Abstract
The Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA-3C) regulates virus and cell genes and is essential for EBV-mediated transformation of primary B lymphocytes. EBNA-3C associates with the cellular DNA sequence-specific transcription factors RBP-Jκ and PU.1 and coactivates the EBV LMP1 promoter with EBNA-2 in BL2 and Raji cells under conditions of restrictive growth. We now find that EBNA-3C is similar to EBNA-LP in coactivating the LMP1 promoter with EBNA-2 in non-EBV-infected Burkitt lymphoma cells under conditions of maximal cell growth, whereas the EBV Cp promoter is repressed under the same conditions. EBNA-3A and EBNA-3B coactivation are at most 40% that of EBNA-3C. The RBP-Jκ binding sites of EBNA-2 and the LMP1 promoter are not required for EBNA-3C coactivation, whereas the PU.1 site in the LMP1 promoter is required for EBNA-2-mediated activation and EBNA-3C coactivation. EBNA-3C amino acids (aa) 365 to 545, including most of the previously identified repression domain (M. Bain, R. J. Watson, P. J. Farrell, and M. J. Allday, J. Virol. 70:2481–2489, 1996), are necessary and sufficient for coactivation with wild-type EBNA-2. EBNA-3C can also coactivate with the EBNA-2 acidic activating domain; this activation does not require aa 343 to 545. These data indicate that there are at least two mechanisms by which EBNA-3C coactivates the LMP1 promoter with EBNA-2. Of the proteins that interact with EBNA-3C in a yeast two-hybrid screen, only the ubiquitin-like proteins SUMO-1 and SUMO-3/hSMT3B map to aa 365 to 545, implicating these molecules in EBNA-3C coactivation. In addition, SUMO-1 associates at a high level with EBNA-3C in lymphoblasts. Promoter coactivation by EBNA-3C is likely to be important in ensuring adequate levels of LMP1, while inhibition of the EBNA-Cp promoter under the same conditions prevents uncontrolled up-regulation of EBNA expression from a positive-feedback loop.
Epstein-Barr virus (EBV) infection causes B-lymphocyte proliferative disorders in immunocompromised patients and is etiologically implicated in Burkitt lymphoma (BL), Hodgkin’s disease, and nasopharyngeal carcinoma. EBV infection of B lymphocytes results in the expression of six EBV-encoded nuclear proteins (EBNA-LP, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-1) and two integral membrane proteins (LMP1 and LMP2) and continuous cell proliferation as lymphoblastoid cell lines (LCLs) (25). In B-lymphocyte infection, EBNA-LP and EBNA-2 are the first EBV genes expressed. EBNA-2 activates transcription of cell and viral genes by self-associating (17), by interacting with the cellular sequence-specific DNA-binding proteins RBP-Jκ/CBF-1 (13, 18) and PU.1/Spi1 (21), and by recruiting TFIIB, TFIIH, TAF40, p100, and p300/CBP to its acidic activating domain (51–53, 60). EBNA-LP is a strong coactivator of transcription mediated by EBNA-2 or the EBNA-2 acidic domain (16, 36). EBNA-2 and EBNA-LP coactivate specific cell and viral promoters, including the viral LMP1 and the Cp and Wp EBNA promoters. EBNA promoter up-regulation results in some EBNA transcripts being spliced to encode EBNA-3A, EBNA-3B, EBNA-3C, or EBNA-1. The EBNA-3 proteins all bind to RBP-Jκ/CBF-1 and can interfere with its association with EBNA-2 and cognate DNA, limiting EBNA-2 up-regulation of viral and cellular promoters (31, 39, 42, 43, 55). Reverse genetic EBV recombinant analyses indicate that EBNA-3C, EBNA-3A, EBNA-LP, EBNA-2, and LMP1 are the critical EBV genes for conversion of B cells into LCLs (25).
EBNA-3C is 992 amino acids (aa) in length and contains an RBP-Jκ-binding domain, a putative leucine zipper, proline- and proline-glutamine-rich domains, and domains which mediate repression or activation when fused to the Gal4 DNA-binding domain (DBD) (Fig. 1A) (3, 31, 39). Several EBNA-3C-interacting proteins have been identified, including TBP, HDAC1, DP103, prothymosin-α, and Nm23-H1 (3, 5, 14, 40, 49). The biological significance of these interaction partners remains to be fully elucidated.
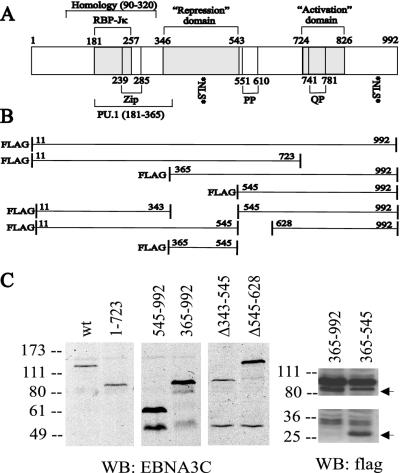
FIG. 1.
Schematic diagram of wild-type EBNA-3C and deletion mutants used in these experiments. (A) aa 90 to 320 have the highest level of identity (22 to 27%) among the EBNA-3 proteins. The RBP-Jκ-binding site (aa 181 to 257), a PU.1-binding region (aa 181 to 365) (64), and a putative leucine zipper (aa 239 to 285) are indicated. The repression (aa 346 to 543) and activation (aa 724 to 826) domains were defined based on their effects on a Gal4-responsive promoter when fused to the Gal4 DBD (3, 31). Repeats containing prolines (PP) and glutamine-proline residues (QP) are noted, as are two potential nuclear localization signal sequences (NLS). (B) EBNA-3C deletion constructs utilized in this study. (C) Immunoblots of lysates from 12 million BJAB cells that had been transfected 40 to 48 h previously with 1 μg of pSG5 expression vector for the indicated EBNA-3C deletion construct. The left panel used the EBNA-3C-specific A10 monoclonal antibody, while the right panel used M2 and M5 monoclonal antibodies against the Flag epitope tag. The top arrow indicates the position of Flag-EBNA-3C aa 363 to 992, which migrates just below a background band present in both lanes, and the bottom arrow indicates the position of Flag-EBNA-3C aa 365 to 545. WB, Western blot; wt, wild type.
EBNA-3C has had direct up-regulatory effects on the cellular CD21 promoter (58), down-regulatory effects on the viral Cp promoter (39), and down- or up-regulatory effects on the viral LMP1 promoter (31, 42, 64). Precise LMP1 promoter regulation is important because LMP1 levels are critical for cell activation, adhesion, autocrine growth factor secretion, and survival, but excessive LMP1 levels can be cytostatic (15, 24, 47, 56, 57). As a consequence of deletion of the EBNA-3C gene, LMP1 levels fall in Raji BL cells when the cells reach saturation, but EBNA-3C expression in Raji cells increases LMP1 levels at cell saturation (1, 2). EBNA-3C also potentiates EBNA-2 activation of the LMP1 promoter in non-EBV-infected BL2 cells under specific cell growth conditions and weakly activates the LMP1 promoter when expressed alone (31). More recently, EBNA-3C and EBNA-2 coactivated an enhancer composed of several PU.1/Spi1 sites in BL2 cells, and EBNA-3C aa 181 to 365 were found to interact in vitro with PU.1/Spi1 (64). However, in BL2 cells under slightly different feeding conditions and in the EBV-negative cell line Louckes, EBNA-3C repressed EBNA-2 activation of the LMP1 promoter (31). These latter effects are attributed to EBNA-3C competition with EBNA-2 for interaction with RBP-Jκ. The experiments reported here further investigate EBNA-3C regulation of the LMP1 and EBNA Cp promoters in the EBV-negative BJAB BL cell line.
MATERIALS AND METHODS
Cells and cell culture.
BJAB is an EBV-negative BL cell line. Cells were cultured in RPMI 1640 medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum (HyClone) and 2 mM glutamine (HyClone) and incubated at 37°C with 5% CO2.
Antibodies.
A fusion of glutathione S-transferase (GST) to EBNA-3C aa 633 to 913 was used to generate rabbit polyclonal antiserum R1.9 (Pocono Rabbit Farm). A10 monoclonal antibody reacts with EBNA-3C (aa 682 to 686) and was previously described (33). M2 and M5 monoclonal antibodies (Sigma) are directed against the Flag epitope tag. PE2-HRP is the EBNA-2-reactive PE2 monoclonal antibody (63) directly conjugated to horseradish peroxidase (HRP).
