus LCV genetic system to generate mutant viruses for use in experimental infections and study of molecular pathogenesis in vivo also requires a thorough understanding of the rhesus LCV genome and its sequence as a starting point.
Primary sequence and genome structure of rhesus LCV.
Six overlapping cosmid and two plasmid viral DNA clones were isolated from the rhesus LCV-infected B-cell line LCL8664 (Fig. 1A). A shotgun cloning and sequencing strategy was used to derive contiguous sequences from these eight viral DNA clones. The complete rhesus LCV sequence was assembled with a sevenfold average redundancy from 1,500 overlapping sequences of 300 to 800 nucleotides.
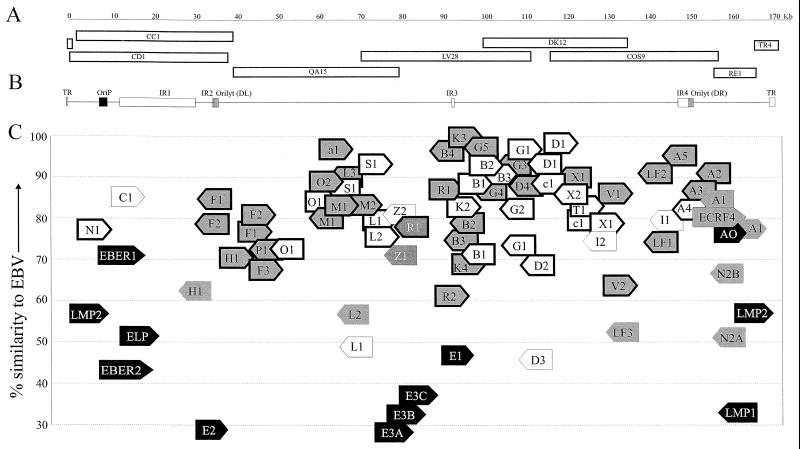
FIG. 1.
Rhesus LCV genome, ORFs, and homology with EBV ORFs. (A) Overlapping cosmid and plasmid DNA clones used to sequence the rhesus LCV genome. Cosmids were identified from the library by hybridization with the EBV BamHI C (CC1, CD1), BamHI Q (QA15), BamHI L (LV28), BamHI D (DK12), and BamHI A (cos9) DNA fragments. RE1 and TR4 are EcoRI and BamHI fragments, respectively, cloned from Hirt DNA. The nucleotide coordinates for each viral DNA clone are as follows: CD1 (140 to 38,206), CC1 (1,785 to 39,553), QA15 (39,641 to 79,990), LV28 (70,760 to 111,969), DK12 (100,417 to 135,417), Cos9 (116,690 to 157,414), RE1 (156,359 to 166,542), and TR4 (166,175 to 783). An 88-bp gap between the CC1 and QA15 cosmid clones was deduced from four PCR clones amplified from rhesus LCV-infected cell DNA using primers from the CC1 and QA15 sequence. (B) Organization of the rhesus LCV genome. Homologues for the EBV lytic and latent origins of replication (ori-p; 7,511 to 9,357), ori-lyt DL (34,141 to 35,138), ori-lyt DR (138,080 to 139,080), major repeat regions IR1 (12,240 to 29,750), IR2 (33,674 to 34,047), IR3 (89,780 to 90,460), and IR4 (135,263 to 137,761), and terminal repeats (TR; 167,326 to 171,106) are identified in the rhesus LCV genome as shown. (C) Rhesus LCV ORFs and amino acid homology with EBV ORFs. The percent amino acid similarity is shown on the y axis. Latent, immediate-early, early, and late lytic ORFs are in black, dark grey, light grey, and white, respectively. Latent infection genes are identified by name (LMPs, EBERs, EBNAs [E], and BARF0 [A0]). Each lytic infection ORF is identified using the EBV nomenclature for BamHI ORFs. The orientations of the ORFs are shown by the direction of the arrow (i.e., right or left). The EBV BamHI fragment is indicated by the letter within the arrow, and the number of the ORF in the EBV BamHI fragment is given last, e.g., the rhesus LCV BCRF1 homologue is indicated by the rightward C1 arrow with approximately 85% amino acid similarity. (The ECRF4 ORF is the only exception to these abbreviations.) ORFs common to other herpesviruses are shown with a bold outline. The initiator codon for each ORF is positioned accurately, but the ORF size is not drawn to scale.
