Abstract
It has been postulated that retroviral recombination, like strong stop template switching, requires the RNase H activity of reverse transcriptase. To address this hypothesis, Moloney murine leukemia virus-based vectors, which were designed to test the recombination-related property of direct repeat deletion, were encapsidated in virions engineered to contain phenotypic mixtures of wild-type and RNase H catalytic site point mutant reverse transcriptase. Integrated provirus titers per milliliter were determined for these phenotypically mixed virions, and vector proviruses were screened to determine what percentage contained repeat deletions. The results revealed a steady decline in direct repeat deletion frequency that correlated with decreases in functional RNase H, with greater than fourfold decreases in repeat deletion frequency observed when 95% of virion reverse transcriptase was RNase H defective. Parallel experiments were performed to address effects of molar excesses of RNase H relative to functional DNA polymerase. These experiments demonstrated that increasing the stoichiometry of RNase H relative to the amount of functional DNA polymerase had minimal effects on direct repeat deletion frequency. DNA synthesis was error prone when directed principally by RNase H mutant reverse transcriptase, suggesting a role for RNase H catalytic integrity in the fidelity of intracellular reverse transcription.
Synthesis of a double-stranded DNA copy of the retroviral genome requires both the DNA polymerase and RNase H activities of reverse transcriptase (RT) (27). The RNase H activity, which is directed by the C-terminal one-third of Moloney murine leukemia virus (M-MuLV) RT, functions to degrade RNA from the RNA-DNA duplexes generated during viral DNA synthesis (1). M-MuLV RT’s DNA polymerase and RNase H active sites both naturally reside in separate domains of a single polypeptide (26). During reverse transcription, some nucleolytic cleavage of the template RNA by the elongating RT probably occurs (16). However, RT’s RNase H is less active than its DNA polymerase, and it is likely that most template degradation does not occur concomitant with DNA synthesis but instead results when preformed RNA-DNA duplexes are bound by nonpolymerizing RT molecules (5, 14). The natural stoichiometry of RT to template RNA in a virion is roughly 50:1 (32).
When two retroviruses coreside in a single cell, the polyproteins they encode can coassemble to generate phenotypically mixed virions with properties of both parents (31). M-MuLV virions engineered to contain phenotypic mixtures of mutant RTs—some with active DNA polymerase but no RNase H and others with defective DNA polymerase but with active RNase H—can synthesize integration-competent DNA at a low level (29). Mutant viruses that lack RNase H activity entirely do not replicate and have been shown to generate minus-strand strong-stop DNA that is retained in RNA-DNA hybrid form (26). This suggests an important role for RNase H in minus-strand strong-stop template switching.
In addition to the required minus- and plus-strand strong-stop template switches that occur during viral DNA synthesis, RT frequently performs nonrequired template switches during viral replication that can lead to genetic recombination (3, 30). If two copackaged RNA molecules are genetically distinct, intermolecular template switching between them may lead to reassortment of genetic traits in progeny proviruses (10, 11). In addition to intermolecular recombination between two copackaged RNAs, intramolecular template switching, which involves transfer of the DNA polymerase growing point from one template position to another position on the same RNA, can also occur. Such switching results in genome rearrangements, such as the deletion of direct repeats (9).
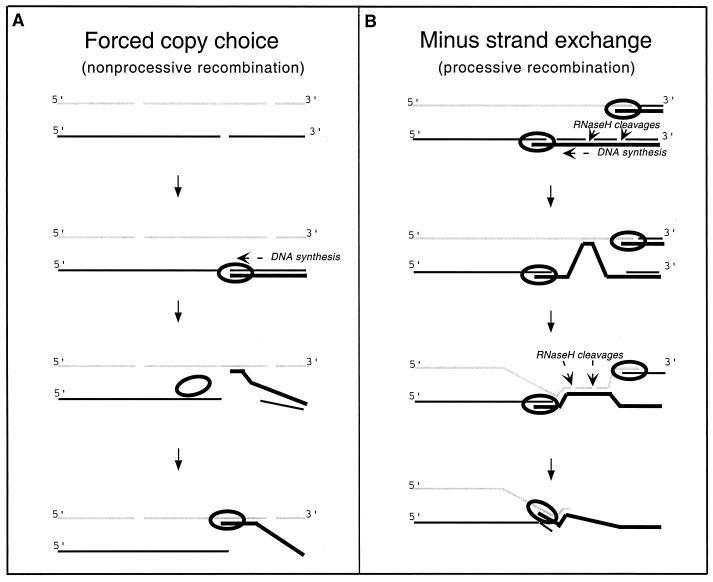
Several models have been proposed for retroviral genetic recombination (2, 15). Some suggest a requirement for RNase H-mediated degradation of the RNA paired with nascent DNA (Fig. 1). The resulting regions of single-stranded DNA are proposed to facilitate acceptor template localization by base pairing with the copackaged RNA molecule, thus allowing primer realignment onto the acceptor template (Fig. 1B) (2). Related models predict that donor template degradation by RNase H should stimulate recombination by rendering interactions between the acceptor template and primer strand more stable than those between donor template and the nascent DNA (Fig. 1A) (4). A prediction of both of these models, as well as of variants of these models, is that decreasing RNase H activity would decrease template switching frequencies.
