Abstract
Most human immunodeficiency virus type 1 (HIV-1) transmissions in sub-Saharan Africa are believed to occur between married adults who are discordant for their HIV-1 infection status; however, no studies to date have investigated the molecular epidemiology of such transmission events. Here we report the genetic characterization of HIV-1 strains from 149 transmission pairs that were identified prospectively in a cohort of discordant couples in Lusaka, Zambia. Subgenomic gag, gp120, gp41, and/or long terminal repeat regions were amplified by PCR analysis of uncultured blood samples from both partners and sequenced without interim cloning. Pairwise genetic distances were calculated for the regions analyzed and compared to those of subtype-specific reference sequences as well as local controls. Sequence relationships were also examined by phylogenetic tree analysis. By these approaches, epidemiological linkage was established for the majority of transmission pairs. Viruses from 129 of the 149 couples (87%) were very closely related and clustered together in phylogenetic trees in a statistically highly significant manner. In contrast, viruses from 20 of the 149 couples (13%) were only distantly related in two independent genomic regions, thus ruling out transmission between the two partners. The great majority (95%) of transmitted viruses were of subtype C origin, although representatives of subtypes A, D, G, and J were also identified. There was no evidence for extensive transmission networks within the cohort, although two phylogenetic subclusters of viruses infecting two couples each were identified. Taken together, these data indicate that molecular epidemiological analyses of presumed transmission pairs are both feasible and required to determine behavioral, virological, and immunological correlates of heterosexual transmission in sub-Saharan Africa with a high level of accuracy.
By the end of the year 2000, an estimated 36 million adults and children were living with human immunodeficiency virus (HIV) infection-AIDS worldwide (39). More than 70% of these individuals resided in sub-Saharan Africa, where the average prevalence of HIV infection is currently 8.8% and transmissions occur predominantly through heterosexual routes or from mother to child (30). One of the African countries with a particularly high prevalence of human immunodeficiency virus type 1 (HIV-1) infection is Zambia, where it is estimated that 20% of all adults harbor HIV-1 and 20% of all cohabitating couples are discordant for their HIV-1 infection status (i.e., one partner is HIV-1 positive and the other is negative) (40). Novel interventions designed to curtail the explosive spread of HIV-1 in Zambia and other high-prevalence countries in sub-Saharan Africa are thus urgently needed but are likely to require detailed knowledge about the factors that influence heterosexual transmission.
The Zambia-UAB HIV Research Project (ZUHRP) was established in 1994 to provide voluntary HIV-1 testing and counseling, long-term monitoring, and health care to cohabitating couples in the capital city of Lusaka (3, 25). To date, 9,569 couples have been tested for HIV-1, of whom 21% were HIV-1 discordant, 26% were concordant HIV-1 positive, and 53% were concordant HIV-1 negative at the time of enrollment. Between February 1994 and October 2000, 1,022 discordant couples (535 with HIV-1-infected men and 487 with HIV-1-infected women) were enrolled into a prospective study of the incidence and predictors of heterosexual transmission and were monitored at 3-month intervals for seroconversion of the seronegative partner. Although testing and counseling prompted substantial risk reduction in this cohort, a seroconversion rate of 8.5 per 100 person years remained, which was similar for male-to-female and female-to-male transmissions (12). Because frequent follow-up visits facilitated blood collection from both the putative donor and the recipient after a transmission event, this cohort has provided a unique setting to examine the incidence, demographics, and behavioral and biological correlates as well as the viral and host determinants of heterosexual transmission of HIV-1. However, a prerequisite for the acquisition of meaningful data, particularly with regard to predictors of contagion in the index seropositive partner, is the ability to confirm, with a high level of confidence, epidemiological linkage of HIV-1 transmission between members of all putative transmission pairs.
Molecular analyses of suspected transmission links have been widely used to characterize localized HIV-1 outbreaks, mother-to-infant transmission, sexual transmission, sharing of contaminated needles, donation of contaminated blood, receipt of contaminated clotting reagent, nosocomial transmissions from health care workers, and intrafamilial contacts (1, 4, 5, 6, 13, 18, 21, 29, 36, 42, 44). In all of these cases, the establishment of epidemiological linkage relied on the documentation of closer genetic relatedness between viruses infecting the suspected transmission pair(s) compared to control viruses isolated from unrelated individuals in the same region. Here we developed a similar approach to confirm (or refute) heterosexual transmission among discordant couples within the ZUHRP cohort.