The rhesus LCV genome contains internal (IR1 to IR4) and terminal repeats (TR) as in EBV (Fig. 1B). The major internal repeat, IR1, contains 5.7 copies of a 3,072-bp motif that is 61.5% homologous to the 3,072-bp BamHI W fragment of the EBV IR1. The rhesus LCV TR consists of a 933-bp motif versus a 538-bp motif in the EBV TR, and there is no significant sequence homology besides a similarly high GC content (75%). Based on 5.7 copies in the major internal repeat and 4 TR copies, the rhesus LCV genome has 171,096 nucleotides (versus 172,231 bp in B95-8 EBV with 11.3 IR1 copies and 4 TR copies), with an overall GC content of 62% (60% for EBV), and 65% overall nucleotide homology with the EBV genome.
ORFs encoded in rhesus LCV.
Eighty open reading frames (ORFs) are identified in the rhesus LCV sequence (Fig. 1C, Table 1). Each of the rhesus LCV ORFs has a homologue in EBV, each is located in a similar relative position as in EBV, and every EBV ORF is represented in the rhesus LCV genome. Thus, the rhesus LCV, in contrast to the recently described New World primate marmoset LCV (5), has the same viral gene repertoire as EBV. The average homology among all EBV and rhesus LCV ORFs is 75.6% compared to an average homology of 47.3% between EBV and marmoset LCV ORFs (5). Because of the overall similarity in repertoire and sequence, we have adopted the EBV nomenclature with the prefix rh to identify the rhesus LCV ORFs.
TABLE 1.
Rhesus LCV genes and amino acid similarity with EBV ORFsa
| Rhesus LCV
|
EBV
|
% aa similarity | Description | |||||
|---|---|---|---|---|---|---|---|---|
| Gene | Exon | Begins (nt) | Stops (nt) | Size (aa) | Gene | Length (aa) | ||
| LMP2A e2 | 138 | 354 | 71 | LMP2A e2 | 71 | 66.2 | ||
| LMP2A e3 | 437 | 535 | 33 | LMP2A e3 | 33 | 51.2 | ||
| LMP2A e4 | 644 | 892 | 83 | LMP2A e4 | 83 | 67.5 | ||
| LMP2A e5 | 974 | 1054 | 27 | LMP2A e5 | 27 | 76.9 | ||
| LMP2A e6 | 1134 | 1289 | 52 | LMP2A e6 | 57 | 50.9 | ||
| LMP2A e7 | 1377 | 1592 | 73 | LMP2A e7 | 72 | 73.6 | ||
| LMP2A e8 | 1673 | 1779 | 35 | LMP2A e8 | 35 | 45.7 | ||
| BNRF1 | 1836 | 5780 | 1,314 | BNRF1 | 1,318 | 77.6 | Tegument protein | |
| LMP2A e9 | 5494 | 5930 | LMP2A e9 | |||||
| EBER1 | 6653 | 6824 | EBER1 | Small RNA | ||||
| EBER2 | 6967 | 7136 | EBER2 | Small RNA | ||||
| BCRF1 | 9920 | 10453 | 177 | BCRF1 | 170 | 84.1 | IL-10 homologue | |
| EBNA-LP | 312 | EBNA-LP | 308 | 53.3 | Nuclear protein | |||