FIG. 1.
Models for genetic recombination during retrovirus minus-strand synthesis. The two models presented here each contain components of more than one previously proposed model. These models include forced copy choice (3), those that require dissociation of the nascent DNA strand from the donor template (17), minus-strand exchange (2), and RNA-DNA-RNA intermediate models (18). The two models presented here describe recombination as processive or nonprocessive, by analogy to terminology used to describe RNA virus recombination mechanisms (12). These models were composed to demonstrate postulated transfer from broken RNA ends and putative facilitating roles of RNase H activity in genetic recombination. (A) Nonprocessive recombination is hypothesized to occur during minus-strand DNA synthesis (thick black line), where a break in the RNA template (thin black line) forces RT to continue DNA polymerization using the homologous region of the copackaged RNA (acceptor template; thin grey line) as a template. RNase H cleavage of RNA from the donor RNA-nascent DNA duplex facilitates template switching by producing a single-stranded region of DNA at the growing point of DNA synthesis which is able to base pair with the other copackaged RNA molecule (acceptor template; thin grey line). (B) During processive recombination, DNA synthesis (thick black line) proceeds farther along on one of the RNA molecules (thin black line) than the other (thin grey line). RNase H cleavage of the associated donor RNA template (thin black line) produces internal regions of single-stranded DNA which can then base pair with the other copackaged RNA molecule (acceptor template; thin grey line). Recombination results from a repositioning of the nascent DNA (primer strand) terminus onto the acceptor template strand. Although, for the sake of clarity, the models as presented describe template switching between copackaged RNAs, these models also pertain to repeat deletion.
To address whether or not retroviral recombinogenic template switching is stimulated by the RNase H activity of RT, phenotypically mixed virions were generated that contained reduced or excess functional RNase H relative to functional DNA polymerase. Template switching mediated by the various RT mixtures was scored by determining deletion frequencies of a 284-bp direct repeat within a lacZ reporter gene. These approaches were used to address the effects of limiting the amounts of functional RT in virions on virus titers and of altering the relative stoichiometry of active RNase H on direct-repeat deletion frequency.
MATERIALS AND METHODS
Plasmid construction.
pLacPuro and derivatives are M-MuLV-based vectors in which viral genes were replaced by the lacZ gene driven by the long terminal repeat promoter, followed by a puromycin resistance gene transcribed from the simian virus 40 promoter. The pLaac-117 and pLaac-284 vectors, which contain direct repeats of 117 and 284 nucleotides within lacZ, respectively, were generated as described by Pfeiffer et al. (20).
RT mutant helper plasmid derivatives were generated using standard approaches by subcloning mutant RT region fragments into the packaging-defective construct pMLV ψ− (20). The pMLV ψ− DNAP+ RH+ construct contained a D524N RNase H catalytic site mutation in the pMLV ψ− background, the pMLV ψ− DNAP− RH+ construct contained the double DD224,225NN mutation within the catalytic core of the DNA polymerase domain, and the pMLV ψ− DNAP− RH− construct contained both the DD224NN and D524N mutations (28, 29).
Cell lines and viruses.
NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum (Gibco). ET-based cell lines were grown in DMEM plus 10% fetal bovine serum (HyClone). ET is a 293T-based line that expresses ecotropic env (20). ET pLacPuro and ET pLaac-284 cells are ET cells that stably express pLacPuro or pLaac-284, respectively (20).
To generate phenotypically mixed virions, ET pLacPuro or ET pLaac-284 cells were transiently cotransfected with mixtures of pMLV ψ−, pMLV ψ− DNAP+ RH−, pMLV ψ− DNAP− RH+, and pMLV ψ− DNAP− RH− plasmid DNAs in various ratios via the calcium phosphate precipitation method as performed previously (20). The total amount of transfected plasmid (8 μg per 6-cm-diameter plate) was the same for all transfection experiments. Harvested virus-containing media were aliquotted and stored at −70°C.
Direct repeat deletion assays and error rate assays.