Plasmids.
pSG5-EBNA-2 contains the entire EBNA-2 open reading frame (ORF) of EBV strain W91 under the control of the simian virus 40 (SV40) early promoter in the vector pSG5 (Stratagene) (16). pSG5-EBNA-2-SS contains point mutations which change aa W319 and W320 to serine residues (61). pGal4-EBNA-2AD is the transcriptional AD (aa 426 to 462) of EBNA-2 fused downstream of the Gal4 DBD (4). pGal4-VP16 contains the VP16 acidic domain fused to the Gal4 DBD (45).
p(−512/+72)LMP1p-Luc (H/S 4) contains two tandem copies of the HinfI-SfiI fragment of the LMP1 promoter ligated into the BglII site of pGL2-Basic (Promega) after blunting all ends with T4 polymerase. This promoter directs the production of a transcript encoding 11 aa of the LMP1 ORF followed by a stop codon, 32 nucleotides of intervening sequence, and the initiating AUG of the luciferase gene in a different reading frame. p(−236/−145)LMP1p-Luc contains three tandem copies of the −236/−145 BamHI fragment from (−236/−145)TKCAT (54) cloned into the BglII site upstream of the TATA box of pGL2-Promoter (Promega). p(−234/+40)LMP1CAT (54) contains the indicated LMP1 promoter sequence cloned into the promoterless chloramphenicol acetyltransferase (CAT) reporter plasmid, pCAT-3M. Mutant p(−234/+40)LMP1CAT reporter constructs have been previously described (21): ΔJκ is mutated at positions −217 to −219 (CAC to AGA), ΔLBF1 is mutated at positions −230 to −228 (CAC to ACA), and ΔPU.1 is mutated at positions −166 to −164 (TTC to GGA). pFR-Luc (Stratagene) contains five copies of the 17-bp Gal4-binding element upstream of a TATA box and the luciferase gene. pLuc-Cp contains eight tandem copies of the RBP-Jκ-binding site derived from the EBV Cp promoter (−430/−330), cloned from a plasmid provided by P. Ling (29) into pGL2-Promoter (Promega).
pSG5-EBNA-3C (42) contains the EBNA-3C ORF cloned into pSG5 (Stratagene). pSG5-flagEBNA-3C was constructed by excising the XbaI-EcoRI fragment from pBS-EBNA-3C, blunting the ends with T4 polymerase, and ligating it into the PmlI site of pSG5-flag. The resulting construct expresses aa 11 to 992 of EBNA-3C with an N-terminal Flag epitope tag. pSG5-flagEBNA-3A, kindly provided by A. Cooper, consists of the entire ORF of EBNA-3A (aa 1 to 944) cloned into pSG5-flag. pSG5-flagEBNA-3B was cloned in multiple steps. pSG5-EBNA-3B was constructed first by cloning the EcoRI-NotI fragment (containing the entire EBNA-3B ORF) from pBS-EBNA-3B into the pSG5 vector digested with EcoRI/NotI. The EcoRI-BlpI fragment was then excised and replaced with partially overlapping oligonucleotides containing compatible ends and an in-frame Flag epitope tag (5′-AATTCACCATGGACTACAAGGACGACGATGACA-3′ and 5′-TGAGCCACGCCTTGTCATCGTCGTCCTTGTAGT-3′).
pSG5-flagEBNA-3C11-723 was created by collapsing the BamHI fragment of pSG5-flagEBNA-3C; the enzyme cleaves in the ORF and downstream of the ORF. pSG5-flagEBNA-3C365-992 was constructed by excising the EcoRI-SpeI fragment of pSG5-flagEBNA-3C, which contains the N-terminal Flag epitope and residues upstream of aa 365, and replacing this with partially overlapping primers containing compatible ends and the Flag epitope tag (5′-AATTCTACCATGGACTACAAGGACGACGATGACAAGA-3′ and 5′-CTAGTCTTGTCATCGTCGTCCTTGTAGTCCATGGTAG-3′). pSG5-flagEBNA-3C545-992 was initially made in the pBS-XS plasmid containing the XbaI-SalI fragment of the EBV genome (genomic coordinates 98398 to 105296) by cutting out the XbaI-AatII fragment (genomic coordinates 98398 to 100079) to delete amino acid residues 1 to 545 of EBNA-3C. Partially overlapping oligonucleotides containing appropriate compatible overhangs, an EcoRI site and a downstream in-frame Flag epitope tag were inserted (5′-CTAGAATTCTACCATGGACTACAAGGACGACGATGACAAGCGACGT-3′ and 5′-CGCTTGTCATCGTCGTCCTTGTAGTCCATGGTAGAATT-3′). This plasmid was then digested with EcoRI and BsiWI (which cuts at genomic site 100328 or aa 628 of EBNA-3C), and this fragment was inserted in place of the EcoRI-BsiWI fragment of pSG5-flagEBNA-3C. To construct pSG5-flagEBNA-3CΔ343-545, PCR was performed by using Pfu polymerase (Gibco), pBS-EBNA-3C as the template, an upstream primer anchored at the BclI site of EBNA-3C (aa 29), and a downstream primer changing the BstBI site at aa 343 in the EBNA-3C ORF to an AatII site (5′-ATAAACCCTGTATTTGTGGACGTCGAAATGCGTTCTGGAT-3′ and 5′-ATGGAATCATTTGAAGGACAGGGGG-3′). Following digestion with BclI/AatII, the PCR product was ligated into pBS-XS from which the BclI-AatII fragment (EBNA-3C residues 29 to 545) had been excised, thereby deleting the nucleotides encoding aa 343 to 545. The BclI-BsiWI fragment of this construct was then used to replace the BclI-BsiWI fragment of pSG5-flagEBNA-3C. To construct pSG5-flagEBNA-3CΔ545-628, PCR was performed by using Pfu polymerase, pBS-EBNA-3C as the template, an upstream primer anchored at the SpeI site (EBNA-3C aa 365), and a downstream primer which altered the AatII site at aa 545 in the EBNA-3C ORF to a BsrGI site (5′-TGGGAGGCGCGGCTCGTTGTACACGTCGGCCTGAACGTTG-3′ and 5′-ACTGGTTTCACAAGACAATCGG-3′). The PCR product was digested with SpeI and BsrGI and ligated into pSG5-flagEBNA-3C11-992 from which the SpeI-BsiWI fragment (encoding EBNA-3C aa 365 to 628) had been removed. pSG5-flagEBNA-3C365-545 was constructed in multiple steps. pSG5-flagEBNA-3C11-992 was digested with SpeI (cuts at EBNA-3C residue 365) and XhoI (site in the vector downstream of the insert), and this fragment was replaced with the SpeI-SalI fragment from pAS-EBNA-3C1-545 (described below), thus creating pSG5-flagEBNA-3C1-545. The EcoRI-SpeI fragment, containing the Flag epitope and the amino acid residues up to position 365, was then replaced with partially overlapping oligonucleotides containing compatible ends and an in-frame Flag epitope tag (5′-AATTCTACCATGGACTACAAGGACGACGATGACAAGA-3′ and 5′-CTAGTCTTGTCATCGTCGTCCTTGTAGTCCATGGTAG-3′).
pAS-EBNA-3C11-992 was constructed by cloning the XbaI-HindIII fragment of pBS-EBNA-3C in frame with the Gal4 DBD of plasmid pAS2-CYH2 (provided by S. Elledge). Deletion mutants of EBNA-3C were constructed in pAS2-CYH2 by PCR amplification using Pfu polymerase and the appropriate upstream and downstream primers, each containing BamHI or SalI sites, respectively. Amplification products were subsequently cleaved with BamHI and SalI and inserted into pAS2-CYH2 at these sites. Primer sequences are as follows, with numbers indicating the first (for upstream primers) or last (for downstream primers) EBNA-3C amino acid encoded by the resulting amplified fragment: upstream primers N1 (5′-CGGGATCCCCATGGAATCATTTGAAGGACAGGGGG-3′), N183 (5′-CAGCCAATCCTGGCGGATCCGATATCGTACAGCAACAC-3′), N365 (5′-AGGTGAACCCAGAGGGATCCCTACTAGTGAAACGAGCAGT-3′), and N545 (5′-CGGCTTTCAACGTTGGATCCGACGTCAAAAACGAGCCG-3′) and downstream primers C183 (5′-GTCGACTCCCATGGGCCAGGATTGGCTGGGA-3′), C365 (5′-CTCATCACTGCTGTCGACACTAGTAGCATCACCTCTGG-3′), C545 (5′-GAGGCGCGGCTCGTCGACGACGTCGGCCTGAACGTTGA-3′), and C713 (5′-GGAATTCGTCGACCGGCTGTGTGGGCGACATGGAACTC-3′).
To create pGEX-SUMO-1, the BglII fragment from pACT-SUMO-1 was excised, blunted with Klenow, and ligated into the SmaI site of pGEX-2TK (Pharmacia). To make pSG5-flagSUMO-1, the BglII fragment was cloned into the HpaI site of pSG5-flag. pGEX-SUMO-3 and pSG5-flagSUMO-3 were created identically but with the BamHI fragment of pACT-SUMO-3.
Yeast two-hybrid screen and assay.
