Abstract
A 2.6-kbp fragment of the pseudorabies virus (PrV) genome was sequenced and shown to contain the homologues of the highly conserved herpesvirus genes UL31 and UL32. By use of a monospecific antiserum, the UL31 gene product was identified as a nuclear protein with an apparent molecular mass of 29 kDa. For functional analysis, UL31 was deleted by mutagenesis in Escherichia coli of an infectious full-length clone of the PrV genome. The resulting virus mutants were deficient in plaque formation, and titers were reduced more than 100-fold from those of wild-type PrV. Ultrastructural analyses demonstrated that capsid maturation and DNA packaging were not affected. However, neither budding at the inner nuclear membrane nor cytoplasmic or extracellular virus particles were observed. These replication defects were similar to those of a UL34 deletion mutant (B. G. Klupp, H. Granzow, and T. C. Mettenleiter, J. Virol. 74:10063–10073, 2000) and could be completely repaired in a cell line which constitutively expresses the UL31 protein. Yeast two-hybrid studies revealed that a UL31 fusion protein specifically interacts with plasmids of a PrV genome library expressing the N-terminal part of UL34. Vice versa, UL34 selected UL31-encoding plasmids from the library. Immunofluorescence studies and immune electron microscopy demonstrated that in cells infected with wild-type PrV, both proteins accumulate at the nuclear membrane, whereas in the absence of UL34 the UL31 protein is dispersed throughout the nucleus. Like the UL34 protein, the UL31 gene product is a component of enveloped virus particles within the perinuclear space and absent from mature virions. Our findings suggest that physical interaction between these two virus proteins might be a prerequisite for primary envelopment of PrV at the inner nuclear membrane and that this envelope is removed by fusion with the outer nuclear membrane.
Pseudorabies virus (PrV, suid herpesvirus 1) is the causative agent of Aujeszky’s disease in pigs (36). It is classified as a member of the Varicellovirus genus within the Alphaherpesvirinae subfamily of Herpesviridae (43). Herpesvirus particles are composed of an icosahedral capsid which encloses the double-stranded DNA genome and which in turn is surrounded by a layer of numerous viral gene products named tegument and a lipid membrane of cellular origin containing mostly glycosylated virus-encoded proteins (36, 43). After DNA replication and encapsidation in the host-cell nucleus, virus egress starts with budding of nucleocapsids at the inner nuclear membrane, leading to enveloped particles in the perinuclear space (44). It has been proposed that these perinuclear virions are released by vesicular transport through the endoplasmic reticulum and Golgi apparatus (26, 51), which would permit processing of viral glycoproteins, but not substitution of envelope or tegument proteins. According to another hypothesis, the primary envelope of herpesviruses is lost by fusion with the outer nuclear membrane, followed by release of naked nucleocapsids into the cytoplasm and a final, secondary envelopment in the trans-Golgi region (54). During the past decade, the latter model was supported by genetic, biochemical, and ultrastructural analyses of different alpha- and betaherpesviruses including PrV (7, 9, 18, 20, 23, 29, 41, 47, 61).
Like other representatives of the Varicellovirus genus, PrV exhibits a type D herpesvirus genome (43) consisting of a unique long region (UL) and a unique short region (US) which is flanked by inverted repeat sequences (IRS, TRS). Major parts of the 150-kbp genome of PrV have been sequenced (36) and shown to contain a set of ca. 70 conserved genes, which exhibit a widely similar arrangement to that found within the genomes of other mammalian alphaherpesviruses, e.g., herpes simplex virus type 1 (HSV-1) (33), varicella-zoster virus (VZV) (13), or equine herpesvirus 1 (EHV-1) (50). However, the UL region of PrV contains an internal inversion encompassing the homologues of the UL27 to UL44 genes of HSV-1 (4, 8). In the present study, sequence analyses were performed to close one of the remaining gaps within this part of the PrV genome. Since this gap was located between the UL30 gene encoding the viral DNA polymerase (5) and the described UL33 to UL35 homologues (29), it was considered to contain the UL31 and UL32 genes of PrV. The corresponding genes of HSV-1 were shown to be essential for virus replication in cell culture (44). UL31 and UL32 are highly conserved among alphaherpesviruses and have homologues in the genomes of beta- and gammaherpesviruses, such as human cytomegalovirus (HCMV) (12) and Epstein-Barr virus (EBV) (2).
The UL31 gene products of HSV-1 and HSV-2 have been identified as nuclear proteins (10, 60), and the UL31 protein of HSV-1 has been demonstrated to be phosphorylated as well as nucleotidylylated in infected cells (6, 10). Pull-down experiments indicated a physical interaction between the UL31 and UL34 gene products of HSV-1 (58), which was somewhat surprising, since UL31 was previously thought to be involved in viral DNA synthesis or encapsidation within the nucleus (11), whereas UL34 was supposed to participate in dynein-mediated transport of incoming nucleocapsids to the nucleus (58). However, recent studies indicated that the UL34 gene product of HSV-1 is required for stabilization of the UL31 protein (57) and for virus envelopment (45). We have previously demonstrated that the UL34 protein of PrV is also required for envelopment of newly synthesized nucleocapsids in the nuclei of infected cells but is not detectable in mature virions and therefore is presumably not involved in virus entry (29). Furthermore, the presence of the PrV UL34 protein within primary enveloped virus particles in the perinuclear space, and its absence from cytoplasmic as well as from released virions, confirmed the hypothesis that envelopment of PrV is a two-step event (23, 54).
In the present study, a monospecific rabbit antiserum was prepared which allowed identification and localization of the UL31 gene product of PrV. Possible interactions between the UL31 and UL34 proteins of PrV were tested by yeast two-hybrid studies (17, 24). Virus recombinants were prepared to investigate whether the UL31 protein of PrV is directly involved in nucleocapsid formation or in virus envelopment. To facilitate mutagenesis of viral DNA, an infectious full-length clone of the PrV genome was generated by insertion of a mini-F plasmid. Such plasmids were shown to be suitable vectors for stable replication of large DNA-fragments in Escherichia coli (39) and have already been used for cloning of numerous herpesvirus genomes including that of PrV (34, 46, 48). For deletion of the UL31 gene, the cloned PrV genome was mutated in E. coli by RecE- and RecT-mediated recombination (46, 59). Subsequently, the bacterial vector sequences were removed to avoid undesired phenotypic effects, and the PrV recombinants were characterized in UL31-expressing and noncomplementing eucaryotic cells.
MATERIALS AND METHODS
Viruses and cells.
All virus recombinants used in this study are based on the laboratory strain PrV-Ka (28). PrV-ΔUL34B and PrV-ΔUS3 have been described recently (29, 30). The newly generated UL31 deletion mutants were derived from the glycoprotein G (gG)-negative, LacZ-expressing recombinant PrVgXβGal (37). Virus was propagated in rabbit kidney (RK13) cells which were grown in minimum essential medium (MEM; Life Technologies) supplemented with 10% fetal calf serum. To generate UL31-expressing cell lines, RK13 cells were transfected with plasmid pIRES-UL31 (Fig. 1B) by calcium phosphate coprecipitation (22), trypsinized after 48 h, diluted in medium containing 500 μg of Geneticin (Life Technologies)/ml, and seeded into microtiter plates. Single resistant cell colonies were tested for constitutive UL31 expression by Western blot analyses. One positive cell clone, named RK-UL31, was selected for propagation of UL31 deletion mutants of PrV.
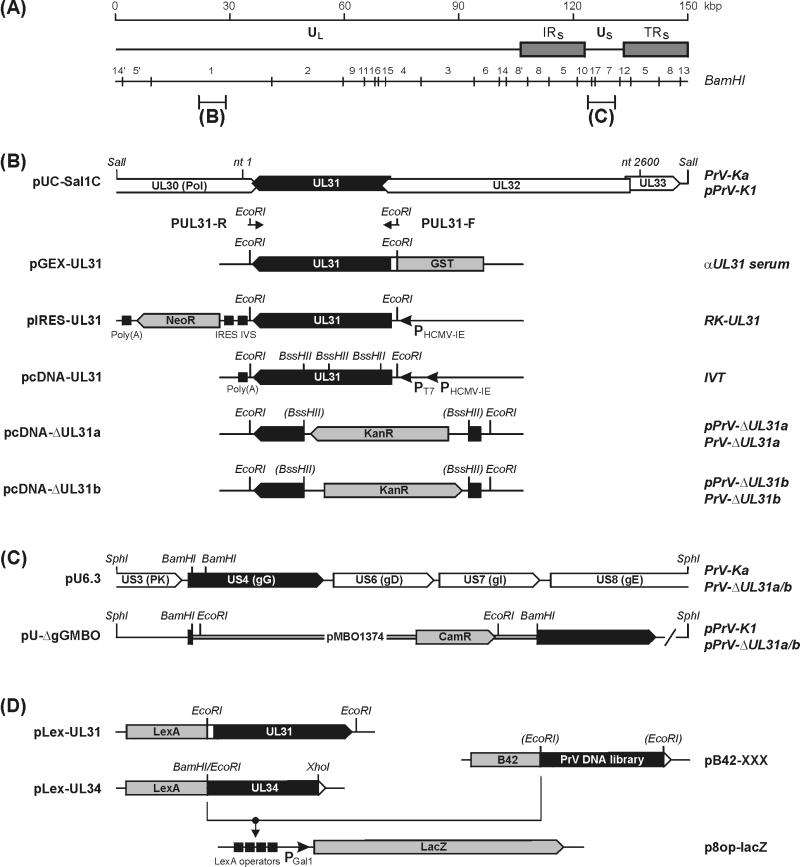
FIG. 1.