Assays to examine rates of tandem repeat deletion were performed as follows. Fifty, 100, or 300 μl of virus-containing medium harvested from transiently transfected ET pLaac-284 cells was incorporated in a total of 750 μl of complete medium containing 0.8 μg of hexadimethrine bromide (Polybrene) (Sigma) per ml and added to 10% confluent 3T3 cells in a 6-cm-diameter dish. After a 2-h incubation period at 37°C, 3 ml of complete medium was added to each dish. Forty-eight hours after infection, virally transduced cells were selected with puromycin, and 10 days postinfection puromycin-resistant colonies were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) using standard protocols (21).
For tandem repeat vectors, deletion frequencies were calculated as the number of blue colonies divided by the total number of puromycin-resistant colonies and were adjusted for lacZ-inactivating mutation rates. The total colony values in Table 1 represent the sum of colonies counted on six plates from each of four independent infection experiments; that is, two separate infection experiments apiece for two separate transfection and virus harvest experiments. The lacZ inactivation rates were determined by dividing the number of white puromycin-resistant colonies by the total number of colonies produced by pLacPuro when mobilized by virions with the indicated protein composition.
TABLE 1.
Effect of limiting RNase H on deletion rates
| Infectiona | Total no. of colonies | Total no. of blue colonies | Adjusted deletion rateb (%) |
|---|---|---|---|
| Mock (no DNA) | |||
| A | 0 | 0 | NA |
| B | 0 | 0 | NA |
| C | 0 | 0 | NA |
| D | 0 | 0 | NA |
| 100% WT | |||
| A | 150 | 53 | 37.7 |
| B | 159 | 58 | 39.0 |
| C | 240 | 93 | 41.4 |
| D | 464 | 203 | 46.8 |
| 50% DNAP+ RH−/50% WT | |||
| A | 304 | 60 | 20.8 |
| B | 291 | 70 | 25.5 |
| C | 174 | 38 | 23.0 |
| D | 520 | 153 | 31.1 |
| 80% DNAP+ RH−/20% WT | |||
| A | 250 | 29 | 12.6 |
| B | 180 | 31 | 18.7 |
| C | 338 | 58 | 18.7 |
| D | 407 | 59 | 15.7 |
| 90% DNAP+ RH−/10% WT | |||
| A | 83 | 12 | 16.0 |
| B | 186 | 27 | 16.0 |
| C | 337 | 34 | 11.1 |
| D | 427 | 57 | 14.7 |
| 95% DNAP+ RH−/5% WT | |||
| A | 50 | 5 | 11.6 |
| B | 98 | 11 | 13.0 |
| C | 52 | 3 | 6.7 |
| D | 293 | 22 | 8.7 |
Data from (A) transfection 1, infection 1; (B) transfection 1, infection 2; (C) transfection 2, infection 1; and (D) transfection 2, infection 2.
Deletion frequency values were adjusted to include rates of lacZ inactivation, determined as described in Materials and Methods. NA, no colonies were observed on mock-infected plates, and thus deletion rates were not applicable.
RESULTS
Generation of phenotypically mixed virions and determination of their titers.
To generate phenotypically mixed virions containing RT mixtures, ET pLaac-284 cells were transfected with various ratios of pMLV ψ−-based helper plasmids containing different RT alleles. ET pLaac-284 cells are 293T-derived cells that stably express ecotropic envelope and a replication-defective retrovirus vector that confers puromycin resistance (20).
The Laac-284 vector that is constitutively expressed by ET pLaac-284 cells is a Ψ+ packaging-competent RNA that becomes encapsidated when viral proteins are provided in trans by helper constructs. The mutant helper constructs contained the following RT alleles: pMLV ψ− DNAP− RH+ contained asparagine substitutions in each of two aspartic acid residues (224 and 225), which are located in the DNA polymerase domain’s highly conserved YxDD motif; the mutant designated DNAP+ RH− contained a substitution (D524N) in a residue essential for RNase H activity; and the DNAP− RH− mutant contained both these mutations (28, 29). When tested in the context of intact proviral clones, all of these RT mutants are replication defective and display no detectable virus spread after prolonged passage of infected or transfected cells (29).
The wild-type plus mutant helper plasmid cotransfection protocol used here should result in the production of virions that contain a phenotypic mixture of viral proteins and have packaged a vector RNA which harbors the puromycin resistance gene, provided assembly of polyproteins is random. This is because we have previously determined that our standard transfection protocol results in cotransfection of each transfected cell by significantly more than 20 plasmids (23), and each retrovirus particle is believed to assemble from nearly 2,000 Gag precursors, randomly selected for coassembly from among all precursors expressed in an individual cell, and to contain roughly 100 molecules of RT (32).