Saccharomyces cerevisiae strain Y190 was transformed with the bait plasmid pAS-EBNA-3C11-992 and selected on tryptophan-deficient media (Bio 101). After confirmation of bait protein expression by immunoblotting (data not shown), yeast cells grown from a single colony were transformed with a cDNA library (generously provided by S. Elledge) derived from an EBV-infected human B-cell line and fused to the Gal4 AD in the plasmid pACT. Transformants were selected on media lacking tryptophan, leucine, and histidine (Bio 101) and containing 25 mM 3-aminotriazole (3-AT) (Sigma). Interaction was scored by transactivation of HIS3, which permits growth on histidine-deficient media with 3-AT, and lacZ, which directs expression of β-galactosidase and was detected by filter lift assay. Plasmids containing cDNAs were isolated and retransformed into yeast bait strains expressing proteins from pAS-EBNA-3C11-992 or pAS-LMP1(CTD) (34) to confirm reproducibility and specificity. Plasmids were subsequently sequenced to identify cDNA inserts (at the core facilities of the Brigham and Women’s Hospital and the Department of Microbiology and Molecular Genetics, Harvard Medical School). Other yeast clones were subsequently subjected to PCR using primers specific for these cDNA inserts to identify plasmids without the need for isolation and sequencing.
For directed two-hybrid interaction assays, strain Y190 was transformed with the appropriate bait plasmids and single yeast colonies were confirmed for protein expression. These yeast cells were then transformed with single pACT clones containing target proteins of interest fused to the Gal4 AD. The resulting transformants were selected and screened as noted above.
Transfections.
Approximately 12 million log-phase BJAB cells were transfected in 0.4 ml of RPMI 1640-10% fetal bovine serum with a Bio-Rad GenePulser at 220 V and 960 μF. All transfected plasmids were purified twice on CsCl gradients. For reporter assays, each transfection contained 10 μg of the appropriate reporter plasmid, the indicated amounts of effector plasmids, and 0.5 μg of pGK-βgal to permit normalization for transfection efficiency. Total plasmid DNA was made constant by the addition of pSG5 vector DNA. After electroporation, cells were resuspended in 13 ml of complete medium and incubated at 37°C for 24 to 48 h.
Reporter assays.
Cells were rinsed in phosphate-buffered saline (PBS) and lysed in reporter lysis buffer (Promega) with one freeze-thaw cycle. For luciferase reporter assays, clarified lysates were measured for luciferase (Luciferase Assay system; Promega) and β-galactosidase (Galacto-Light; Tropix) activities using an Optocomp I luminometer (MGM Instruments). β-Galactosidase activity was used to standardize all results for transfection efficiency. For CAT assays, lysates were assayed for CAT activity as described previously (54). Percent acetylation was determined using a PhosphorImager and ImageQuant software (Molecular Dynamics). All reporter assay results presented are from single experiments representative of multiple independent trials or are averages derived from multiple independent repetitions.
Immunoprecipitations.
Transiently transfected BJAB cells were lysed in IP buffer (150 mM NaCl, 1% NP-40, 50 mM Tris [pH 7.4], 2 mM EDTA) containing protease inhibitors (10 μg of aprotinin per ml, 0.5 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 1 μM leupeptin) by vortexing, incubated on ice for 30 min, and centrifuged to pellet debris. Six microliters of EBNA-3C-reactive polyclonal rabbit serum R1.9 was incubated with the lysate for 1 h at 4°C, followed by binding to Gamma-Bind Sepharose beads (Amersham Pharmacia Biotech) for another hour at 4°C. The beads were washed with 1 ml of IP buffer five times. Proteins were eluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, electrophoresed through SDS-8% PAGE minigels, and analyzed by immunoblotting.
Immunoblots.
Whole-cell lysates or other samples were prepared by solubilization in SDS sample buffer and heating at 95°C for 10 min. Samples were separated on SDS-PAGE gels, transferred to 0.4-μm-pore-size nitrocellulose filters (Bio-Rad), and probed with the previously noted antibodies. EBNA-3C-reactive monoclonal antibody A10 from tissue culture cell supernatant was used undiluted, anti-Flag antibodies M2 and M5 were mixed and used at a concentration of 2 μg/ml (each), and EBNA-2-reactive PE2-HRP was used at a concentration of 0.28 μg/ml in PBS containing 5% milk. As needed, blots were subsequently incubated with HRP-conjugated anti-mouse immunoglobulin G antibodies (Amersham) at a 1:5,000 dilution in PBS containing 0.5% milk. Immunoreactive proteins were visualized by chemiluminescent detection (NEN).
GST-binding assays.
EBNA-3C was transcribed and translated from pSG5-EBNA-3C in vitro by using the TNT Coupled Reticulocyte Lysate system (Promega) in reaction mixtures containing [35S]-Express (NEN), and samples were precleared by binding to Sepharose beads (Sigma). GST-SUMO-1 and GST-SUMO-3/hSMT3B were purified on glutathione-Sepharose 4B beads (Pharmacia) following solubilization with N-lauroylsarcosine (10). Labeled EBNA-3C was incubated with bead-bound GST fusion protein in binding buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 10% glycerol, 0.1% NP-40, 0.5 mM dithiothreitol) containing protease inhibitors (10 μg of aprotinin per ml, 0.5 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 1 μM leupeptin) at 4°C for 2 h. Beads were washed five times in binding buffer, and proteins were eluted in SDS-PAGE loading buffer and separated in SDS-12% PAGE minigels. Gels were stained with Coomassie blue to visualize input GST protein levels, treated with Amplify (Amersham), and subsequently used to expose film at −70°C.
RESULTS
EBNA-3C potentiates EBNA-2 activation of the LMP1 promoter in rapidly growing lymphoblasts.
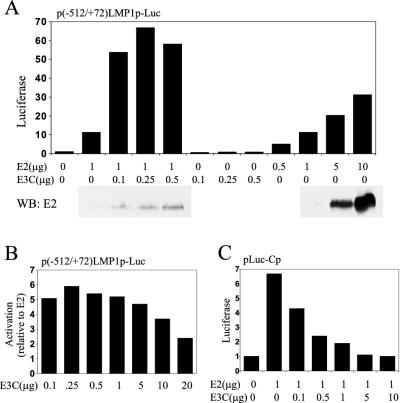
To further investigate the dependence of EBNA-3C potentiation of EBNA-2 activation on cell growth conditions, EBV-negative BJAB BL cells in log-phase growth were cotransfected with a −512/+72 LMP1 promoter fused to a luciferase reporter, pSG5-EBNA-2, and various amounts of pSG5-EBNA-3C (Fig. 2A). While 1 μg of pSG5-EBNA-2 activated the promoter 11-fold, the addition of as little as 0.1 μg of cotransfected pSG5-EBNA-3C increased luciferase activity to 50-fold. EBNA-3C alone had no activating effects (Fig. 2A). EBNA-3C did not significantly increase EBNA-2 levels (Fig. 2A, bottom panel). Even 10 μg of pSG5-EBNA-2 did not induce luciferase levels equal to those induced by 1 μg of pSG5-EBNA-2 and 0.1 μg of pSG5-EBNA-3C. Therefore, EBNA-3C has a specific synergistic effect on EBNA-2 in activating the LMP1 promoter in log-phase BJAB cells which is similar to that of EBNA-LP.
FIG. 2.
EBNA-3C potentiates EBNA-2 activation of the LMP1 promoter in BJAB, a non-EBV-infected BL cell line. (A) BJAB cells were transfected with reporter plasmid p(−512/+72)LMP1p-Luc, which has two tandem copies of the −512/+72 LMP1 promoter upstream of the luciferase gene, in addition to the indicated amounts of the pSG5-EBNA-2 (E2) and pSG5-EBNA-3C (E3C) expression vectors. Fold increase in luciferase activity is shown on the left. The panels below the graph show corresponding immunoblots using a monoclonal antibody against EBNA-2 (PE2). The results are representative of at least two independent experiments. WB, Western blot. (B) Response of p(−512/+72)LMP1p-Luc activity to 1 μg of pSG5-EBNA-2 and various increasing amounts (0.1 to 20 μg) of pSG5-EBNA-3C. The values shown are representative ratios of observed luciferase activity relative to that of EBNA-2 alone from at least two experiments. (C) Representative reporter assay using BJAB cells transfected with a reporter construct (pLuc-Cp) containing eight copies of the RBP-Jκ-binding site from the Cp promoter, 1 μg of pSG5-EBNA-2, and various amounts of pSG5-EBNA-3C. The relative luciferase values, indicated on the left in all experiments, were normalized to the β-galactosidase activity of a cotransfected plasmid, pGK-βgal.
Low levels (0.1 μg) of pSG5-EBNA-3C resulted in near-maximal coactivation with EBNA-2 (Fig. 2A and 2B), whereas amounts of pSG5-EBNA-3C in excess of 5 μg resulted in less activity (Fig. 2B). The progressive decrease with high-level EBNA-3C expression is similar to previously described squelching effects.