Structure of the PrV genome and cloning strategies. (A) A schematic map of the PrV genome shows the long (UL) and short (US) unique regions, the inverted repeat sequences (IRS, TRS), and the positions of BamHI restriction sites. (B) The sequenced genome region of PrV-Ka (nt 1 to 2600; GenBank accession no. AJ319028) is part of plasmid pUC-SalIC. Primers PUL31-F and PUL31-R contain artificial EcoRI sites and were used to amplify the UL31 ORF for cloning in expression vectors. A bacterial GST fusion protein expressed from pGEX-UL31 was used for rabbit immunization (αUL31 serum). Plasmid pIRES-UL31 was used to generate a cell line (RK-UL31) expressing UL31 together with the neomycin resistance gene (neoR) under the control of the HCMV immediate-early gene promoter (PHCMV-IE) from a bicistronic mRNA which contains an intron (IVS) and an internal ribosomal entry site (IRES) between the two genes, and a polyadenylation signal [poly(A)] at the end. The T7 promoter (PT7) of pcDNA-UL31 permitted in vitro transcription and translation (IVT). In plasmids pcDNA-ΔUL31a and pcDNA-ΔUL31b, a major portion of UL31 was replaced by a kanamycin resistance gene (kanR) inserted in either orientation (pcDNA-ΔUL31a and -b). The modified gene was amplified by PCR with primers PUL31-F and PUL31-R and used for RecE- and RecT-mediated mutagenesis of pPrV-K1 (see panel C) in E. coli. (C) The mini-F plasmid pMBO1374 containing a chloramphenicol resistance gene (camR) was inserted into the cloned gG gene of PrV-Ka in plasmid pU6.3. The plasmid obtained, pU-ΔgGMBO, was used for generation of an infectious full-length clone of the PrV genome (pPrV-K1). After mutagenesis of the UL31 gene (pPrV-ΔUL31a and -b; see panel B), the vector insertion was removed by EcoRI digestion and cotransfection of cells with plasmid pU6.3. Thus, the resulting virus mutants (PrV-ΔUL31a and -b) contain an intact gG gene. (D) Plasmids pLex-UL31 and pLex-UL34 were utilized for yeast two-hybrid screening of a PrV genome library (pB42-XXX). Interactions between the fusion proteins were detected by LacZ expression from p8op-lacZ (Clontech), which is induced by binding of LexA to the LexA operator elements preceding the Gal 1 minimal promoter (PGal 1), and subsequent activation of transcription by the B42 protein part. Only relevant restriction sites are included in the maps in panels B to D. ORFs are drawn as pointed rectangles, and a slash indicates that the DNA fragment is not plotted to scale.
DNA sequencing.
The genomic BamHI fragment 1 of PrV-Ka (Fig. 1A) was digested with SalI and subcloned into pUC19. Two independently isolated plasmids containing the 3,320-bp SalI subfragment 1C were seqenced bidirectionally as described elsewhere (19) by primer walking. PrV-specific primers were deduced sequentially starting from the published DNA sequences of the UL30 gene (5; GenBank accession no. L24487) and the UL33 gene (29; GenBank accession no. AJ276165). The sequences obtained were assembled and analyzed with the Wisconsin software package (Genetics Computer Group) (14) in Unix, version 10.2.
Construction of UL31 expression and deletion plasmids.
The UL31 open reading frame (ORF) was amplified from cloned PrV DNA by PCR with Pfx DNA polymerase (Life Technologies) using primers PUL31-F (CTGAATTCACACGCTCGGCAGCTATGTTTG [initiation codon underlined]) and PUL31-R (CTGAATTCTTCGCGGCGCTCACGG [reverse of termination codon underlined]). The 857-bp amplification product contains artificial EcoRI sites at both ends (printed in italics), allowing insertion into the EcoRI-digested expression plasmids pGEX-4T-1 (Amersham Pharmacia), pLexA (Clontech), pcDNA3 (Invitrogen), and pIRES1neo (Clontech). Whereas the obtained plasmids pGEX-UL31, pLex-UL31, and pIRES-UL31 (Fig. 1B and D) were used for protein expression in E. coli, yeast, and RK13 cells, respectively, pcDNA-UL31 was utilized for in vitro transcription and translation of UL31.
To generate UL31 deletion constructs from pcDNA-UL31 (Fig. 1B), a part of the vector sequence containing an unwanted BssHII site was removed by double digestion with EcoRV and BstBI, treatment with Klenow polymerase, and religation. The resulting plasmid was cleaved with BssHII to delete two 342- and 116-bp fragments representing codons 23 to 174 of UL31. After Klenow treatment, the PCR-amplified kanamycin resistance gene (kanR) consisting of nucleotides 1800 to 2769 of pACYC177 (GenBank accession no. X06402) was inserted in either orientation (pcDNA-ΔUL31a and -b [Fig. 1B]).
Generation of UL31 deletion mutants of PrV.
For cloning of the PrV genome we used the mini-F plasmid pMBO1374 (48), which was provided with the permission of M. O’Connor (University of California, Irvine) by G. A. Smith (Princeton University, Princeton, N.J.). This vector was inserted into plasmid pU6.3 (27) with concomitant deletion of a 196-bp BamHI-fragment from the gG gene (Fig. 1C). The resulting plasmid, pU-ΔgGMBO, was used for cotransfection (22) of RK13 cells together with virion DNA of a previously described LacZ-expressing gG gene deletion mutant (37). Blue plaque assays (37) of progeny virus were performed to isolate the desired β-galactosidase-negative recombinants. To obtain circular monomers of the F plasmid-containing PrV genome, RK13 cells were infected at a multiplicity of infection (MOI) of 5 and incubated for 6 h at 37°C in the presence of 100 μg of cycloheximide/ml. Viral DNA was prepared (25) and used for electroshock transformation of E. coli DH10B cells (Life Technologies) as described recently (46). Chloramphenicol-resistant clones were tested for integrity of the PrV genome by DNA analyses and by retransfection of RK13 cells.
One infectious full-length clone (pPrV-K1 [Fig. 1C]) was used for RecE- and RecT-mediated mutagenesis in E. coli (46, 59). For this purpose, the bacteria were additionally transformed with plasmid pGETrec (38), providing the recombination factors, and linear PCR products of kanR-containing UL31 deletion plasmid pcDNA-ΔUL31a or -b (Fig. 1B) obtained after amplification with primers PUL31-F and PUL31-R (see above). DNA of chloramphenicol and kanamycin double-resistant clones (pPrV-ΔUL31a/b) was characterized by blot hybridization and sequencing of the UL31 gene region.
To eliminate unwanted effects of the vector insertion on virus replication, pPrV-ΔUL31a and -b were digested with EcoRI and repaired with plasmid pU6.3 (Fig. 1C) by cotransfection of RK-UL31 cells. Virus progeny was plaque purified and tested for indirect immunofluorescence reaction with a rabbit antiserum produced against a bacterial gG fusion protein. Two gG-expressing virus isolates named PrV-ΔUL31a and PrV-ΔUL31b (Fig. 1C) were further propagated and again characterized by DNA analyses including PCR amplification and DNA sequencing to confirm complete removal of the mini-F plasmid.
One-step growth analysis.
Confluent monolayers of RK13 and RK-UL31 cells were infected at an MOI of 10 with PrV-Ka or UL31 deletion mutants and incubated on ice for 1 h. Then prewarmed medium was added, and incubation was continued at 37°C. After 1 h, nonpenetrated virus was inactivated by low-pH treatment (35), and at 3, 6, 9, 12, 24, and 48 h after the temperature shift, cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). Titers of progeny virus were determined by plaque assays in RK-UL31 cells overlaid with MEM containing 5% fetal calf serum and 6 g of methyl cellulose/liter.
Preparation of a UL31-specific rabbit antiserum.
In pGEX-UL31 the complete UL31 ORF, preceded by several nucleotides of originally noncoding PrV DNA, was fused in frame to the glutathione S-transferase (GST) gene (Fig. 1B). A 55-kDa GST fusion protein was expressed in E. coli strain DH5α (Amersham Pharmacia) and purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (32) followed by electroelution into ultrafiltration units (Amicon). After concentration and repeated washes with phosphate-buffered saline (PBS), 100 μg of the eluted protein was emulsified in mineral oil and injected intramuscularly into a rabbit four times at 3-week intervals. Sera collected before and 3 weeks after each immunization were analyzed.