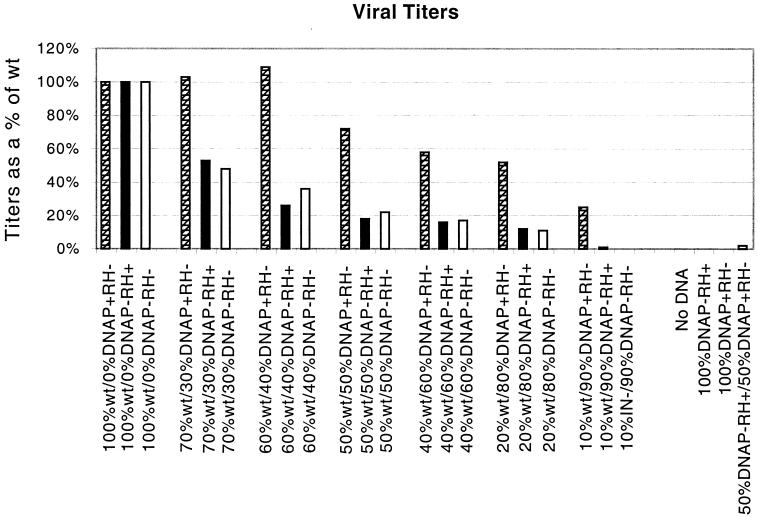
Virions harvested from cotransfected cells were used to infect 3T3 cells. Infected cells were selected with puromycin, resulting colonies were counted, and virus titers per milliliter of virus-containing medium were determined and expressed as a percentage of the titer for virions with 100% wild-type RT (Fig. 2). Virions containing only wild-type RT yielded titers of approximately 2,000 puromycin-resistant colonies per ml, while medium from mock-infected cells or media containing virions with only DNAP− RH−, DNAP− RH+, or DNAP+ RH− RTs were apparently unable to synthesize viral DNA. (Because a single colony either was or was not observed when 100 or 300 μl was assayed, values ranged from ≈10 per ml to <3 per ml.)
FIG. 2.
Virus titers (CFU per milliliter) of phenotypically mixed and unmixed virions expressed as a percentage of titers of virions containing 100% wild-type (wt) RT. See Materials and Methods for descriptions of the transfection and infection approaches. Hatched bars represent values for phenotypically mixed virions that include wild-type and DNAP+ RH− RT, solid bars represent values for virions with wild-type and DNAP− RH+ RT, and open bars represent wild-type plus DNAP− RH− RT.
When phenotypically mixed virions engineered to contain equal amounts of DNAP+ RH− and DNAP− RH+ RT were tested, low puromycin-resistant colony titers were observed that were reproducibly about 10 times higher than those of virions containing DNAP+ RH− or DNAP− RH+ RT alone. That is, within each infection experiment, values for the combined mutants were about 10-fold higher than titers for mock-infected cells, with an average of roughly 30 colonies per ml for virions with equal amounts of DNAP+ RH− and DNAP− RH+ RT (see values at the far right in Fig. 2. Note that 50%/50% values presented further to the left in this figure are for mixtures of one RT mutant with wild-type RT. Note that 50%/50% values presented further to the left in this figure are for mixtures of one RT mutant with wild-type RT.
This result confirms previous findings that certain pairs of defective M-MuLV RTs can complement and synthesize integration-competent DNAs at a low level and is consistent with complementation experiments recently performed with human immunodeficiency virus type 1 (HIV-1) RT mutants (13, 29). However, the colony-forming titers of the DNAP+ RH− plus DNAP− RH+ mixed virions were too low to allow accurate assessment of template switching properties. Thus, subsequent experiments designed to test template switching effects of RNase H limitation were performed using phenotypically mixed virions with mixtures of wild-type and RNase H and/or DNA polymerase mutant RTs.
Results of experiments that addressed titer effects of reducing the amount of functional RNase H or DNA polymerase activity by coexpression of wild-type and mutant RTs are also displayed in Fig. 2. Values shown are averages from two or more independent transfection and infection experiments for each wild-type to mutant helper ratio. Virions containing 50% wild-type and 50% DNAP+ RH− RT produced titers only modestly reduced relative to those with 100% wild-type RT. The apparent modest titer stimulation values for virions with 70 or 60% wild-type plus 30 or 40% DNAP+ RH− RT were within 1 standard deviation of the wild-type value and thus presumably represent random sampling variation. As the ratio of mutant RT to wild-type RT was increased so that more than half the RT was mutant, titers decreased further, with more severe titer reductions observed for mixed virions with limited DNA polymerase activity than for virions limited in RNase H.