The effects of EBNA-3C and EBNA-2 in log-phase BJAB cells on an enhancer composed of eight RBP-Jκ sites positioned upstream of a minimal promoter and a luciferase reporter were quite different from the effects on the LMP1 promoter. EBNA2 expressed from 1 μg of pSG5-EBNA-2 resulted in 6.5-fold activation (Fig. 2C). As little as 0.1 μg of pSG5-EBNA-3C reduced the EBNA2 effect by 45% (4.3-fold activation). Larger amounts of cotransfected plasmid further inhibited EBNA-2 activation, and 5 μg of pSG5-EBNA-3C completely blocked EBNA-2 activity. In other experiments, similar results were seen with a −1425/+6 viral Cp promoter and luciferase reporter (data not shown). EBNA-3C alone had no effects on the eight RBP-Jκ-sites or −1425/+6 Cp promoter reporters (data not shown). EBNA-3C alone has been previously reported to have repressive effects on a −1425/+1 Cp promoter in DG75 EBV-negative BL cells (39). These data indicate that EBNA-3C does not activate alone or coactivate with EBNA-2 through RBP-Jκ sites under conditions in which EBNA-3C and EBNA-2 coactivate the LMP1 promoter. EBNA-3C therefore differs from EBNA-LP, which coactivates the Cp promoter or a multimerized RBP-Jκ enhancer as well as the LMP1 promoter.
EBNA-3C coactivation with EBNA-2 of the LMP1 promoter is dependent on the PU.1 but not the RBP-Jκ or AML1 site.
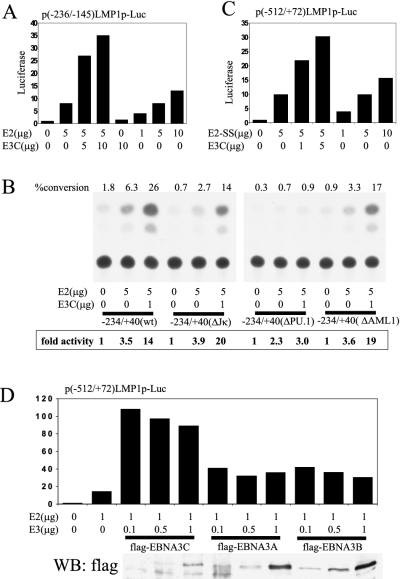
The minimal EBNA-2-responsive LMP1 promoter element (−236/−145) was activated 8-fold by 5 μg of pSG5-EBNA-2, and cotransfection with 5 μg of pSG5-EBNA-3C resulted in 27- to 35-fold activation (Fig. 3A). The EBNA-3C effect could not be reproduced by higher levels of EBNA-2 expression. Similar results were obtained with a −234/+40 LMP1 promoter CAT reporter (Fig. 3B): −234/+40 was 3.5-fold responsive to EBNA-2 and 14-fold responsive to EBNA-2 and EBNA-3C. Mutation of the RBP-Jκ-binding site (ΔJκ) did not decrease EBNA-2 activation (3.9-fold) or EBNA-3C coactivation (20-fold). Mutation of the AML1 site (ΔAML1) resulted in 3.6-fold activation with EBNA-2 and 19-fold coactivation with EBNA-2 and EBNA-3C. Consistent with previous results in BL2 cells, mutation of the PU.1 site (ΔPU.1) eliminated EBNA-2 responsiveness and EBNA-3C coactivation with EBNA-2 (Fig. 3B). These results indicate that EBNA-3C coactivation with EBNA-2 is dependent on the PU.1 site in the LMP1 promoter but not the RBP-Jκ or AML1 site.
FIG. 3.
EBNA-3C coactivation of the LMP1 promoter element is independent of RBP-Jκ. (A) Representative assay (of two repetitions) using BJAB cells transfected with the reporter plasmid p(−236/−145)LMP1p-Luc containing the −236/−145 LMP1 promoter element upstream of a minimal SV40 early promoter and the luciferase ORF, along with the indicated amounts of pSG5-EBNA-2 (E2) and pSG5-EBNA-3C (E3C). Fold increase in luciferase activity is indicated. (B) BJAB cells were transfected with CAT reporter plasmids containing wild-type (wt) or mutant LMP1 promoter sequences and the indicated amounts of pSG5-EBNA-2 and pSG5-EBNA-3C. Results are representative of two independent repetitions. Percent conversion was determined using ImageQuant software and a PhosphorImager. Fold activity is relative to that of the vector-only control, which was assigned a value of 1. (C) BJAB cells were transfected with p(−512/+72)LMP1p-Luc and the indicated amounts of pSG5-EBNA-2-SS (E2-SS) and pSG5-EBNA-3C. Results are representative of independent duplicate experiments. Fold activation of luciferase is indicated. (D) BJAB cells were transfected with p(−512/+72)LMP1p-Luc and the indicated amounts of pSG5-EBNA-2 and pSG5-flagEBNA-3C, pSG5-flagEBNA-3A, or pSG5-flagEBNA-3B. The results are representative of two independent experiments. Relative luciferase activity is indicated. The panel below the graph shows a corresponding immunoblot with monoclonal antibodies directed against the Flag epitope tag. WB, Western blot.
To test whether EBNA-3C coactivation with EBNA-2 is mediated by the high-level interaction of both proteins with RBP-Jκ, an EBNA-2 mutant incapable of binding RBP-Jκ, EBNA-2-SS, was tested for the ability to activate the LMP1 promoter and to be coactivated by EBNA-3C. The −512/+72 LMP1 promoter and luciferase reporter were activated 10-fold with 5 μg of pSG5-EBNA-2-SS and were only activated 15-fold by 10 μg of pSG5-EBNA-2-SS, whereas 5 μg of pSG5-EBNA-2-SS and 5 μg of pSG5-EBNA-3C activated the LMP1 promoter 30-fold (Fig. 3C). These data indicate that EBNA-2 association with RBP-Jκ is not critical for EBNA-3C coactivation of the LMP1 promoter.
EBNA-3C is much stronger than EBNA-3A and EBNA-3B in coactivation of the LMP1 promoter.
To determine whether EBNA-3A and EBNA-3B are also able to coactivate the LMP1 promoter with EBNA-2, BJAB cells were transfected with the −512/+72 LMP1 promoter reporter, 1 μg of pSG5-EBNA-2, and increasing amounts of expression constructs encoding Flag epitope-tagged EBNA-3A, EBNA-3B, and EBNA-3C (Fig. 3D). EBNA-2 activated the LMP1 promoter 14-fold, and pSG5-flagEBNA-3C increased EBNA-2 activation a further 7.5-fold, whereas pSG5-flagEBNA-3A or pSG5-flagEBNA-3B had, at most, a 3-fold effect. Similar levels of Flag-tagged EBNA-3A, EBNA-3B, and EBNA-3C were expressed (Fig. 3D, bottom panel). These experiments indicate that EBNA-3A and EBNA-3B can have similar but weaker coactivating effects on the LMP1 promoter.
EBNA-3C aa 365 to 545 strongly coactivate the LMP1 promoter with EBNA-2.
Parts of EBNA-3C have been implicated in transcriptional activation or repression based on their effects on a minimal promoter when fused to the Gal4 DBD or by their interaction with PU.1. EBNA-3C mutants were made to evaluate the significance of these domains in EBNA-3C coactivation (Fig. 1B). Immunoblotting of transiently transfected cell lysates indicated that the mutant proteins were expressed at levels equal to or greater than those of wild-type EBNA-3C (Fig. 1C).
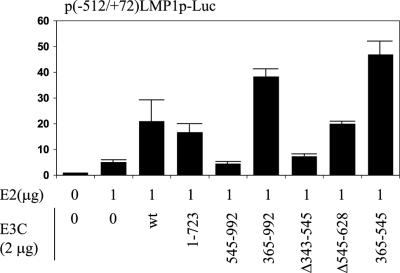
Plasmids expressing wild-type or mutant EBNA-3C were transfected into BJAB cells with pSG5-EBNA-2 and the −512/+72 LMP1 promoter reporter (Fig. 4). EBNA-2 increased luciferase activity 5-fold. Adding wild-type EBNA-3C resulted in 21-fold overall activation. EBNA-3C aa 1 to 723, from which the putative AD had been deleted (31) (Fig. 2A), had 73% wild-type EBNA-3C activity, indicating that the previously identified AD is a small component of the coactivation effect. Surprisingly, EBNA-3C aa 365 to 992, from which the N-terminal amino acids implicated in interaction with RBP-Jκ and PU.1 had been deleted, coactivated the LMP1 promoter with EBNA-2 nearly twice as effectively as wild-type EBNA-3C. Further N-terminal deletion of residues 365 to 545, which constitute most of a repression domain, resulted in complete loss of coactivation (EBNA-3C aa 545 to 992) (Fig. 4). The importance of EBNA-3C aa 343 to 545 for coactivation was also evident in experiments in which EBNA-3C aa 343 to 545 had been deleted, in which case there was almost no coactivating effect (Fig. 4). In contrast, the deletion of aa 545 to 628 resulted in 95% wild-type coactivation. These data show that aa 365 to 545 of EBNA-3C are necessary for coactivation of EBNA-2 at the LMP1 promoter and that aa 1 to 365 and 724 to 992 have small negative and positive effects, respectively.