Western blot analysis.
For Western blotting, RK13 cells were infected with PrV at an MOI of 5 and incubated at 37°C for 2 to 24 h. Samples of infected and noninfected cells, as well as PrV virions, were prepared as described previously (15), separated by SDS-PAGE, and electrotransferred to nitrocellulose membranes (Schleicher & Schuell). Blots were blocked with 5% low-fat milk in PBS and incubated for 1 h with anti-UL31, anti-UL34 (29), and anti-UL49 (7) rabbit sera at dilutions of 1:100,000. Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
Indirect immunofluorescence and confocal microscopy.
Cells were grown on coverslips and infected at an MOI of ca. 0.001 with wild-type or mutant PrV. After 24 or 48 h, the cells were fixed for 10 min with a 1:1 mixture of methanol and acetone, dried, and subsequently incubated with a gC-specific monoclonal antibody (B16-c8 [31]) or rabbit antisera (anti-UL31; anti-UL34) at dilutions of 1:200 and Alexa 488- or tetramethyl rhodamine isocyanate (TRITC)-conjugated secondary antibodies (Molecular Probes; Dako). After each step the slides were washed repeatedly with PBS, and finally they were preserved with a 9:1 mixture of glycerol and PBS, containing 25 mg of 1,4-diazabicyclooctane/ml and 1 μg of propidium iodide/ml. Fluorescence was recorded in a confocal laser scan microscope (LSM 510; Zeiss).
Electron microscopy and immunolabeling.
RK13 and RK-UL31 cells were infected at an MOI of 1 with either PrV-Ka, PrV-ΔUL31a, or PrV-ΔUS3 (30). After 1 h on ice and an additional hour at 37°C, the inoculum was replaced by fresh medium, and incubation was continued for 13 h at 37°C. Fixation and embedding for routine microscopy, as well as for intracellular immunolabeling of viral proteins, were done as described recently (29). The reaction of the anti-UL31 serum was visualized by binding of gold-tagged anti-rabbit antibodies (British BioCell International), and ultrathin sections of the infected cells were examined with an electron microscope (400T; Philips).
Yeast two-hybrid analysis.
For investigation of interactions between PrV proteins, the Matchmaker LexA two-hybrid system (Clontech) was used according to the detailed protocols of the manufacturer. To generate an expression library of PrV proteins fused to a transcription-activating peptide (B42), DNA was prepared from sucrose gradient-purified virions of the Ka strain (3). The PrV DNA obtained was disrupted by ultrasonic treatment, and fragments ranging from 500 to 1,000 bp were isolated from an agarose gel by using DEAE membranes (16). To generate blunt ends, the isolated DNA fragments were incubated with Klenow polymerase and ligated to the EcoRI-digested, Klenow fragment- and phosphatase-treated vector pB42AD (Fig. 1D). After electroshock transformation of E. coli DH10B cells (46), a library of ca. 2 × 105 plasmid clones was obtained and further amplified for DNA preparation (Plasmid Kit; Qiagen). The UL31 and UL34 gene products of PrV were used as baits, and for that purpose they were fused to a sequence-specific DNA-binding protein (LexA). Whereas the respective expression plasmid pLex-UL31 contains the entire UL31 ORF of PrV (Fig. 1D), pLex-UL34 was obtained by recloning of a 669-bp EcoRI-XhoI fragment from a previously described bacterial expression plasmid which contained codons 1 to 220 of UL34 (29). To permit in-frame fusion with LexA, the vector was digested with BamHI and XhoI, and noncompatible single-stranded overhangs were filled in by using Klenow polymerase (Fig. 1D).
Yeast cells were subsequently transformed with the reporter plasmid p8op-lacZ (Clontech), the chosen bait construct, and the PrV expression library (Fig. 1D). In several yeast clones, interaction between bait and library proteins caused trans-activation of the lacZ reporter gene, leading to blue colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-containing agar plates. DNA was prepared from these clones, and the library plasmids were recloned in E. coli strain KC8 (Clontech), tested again for specific yeast two-hybrid interactions with the respective bait construct, and characterized by DNA sequencing of the insert fragment with vector-specific primers.
Nucleotide sequence accession number.
The PrV DNA sequence determined in this study has been deposited in GenBank under accession no. AJ319028.
RESULTS
DNA sequences of the UL31 and UL32 genes of PrV.
A 2,600-bp fragment of genomic DNA of PrV-Ka, which closes a gap between two characterized parts of the UL region of the virus genome, was analyzed. The fragment contains the complete UL31 (272 codons) and UL32 (471 codons) genes (Fig. 1B). At its left end, the sequence determined overlaps with the DNA polymerase gene UL30 (5) by 204 bp, and the last 471 bp correspond to the first nucleotides of the previously described UL33-to-UL35 gene region (29). Since all these DNA sequences originate from the same PrV strain, it was not surprising that no differences were found within the overlapping parts. The positions of putative TATA and polyadenylation signals indicate that UL31 and UL32 presumably form a 3′-coterminal transcription unit (Table 1). Remarkably, both ORFs overlap by several nucleotides with each other as well as with the adjacent UL30 and UL33 genes (Fig. 1B). The deduced PrV gene products of UL31 and UL32 are highly conserved in other alphaherpesviruses and also possess detectable homology to corresponding beta- and gammaherpesvirus proteins, e.g., UL53 and UL52 of HCMV and BFLF2 and BFLF1 of EBV (Table 1).
TABLE 1.
Properties of the identified PrV genes
| Property | UL31 | UL32 |
|---|---|---|
| Gene | ||
| Coding sequence | nt 943–128a | nt 2348–936 |
| TATA box | nt 1069–1063 | nt 2536–2530 |
| Polyadenylation signal | nt 75–70 | nt 75–70 |
| Deduced protein | ||
| Size | 271 aa | 470 aa |
| Isoelectric point | 9.00 | 6.96 |
| Expected molecular mass | 30.41 kDa | 51.58 kDa |
| Apparent molecular mass | 29 kDa | —b |
| Homologous proteinsc | ||
| Alphaherpes viruses | ||
| EHV-1 | 76.2 (67.9) | 66.2 (59.8) |
| VZV | 72.1 (62.5) | 59.5 (51.0) |
| HSV-1 | 69.3 (57.7) | 60.4 (53.3) |
| Betaherpesvirus (HCMV) | 38.2 (27.5) | 37.7 (28.7) |
| Gammaherpesvirus (EBV) | 33.1 (23.8) | 38.2 (30.5) |
All ORFs and putative transcription signals are located at the reverse strand of the determined DNA sequence (GenBank accession no. AJ319028), and nucleotide positions (nt) are therefore numbered from right to left.
The UL32 protein of PrV has not been identified.
Values are percent similar residues (percent identical residues) and were determined using the Genetics Computer Group program “gap”. The amino acid sequences of the homologous alpha-, beta-, and gammaherpesvirus proteins were deduced from genomic DNA sequences (2, 12, 13, 33, 50).
The UL31 gene product of HSV-1 has been described as a phosphorylated and nucleotidylylated nuclear protein (6, 10). Like its homologue, the predicted UL31 protein of PrV is moderately basic, and exhibits a highly hydrophilic N-terminal part containing short clusters of arginine and lysine residues (positions 4 to 6, 9 to 12, and 17 to 19), which might serve as nuclear localization signals (21). The PrV protein also contains possible phosphorylation sites of casein kinase II, protein kinase C, and cyclic AMP (cAMP)-dependent protein kinases, but the consensus motif (R/P)RA(P/S)R of nucleotidylylated HSV proteins (6) was not found.
Identification of the UL31 protein of PrV.
For identification of the UL31 gene product, the entire ORF was cloned in a procaryotic expression vector (pGEX-UL31 [Fig. 1B]) and the GST fusion protein isolated from transformed bacteria was used for rabbit immunization. The antiserum obtained specifically reacted in Western blot analyses with a 29-kDa protein in lysates of the neomycin-resistant cell line RK-UL31 (Fig. 2A), which was isolated after transfection of RK13 cells with a plasmid containing UL31 and the resistance gene as a bicistronic transcription unit under the control of the HCMV immediate-early gene promoter (pIRES-UL31 [Fig. 1B]). A protein of similar size was also detected after in vitro transcription and translation of pcDNA-UL31 (data not shown) as well as in RK13 cells infected with wild-type PrV-Ka (Fig. 2A). This indicates that stable constitutive UL31 expression does not require any other viral protein and that posttranslational modifications by viral or cellular enzymes, if present at all, have no visible effect on electrophoretic mobility. Consistently, the UL31 gene product of PrV was not detectable after immunoprecipitation if infected cells were labeled with [32P]orthophosphate, whereas the UL31 protein was efficiently precipitated by the respective antiserum from infected cells labeled with [35S]methionine (data not shown). Thus, unlike the homologous HSV-1 protein (10), the UL31 gene product of PrV is apparently not phosphorylated.