Effect of limiting RNase H on template switching rates.
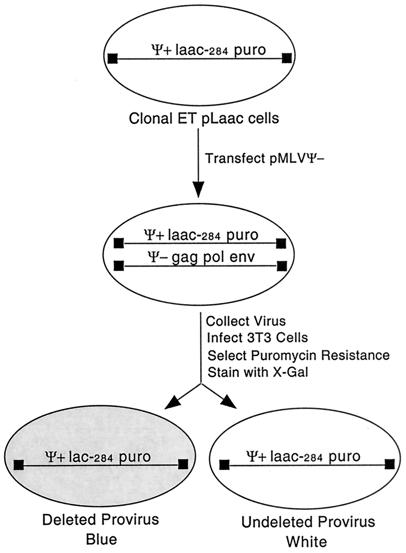
The tandem repeat deletion assay introduced in Fig. 3 was used to study the effects of limiting functional RNase H on template switching rates. This assay uses a member of the pLaac series of retrovirus vectors, which are derivatives of the pLacPuro vector that contain internal direct repeats within a lacZ cassette (20. If the repeat within laacZ (so named to indicate the internal repeat) remains undeleted during reverse transcription, transduced cells will remain unstained when incubated with X-Gal. In contrast, precise deletion of the direct repeat within laacZ during reverse transcription causes transduced cells to stain blue in the presence of X-Gal. Thus, repeat deletion frequency can be estimated by determining the ratio of blue to total puromycin-resistant colonies (Fig. 3). The repeat within lacZ studied here was 284 nucleotides long.
FIG. 3.
Schematic overview of the experimental approach and the assay used to assess direct repeat deletion frequencies that is described in the text.
Phenotypically mixed virions were prepared by cotransfecting ET pLaac-284 vector-expressing cells with a mixture of the wild-type RT helper, pMLV ψ−, and pMLV ψ− DNAP+ RH− helper plasmids. Virus was harvested and used to infect fresh cells. Ten days postinfection, puromycin-resistant colonies were stained, and the number of blue and unstained colonies was counted.
Data from several experimental repetitions of assays to determine Laac-284 repeat deletion frequencies, performed as described in the Materials and Methods section, are presented in Table 1. Virions with mixtures of wild-type and RNase H mutant RTs were observed to generate blue-staining colonies less frequently than virions that contained only wild-type RT. For virions containing 50, 20, 10, and 5% wild-type RT, with the remainder of the RT molecules in those virions containing the RNase H-inactivating mutation, blue colonies were observed at 61.5, 39.5, 34.4, and 22.9%, respectively, the frequency that they were observed for virions with 100% wild-type RT. Note that different volumes of each mixed-virion-containing media, empirically determined to yield well-separated colonies, were used to infect cells and generate the data in Table 1. Thus, relative titers cannot be inferred from the colony counts reported here; that information is reported in Fig. 2.
Direct repeat deletion rates were adjusted to account for the frequency of lacZ gene inactivation, because such mutations might lead to underestimation of repeat deletion frequencies. lacZ inactivation rates were determined by cotransfecting ET pLacPuro cells, which express a vector containing nondisrupted lacZ, with the same ratios of pMLV ψ− and the RNase H mutant derivative as above. Following puromycin selection, transduced colonies were stained with X-Gal. Most cells transduced with LacPuro stain blue, but the fraction of total puromycin-resistant colonies that did not stain blue was used as an estimate of how frequently the lacZ gene was mutationally inactivated during reverse transcription. Virions packaged with 100, 50, 20, 10, and 5% wild-type RT, with the remainder of the virion RT consisting of the RNase H mutant, generated nonstaining colonies at frequencies of 6.8, 5.7, 8.6, 10.2, and 16%, respectively (Fig. 4B).