FIG. 4.
Amino acid residues 365 to 545 of EBNA-3C are necessary and sufficient for coactivating activity with EBNA-2. Reporter assay in BJAB cells using p(−512/+72)LMP1p-Luc, 1 μg of pSG5-EBNA-2 (E2), and 2 μg of either pSG5-EBNA-3C (E3C) or the indicated EBNA-3C deletion construct. Relative luciferase activity is indicated. The results are averages of duplicate samples and are representative of at least three independent repetitions. Error bars indicate standard deviations.
The importance of EBNA-3C aa 365 to 545 alone was further evaluated in a coactivation assay with the −512/+72 LMP1 promoter and luciferase reporter. Surprisingly, EBNA-3C aa 365 to 545 had a twofold-higher coactivating effect than wild-type EBNA-3C (Fig. 4). Thus, EBNA-3C aa 365 to 545 are both necessary and sufficient for coactivation of the LMP1 promoter with EBNA-2.
EBNA-3C can coactivate with the EBNA-2 acidic AD (EBNA-2AD).
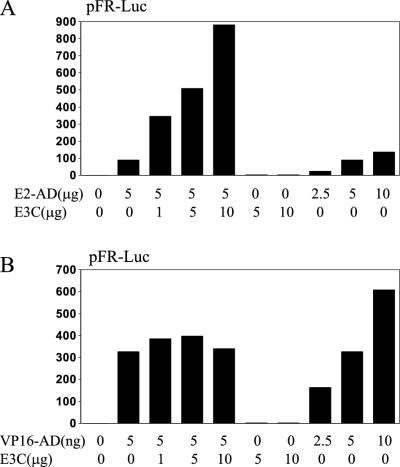
Since EBNA-LP can coactivate wild-type EBNA-2 or the EBNA-2AD (EBNA-2 residues 426 to 462), we evaluated whether EBNA-3C can also coactivate the EBNA-2AD. BJAB cells were transfected with an expression vector for the EBNA-2AD fused to the Gal4 DBD, the pFR-Luc reporter plasmid, which contains five copies of the Gal4 DNA-binding site upstream of the luciferase reporter, and indicated amounts of pSG5-EBNA-3C (Fig. 5A). While 5 μg of pSG5-Gal4-EBNA-2AD resulted in 90-fold transactivation compared to vector alone, increasing amounts of cotransfected pSG5-EBNA-3C increased reporter activity more than 9-fold relative to that of the EBNA-2AD alone. EBNA-3C alone had no effect on luciferase activity. Transfections with more pSG5-Gal4-EBNA-2AD showed that 5 μg was within the linear-response range for the effects of the EBNA-2AD on luciferase expression. EBNA-3C coactivation of the EBNA-2AD was not limited to BJAB cells and was also seen in HeLa and 293 cells (data not shown). These data indicate that EBNA-3C can coactivate with the EBNA-2AD, although this differs from coactivation with wild-type EBNA-2 in that the latter was only evident in lymphocytes (9, 59).
FIG. 5.
EBNA-3C can enhance transcriptional activity of the EBNA-2AD (aa 426 to 462) but not the VP16AD. (A) BJAB cells were transfected with reporter plasmid pFR-Luc, which contains five Gal4-binding sites, and the indicated amounts of pGal4-EBNA-2AD (E2-AD) and pSG5-EBNA-3C (E3C). The results from a representative experiment are depicted. (B) BJAB cells were transfected with pFR-Luc and the indicated amounts of pGal4-VP16 (VP16-AD) and pSG5-EBNA-3C. The results from a representative experiment are depicted. Luciferase activities are shown relative to those of reporter and empty expression vector control.
EBNA-3C coactivation was not mediated by the Gal4 DBD part of the fusion protein and specifically required the EBNA-2AD, since EBNA-3C did not coactivate the Gal4 DBD fused to the VP16 acidic AD (Fig. 5B). Transfections with increasing amounts of Gal4-VP16AD resulted in parallel increases in luciferase expression, indicating that the amount of Gal4-VP16AD expression plasmid used in these experiments was within the linear dose-responsive range. Thus, EBNA-3C is also similar to EBNA-LP in specifically potentiating the EBNA-2AD and not the VP16AD.
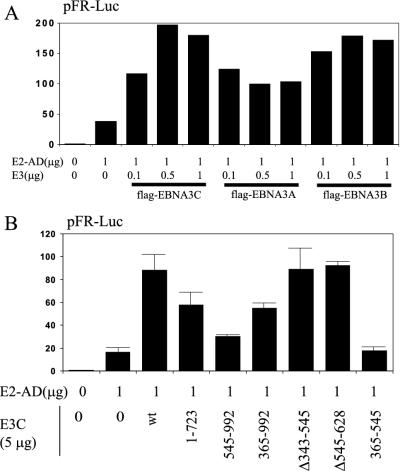
In comparison to coactivation with wild-type EBNA-2 on the LMP1 promoter, for which EBNA-3A and EBNA-3B had threefold-lower effects than EBNA-3C (Fig. 3D), flag-EBNA-3A was only twofold less active than flag-EBNA-3C in coactivation with the EBNA-2AD and flag-EBNA-3B was almost as active as flag-EBNA-3C (Fig. 6A). Thus, EBNA-3 coactivation with the EBNA-2AD appears to be different than coactivation with wild-type EBNA-2.
FIG. 6.
EBNA-3C coactivation of EBNA-2 and the EBNA-2AD are distinct activities. (A) Typical results from a reporter assay in BJAB cells using the pFR-Luc reporter construct and the indicated amounts of pGal4-EBNA-2AD (E2-AD) and pSG5-flagEBNA-3C (flag-EBNA3C), pSG5-flagEBNA-3A (flag-EBNA3A), or pSG5-flagEBNA-3B (flag-EBNA3B). (B) Reporter assay in BJAB cells using pFR-Luc, 1 μg of pGal4-EBNA-2AD, and 5 μg of either pSG5-EBNA-3C (E3C) or the indicated EBNA-3C deletion construct. The results are averages of duplicate samples and are representative of at least three independent repetitions. Error bars indicate standard deviations. Luciferase activities are shown relative to those of reporter and empty expression vector control.
EBNA-3C coactivation with the EBNA-2AD does not require aa 343 to 545, which are critical for coactivation with wild-type EBNA-2.
The EBNA-3C amino acids important for coactivation with the EBNA-2AD were identified by transfection of BJAB cells with expression vectors for wild-type EBNA-3C or deletion mutants (Fig. 6B). Full-length EBNA-3C resulted in fivefold-higher luciferase activity than did Gal4-EBNA-2AD alone. EBNA-3C aa 1 to 723 had 58% of wild-type EBNA-3C activity, and EBNA-3C with a deletion of aa 545 to 628 was similar to wild-type EBNA-3C, reminiscent of how these mutants affected coactivation of the LMP1 promoter with wild-type EBNA-2 (Fig. 6B and 4). EBNA-3C aa 365 to 992 had 54% of wild-type EBNA-3C activity in the EBNA-2AD coactivation, whereas these residues had more of an effect than wild-type EBNA-3C in coactivation with EBNA-2 at the LMP1 promoter. EBNA-3C aa 545 to 992 had 17% activity, similar to their nearly null effect in coactivation of the LMP1 promoter. However, EBNA-3C aa 343 to 545 could be deleted with minimal effect on coactivation with the EBNA-2AD, and EBNA-3C aa 365 to 545 had almost no effect alone. Thus, most of EBNA-3C, with the exception of residues 343 to 545 and 545 to 628, contribute to the EBNA-2AD coactivation. This was in marked contrast to coactivation of wild-type EBNA-2 at the LMP1 promoter, for which EBNA-3C aa 365 to 545 were both essential and sufficient. Therefore, EBNA-3C coactivates with EBNA-2 through at least two different mechanisms.
EBNA-3C aa 365 to 545 are important for binding ubiquitin-like proteins, SUMO-1 and SUMO-3.
A yeast two-hybrid screen was carried out to identify cellular proteins that interact with EBNA-3C, using EBNA-3C aa 11 to 992 fused in frame to the Gal4 DBD. Of 20 million S. cerevisiae strain Y190 cells transformed with expression plasmids for the EBNA-3C bait protein and B-lymphocyte cDNAs fused to the Gal4 acidic domain, 432 clones were picked based on selection for growth on histidine-deficient media containing 25 mM 3-AT. The lymphocyte cDNA sequences in the 156 clones that were β-galactosidase-positive in 1 h, as determined by a filter-lift assay, were identified (Table 1). Nine of these encoded the previously known EBNA-3C-interacting protein RBP-Jκ. Three clones were the ubiquitin-like protein SUMO-1, and three others were the related protein SUMO-3/hSMT3B.