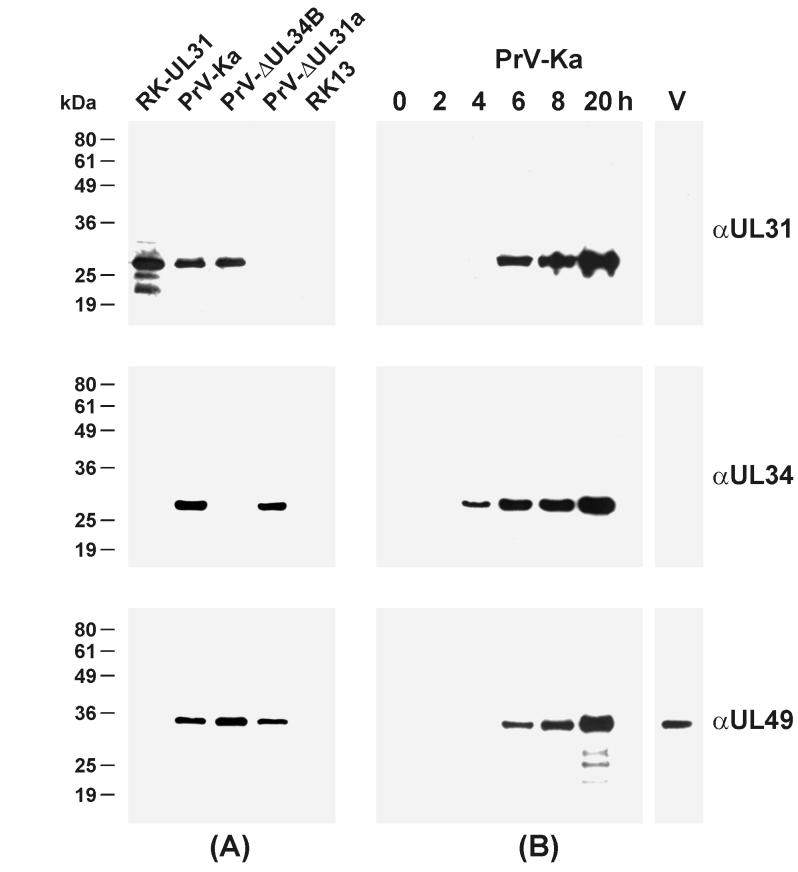
FIG. 2.
Identification of the UL31 protein of PrV. RK13 cells were infected with either PrV-Ka, PrV-ΔUL31a, or PrV-ΔUL34B (29) at an MOI of 5 and incubated for 8 h or the indicated times at 37°C. Lysates of infected cells, of noninfected RK13 and RK-UL31 cells (105 cells/lane), and of purified wild-type virions (V; 3 μg of protein/lane) were separated on SDS-12% polyacrylamide gels. Western blots were analyzed with monospecific rabbit antisera against UL31, UL34, and UL49. The digitally scanned images shown in panel A indicate that expression of UL31 and UL34 is not interdependent, whereas panel B illustrates the expression kinetics and the absence of the UL31 and UL34, but not of the UL49 proteins, from mature virions
Western blot analyses further revealed that the amount of UL31 protein continuously increased from 6 to 20 h after infection of RK13 cells with wild-type virus (Fig. 2B), indicating that UL31 of PrV, like its HSV-1 homologue, presumably represents a late virus gene (44). Remarkably, the UL31 gene product of PrV, like the previously described UL34 protein (29), could not be detected in gradient-purified extracellular virions, whereas the tegument protein encoded by UL49, as expected (7), was present in mature virus particles (Fig. 2B).
Construction and in vitro characterization of UL31 deletion mutants.
An infectious clone of the PrV genome containing a mini-F plasmid vector within the nonessential gG gene (pPrV-K1 [Fig. 1C]) was used for deletion of UL31 and concomitant insertion of the selectable kanamycin resistance gene in either orientation (Fig. 1C) by RecE- and RecT-mediated recombination in E. coli. Because of overlaps with the adjoining UL30 and UL32 genes, the UL31 ORF was not completely removed from the genomic clones generated, pPrV-ΔUL31a and -b (Fig. 1B). However, the remaining first 22 codons of UL31 should be insufficient to form a functional protein, and the 3′-terminal fragment lacks the preceding promoter. To avoid unwanted side effects of the mini-F plasmid insertion at the gG gene locus, pPrV-ΔUL31a and -b were rescued by digestion at unique EcoRI sites within the bacterial vector and subsequent cotransfection of eucaryotic cells together with a second plasmid containing the gG gene region of the parental PrV strain Ka (Fig. 1C). Thus, as verified by DNA analyses (data not shown), the resulting recombinants PrV-ΔUL31a and -b differed from wild-type virus only by their mutations within the UL31 gene. Western blot analyses further demonstrated that neither the full-length UL31 protein nor truncated gene products were expressed in cells infected with either of the deletion mutants (Fig. 2A), whereas expression of other capsid or tegument proteins, such as UL19, UL46, UL48 (data not shown), and UL49 (Fig. 2A) was not affected.
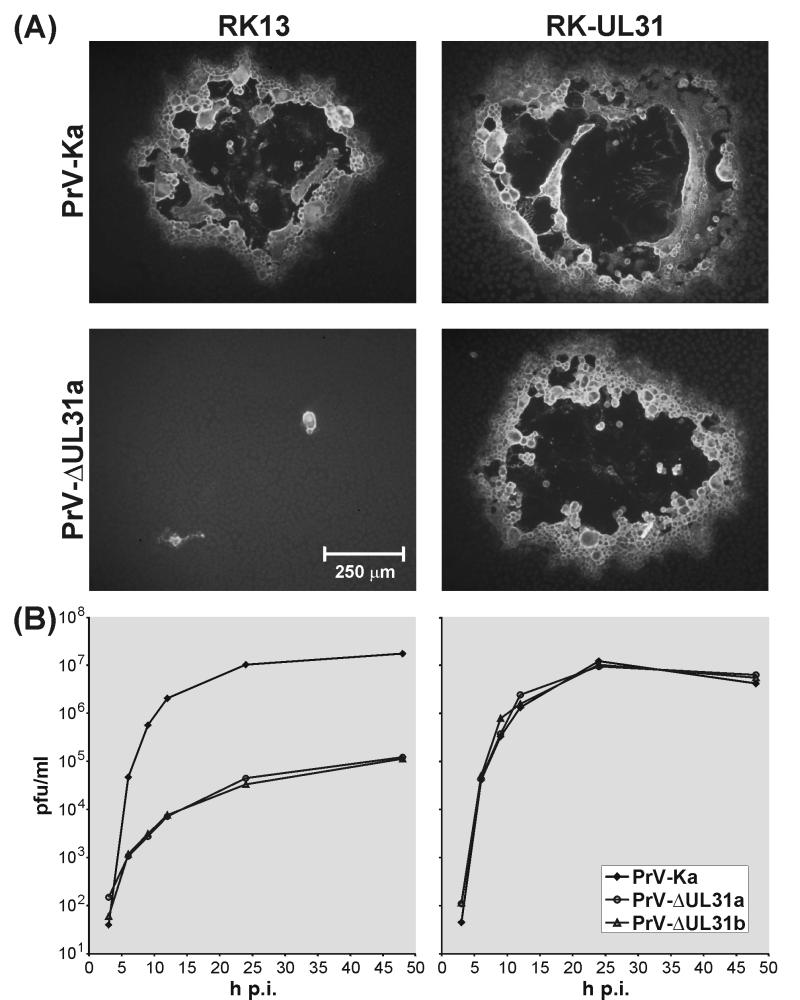
Growth properties of the UL31 deletion mutants were analyzed in RK13 cells and in UL31-expressing RK-UL31 cells (Fig. 3). Whereas in trans-complementing RK-UL31 cells the plaques formed by PrV-ΔUL31a or -b were indistinguishable from those of PrV-Ka, plaque formation of the UL31 deletion mutants was almost completely inhibited in noncomplementing RK13 cells, and only small foci of infected cells were observed (Fig. 3A). One-step growth kinetics analysis of RK13 cells revealed that maximum virus titers of PrV-ΔUL31a or -b (ca. 105 PFU/ml) are more than 100-fold lower than those of wild-type PrV (Fig. 3B). The replication defect of UL31 deletion mutants was completely repaired in trans-complementing RK-UL31 cells (Fig. 3B).
FIG. 3.
In vitro growth properties of UL31 deletion mutants. (A) RK13 and RK-UL31 cells were fixed 48 h after infection under plaque assay conditions with PrV-Ka or PrV-ΔUL31a. Plaques were visualized by indirect immunofluorescence with a gC-specific monoclonal antibody. (B) One-step growth kinetics of PrV-Ka and of PrV-ΔUL31a and -b were determined in RK13 and RK-UL31 cells. At the indicated times after infection at an MOI of 10, the total amount of infectious intracellular and released progeny virus was determined by plaque assays on RK-UL31 cells.