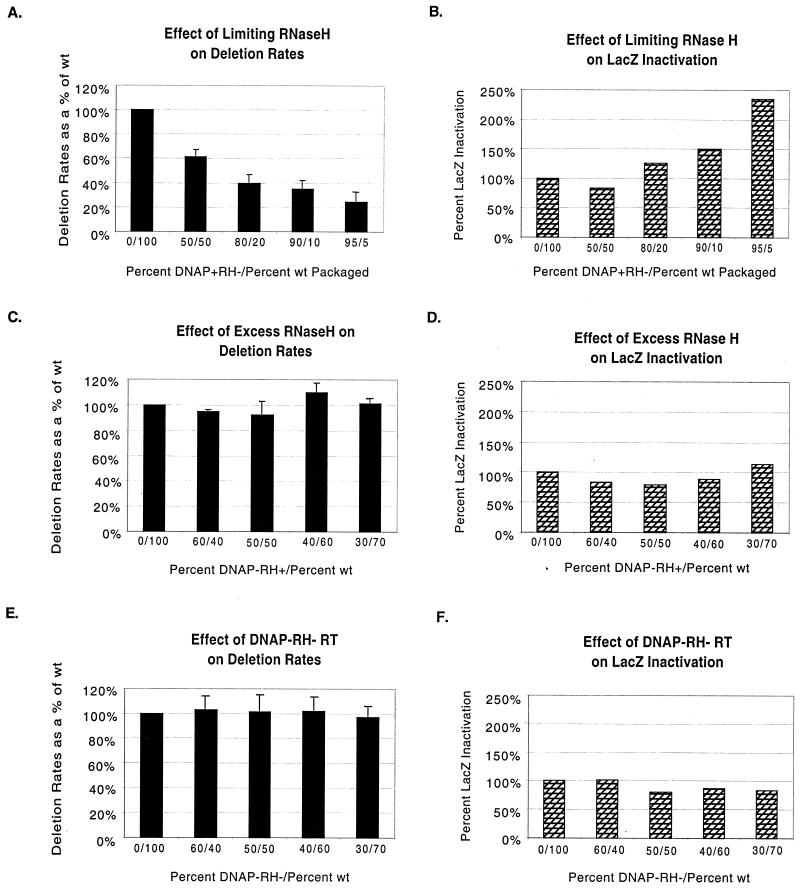
FIG. 4.
Fidelity of DNA synthesis and direct repeat deletion frequencies for phenotypically mixed virions. (A and B) Effects of limited RNase H on repeat deletion and replication fidelity. Phenotypically mixed virions with the indicated ratios of DNAP+ RH− mutant and wild-type RTs were prepared as described in the text. (A) Repeat deletion frequencies of phenotypically mixed virions. Values given are the averages of the values determined for each of the four separate infection experiments whose data are provided in Table 1, adjusted for lacZ inactivation (see text and panel B). (B) lacZ inactivation rates. Assays were performed as described in the Materials and Methods section. (C and E) Normalized frequencies of direct repeat for the indicated mixed virions; (D and F) lacZ inactivation rates for the indicated mixed virions. All values for phenotypically mixed virions are provided as percentages of the rate, set to 100%, that was determined for virions with 100% wild-type RT within an individual experiment. The indicated standard deviations for mixed virions were calculated by comparing percent wild-type deletion values among experimental repetitions, as opposed to comparing the absolute deletion values as reported in Table 1.
Average adjusted deletion values from the four infection experiments reported in Table 1 are presented in Fig. 4A. These values were derived by first determining blue to total (stained plus unstained) colony ratios and then dividing these values by the percentage of colonies that retained functional lacZ when assayed with LacPuro. For example, because the 100% wild-type virions above displayed lacZ inactivation rates of 6.8%, observed blue colony frequencies were divided by 93.2%. Factoring in lacZ inactivation differences in this way, the direct repeat deletion rates for the mixed virions with reduced functional RNase H were adjusted to 61.4, 39.8, 35.0, and 24.5% of the wild-type rate (Fig. 4A). Thus, reducing the amount of functional RNase H in virions caused a marked decrease in tandem repeat deletion frequency.
Effect of excess RNase H on template switching rates.
Tandem repeat deletion assays were also used to determine the effects of excess RNase H on template switching. Phenotypically mixed virions were prepared as above, here by transiently cotransfecting ET pLaac-284 cells with various ratios of pMLV ψ− and pMLV ψ− DNAP− RH+ helper plasmids. Virus harvested from cotransfected cells was used to infect 3T3 cells, and deletion rates were determined by puromycin selection and LacZ staining, and adjusted for lacZ inactivation rates (Fig. 4D) as above. As shown in Fig. 4C, virions with 30, 40, 50, and 60% DNAP− RH+ RT displayed direct repeat deletion frequencies of 101.5, 110.0, 92.2, and 95.1%, respectively, that of virus with 100% wild-type RT. Thus, there were no consistent trends, and incorporating excess RNase H relative to functional DNA polymerase did not appear to affect rates of tandem repeat deletion.
In parallel experiments, ET pLaac-284 cells were cotransfected with pMLV ψ− and pMLV ψ− DNAP− RH− helper plasmids. Relative titers for these mixed virions are presented in Fig. 2. This cotransfection experiment was performed to address whether or not template switching rates would change when reduced amounts of wild-type RT synthesized DNA in the presence of doubly defective RT. Figure 4 presents data which demonstrate that virions packaged with 30, 40, 50, and 60% RT defective in both DNA polymerase and RNase H activities performed direct repeat deletion at frequencies 97.0, 101.8, 101.3, and 103.1%, respectively, that of virus with wild-type RT. These frequencies are similar to those obtained for virions assembled with excess RNase H as well as for virions containing 100% wild-type RT.