TABLE 1.
Yeast two-hybrid screen results using EBNA-3C aa 11 to 992 as bait
| cDNA identity | No. of times represented/156 clones | Descriptive notes |
|---|---|---|
| PK/THBP | 45 | Pyruvate kinase-thyroid hormone binding protein |
| DEEPEST | 43 | Mitotic spindle coiled coil-related protein |
| eIF-3 (47-kDa subunit) | 11 | Translation initiation factor subunit |
| RINT-1 | 10 | Rad50-interacting protein 1 |
| RBP-Jκ | 9 | Transcription factor, known EBNA-3C-interacting protein |
| hNOT-2 | 6 | Transcriptional repressor homolog |
| DP103 (DEAD-box) | 4 | Putative RNA helicase, known EBNA-2 and EBNA-3C interactor |
| RBP-MS | 4 | RNA-binding protein |
| SODD | 4 | Suppressor of death domains |
| SUMO-1 | 3 | Ubiquitin-like protein |
| SUMO-3/hSMT3B | 3 | Ubiquitin-like protein |
| HRMT1L1 | 2 | Arginine methyltransferase homolog |
| TERF2 | 2 | Telomeric repeat binding factor 2 |
| HIC p40 | 1 | I-mfa domain-containing protein regulator of gene expression |
| hTid1 | 1 | DnaJ homolog |
| CPAP | 1 | Centrosomal P4.1-associated protein |
| Unknown | 5 | 3 cDNAs of unknown function |
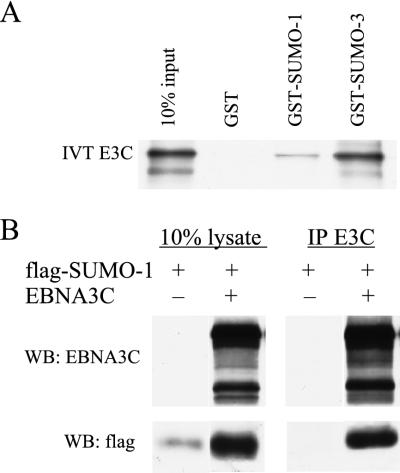
To confirm the interaction of EBNA-3C with SUMO-1 and SUMO-3, the binding of EBNA-3C to GST-SUMO and in vivo association of EBNA-3C with SUMO-1 were assessed. First, GST and GST fusions with SUMO-1 and SUMO-3 were produced in bacteria, purified on glutathione-coupled beads, and tested for binding of in vitro-translated EBNA-3C. EBNA-3C did not bind to GST alone. Approximately 1 to 2% of input EBNA-3C bound to GST-SUMO-1, while 10% of input EBNA-3C bound to GST-SUMO-3 (Fig. 7A). Expression vectors encoding Flag-tagged SUMO-1 and SUMO-3 were constructed. Flag-SUMO-1 was detected with Flag antibody in whole-cell lysates (data not shown) or in NP-40 extracts. Cotransfection of Flag-SUMO-1 and EBNA-3C expression vectors into BJAB cells resulted in increased accumulation of Flag-SUMO-1 (Fig. 7B). However, Flag-SUMO-3 expression was difficult to detect despite many attempts. Cotransfection of expression vectors for Flag-SUMO-3 and EBNA-3C did not enable Flag-SUMO-3 detection. Immunoprecipitation of EBNA-3C from lysates of BJAB cells cotransfected with Flag-SUMO-1 and EBNA-3C expression vectors resulted in precipitation of about 15% of the EBNA-3C and about 15% of the Flag-SUMO-1, based on comparisons with the cell lysate input lanes (Fig. 7B). Reciprocal immunoprecipitations were not interpretable because of a high EBNA-3C background with Flag antibody immunoprecipitation. Thus, most of the free SUMO-1 in overexpressing BJAB cells is associated with EBNA-3C, and EBNA-3C stabilizes SUMO-1.
FIG. 7.
EBNA-3C, which binds ubiquitin-like proteins SUMO-1 and SUMO-3/hSMT3b in yeast, can interact with these proteins in vitro and in cells. (A) GST-binding assay with the indicated GST fusion protein and in vitro-transcribed-translated [35S]-labeled EBNA-3C (IVT E3C). The first lane contains 10% of the amount of input EBNA-3C applied to each binding reaction, and subsequent lanes show the results of binding to the indicated GST fusion proteins. Coomassie staining confirmed that the GST control protein was used at a >5-fold excess relative to the GST fusion proteins (data not shown). (B) BJAB cells were transiently transfected with 10 μg of pSG5-EBNA-3C (EBNA3C) and/or pSG5-flagSUMO-1 (flag-SUMO-1) as indicated. Immunoprecipitations (IP) from these cell lysates were performed using rabbit antiserum reactive against EBNA-3C (R1.9), followed by immunoblotting with EBNA-3C-reactive monoclonal antibody (A10) and anti-Flag monoclonal antibodies (M2/M5). WB, Western blot.
To identify the EBNA-3C residues responsible for interaction with SUMO-1 and SUMO-3, EBNA-3C deletion mutant ORFs were fused in frame with the Gal4 DBD for use in yeast two-hybrid assays. Yeast clones expressing wild-type or mutant EBNA-3C proteins were transformed with plasmids containing SUMO-1 or SUMO-3 fused in frame with the Gal4 AD. The outcomes of the resulting β-galactosidase assays are shown in Table 2. SUMO-1 and SUMO-3 interacted with wild-type EBNA-3C and with EBNA-3C residues 1 to 545, 1 to 713, 183 to 713, or 365 to 713. Interactions were not evident with EBNA-3C residues 1 to 365 or 545 to 713. All of the EBNA-3C constructs which contain EBNA-3C aa 181 to 257 interacted strongly with RBP-Jκ, demonstrating the appropriate protein production and folding for at least that portion of the mutant proteins. These data indicate that EBNA-3C aa 365 to 545 are necessary for interaction with SUMO-1 and SUMO-3. Similar identification of the EBNA-3C site of interaction with each of the other cDNAs listed in Table 1 (data not shown) indicated that SUMO-1 and SUMO-3 are unique in their dependence on EBNA-3C residues 365 to 545 for interaction.
TABLE 2.
Directed yeast two-hybrid assays
| Target protein | Encoded amino acids of bait plasmid pAS-EBNA-3Ca | Result of reporter gene assay forb:
|
|
|---|---|---|---|
| His-deficient growth | β-Gal activity | ||
| RBP-Jκ | 1–183 | − | − |
| 1–365 | + | + | |
| 1–545 | + | + | |
| 1–713 | + | + | |
| 11–992 | + | + | |
| 183–713 | + | + | |
| 365–713 | − | − | |
| 545–713 | − | − | |
| SUMO-1 | 1–183 | − | − |
| 1–365 | − | − | |
| 1–545 | + | + | |
| 1–713 | +/− | + | |
| 11–992 | + | + | |
| 183–713 | +/− | + | |
| 365–713 | + | + | |
| 545–713 | − | − | |
| SUMO-3 | 1–183 | + | − |
| 1–365 | − | − | |
| 1–545 | +/− | − | |
| 1–713 | + | +/− | |
| 11–992 | + | + | |
| 183–713 | + | + | |
| 365–713 | + | + | |
| 545–713 | − | − | |
Y190 yeast cells were confirmed for expression of the EBNA-3C polypeptides fused to the Gal4 DBD by immunoblotting (data not shown) and subsequently transformed with the listed target proteins (fused to the Gal4 AD).
Reporter gene activities were determined as described in Materials and Methods. His-deficient growth indicates growth in the absence of histidine and in the presence of 25 mM 3-AT. β-Gal activity indicates β-galactosidase activity as determined by filter-lift assays.
DISCUSSION
Recombinant EBV reverse genetic experiments indicated that EBNA-3C is critical for transformation of human B lymphocytes into LCLs, and the experiments reported here provide new insight into the role of EBNA-3C in regulating transcription in human B lymphoblasts. Based on previous experiments and these new data, we propose that EBNA-3C is distinguished among EBNA-3A, EBNA-3B, and EBNA-3C in the strength of coactivation of the LMP1 promoter with EBNA-2. Importantly, even small amounts of EBNA-3C are effective in coactivation of the LMP1 promoter with EBNA-2. The greater potency of EBNA-3C in coactivation may explain why EBNA-3A and EBNA-3B cannot substitute for EBNA-3C in reverse genetic studies. Although distinctive in coactivation, EBNA-3C is similar to EBNA-3A and EBNA-3B in cooperatively down-modulating EBNA-2-mediated activation of the Cp promoter.