| EBNA-LP C1 | 11765 | 11771 | EBNA-LP C1 | |||||
| EBNA-LP C2 | 11930 | 11959 | EBNA-LP C2 | |||||
| EBNA-LP W1 | 15018 | 15078 | 21 | EBNA-LP W1 | 21 | 76.2 | ||
| EBNA-LP W2 | 15160 | 15294 | 45 | EBNA-LP W2 | 44 | 52.3 | ||
| EBNA-LP Y1 | 29839 | 29871 | 11 | EBNA-LP Y1 | 11 | 45.5 | ||
| EBNA-LP Y2 | 29956 | 30060 | 34 | EBNA-LP Y2 | 34 | 44.1 | ||
| EBNA2 | 30612 | 32429 | 606 | EBNA-2 | 490 | 29.8 | Nuclear protein | |
| BHLF1 | 33940 | 33089 | 283 | BHLF1 | 660 | 63.6 | ||
| BHRF1 | 35902 | 36477 | 190 | BHRF1 | 191 | 72.8 | be1-2 homologue | |
| BFLF2 | 38519 | 37566 | 318 | BFLF2 | 318 | 79.6 | ||
| BFLF1 | 40106 | 38532 | 525 | BFLF1 | 525 | 85.5 | Glycoprotein | |
| BFRF1 | 40465 | 41454 | 328 | BFRF1 | 336 | 78.4 | Tegument protein | |
| BFRF2 | 41358 | 43169 | 603 | BFRF2 | 591 | 80.4 | ||
| BFRF3 | 43093 | 43605 | 169 | BFRF3 | 176 | 69.2 | Capsid protein | |
| BPLF1 | 52963 | 43646 | 3,106 | BPLF1 | 3,149 | 74.3 | Tegument protein | |
| BORF1 | 56698 | 57789 | 362 | BORF1 | 364 | 86.5 | DNA maturation | |
| BOLF1 | 56699 | 53013 | 1,224 | BOLF1 | 1,239 | 71.1 | Tegument protein | |
| BORF2 | 57852 | 60320 | 821 | BORF2 | 826 | 87.5 | Ribonucleotide reductase | |
| BaRF1 | 60333 | 61241 | 300 | BaRF1 | 302 | 96.0 | Ribonucleotide reductase | |
| BMRF1 | 61334 | 62548 | 404 | BMRF1 | 404 | 85.9 | DNA replication | |
| BMRF2 | 62553 | 63626 | 357 | BMRF2 | 357 | 86.0 | Membrane protein | |
| BMLF1 | 65658 | 64261 | 464 | BMLF1 | 438 | 80.4 | Transactivator | |
| BSLF1 | 68479 | 65855 | 875 | BSLF1 | 874 | 86.4 | Helicase complex | |
| BSRF1 | 68522 | 69187 | 218 | BSRF1 | 218 | 89.4 | ||
| BLLF3 | 70089 | 69250 | 276 | BLLF3 | 278 | 87.3 | dUTPase | |
| BLRF1 | 70163 | 70451 | 102 | BLRF1 | 102 | 74.5 | Glycoprotein N, gp15 | |
| BLRF2 | 70542 | 71033 | 162 | BLRF2 | 162 | 76.5 | ||
| BLLF2 | 71609 | 71190 | 139 | BLLF2 | 148 | 56.0 | ||
| gp350 | 73401 | 71050 | 783 | gp350 | 886 | 49.3 | Glycoprotein, gp350 | |
| EBNA3A | 955 | EBNA3A | 925 | 29.4 | Nuclear protein | |||
| EBNA3A e1 | 73534 | 73845 | 117 | EBNA3A e1 | 117 | 37.6 | ||
| EBNA3A e2 | 73933 | 76446 | 838 | EBNA3A e2 | 808 | 28.0 | ||
| EBNA3B | 928 | EBNA3B | 938 | 30.5 | Nuclear protein | |||
| EBNA3B e1 | 76628 | 76993 | 122 | EBNA3B e1 | 121 | 37.7 | ||
| EBNA3B e2 | 77075 | 79495 | 806 | EBNA3B e2 | 817 | 31.8 | ||
| EBNA3C | 1,157 | EBNA3C | 1,069 | 31.2 | Nuclear protein | |||