DISCUSSION
Retrovirus assembly results in the inclusion of roughly 100 RT molecules per virion (32). Most virions probably contain only two genomic RNAs, and thus, retroviruses contain a significant molar excess of reverse transcriptase relative to viral RNA (3). Here, the properties of phenotypically mixed virions with mixtures of wild-type and mutant RTs demonstrated that viral replication could occur, albeit at moderately impaired levels, with less than the normal complement of functional RT. DNA polymerase activity, more than RNase H activity, became limiting in virions when amounts of functional RT were reduced. This was evident from the finding that titer reductions of mixed virions containing wild-type and DNA polymerase mutant RTs were reduced more than were titers of RNase H mutant mixed virions with the same ratio of mutant to wild-type RT (Fig. 2). These findings are consistent with those reported by Julias and colleagues, who studied the effects of limiting functional DNA polymerase or RNase H on titers for HIV-1 (13). Interestingly, HIV-2 RT naturally possesses only about 10% of the basal RNase H activity of HIV-1 RT (8), further supporting the notion that at least some retroviruses possess more RNase H activity than is necessary for viral replication.
When RNase H was limited, fewer proviral products contained repeat deletions than when wild-type levels of RNase H were present. This finding is consistent with the models for retrovirus recombination introduced above (2, 4). M-MuLV RNase H mutants that perform template switching at decreased frequencies have been reported previously (19, 25). However, it is unclear whether these mutants, which retain some infectivity, display reduced template switching frequencies due to decreases in levels of RNase H activity or if some property of the DNA polymerase domain or of the enzyme as a whole contributes to template switching impairment. Both the present study and the earlier replication-competent mutant work (19, 25) demonstrate the importance of RT’s RNase H domain to template switching.
Here, increasing the ratio of functional RNase H to DNA polymerase above its natural level did not augment repeat deletion rates. One possible reason for this is that virions may be saturated for RNase H activity at the normal DNA polymerase to RNase H ratio. However, deletion frequency is not maximal under native conditions. We previously reported that repeat deletion frequencies can be increased by increasing the amount of time required to synthesize viral DNA (20). One possible explanation of those earlier findings was that increasing the time spent synthesizing DNA might allow further unmasking of single-stranded DNA—which is postulated to be required for template switching—by the RNase H activity of surplus RT molecules over time. The current findings of no increase in repeat deletion when excess RNase H was present may weaken arguments favoring that explanation.
Another possible reason for why additional RNase H failed to stimulate template switching here is that RNase H in the context of a catalytically inactive DNA polymerase may be unable to contribute to replication. Possibly consistent with this interpretation are the findings of this study that there were no significant differences in titer, repeat deletion, or fidelity between phenotypically mixed virions whose mutant RT was DNAP− RH+ and those whose mutant was DNAP− RH− (Fig. 2 and 4). Although a possible argument against this interpretation lies in observations that DNAP− RH+ and DNAP+ RH− virions can complement to generate viral DNA, it may be significant that the titers observed for such mixed virions were only marginally higher than the titer observed for 100% DNAP+ RH− virions.
Because recombination rapidly reverts virions with both DNAP− RH+ and DNAP+ RH− RT gene-containing intact genomes to wild type (29), it would be difficult to assess whether or not this low ability to synthesize DNA would be sufficient to support multiple replication rounds. Although RTs with inactivated DNA polymerase domains have been shown to display RNase H activity in purified reactions, numerous studies have reported examples of functional coupling between RT’s DNA polymerase and RNase H domains (22, 24, 28). It is possible that a catalytic site mutation in the DNA polymerase domain was sufficient to block normal functioning of the RNase H domain and that the DNAP− RH+ functioned as if it were RNase H and DNA polymerase defective.
This study revealed one striking example—the reduced fidelity of DNA synthesis by the DNAP+ RH− form of RT—of an apparent functional interaction between M-MuLV RT’s RNase H and DNA polymerase domains. Compared to the replicative fidelity of previously described M-MuLV RT mutants, including structure-based DNA polymerase domain mutants, the nearly 2.5-fold-increased frequency of lacZ inactivation observed for phenotypically mixed virions containing 95% DNAP+ RH− plus 5% wild-type RT is high (19). The nature of the mutations in these proviruses was not determined. However, these findings may suggest that, at least in the context of viral replication, introducing the RNase H domain single D524N catalytic residue substitution was sufficient to generate a highly error-prone RT (7).