Blood samples were collected between 1996 and 2000 from both partners of 149 (of a total of 162) discordant couples in whom seroconversion had been documented. The time period between the last negative and the first positive blood tests for the seroconvertor (which in most cases was also the blood sample used for linkage analysis) was 4.9 months on average, but in some cases it extended up to 4 years. High-molecular-weight DNA was extracted from whole blood or Ficoll gradient-purified peripheral blood mononuclear cells by using the QIAamp Blood Kit (Qiagen, Valencia, Calif.). For a small number of couples, DNA was extracted from dried blood spots (9). Because of the known variability of HIV-1, different regions of the HIV-1 genome were targeted for PCR amplification, resulting in comparisons of gag, gp120, gp41, and/or long terminal repeat (LTR) regions as shown in Fig. 1. Although the gp41 primers were by far the most cross-reactive, the suitability of this primer set was discovered only after alternative genomic regions from a number of transmission pairs had already been analyzed (43). LTR, gp120, and gp41 primers and amplification conditions have been described previously (15, 16). The primers that were used to amplify sequences within gag were cgagA 5′-TGATAAAACCTCCAATTCCCCCTAT-3′ and PBS1A 5′-TTTGCCTGTACTGGGTCTCTCTGGTT-3′ in the first round and cgagB 5′-AATACTGTATCATCTGCTCCTGTATC-3′ and PBS1B 5′-GCTTAAGCCTCAATAAAGCTTGCCTT-3′ in the second round. PCR products were sequenced directly, using cycle sequencing and dye terminator methodologies, on an automated DNA sequencer (model 377A; Applied Biosystems, Inc., Foster City, Calif.). Both strands of the PCR products were sequenced (sequences are available under GenBank accession numbers AF404868 through AF405203, AF406742, and AF406743). Although population-based sequencing was used to allow analysis of the predominant viral form in each individual, the number of ambiguous base pairs in the entire data set was <0.3%.
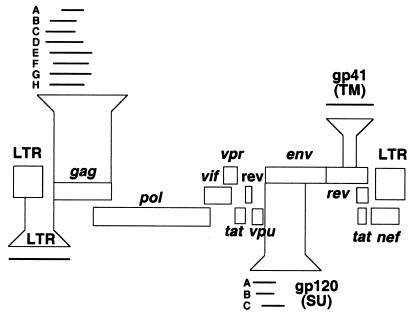
FIG. 1.
HIV-1 subgenomic regions utilized for epidemiological linkage analysis. A schematic representation of the HIV-1 genome is shown, with brackets denoting the subgenomic gag, gp120, gp41, and LTR fragments that were amplified for diagnostic PCR analysis. Overlapping gag and gp120 regions are denoted by capital letters (gagA to -H; gp120A to -C) and are referred to in Tables 2 to 4.
To establish suitable linkage criteria for HIV-1 strains infecting the Zambian couples, amplified viral sequences were first subjected to preliminary phylogenetic tree analyses to identify all circulating HIV-1 group M subtypes (not shown). Full-length and nonrecombinant reference sequences representing these subtypes were then obtained from the Los Alamos HIV Sequence Database (Table 1) and subjected to pairwise sequence comparisons in the genomic regions corresponding to the PCR amplification products. Eight partially overlapping regions in gag, three in gp120, one in gp41, and one in the LTR were used for analysis (Fig. 1). Uncorrected nucleotide sequence distances were then calculated for each transmission pair and compared to the mean sequence distances calculated for the reference sequence set in the corresponding genomic region. The latter minus two standard deviations (SDs) was arbitrarily assigned as the cutoff value for epidemiologically linked sequence pairs (Table 2). Transmission pairs were tentatively classified as epidemiologically linked when their pairwise sequence distances fell below this limit. Conversely, transmission pairs were tentatively classified as unlinked when their pairwise distances exceeded this limit. For the subtype C reference set, only single representatives from India and Brazil were included, so as to not skew results due to the more recent introduction of HIV-1 into these countries. Mean distances for the gp41 region of subtype J and the LTR region of subtype C could not be calculated because of a lack of sufficient reference sequences.
TABLE 1.
Subtype-specific full-length reference sequences from the HIV Sequence Database
| Subtype_sequence name | Accession no. | Reference |
|---|---|---|
| A_Q2317 | AF004885 | 31 |
| A_SE8891 | AF069673 | 7 |
| A_SE8538 | AF069669 | 7 |
| A_SE6594 | AF069672 | 7 |
| A_SE7535 | AF069671 | 7 |
| A_SE7253 | AF069670 | 7 |
| A_SE8131 | AF107771 | 7 |
| A_U455 | M62320 | 28 |
| A_92UG037.1 | U51190 | 14 |
| C_96BW04.07 | AF110963 | 26 |
| C_96BW11B01 | AF110971 | 26 |
| C_96BW15C02 | AF110974 | 26 |
| C_96BW05.04 | AF110968 | 26 |
| C_96BW16.26 | AF110978 | 26 |
| C_96BW12.10 | AF110972 | 26 |
| C_96BW17A09 | AF110979 | 26 |
| C_96BW01B21 | AF110960 | 26 |
| C_C2220 | U46016 | 35 |
| C_94IN11246 | AF067159 | 24 |
| C_98BR004 | AF286228 | 33 |
| C_98IS002.5 | AF286233 | 33 |
| C_98TZ013.10 | AF286234 | 33 |
| C_98TZ017.2 | AF286235 | 33 |
| C_97ZA012.1 | AF286227 | 33 |
| D_94UG114.1 | U88824 | 14 |
| D_84ZR085 | U88822 | 14 |
| D_NDK | M27323 | 37 |
| D_ELI | K03454 | 2 |
| D_Z2Z6 | M22639 | 38 |
| G_DRCBL | AF084936 | 27 |
| G_HH8793 | AF061641 | 8 |
| G_92NG083.2 | U88826 | 14 |
| G_SE6165 | AF061642 | 8 |
| J_SE91733 | AF082395 | 23 |
| J_SE92809 | AF082394 | 23 |
TABLE 2.