| EBNA3C e1 | 79626 | 79970 | 117 | EBNA3C e1 | 117 | 32.5 | ||
| EBNA3C e2 | 80050 | 83184 | 1,040 | EBNA3C e2 | 952 | 30.1 | ||
| BZLF2 | 83897 | 83232 | 222 | BZLF2 | 223 | 77.6 | Glycoprotein, gp42 | |
| BZLF1 | 248 | BZLF1 | 245 | 71.3 | Transactivator | |||
| BZLF1 e3 | 84118 | 83997 | 41 | BZLF1 e3 | 42 | 80.5 | ||
| BZLF1 e2 | 84335 | 84227 | 36 | BZLF1 e2 | 36 | 88.9 | ||
| BZLF1 e1 | 85093 | 84581 | 171 | BZLF1 e1 | 167 | 60.5 | ||
| BRLF1 | 87109 | 85301 | 603 | BRLF1 | 605 | 76.3 | Transactivator | |
| BRRF1 | 87108 | 88037 | 310 | BRRF1 | 310 | 85.5 | ||
| BRRF2 | 88226 | 89731 | 500 | BRRF2 | 537 | 60.6 | ||
| EBNA1 | 89767 | 91302 | 512 | EBNA1 | 641 | 46.3 | Episomal maintenance | |
| BKRF2 | 91383 | 91796 | 136 | BKRF2 | 137 | 81.6 | Glycoprotein L, gp25 | |
| BKRF3 | 91778 | 92545 | 255 | BKRF3 | 255 | 96.9 | Uracyl DNA glucosidase | |
| BKRF4 | 92556 | 93275 | 239 | BKRF4 | 217 | 69.0 | ||
| BBRF1 | 95691 | 97535 | 615 | BBRF1 | 613 | 92.5 | Capsid protein | |
| BBLF4 | 95746 | 93317 | 810 | BBLF4 | 809 | 93.4 | Helicase complex | |
| BBRF2 | 97438 | 98274 | 279 | BBRF2 | 278 | 91.4 | ||
| BBLF3 | 98846 | 98271 | 205 | BBLF3 | 201 | 73.6 | Helicase complex | |
| BBLF2 | 100552 | 98945 | 514 | BBLF2 | 522 | 72.8 | Helicase complex | |
| BBRF3 | 100652 | 101872 | 407 | BBRF3 | 405 | 90.4 | Glycoprotein M, gp84/113 | |
| BBLF1 | 102535 | 102311 | 75 | BBLF1 | 75 | 72.0 | Myristylated tegument protein | |
| BGLF5 | 103902 | 102490 | 470 | BGLF5 | 470 | 94.0 | Alkaline exonuclease | |
| BGLF4 | 105178 | 103889 | 452 | BGLF4 | 455 | 89.6 | Kinase | |
| BDRF1 | 690 | BDRF1 | 690 | 92.9 | Packaging protein | |||
| BDRF1 e1 | 106502 | 107437 | 312 | BGRF1 | 311 | 91.6 | ||
| BGLF3 | 106503 | 105505 | 332 | BGLF3 | 332 | 88.3 | ||
| BGLF2 | 108437 | 107427 | 336 | BGLF2 | 336 | 87.8 | ||
| BGLF1 | 109911 | 108415 | 498 | BGLF1 | 507 | 74.7 | ||
| BDLF4 | 110597 | 109881 | 238 | BDLF4 | 225 | 86.2 | ||
| BDRF1 e2 | 110794 | 111930 | 378 | BDRF1 | 379 | 94.7 | ||
| BDLF3 | 112748 | 111969 | 260 | BDLF3 | 234 | 46.6 | Glycoprotein, gp150 | |
| BDLF2 | 114024 | 112813 | 403 | BDLF2 | 420 | 68.5 | ||
| BDLF1 | 114939 | 114034 | 302 | BDLF1 | 301 | 96.7 | Capsid protein | |
| BcLF1 | 119097 | 114955 | 1,381 | BcLF1 | 1,381 | 95.8 | Capsid protein | |
| BcRF1 | 119641 | 121374 | 578 | BcRF1 | 575 | 83.0 | ||
| BTRF1 | 121361 | 122572 | 425 | BTRF1 | 425 | 86.6 | ||