Coactivation of the LMP1 promoter and partial down-modulation of the repression of the Cp enhancer requires 10-fold less EBNA-3C than EBNA-2 expression vector for near-maximal effects. Because the SV40 early promoter effected expression of both genes, lower levels of EBNA-3C expression vector probably result in differentially less EBNA-3C transcript. EBNA-3C mRNAs are among the least abundant viral messages (25), and the lower ratio of EBNA-3C to EBNA-2 likely mimics the relative expression levels of EBNA-3C and EBNA-2 in EBV LCLs.
In initial EBV infection of B lymphocytes, EBNA-2 and EBNA-LP are the first viral genes expressed (25). EBNA-2 associates with RBP-Jκ and PU.1 to activate the LMP1 and LMP2 promoters (21) and with RBP-Jκ and AUF1 to activate the Cp promoter (11). EBNA-LP strongly coactivates with EBNA-2 on both promoters, and EBNA-LP and EBNA-2 coactivation maps to the EBNA-LP repeat domain (16, 36, 37) and the EBNA-2 acidic domain (16). Since EBNA-LP increases the activity of the EBNA-2 acidic domain, EBNA-LP is likely to globally coactivate EBNA-2 at viral as well as cell promoters, including the c-myc (22), CD21 (58), and CD23 (59) promoters. Up-regulation of the EBNA Cp and Wp promoters leads to higher-level transcription extending through the EBNA-2 polyadenylation site to include the DNA encoding EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-1. These longer transcripts are differentially spliced to yield EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-1 mRNAs and proteins. Previous experiments and the data described here indicate that EBNA-3C, and to a lesser extent EBNA-3A and EBNA-3B, contribute to EBNA-2-mediated activation of the LMP1 promoter (31, 64). Since the LMP2A and B promoters appear to be similar to LMP1, EBNA-3C, EBNA-3A, and EBNA-3B are likely to have similar effects on these promoters (27). EBNA-1 may also be involved in further up-regulation of the Cp and LMP1 promoters (38, 41, 50).
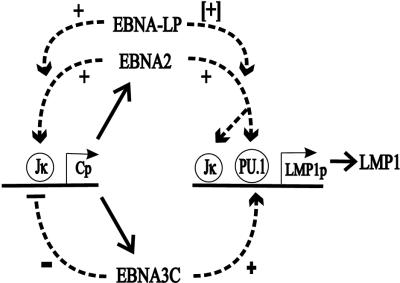
In up-regulating the LMP1 promoter and down-regulating the Cp promoter, EBNA-3C, EBNA-3A, and EBNA-3B differ from EBNA-LP and EBNA-1, which have positive effects on both promoters. LMP1, the major EBV oncogene, is a key mediator of viral effects on cell transformation, activation, adhesion, and survival; and appropriate LMP1 levels are critical to these effects (56, 57). While EBNA-3C, EBNA-LP, and EBNA-1 may enhance EBNA-2 activation of the LMP1 promoter to ensure adequate levels of LMP1, excessive levels of LMP1 are cytostatic (15, 24, 47, 56). Because the Cp promoter transcribes all of the EBNAs including EBNA-2 and EBNA-LP, coordinated repression of the Cp promoter by EBNA-3A, EBNA-3B, and EBNA-3C indirectly down-modulates LMP1 promoter activity. Therefore, through differential effects on EBNA-2-mediated transactivation, EBNA-3A, EBNA-3B, and EBNA-3C provide fine-tuning of LMP1 levels (Fig. 8).
FIG. 8.
Proposed roles of EBNA-3C in regulating EBNA-2 transactivation activity in EBV-infected B lymphocytes. The Wp EBNA promoter directs initial transcription of EBNA-2 and EBNA-LP mRNAs, which are polyadenylated at a site downstream of the EBNA-2 ORF. EBNA-2 activates the Cp promoter and probably also up-regulates the Wp promoter, resulting in higher-level transcription through EBNA-LP and EBNA-2 and transcription of EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-1 mRNAs. EBNA-2, EBNA-LP, and EBNA-3C collaborate to coactivate the LMP1 promoter and to sustain LMP1 expression at the appropriate level under all conditions of cell growth. LMP1 has a key role in LCL transformation, growth, and survival. EBNA-2, EBNA-LP, and EBNA-3C could cause runaway positive feedback up-regulation of the EBNA Cp and Wp promoters, but EBNA-3C, EBNA-3A, and EBNA-3B all contribute to repressing EBNA-2 and EBNA-LP coactivation of these promoters. Similar effects of EBNA-2, EBNA-LP, EBNA-3C, EBNA-3A, and EBNA-3B are presumed to govern transcription at other viral and cell promoters in latent EBV infection.
Several lines of evidence indicate that EBNA-3C coactivation of the LMP1 promoter may be critical for sustaining LMP1 levels in LCLs under specific cell growth conditions. First, EBNA-3C coactivation with EBNA-2 of the LMP1 promoter is evident when EBV-negative BL2 cells are fed 2 days, but not 1 day, before transfection (31). Second, LMP1 levels fall in the EBV-positive BL cell line Raji, which has a deletion of EBNA-3C, when cells are allowed to grow to maximal density. Expression of EBNA-3C in Raji cells restores LMP1 expression under these restrictive growth conditions (1, 2), indicating a unique role for EBNA-3C in sustaining LMP1 levels. Since Raji cells express normal levels of EBNA-2 and EBNA-LP, one important physiologic role of EBNA-3C may be to substitute functionally for EBNA-LP under conditions of growth restriction, when EBNA-LP coactivation with EBNA-2 may be inhibited. Such an inhibition could be envisioned to occur through several possible mechanisms. The EBNA-2AD is associated with p100, a scaffolding protein for PIM-1, and PIM-1 could connect cytokine signal transduction to the regulation of EBNA-2 activity (6, 28, 52). Alternatively, EBNA-LP and EBNA-2 coactivation of the LMP1 promoter may be regulated through differential phosphorylation of EBNA-LP (62). EBNA-LP undergoes cell cycle-specific phosphorylation and can be phosphorylated in vitro by p34(cdc2) (26). The precise mechanisms that result in down-regulation of LMP1 in growth-arrested, EBNA-3C-deficient cells remain to be established. Whatever those mechanisms are, EBNA-3C appears to be unable to fully replace EBNA-LP in coactivation, since EBNA-LP and EBNA-2 coactivate LMP1 expression from the endogenous EBV episome of Akata cells (36), whereas EBNA-3C and EBNA-2 appear to be unable to do so (data not shown).
Previous work with the LMP1 promoter has shown that the RBP-Jκ-binding sites contribute significantly to the effects of EBNA-2 but are not required, whereas the PU.1 site is essential for EBNA-2 responsiveness (21). EBNA-2 interaction with RBP-Jκ is also not essential for coactivation, because EBNA-2-SS does not associate with RBP-Jκ (61) but was readily coactivated by EBNA-3C (Fig. 3C). Further, EBNA-3C aa 365 to 992, which lack the RBP-Jκ-interacting site (aa 181 to 257), strongly coactivated the LMP1 promoter with EBNA-2, indicating that the RBP-Jκ site is not required for strong coactivation with EBNA-2 at the LMP1 promoter. EBNA-3C coactivation with EBNA-2 depends on the PU.1 site in BL2 cells, and a multimerized element from the LMP1 promoter (−181/−141) encompassing the PU.1 site sufficed for EBNA-3C-mediated coactivation with EBNA-2 (64). The data presented here confirm that EBNA-3C coactivation with EBNA-2 is dependent on the PU.1 site but not on the RBP-Jκ or AML1 site. Interestingly, PU.1 binds in vitro to EBNA-3C aa 181 to 365 (64), whereas aa 181 to 365 are not important for strong coactivation of the LMP1 promoter with EBNA-2 in transient transfection assays. Perhaps PU.1 can also interact with EBNA-3C residues located on the C-terminal side of aa 365.
Our observation that EBNA-3C coactivates the LMP1 promoter in actively growing BJAB cells has facilitated identification of the EBNA-3C components that mediate these effects. Interestingly, EBNA-3C aa 724 to 628, described as a putative AD based on their ability to activate transcription when fused to the Gal4 DBD (31), are dispensable for the majority of EBNA-3C activity in coactivation of the LMP1 promoter (Fig. 4). Therefore, a simple model proposing the recruitment of an EBNA-3C AD to the EBNA-2 complex to effect further activation of the LMP1 promoter seems unlikely.