Acknowledgments
We thank John Julias and Steve Hughes for sharing results prior to publication and Tammy Wood for technical assistance during early stages of this project.
This work was supported by American Cancer Society grant RPG-95-058-06-MBC to A.T., a Rackham Predoctoral Fellowship to J.K.P., and NIH grant T32 GM 07544 to J.L.B.
REFERENCES
- 1.Champoux, J. J. 1993. Roles of ribonuclease H in reverse transcription, p. 103–117. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 2.Coffin, J. 1996. Retroviridae: the viruses and their replication. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott, Philadelphia, Pa.
- 3.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1–26. [DOI] [PubMed] [Google Scholar]
- 4.Delviks, K. A., and V. K. Pathak. 1999. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J. Virol. 73:7923–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeStefano, J. J., R. G. Buiser, L. M. Mallaber, T. W. Myers, R. A. Bambara, and P. J. Fay. 1991. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency and Moloney murine leukemia viruses are functionally uncoupled. J. Biol. Chem. 266:7423–7431. [PubMed] [Google Scholar]
- 6.Gao, H.-Q., P. L. Boyer, E. Arnold, and S. H. Hughes. 1998. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol. 277:559–572. [DOI] [PubMed] [Google Scholar]
- 7.Halvas, E. K., E. S. Svarovskaia, and V. K. Pathak. 2000. Development of an in vivo assay to identify structural determinants in murine leukemia virus reverse transcriptase important for fidelity. J. Virol. 74:312–319. [PMC free article] [PubMed] [Google Scholar]
- 8.Hizi, A., R. Tal, M. Shaharabany, and S. Loya. 1991. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J. Biol. Chem. 266:6230–6239. [PubMed] [Google Scholar]
- 9.Hu, W.-S., E. H. Bowman, K. A. Delviks, and V. K. Pathak. 1997. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J. Virol. 71:6028–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, >W.-S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter, E. 1978. The mechanism for genetic recombination in the avian retroviruses. Curr. Top. Microbiol. Immunol. 79:295–309. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis, T. C., and K. Kirkegaard. 1991. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 7:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed HIV-1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988–25997. [PubMed] [Google Scholar]
- 15.Negroni, M., and H. Buc. 2001. Retroviral recombination: what drives the switch? Nat. Rev. Mol. Cell. Biol. 2:151–155. [DOI] [PubMed] [Google Scholar]
- 16.Oyama, F., R. Kikuchi, R. J. Crouch, and T. Uchida. 1989. Intrinsic properties of reverse transcriptase in reverse transcription: associated RNase H is essentially regarded as an endonuclease. J. Biol. Chem. 264:18808–18817. [PubMed] [Google Scholar]
- 17.Pathak, V. K., and W.-S. Hu. 1997. “Might as well jump!”: template switching by retroviral reverse transcriptase, defective genome formation, and recombination. Semin. Virol. 8:141–150. [Google Scholar]
- 18.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112–1118. [DOI] [PubMed] [Google Scholar]
- 19.Pfeiffer, J. K., M. M. Georgiadis, and A. Telesnitsky. 2000. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct repeat deletion frequencies. J. Virol. 74:9629–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer, J. K., R. Topping, N.-H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 3:8441–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price, J., D. Turner, and C. Cepko. 1987. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. USA 84:156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz, S., and J. Champoux. 1996. RNase H domain of Moloney murine leukemia virus reverse transcriptase retains activity but requires the polymerase domain for specificity. J. Virol. 70:8630–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, N. H., D. Hartigan-O’Connor, J. K. Pfeiffer, and A. Telesnitsky. 2000. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J. Virol. 74:2694–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, J., K. Gristman, and M. J. Roth. 1994. Contributions of DNA polymerase subdomains to the RNase H activity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 68:5721–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svarovskaia, E. S., K. A. Delviks, C. K. Hwang, and V. K. Pathak. 2000. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74:7171–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanese, N., and S. P. Goff. 1988. Domain structure of the Moloney MuLV reverse transcriptase: mutational analysis and separate expression of the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 85:1777–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121–160. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 28.Telesnitsky, A., and S. P. Goff. 1993. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc. Natl. Acad. Sci. USA 90:1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telesnitsky, A., and S. P. Goff. 1993. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 12:4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temin, H. M. 1993. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. USA 90:6900–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt, P. K. 1967. Phenotypic mixing in the avian tumor virus group. Virology 32:708–717. [DOI] [PubMed] [Google Scholar]
- 32.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27–69. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]