| BXLF2 | 124692 | 122569 | 708 | BXLF2 | 706 | 85.1 | Glycoprotein H, gp85 | |
| BXLF1 | 126517 | 124694 | 608 | BXLF1 | 607 | 89.1 | Thymidine kinase | |
| BXRF1 | 126516 | 127265 | 250 | BXRF1 | 248 | 80.6 | ||
| BVRF1 | 127075 | 128781 | 566 | BVRF1 | 570 | 84.6 | Tegument protein | |
| BVRF2 | 129594 | 131453 | 623 | BVRF2 | 605 | 66.4 | Capsid protein | |
| BILF2 | 132248 | 131487 | 249 | BILF2 | 248 | 75.0 | Glycoprotein, gp78 | |
| LF3 | 137831 | 135159 | 905 | LF3 | 924 | 42.7 | ||
| LF2 | 146382 | 145093 | 429 | LF2 | 429 | 92.5 | ||
| LF1 | 147596 | 146343 | 417 | LF1 | 422 | 75.3 | ||
| BILF1 | 148684 | 147746 | 312 | BILF1 | 312 | 80.4 | Glycoprotein, gp64 | |
| ECRF4 | 151083 | 151946 | 289 | ECRF4 | 289 | 79.3 | ||
| BALF5 | 152230 | 149283 | 1,015 | BALF5 | 1,015 | 94.8 | DNA polymerase | |
| BALF4 | 154927 | 152333 | 865 | BALF4 | 857 | 85.6 | Glycoprotein B, gp110 | |
| BARF0 | 156117 | 156602 | 165 | BARF0 | 175 | 77.0 | Nuclear protein | |
| BALF3 | 156968 | 154914 | 684 | BALF3 | 789 | 85.7 | Transport protein | |
| BALF2 | 160385 | 156984 | 1,134 | BALF2 | 1,128 | 90.3 | DNA binding protein | |
| BALF1 | 161020 | 160472 | 183 | BALF1 | 220 | 84.1 | bcl-2 homologue | |
| BARF1 | 161120 | 161782 | 220 | BARF1 | 221 | 75.0 | CSF1R homologue | |
| LMP2A | 495 | LMP2A | 498 | 57.0 | Membrane protein | |||
| LMP2A e1 | 162521 | 162879 | 120 | LMP2A | 119 | 31.9 | ||
| BNLF2B | 163256 | 162960 | 94 | BNLF2b | 101 | 68.1 | ||
| BNLF2A | 163444 | 163265 | 60 | BNLF2a | 60 | 51.7 | ||
| LMP1 e3 | 165723 | 164303 | 474 | LMP1 e3 | 267 | 52.3 | ||
| LMP1 e2 | 165894 | 165808 | 28 | LMP1 e2 | 32 | 39.3 | ||
| LMP1 e1 | 166242 | 165981 | 88 | LMP1e1 | 88 | 30.7 | ||
| LMP1 | 588 | LMP1 | 386 | 32.4 | Transforming gene | |||
| LMP2B e1 | 166602 | 166718 | LMP2B e1 | |||||
Descriptions as defined for EBV ORFs. EBV ORFs derived by combining B95-8 sequence (2) (GenBank accession no. NC 001345) and Raji sequence (23) (GenBank accession no. M35547). Criteria used to evaluate rhesus LCV ORFs were as follows: >150 amino acids (aa), no more than 30% overlap with a known ORF. Shorter ORFs were identified only if homologues were identified in other herpesviruses. The complete rhesus LCV genome sequence has been deposited in GenBank (accession no. AY037858). Nucleotide homology between EBER-1 and rh EBER-1, 70.5%. Nucleotide homology between EBER-2 and rh EBER-2, 42.4%.