These findings demonstrate that UL31 is important for replication of PrV. However, productive virus replication of UL31 deletion mutants at lower levels was also observed in noncomplementing cells (Fig. 3B). This was confirmed by detection of progeny virus after transfection of RK13 or porcine kidney (PSEK) cells with the E. coli-derived infectious plasmid pPrV-ΔUL31a or -b. The virus obtained could be serially passaged in these noncomplementing cells. Moreover, infectivity was demonstrated not to be cell associated, since it was resistant to freezing and thawing, remained in the supernatant after low-speed centrifugation, and could be filtered through 0.2-μm-pore-size nitrocellulose filters (data not shown). Thus, UL31 is not absolutely essential for replication of PrV in cultured cells.
The UL31 gene product is involved in primary nucleocapsid envelopment.
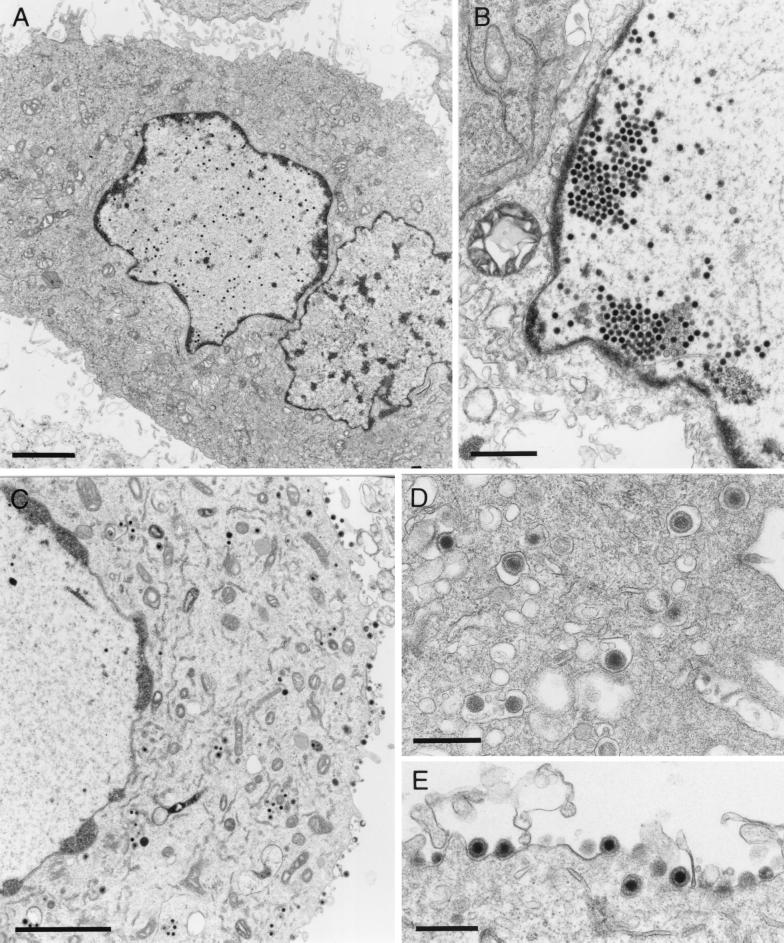
Electron microscopy of RK13 cells which were fixed 14 h after infection with either of the UL31 deletion mutants of PrV revealed that capsid formation and DNA encapsidation occurred but that nucleocapsids apparently were unable to exit the nucleus (Fig. 4A and B). Sometimes pseudocrystalline aggregates of filled capsids could be found in the vicinity of the inner nuclear membrane (Fig. 4B), but no unequivocal budding stages, and consequently no cytoplasmic or extracellular virions, were detectable (Fig. 4A). Electron microscopy also confirmed that these replication defects of PrV-ΔUL31a and -b could be corrected in UL31-expressing RK-UL31 cells (Fig. 4C through E). As in normal cells infected with wild-type PrV (23), only a few enveloped virus particles were found in the perinuclear space, but naked nucleocapsids as well as vesicles containing enveloped particles were present in the cytoplasm (Fig. 4D), and numerous released virions were visible (Fig. 4E).
FIG. 4.
Electron microscopy of cells infected with PrV-ΔUL31a. Noncomplementing RK13 cells (A and B) and trans-complementing RK-UL31 cells (C through E) were fixed 14 h after infection at an MOI of 1 and embedded in glycid ether 100. Ultrathin sections were stained with uranyl acetate and lead salts. In the absence of UL31, nucleocapsids are retained in the nucleus (A and B), whereas virus maturation in the cytoplasm (C and D) and release (C and E) are restored in UL31-expressing cells. Bars, 2.5 μm (A and C), 1 μm (B), or 500 nm (D and E).
Yeast two-hybrid interactions between the UL31 and UL34 gene products of PrV.
To investigate physical interactions of the UL31 gene product with other proteins, the entire ORF, preceded by 5 artificial codons, was 3′-terminally fused to the LexA gene (pLex-UL31 [Fig. 1D]) and expressed as a DNA-binding bait protein. This construct was used to screen an expression library of randomly cleaved genomic PrV DNA, fused to the gene of a transcriptional activator (B42). Interactions between bait and library proteins in yeast cells were detected by trans-activation of a lacZ reporter gene regulated by LexA operator elements (Fig. 1D). Single library plasmids were isolated from yeast clones forming blue colonies on X-Gal-containing agar plates, sequenced, and tested again for specific two-hybrid interactions with pLex-UL31, including the empty vectors as controls (Table 2). Of six positive library plasmids tested, five harbored fragments of the UL34 gene of PrV, and four of the five pB42-UL34 plasmids contained different inserts. Interestingly, all of them started in frame with, but upstream of, the original UL34 initiation codon and extended at least to position 162 of the 262-codon ORF (Table 2). This finding indicates that the domain responsible for interaction with the UL31 protein is located in the predicted nucleoplasmic N-terminal part of the putative type II membrane protein UL34 (29).
TABLE 2.
Yeast two-hybrid interactions between the UL31 and UL34 proteins of PrV
| Library plasmid | Interactiona with the following bait plasmid:
|
||
|---|---|---|---|
| pLexA | pLex-UL31, (−5)–271* | pLex-UL34, (−2)–220 | |
| pB42AD | No | No | No |
| pB42-UL31 | |||
| 32–271* | No | NT | Yes |
| 42–271* | No | NT | Yes |
| pB42-UL34 | |||
| (−20)–216 | No | Yes | NT |
| (−19)–193 | No | Yes | NT |
| (−16)–162 | No | Yes | NT |
| (−8)–193 | No | Yes | NT |
Two-hybrid interactions in yeast clones containing reporter, bait, and library plasmids were detected by X-Gal staining. Yes, positive; No, negative; NT, not tested. The codons of the PrV UL31 and UL34 genes expressed as LexA or B42 fusion proteins are indicated. The presence of the authentic stop codon is indicated by an asterisk, and artificial codons preceding the original initiation site are enclosed in parentheses.
To further confirm the specificity of this interaction between the two viral proteins, the 5′-terminal part of UL34 (codons 1 to 220) was cloned in pLexA (Fig. 1D) and expressed together with the PrV library. After screening for LacZ-expressing yeast colonies, two positive clones were identified. Both contained library plasmid inserts permitting expression of slightly different parts of the UL31 gene product (Table 2), including the complete C terminus of the viral protein, but lacking the hydrophilic N-terminal part with the putative nuclear localization signals.
Subcellular localization of the UL31 protein depends on the UL34 gene product.
In Western blot analyses, the UL31 protein expressed by the UL34 deletion mutant PrV-ΔUL34B (29) was indistinguishable from that of wild-type PrV (Fig. 2A), confirming that the UL34 gene product is not required for stable expression of UL31. However, whereas 8 h after infection of RK13 cells with either PrV-ΔUL34B or wild-type virus, UL31 and UL49 were expressed at comparable levels (Fig. 2A), the relative amount of UL31 protein declined at later times after infection with PrV-ΔUL34B (data not shown). Conversely, deletion of the UL31 gene of PrV does not affect the apparent size or amount of the UL34 protein (Fig. 2A).