Most intriguingly, EBNA-3C residues 365 to 545 are both necessary and sufficient for strong coactivation with EBNA-2 at the LMP1 promoter. These residues correspond closely with aa 346 to 543, which constitute a repression domain when fused to the Gal4 DBD (3). That this domain exhibits strong coactivation with EBNA-2 raises the possibility that coactivation could be due to the squelching of a highly specific inhibitor that is otherwise active at sites of EBNA-2 transcription complexes. The binding of inhibitor by this domain of EBNA-3C when it is artificially tethered to DNA results in a repressive effect on transcription. RBP-Jκ can repress transcription (8, 19) and can associate with repressor complexes containing SMRT, HDAC1, CIR, SAP30, and HDAC2 (20, 23), and these components may be targets of EBNA-3C. Alternatively, EBNA-3C aa 365 to 545 might interact biochemically with EBNA-2 or EBNA-2-PU.1 complexes, even though this is exclusive of the previously described PU.1-interacting region (aa 181 to 365). EBNA-3C aa 365 to 545 lie outside of the region of EBNA-3C with the highest level (22 to 27%) of identity among the EBNA-3 proteins (aa 90 to 320) (Fig. 1)A, and a manual alignment showed that this domain lacks obvious similarities with the comparable regions of EBNA-3A and EBNA-3B (data not shown). This may account for the greater ability of EBNA-3C to coactivate the LMP1 promoter with EBNA-2 in our assays.
EBNA-3C has some similarity with EBNA-LP in coactivating transcription of both wild-type EBNA-2 and the EBNA-2AD (aa 426 to 462). This latter activity is clearly separable from the strong coactivating effects of EBNA-3C aa 365 to 545 with wild-type EBNA-2 on the LMP1 promoter, since EBNA-3C aa 365 to 545 are not important in coactivation of the EBNA-2AD. Thus, EBNA-3C coactivates with EBNA-2 through at least two different mechanisms: one involves most of EBNA-3C except for aa 365 to 545 and the EBNA-2AD, whereas the other is primarily mediated by EBNA-3C aa 365 to 545, which have no effect through the EBNA-2AD but strongly coactivate with wild-type EBNA-2. Most likely, stable regulation of the LMP1 promoter in LCLs involves both mechanisms and consequently requires most of the EBNA-3C ORF.
Based on the critical role of EBNA-3C aa 365 to 545 in LMP1 promoter coactivation with EBNA-2, we screened a panel of EBNA-3C-interacting proteins obtained from a yeast two-hybrid screen for dependence on this region and identified the small ubiquitin-like modifiers SUMO-1 and SUMO-3/hSMT3B. EBNA-3C increased the accumulation of coexpressed SUMO-1 and associated with SUMO-1 at very high levels. SUMO-1 is covalently conjugated to a number of proteins and alters their intracellular localization and protein interactions. SUMO-1 is linked to RanGAP1 (30, 32) and PML (35, 48, 65), protects IκBα from degradation (7), and regulates the transcriptional activity of p53 (12, 44). EBNA-3C and EBNA-2 lack identifiable consensus sites for SUMO-1 modification, and we have been unable to detect SUMO-1 modified forms. Although overexpression of SUMO-1 in BJAB cells has not altered EBNA-3C-mediated LMP1 promoter coactivation (data not shown), adequate levels of endogenous SUMO-1 may already exist. EBNA-3C may be a scaffolding protein for bringing SUMO-1 to specific substrate(s) or conjugating enzyme(s), and this may be integrally involved in the effects of EBNA-3C aa 365 to 545 on transcription. Though little is known about SUMO-3, which appears to be functionally distinct from SUMO-1 (46), EBNA-3C may also function to regulate its activity. Further investigation of the roles of SUMO-1 and SUMO-3 in EBNA-3C coactivation is appropriate, since EBNA-3C aa 365 to 545 may coactivate the LMP1 promoter by effecting sumoylation or desumoylation.
Other groups have reported EBNA-3C-interacting proteins that may have potential roles in mediating transcriptional regulation by EBNA-3C. DP103, a putative RNA helicase, interacts with EBNA-2 and EBNA-3C aa 534 to 778 (14). HDAC1 interacts with residues 146 to 565 of EBNA-3C, and this was proposed to contribute significantly to the EBNA-3C-mediated repression of the Cp promoter (40). More recently, prothymosin-α interaction has been mapped to EBNA-3C aa 365 to 393 (5), and the human metastatic repressor protein Nm23-H1 has been mapped to EBNA-3C aa 365 to 992 (49). Thus, prothymosin-α and Nm23-H1 are also candidates for involvement in the EBNA-3C aa 365 to 545 coactivation with EBNA-2 at the LMP1 promoter.
In addition to regulating the EBV LMP1 and Cp promoters, EBNA-3C probably positively affects transcription of some cellular genes and negatively affects others through interactions with RBP-Jκ and PU.1. EBNA-3C has EBNA-2-independent effects on CD21 expression, and these may be mediated by RBP-Jκ or PU.1. Effects of EBNA-3C on cellular gene transcription are likely important components of the role of EBNA-3C in the transformation of B cells into LCLs. Reverse genetic studies are necessary to evaluate the significance of EBNA-3C domains, including aa 365 to 545, in EBV-mediated LCL outgrowth.
Acknowledgments
These investigations were supported by grant CA-47006 from the National Cancer Institute of the U.S. Public Health Service. E.J. received support from grant 5K12 CA77845-03 from the National Institutes of Health.
We are grateful to Steve Elledge for the yeast two-hybrid reagents, Frederick Wang for the HRP-conjugated PE2, and Andrew Cooper for the pSG5-flagEBNA-3A plasmid.
REFERENCES
- 1.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361–369. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., and E. Kieff. 1991. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J. Virol. 65:5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dash, A. B., F. C. Orrico, and S. A. Ness. 1996. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 10:1858–1869. [DOI] [PubMed] [Google Scholar]
- 7.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233–239. [DOI] [PubMed] [Google Scholar]
- 8.Dou, S., X. Zeng, P. Cortes, H. Erdjument-Bromage, P. Tempst, T. Honjo, and L. D. Vales. 1994. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol. Cell. Biol. 14:3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahraeus, R., A. Jansson, A. Sjoblom, T. Nilsson, G. Klein, and L. Rymo. 1993. Cell phenotype-dependent control of Epstein-Barr virus latent membrane protein 1 gene regulatory sequences. Virology 195:71–80. [DOI] [PubMed] [Google Scholar]
- 10.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179–187. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes-Pananá, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136–19144. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada, S., R. Yalamanchili, and E. Kieff. 2001. Epstein-Barr virus nuclear protein 2 has at least two N-terminal domains that mediate self-association. J. Virol. 75:2482–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92–95. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J. J., and S. D. Hayward. 1995. Masking of the CBF1/RBP-Jκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science 268:560–563. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaykas, A., and B. Sugden. 2000. The amino-terminus and membrane-spanning domains of LMP-1 inhibit cell proliferation. Oncogene 19:1400–1410. [DOI] [PubMed] [Google Scholar]
- 25.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511–2573. In D. Knipe, P. Howley, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Kitay, M. K., and D. T. Rowe. 1996. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J. Virol. 70:7885–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Roux, A., B. Kerdiles, D. Walls, J. F. Dedieu, and M. Perricaudet. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596–602. [DOI] [PubMed] [Google Scholar]
- 28.Leverson, J. D., P. J. Koskinen, F. C. Orrico, E. M. Rainio, K. J. Jalkanen, A. B. Dash, R. N. Eisenman, and S. A. Ness. 1998. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell 2:417–425. [DOI] [PubMed] [Google Scholar]
- 29.Ling, P. D., J. J. Ryon, and S. D. Hayward. 1993. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J. Virol. 67:2990–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maunders, M. J., L. Petti, and M. Rowe. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3C-specific monoclonal antibody. J. Gen. Virol. 75:769–778. [DOI] [PubMed] [Google Scholar]
- 34.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389–399. [DOI] [PubMed] [Google Scholar]
- 35.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puglielli, M. T., M. Woisetschlaeger, and S. H. Speck. 1996. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 70:5758–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisman, D., and B. Sugden. 1986. Transactivation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 6:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBP-Jκ. J. Virol. 70:3068–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563–564. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258. [DOI] [PubMed] [Google Scholar]
- 47.Sandberg, M. L., A. Kaykas, and B. Sugden. 2000. Latent membrane protein 1 of Epstein-Barr virus inhibits as well as stimulates gene expression. J. Virol. 74:9755–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23–H1: a molecular link to cancer metastasis. Nat. Med. 7:350–355. [DOI] [PubMed] [Google Scholar]
- 50.Sugden, B., and N. Warren. 1989. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 63:2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong, X., R. Drapkin, R. Yalamanchili, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15:4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang, S. F., F. Wang, K. M. Izumi, and E. Kieff. 1991. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J. Virol. 65:6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ–EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J. Virol. 70:5909–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840. [DOI] [PubMed] [Google Scholar]
- 57.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, F., C. D. Gregory, M. Rowe, A. B. Rickinson, D. Wang, M. Birkenbach, H. Kikutani, T. Kishimoto, and E. Kieff. 1987. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 84:3452–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634–641. [DOI] [PubMed] [Google Scholar]
- 62.Yokoyama, A., M. Tanaka, G. Matsuda, K. Kato, M. Kanamori, H. Kawasaki, H. Hirano, I. Kitabayashi, M. Ohki, K. Hirai, and Y. Kawaguchi. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): ability of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 75:5119–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O’Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080–1085. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2752. [PubMed] [Google Scholar]