Rhesus LCV latent infection genes.
Homologues for the rhesus LCV EBV-encoded small RNAs (EBERs), EBNA-LP, two types of EBNA-2, EBNA-1, EBNA-3A, -3B, and -3C, LMP1, LMP2A, and LMP2B have been reported previously (3, 6, 8, 15, 24, 26, 27). The complete rhesus LCV sequence shows that there is also a homologue for the EBV BARF0 open reading frame, with 77% homology, suggesting that the family of EBV BamHI A transcripts expressed during latent infection are also likely to be conserved in the rhesus LCV. The latent infection genes are generally the least well conserved among all rhesus LCV genes (Fig. 1C, Table 1).
Conservation of LCV lytic infection genes.
Most of the EBV lytic infection genes have homologues in other herpesviruses due to the conserved mechanisms for herpesvirus replication. These 56 ORFs (24 late, 32 early, and 1 immediate-early lytic infection viral gene product) have an average homology of 82.8% with the rhesus LCV homologues (Fig. 1C, ORFs with bold outline). Fifteen EBV lytic infection ORFs do not have homologues in other herpesviruses, i.e., they are restricted to gamma-1 herpesviruses, and the rhesus LCV homologues for these ORFs have an average homology of 60.3%. These genes have presumably evolved more recently and are generally less well conserved between EBV and rhesus LCV.
ORFs encoding homologues of cellular proteins.
Four EBV lytic infection genes are cell gene homologues likely to have been captured because they provide a biologic advantage during EBV infection. These include a viral interleukin-10 (vIL-10; BCRF1), two bcl-2 homologues (BHRF1 and BALF1), and a colony-stimulating factor 1 receptor (CSF-1R) homologue (BARF1) (14, 18, 19, 32). These viral genes are not essential for EBV-induced transformation of B-cell growth and for EBV replication in vitro (7, 18, 33). Rhesus LCV has captured an identical repertoire of cellular homologues. Conservation of these ORFs in the rhesus LCV (73 to 84% homology relative to the EBV proteins) indicates that these cellular homologues provide biologic advantages that are common to both EBV and rhesus LCV infection in their natural hosts.
Viral membrane proteins.
Viral membrane proteins are important for cell tropism, as targets for the host immune response, and for pathogenesis of infection in vivo. All 10 EBV ORFs known to encode viral membrane glycoproteins are positionally conserved in the rhesus LCV. Five of these glycoproteins are conserved in all herpesviruses (gB, gH, gL, gM, and gN) and are important for herpesvirus virus assembly, egress, and cell fusion (13, 16, 17, 21). These glycoproteins are well conserved in the rhesus LCV (74 to 90% homology with the EBV glycoproteins). Five glycoproteins are restricted to gammaherpesviruses, and these viral gene products are likely to be important for LCV biology and pathogenesis. Among these, gp350 and gp150 have the lowest degree of homology between EBV and the rhesus LCV, 49.3 and 46.6% homology, respectively. gp350 is the major viral membrane glycoprotein that binds to CR2/CD21 and is a major determinant for EBV’s B-cell tropism (22, 34). gp150 is not essential for EBV replication and infection in vitro (4), suggesting an important role for this glycoprotein during human and rhesus LCV infection in vivo.