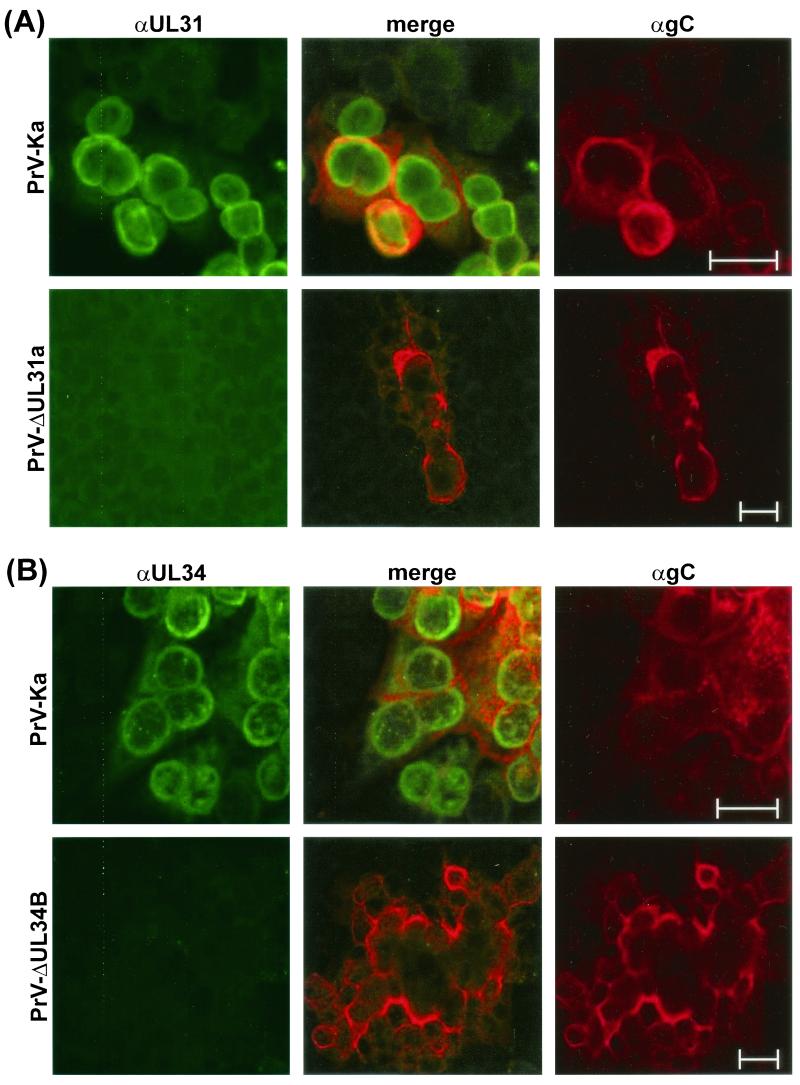
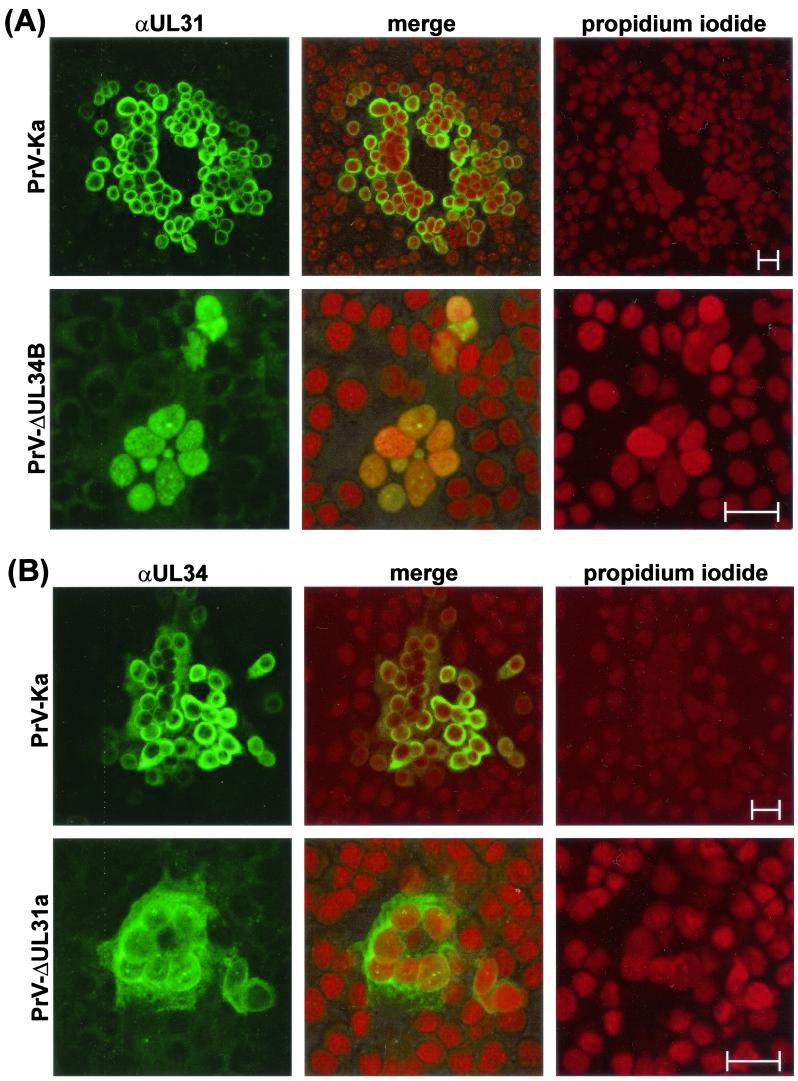
A more pronounced effect of interdependence between UL31 and UL34 was detected by indirect immunofluorescence reactions of the monospecific antisera, which revealed a similar localization of both proteins in PrV-Ka-infected cells (Fig. 5 and 6). As expected, no specific reactivity of the anti-UL31 serum (Fig. 5A) or of the anti-UL34 serum (Fig. 5B) was observed in noncomplementing RK13 cells infected with the respective deletion mutant PrV-ΔUL31a or PrV-ΔUL34B, although virus replication could be demonstrated by detection of the virion gC. In cells infected with PrV-Ka, confocal microscopy showed that the majority of gC was localized at cytoplasmic membranes and microsomes (Fig. 5, red fluorescence), whereas the UL31- and UL34-specific antisera predominantly labeled clearly distinct perinuclear structures (Fig. 5, green fluorescence). An occasional merged yellow fluorescence indicative of “colocalization” of the investigated proteins with gC was most likely caused by disintegration of cell compartments or cell contraction during the late phase of infection. Counterstaining of chromatin with propidium iodide (Fig. 6) also indicated that the bulk of the UL31 and UL34 proteins are associated either with the lamina or with the nuclear membrane. The latter localization was confirmed by immune electron microscopy for UL34 (29) as well as for UL31 (see below).
FIG. 5.
Subcellular localization of the UL31 and UL34 proteins of PrV. Infected RK13 cells were incubated for 24 h (PrV-Ka) or 48 h (PrV-ΔUL31a and PrV-ΔUL34B) and fixed with a 1:1 mixture of methanol and acetone. Binding of αUL31 (A) or αUL34 (B) sera was detected by Alexa 488-conjugated secondary antibodies. For comparison, gC was visualized by a monoclonal antibody and TRITC-conjugated anti-mouse immunoglobulins. Green, red, and merged fluorescence reactions were analyzed by confocal laser scanning microscopy. Bars, 25 μm.
FIG. 6.
Nuclear localization of the UL31 and UL34 proteins. PrV-infected RK13 cells were fixed after 24 h (PrV-Ka) or 48 h (PrV-ΔUL31a, PrV-ΔUL34B) with a 1:1 mixture of methanol and acetone. Indirect immunofluorescence reactions of UL31- (A) and UL34 (B)-specific antisera and Alexa 488-conjugated secondary antibodies were analyzed by confocal laser scanning microscopy after chromatin counterstaining with propidium iodide. Green, red, and merged fluorescence reactions are imaged separately. Bars, 25 μm.
In the absence of the UL34 protein, i.e., in cells infected with PrV-ΔUL34B (Fig. 6A) or in uninfected RK-UL31 cells (data not shown), the UL31 protein was no longer found at nuclear membranes but colocalized with chromatin throughout the nucleus, resulting in a merger of green and red fluorescence (Fig. 6A). In contrast, localization of the UL34 protein at the nuclear membrane was not abolished in the absence of the UL31 protein. However, in RK13 cells transfected with an UL34 expression plasmid (29), as well as in cells infected with PrV-ΔUL31a or -b, a larger proportion of the UL34 gene product was found to be dispersed within the cytoplasm compared to the pattern in cells infected with wild-type PrV (Fig. 6B). Since UL31 and UL34 deletion mutants were drastically impaired in cell-to-cell spread, different time points after infection with these mutants (48 h) and with PrV-Ka (24 h) were chosen to achieve comparable cytopathic effects. However, in the single infected cells observed 24 h after infection with PrV-ΔUL31a, the UL34 protein was also found predominantly in the cytoplasm, and the UL31 protein was never detectable at the nuclear rims of cells analyzed 24 h after infection with PrV-ΔUL34B (data not shown). Thus, association of the two proteins with the nuclear membrane is indeed interdependent.