The rhesus LCV is only the second completely sequenced genome from the oncogenic LCV genera. The EBV B95-8 strain was the first gamma-1 herpesvirus fully sequenced (2). Portions of several other EBV strains have been sequenced (23, 29). Analysis of an 11-kb DNA sequence from Raji EBV demonstrated that the B95-8 strain is a deletion mutant, missing a duplicated ori-lyt sequence (DR) at the right-hand side of the genome (10, 23, 25). Thus, the rhesus LCV genome is the first complete sequence derived from a prototypical LCV genome.
The identical repertoire of lytic and latent infection genes between EBV and the rhesus LCV demonstrates the close genetic relationship between these two viruses and provides genetic validation that the rhesus LCV is an accurate model for studying EBV pathogenesis. The conservation of a type 1 latency EBNA-1 promoter (28) and the existence of two different rhesus LCV types similar to type 1 and 2 EBV (6) provide further evidence of the biologic and genetic similarities between EBV and the rhesus LCV. Thus, Old World LCVs, such as the rhesus LCV, appear to have evolved very closely in parallel with EBV, whereas New World LCVs, such as the marmoset LCV, appear to have evolved somewhat differently despite the overt biologic similarities (5). Thus, the evolutionary distances between human, Old World, and New World LCVs are similar to the relationships between New World, Old World, and human hosts. Studies in both New and Old World model systems may provide a better understanding of how various viral genes contribute to successful EBV infection and pathogenesis in vivo.
Primates have been touted as important animal model systems for studying human virus infection because of the strong similarities in both the viruses and the natural hosts. To our knowledge, only two other herpesviruses naturally infecting Old World nonhuman primate species have been completely sequenced, rhesus rhadinovirus (RRV) and simian varicella virus (SVV) (1, 9, 30), and both have been proposed as animal models for human herpesvirus infections (31, 36). However, the viral gene repertoire from these two viruses is not identical to their human counterparts, Kaposi’s sarcoma herpesvirus (KSHV) and varicella-zoster virus (VZV). RRV does not encode homologues for the KSHV K3, K5, K7, K8, K8.1, and K12 ORFs (1, 30). In addition, the dihydrofolate reductase (DHFR) homologue is encoded in a different location and the copy numbers of macrophage inflammatory protein and viral interferon regulatory factor homologues are different.
Similarly, the SVV and VZV genomes do not have an identical gene repertoire. SVV does not encode a homologue for the VZV ORF2, a gene with unknown function, and SVV encodes for a novel ORF A that is a truncated form of VZV ORF4 (9). In addition, the average homology of RRV and SVV ORFs with their human herpesvirus counterparts is relatively low, approximately 55%, versus 75% between rhesus LCV and EBV. Thus, the identical gene repertoire and high overall sequence homology make the rhesus LCV a uniquely accurate animal model for studying EBV pathogenesis.
Acknowledgments
We acknowledge members of the Massachusetts General Hospital DNA Sequencing Core (Harry Orf, Brian Seed, Dan Stetson, and David Levin) and the Brigham & Women’s Hospital Genetics Core (David Beier) for assistance with and performance of high-throughput DNA sequencing. We thank Elliott Kieff for valuable advice.
This work was supported by grants from the U.S. Public Health Service (CA68051) and the American Heart Association.
REFERENCES
- 1.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211. [DOI] [PubMed] [Google Scholar]
- 3.Blake, N. W., A. Moghaddam, P. Rao, A. Kaur, R. Glickman, Y. G. Cho, A. Marchini, T. Haigh, R. P. Johnson, A. B. Rickinson, and F. Wang. 1999. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J. Virol. 73:7381–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borza, C. M., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus recombinant lacking expression of glycoprotein gp150 infects B cells normally but is enhanced for infection of epithelial cells. J. Virol. 72:7577–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, Y., J. Ramer, P. Rivailler, C. Quink, R. L. Garber, D. R. Beier, and F. Wang. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc. Natl. Acad. Sci. USA 98:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, Y. G., A. V. Gordadze, P. D. Ling, and F. Wang. 1999. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J. Virol. 73:9206–9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., and K. Lekstrom. 1999. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J. Virol. 73:7627–7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franken, M., O. Devergne, M. Rosenzweig, B. Annis, E. Kieff, and F. Wang. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, W. L., B. Starnes, M. W. White, and R. Mahalingam. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123–130. [DOI] [PubMed] [Google Scholar]
- 10.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427–433. [DOI] [PubMed] [Google Scholar]
- 11.Heller, M., P. Gerber, and E. Kieff. 1982. DNA of herpesvirus pan, a third member of the Epstein-Barr virus-herpesvirus papio group. J. Virol. 41:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller, M., and E. Kieff. 1981. Colinearity between the DNAs of Epstein-Barr virus and herpesvirus papio. J. Virol. 37:821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 70:2049–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu, D. H., R. de Waal Malefyt, D. F. Fiorentino, M. N. Dang, P. Vieira, J. de Vries, H. Spits, T. R. Mosmann, and K. W. Moore. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830–832. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, H., Y. G. Cho, and F. Wang. 2000. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J. Virol. 74:5921–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini, A., B. Tomkinson, J. I. Cohen, and E. Kieff. 1991. BHRF1, the Epstein-Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J. Virol. 65:5991–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, W. L., C. Yim, E. Gustafson, T. Graf, D. R. Sage, K. Hanify, L. Williams, J. Fingeroth, and R. W. Finberg. 1999. Epstein-Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J. Virol. 73:5181–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moghaddam, A., M. Rosenzweig, D. Lee-Parritz, B. Annis, R. P. Johnson, and F. Wang. 1997. An animal model for acute and persistent Epstein-Barr virus infection. Science 276:2030–2033. [DOI] [PubMed] [Google Scholar]
- 21.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339–346. [DOI] [PubMed] [Google Scholar]
- 24.Peng, R., A. V. Gordadze, E. M. Fuentes Panana, F. Wang, J. Zong, G. S. Hayward, J. Tan, and P. D. Ling. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raab-Traub, N., T. Dambaugh, and E. Kieff. 1980. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell 22:257–267. [DOI] [PubMed] [Google Scholar]
- 26.Rao, P., H. Jiang, and F. Wang. 2000. Cloning of the rhesus lymphocryptovirus viral capsid antigen and epstein-barr virus-encoded small RNA homologues and use in diagnosis of acute and persistent infections. J. Clin. Microbiol. 38:3219–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivailler, P., C. Quink, and F. Wang. 1999. Strong selective pressure for evolution of an Epstein-Barr virus LMP2B homologue in the rhesus lymphocryptovirus. J. Virol. 73:8867–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruf, I. K., A. Moghaddam, F. Wang, and J. Sample. 1999. Mechanisms that regulate Epstein-Barr virus EBNA-1 gene transcription during restricted latency are conserved among lymphocryptoviruses of Old World primates. J. Virol. 73:1980–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, and A. Rickinson. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soike, K. F. 1992. Simian varicella virus infection in African and Asian monkeys: the potential for development of antivirals for animal diseases. Ann. N. Y. Acad. Sci. 653:323–333. [DOI] [PubMed] [Google Scholar]
- 32.Strockbine, L. D., J. I. Cohen, T. Farrah, S. D. Lyman, F. Wagener, R. F. DuBose, R. J. Armitage, and M. K. Spriggs. 1998. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 72:4015–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaminathan, S., R. Hesselton, J. Sullivan, and E. Kieff. 1993. Epstein-Barr virus recombinants with specifically mutated BCRF1 genes. J. Virol. 67:7406–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203–213. [DOI] [PubMed] [Google Scholar]
- 35.Wang, F., P. Rivailler, P. Rao, and Y. Cho. 2001. Simian homologues of Epstein-Barr virus. Phil. Trans. R. Soc. Lond. B Biol. Sci. 356:489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi’s sarcoma-associated herpesvirus. J. Exp. Med. 190:827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]