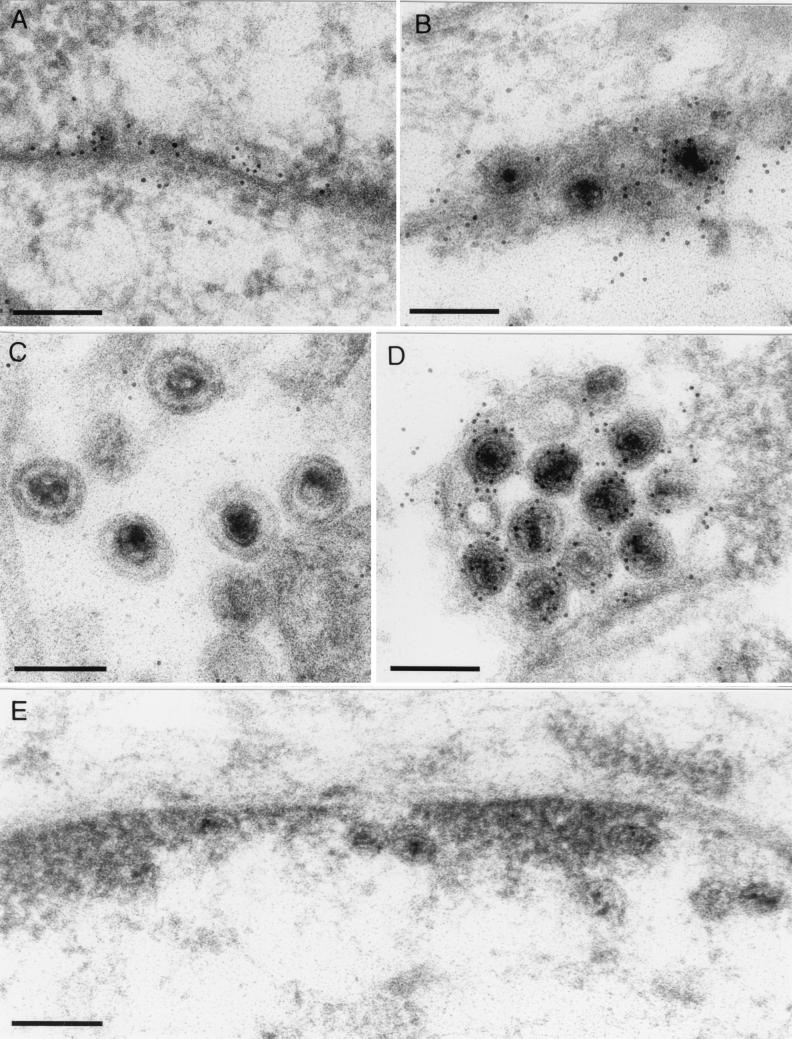
The UL31 protein is incorporated into primary enveloped virions.
Although the UL34 gene product of PrV was shown to be absent from mature virions, it could be detected in enveloped virus particles within the perinuclear space (29). To test whether this is also the case for the UL31 protein, RK13 cells infected with wild-type PrV-Ka, PrV-ΔUL31a, or PrV-ΔUS3 (30) were analyzed by immune electron microscopy (Fig. 7). The latter virus mutant was chosen because in wild-type PrV-infected cells, transit through the nuclear membrane appears to be a rapid process, and only a very few enveloped virus particles are usually found in the perinuclear space (Fig. 7B). In contrast, PrV recombinants lacking the US3 protein kinase reproducibly accumulate enveloped nucleocapsids in the perinuclear space (Fig. 7D) (30, 53). As visualized with an anti-UL31 serum and gold-tagged secondary antibodies, the UL31 gene product is present in these primary enveloped virions of both PrV-Ka (Fig. 7B) and PrV-ΔUS3 (Fig. 7D) either as a membrane-associated protein or as a constituent of tegument-like structures. In accordance with previous Western blot analyses (see above), released virions (Fig. 7C), and enveloped particles in cytoplasmic vesicles (data not shown) were not labeled. Correlating with the nuclear rim staining observed in immunofluorescence studies (see above), the UL31 protein was detected at the nuclear membranes of wild-type PrV-infected cells, including sites where no budding virions were present (Fig. 7A). In contrast, no specific labeling of nuclear membranes, nucleocapsids, or other structures was found in RK13 cells infected with PrV-ΔUL31 (Fig. 7E).
FIG. 7.
Localization of the UL31 protein within primary enveloped virus particles. RK13 cells were infected with PrV-Ka (A through C), PrV-ΔUS3 (D), or PrV-ΔUL31a (E) at an MOI of 1, fixed after 14 h, and embedded in Lowicryl K4M. After subsequent incubation with anti-UL31 serum and gold-tagged secondary antibodies, ultrathin sections were counterstained with uranyl acetate and analyzed by electron microscopy. The UL31 protein of PrV was detected along the nuclear membrane (A) and in primary enveloped virus particles in the perinuclear space (B and D) but not in released virions (C). In cells infected with PrV-ΔUL31a (E), no specific labeling was found. Bars, 200 nm.
DISCUSSION
We analyzed a 2.6-kbp fragment originating from the UL region of the PrV genome which contains two highly conserved herpesvirus genes corresponding to the UL31 and UL32 ORFs of HSV-1 (33). The deduced amino acid sequences of both identified PrV genes exhibit identities of more than 50% to homologous proteins of other mammalian alphaherpesviruses. Early investigations indicated that gene layout within the UL genome region of PrV is distinct from that of other alphaherpesviruses, and sequence analyses demonstrated an internal inversion bordered by the gB (UL27) and gC (UL44) genes (4, 8). Our present studies confirm that the gene arrangement within the left part of this inversion, including the PrV homologues of UL27 (42), UL28 (40), UL29 (55), UL30 (5), UL31, UL32, and UL33 to UL35 (29), is perfectly colinear with that of HSV-1 (33), VZV (13), and EHV-1 (50).
For functional characterization of the UL31 gene, an infectious full-length clone of the genome of PrV-Ka was generated by insertion of a mini-F plasmid vector (39) at the nonessential (37) gG gene locus. Propagation of the PrV genome as a bacterial artificial chromosome permitted efficient mutagenesis by RecE- and RecT-mediated recombination in E. coli (59). However, even at nonessential sites of the genome, bacterial vector insertions may affect in vitro or in vivo replication of reconstituted herpesviruses, and should be removed prior to characterization of the virus mutants generated (1, 49). Our infectious clone pPrV-K1 also exhibited subtle but reproducible replication deficiencies in cultured cells. Ultrastructural analyses of virus morphogenesis revealed that pPrV-K1 and its derivatives, like US3 deletion mutants of PrV (30, 53), accumulated enveloped virus particles in the perinuclear space, and Western blot analyses demonstrated significantly reduced expression of the US3 protein kinase (data not shown). Presumably, the F-plasmid insertion within the gG gene (US4) interferes with the US3 mRNA, since the transcripts of US3 and US4 are 3′ coterminal (52). Therefore, the US3 and gG genes of the UL31 deletion mutants of PrV were repaired, and vector sequences were completely removed after mutagenesis in E. coli.
Independent of the presence or absence of bacterial vector sequences, the UL31 deletion mutants exhibited substantial replication defects in noncomplementing cells. However, although plaque formation was almost completely abolished, virus yields of ca. 105 PFU/ml, which were not caused by phenotypic complementation or genotypic reversion, were reproducibly obtained. Thus, the UL31 gene of PrV is, strictly speaking, not essential for virus replication in vitro. In contrast, the homologous UL31 of HSV-1 was designated an essential gene (44, 57), although a respective deletion mutant also replicated at low levels in noncomplementing cells (11). Electron microscopic studies of cells infected with this UL31 null mutant of HSV-1 revealed the absence of cytoplasmic and extracellular virions, but also an inefficient cleavage and encapsidation of viral DNA in the nucleus, which was confirmed by comparison of the amounts of monomeric and concatemeric genomes (11). PrV-ΔUL31 was also impaired in exit from the nucleus of noncomplementing cells but produced large amounts of full capsids. Thus, although quantitative DNA analyses have yet to be performed, our ultrastructural studies indicate that the UL31 protein of PrV is presumably not required for nucleocapsid formation but functions in a subsequent step of virus egress which leads to envelopment at the inner nuclear membrane. This function could be assigned to the deleted gene by trans-complementation studies, which demonstrated that PrV-ΔUL31 exhibits wild-type-like plaque formation, growth kinetics, and ultrastructural replication characteristics in cells expressing UL31.
Like the PrV-ΔUL31 recombinants, previously described UL34 deletion mutants of PrV (29) and HSV-1 (45) were also characterized by retention of unenveloped nucleocapsids in the host cell nucleus. Coinciding with these findings, our yeast two-hybrid studies revealed a physical interaction between the UL31 and UL34 gene products of PrV, as has also been indicated by GST pull-down experiments for the respective proteins of HSV-1 (58). This concordance, as well as the high degrees of homology between the deduced UL31 and UL34 proteins of HSV-1 and PrV, suggests that the protein interactions detected in different assay systems are functionally relevant. It has been demonstrated that in cells infected with a UL34 deletion mutant of HSV-1, the UL31 protein was also hardly detectable, presumably due to rapid degradation by proteases (57). In noncomplementing cells infected with PrV-ΔUL34B, the amount of the UL31 gene product also declined at late times after infection (data not shown). However, during the productive phase of virus replication, the UL31 protein was detected at similar levels in wild-type- and PrV-ΔUL34B-infected cells. Furthermore, considerable amounts of the viral protein were found in uninfected UL31-expressing cell lines. Thus, in the case of PrV, protection of the UL31 gene product is more likely a side effect than the main function of the supposed interaction with the UL34 protein. There is also no evidence that interaction of the UL31 and UL34 gene products of PrV is required for correct processing, since the electrophoretic mobility of each protein was similar in the presence or absence of its partner. Moreover, the molecular mass of the UL31 protein of PrV appeared identical not only in infected and constitutively expressing cells, but also after translation in a cell-free system. Unlike the homologous protein of HSV-1, which has been shown to be nucleotidylylated and phosphorylated (6, 10), the UL31 gene product of PrV lacks the conserved amino acid sequence motif of nucleotidylylated proteins, and phosphate labeling was also not detectable (data not shown).
The UL31 and UL34 proteins of PrV apparently are not covalently linked, since only monomers were found in Western blot analyses of wild-type PrV-infected cell lysates, even if they were prepared and separated in the absence of β-mercaptoethanol. However, immunofluorescence reactions of monospecific antisera against either of the proteins revealed colocalization at the nuclear membranes of cells infected with wild-type PrV. The UL34 gene product of PrV was predicted to represent a type II membrane protein with a hydrophobic anchor domain in its C-terminal part (29). Since the UL31 gene product lacks extended stretches of nonpolar amino acids, it is most likely attached to the nuclear membrane via its interaction with the UL34 protein. Transient coexpression studies of the homologous proteins of HSV-2 indicated that the UL31 gene product might be required for relocation of the UL34 protein to the inner nuclear membrane (56). Our investigations of cells infected with PrV-ΔUL31 mutants confirmed that accumulation of the UL34 protein at the nuclear membrane is indeed reduced in favor of a more cytoplasmic distribution. However, as in noninfected UL34-expressing cells (29), the protein was still detectable at the inner and outer nuclear membranes, indicating that UL31 might function in retention more than in targeting of the UL34 gene product of PrV. On the other hand, the UL34 gene product of PrV seems to be more important for proper localization of the UL31 protein, because in cells infected with PrV-ΔUL31B, as in noninfected RK-UL31 cells, the expressed protein is excluded from the nuclear membrane region but is dispersed throughout the nucleus. Interestingly, a diffuse intranuclear distribution of the UL31 proteins of HSV-1 and HSV-2 was described even in cells infected with the respective wild-type viruses (10, 60). All identified UL31 proteins exhibit basic amino acid sequence motifs which might serve as nuclear localization signals (21) and are efficiently transported into the nucleus, but only the PrV protein was clearly shown to be targeted to the nuclear membrane in the presence of UL34.
A common function of the UL31 proteins of different alphaherpesviruses might be involvement in transport of nucleocapsids to the inner nuclear membrane. The interacting UL31 and UL34 proteins might also contribute to disintegration of the nuclear lamina or might directly mediate the budding process initiating virus egress. The latter hypothesis was supported by the observation that enveloped virus particles located in the perinuclear space contain considerable amounts of the UL34 (29) as well as the UL31 protein. Whereas the UL34 gene product presumably represents a primary envelope protein, the UL31 gene product might be a component of the primary tegument. Remarkably, the UL31 and UL34 proteins of PrV were not found in mature virions. In contrast, the UL49 gene product, representing a major tegument protein of released virus particles, was not detectable in perinuclear virions (29). Thus, our results indicate that the envelope and presumably also the tegument of perinuclear virus particles are lost and that cytoplasmic nucleocapsids are reenveloped in the trans-Golgi region prior to release from host cells. This two-step model of herpesvirus egress has long been proposed for PrV (7, 18, 23, 54) and VZV (20, 61), and more recent results demonstrate a similar process in cells infected with HSV-1 (9, 23, 47).
An unresolved question is why the UL31 and UL34 proteins of PrV and HSV-1, although crucial for virion formation, are not absolutely essential for productive virus replication in cultured cells (11, 29, 45, 57). Possibly, an independent but less efficient mechanism of nucleocapsid transport through the nuclear membrane is mediated by other viral or cellular proteins. Alternatively, virus induced or cell cycle dependent disintegration of the nuclear membrane might permit direct release of newly synthesized nucleocapsids to the cytoplasm, where they could obtain an envelope.
ADDENDUM
During revision of the present report, a study appeared which indicates that the subcellular localization and major functions of the conserved UL31 and UL34 gene products of PrV and HSV-1 are very similar (41a).
Acknowledgments
The present study was supported by a grant from the DFG (Me854/5-1).
We are obliged to G. A. Smith, L. W. Enquist, and P. Ioannou for providing plasmids and helpful comments required for BAC cloning and mutagenesis. We also thank E. Mundt for help with antiserum preparation. The excellent technical assistance of C. Ehrlich and P. Meyer is greatly appreciated.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211. [DOI] [PubMed] [Google Scholar]
- 3.Ben Porat, T., J. M. Demarchi, and A. S. Kaplan. 1974. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology 61:29–37. [DOI] [PubMed] [Google Scholar]
- 4.Ben Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus type 1 and pseudorabies virus. Virology 127:194–204. [DOI] [PubMed] [Google Scholar]
- 5.Berthomme, H., S. J. Monahan, D. S. Parris, B. Jacquemont, and A. L. Epstein. 1995. Cloning, sequencing, and functional characterization of the two subunits of the pseudorabies virus DNA polymerase holoenzyme: evidence for specificity of interaction. J. Virol. 69:2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401–17410. [PubMed] [Google Scholar]
- 7.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29–40. [DOI] [PubMed] [Google Scholar]
- 9.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus type 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 67:6348–6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 13.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816. [DOI] [PubMed] [Google Scholar]
- 14.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dretzen, G., M. Bellard, P. Sassone-Corsi, and P. Chambon. 1981. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal. Biochem. 112:295–298. [DOI] [PubMed] [Google Scholar]
- 17.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 80:2173–2182. [DOI] [PubMed] [Google Scholar]
- 20.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Görlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271:1513–1518. [DOI] [PubMed] [Google Scholar]
- 22.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. [DOI] [PubMed] [Google Scholar]
- 23.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791–803. [DOI] [PubMed] [Google Scholar]
- 25.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jöns, A., and T. C. Mettenleiter. 1997. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. J. Virol. Methods 66:283–292. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. [DOI] [PubMed] [Google Scholar]
- 29.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein kinase on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363–2371. [DOI] [PubMed] [Google Scholar]
- 31.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531–1574. [DOI] [PubMed] [Google Scholar]
- 34.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. [DOI] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C. 2000. Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99–115. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C., and I. Rauh. 1990. A glycoprotein gX-beta-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J. Virol. Methods 30:55–65. [DOI] [PubMed] [Google Scholar]
- 38.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6:442–447. [DOI] [PubMed] [Google Scholar]
- 39.O’Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 244:1307–1312. [DOI] [PubMed] [Google Scholar]
- 40.Pederson, N. E., and L. W. Enquist. 1989. The nucleotide sequence of a pseudorabies virus gene similar to ICP18.5 of herpes simplex virus type 1. Nucleic Acids Res. 17:3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radsak, K., M. Eickmann, T. Mockenhaupt, E. Bogner, H. Kern, A. Eis-Hubinger, and M. Reschke. 1996. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch. Virol. 141:557–572. [DOI] [PubMed] [Google Scholar]
- 41a.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins, A. K., D. J. Dorney, M. W. Wathen, M. E. Whealy, C. Gold, R. J. Watson, L. E. Holland, S. D. Weed, M. Levine, J. C. Glorioso, and L. W. Enquist. 1987. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J. Virol. 61:2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roizman, B. 1996. Herpesviridae, p. 2221–2230. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 44.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231–2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 45.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088–11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 75:5697–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304–316. [DOI] [PubMed] [Google Scholar]
- 51.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studied by fracture label. J. Virol. 66:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747–1755. [DOI] [PubMed] [Google Scholar]
- 53.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851–1859. [DOI] [PubMed] [Google Scholar]
- 54.Whealy, M. E., J. P. Card, R. P. Meade, A. K. Robbins, and L. W. Enquist. 1991. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J. Virol. 65:1066–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, S. L., C. Y. Hsiang, T. Y. Ho, and T. J. Chang. 1998. Identification, expression, and characterization of the pseudorabies virus DNA-binding protein gene and gene product. Virus Res. 56:1–9. [DOI] [PubMed] [Google Scholar]
- 56.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423–1428. [DOI] [PubMed] [Google Scholar]
- 57.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus type 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123–128. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, H. Y., H. Yamada, Y. M. Jiang, M. Yamada, and Y. Nishiyama. 1999. Intracellular localization of the UL31 protein of herpes simplex virus type 2. Arch. Virol. 144:1923–1935. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, C. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 69:7951–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]