Genetic diversity in different subgenomic regions for two sets of reference sequences
| Subgenomic regiona | Sequence subtype | Reference sequencesb
|
Zambian donor sequencesb
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Mean distance | SD | Cutoff value | n | Mean distance | SD | Cutoff value | ||
| gagA | C | 15 | 5.5 | 1.5 | 2.5 | 6 | 5.5 | 1.4 | 2.7 |
| gagB | C | 15 | 6.9 | 1.3 | 4.3 | 12 | 5.6 | 1.3 | 3.0 |
| gagC | C | 15 | 7.7 | 1.6 | 4.5 | 7 | 6.3 | 1.3 | 3.7 |
| gagD | A | 9 | 6.7 | 1.8 | 3.1 | 3 | 13.1 | 1.6 | 9.9 |
| gagD | C | 15 | 7.6 | 1.5 | 4.6 | 6 | 8.4 | 2.5 | 3.4 |
| gagE | C | 15 | 6.1 | 1.3 | 3.5 | 7 | 5.3 | 2.1 | 1.1 |
| gagF | C | 15 | 6.0 | 1.4 | 3.2 | 8 | 4.2 | 0.8 | 2.6 |
| gagG | C | 15 | 6.2 | 1.3 | 3.6 | 9 | 5.4 | 0.6 | 4.2 |
| gagH | C | 15 | 6.3 | 1.3 | 3.7 | 4 | 7.9 | 1.0 | 5.9 |
| gp120A | C | 15 | 8.6 | 1.8 | 5.0 | 4 | 6.5 | 1.1 | 4.3 |
| gp120B | C | 15 | 7.0 | 1.9 | 3.2 | 11 | 5.9 | 1.1 | 3.7 |
| gp120C | C | 15 | 13.1 | 2.4 | 8.3 | 3 | 13.0 | 1.9 | 9.2 |
| gp41 | C | 15 | 8.4 | 1.5 | 5.4 | 81 | 9.3 | 1.9 | 5.5 |
| gp41 | D | 5 | 5.4 | 1.4 | 2.6 | 1 | NA | NA | NA |
| gp41 | G | 4 | 6.8 | 1.5 | 3.8 | 3 | 5.6 | 2.0 | 1.6 |
| gp41 | J | 2 | NA | NA | NA | 1 | NA | NA | NA |
| LTR | C | NA | NA | NA | NA | 3 | 3.2 | 0.4 | 2.4 |
The subgenomic region was analyzed as shown in Fig. 1.
Full-length (nonmosaic) subtype-specific reference sequences were obtained from the Los Alamos Sequence Database and are listed in Table 1. Mean distance is the mean percent sequence difference in the analyzed genomic region. n, number of sequences compared. The cutoff value was 2 SD below the mean. NA, not available.
Although the HIV-1 epidemic in Zambia is believed to be longstanding and mature (33), we examined the extent of genetic diversity of HIV-1 strains infecting all putative Zambian donors to exclude the possibility of a recent founder effect within this cohort. As shown in Table 2, pairwise comparison of all Zambian donor sequences yielded mean distance values, SDs, and cutoff values that were very similar to those obtained for the reference sequences. This indicated that the selected reference sequences were indeed representative of the viruses infecting the cohort. There was no evidence for an unusually high degree of genetic relatedness among the Zambian donor viruses that could have confounded the linkage analysis. Instead, the results suggested that the viruses circulating within the heterosexual transmission cohort were representative of the viruses circulating in the country at large.
Having established suitable reference sequence sets, we next used the linkage criteria (Table 2) to tentatively classify the 149 transmission pairs as either likely linked or unlinked. Table 3 lists the identification number, dates of blood collection from donor and recipient (identical unless indicated otherwise), genomic region analyzed, and viral subtype for 129 transmission pairs whose uncorrected pairwise distances fell below the cutoff value of the reference sequences (compare with Table 2). Only one transmission pair (couple 136) yielded a pairwise distance (2.7%) that was slightly above the reference cutoff limit (2.6%). However, this pair was included as a likely linked transmission event after inspection of the two sequences revealed G-to-A hypermutation (41) as the cause of 9 of 10 sequence changes between donor and recipient virus. G-to-A hypermutation was also identified as a reason for increased genetic diversity in four other pairs (couples 65, 132, 138, and 149), although in these instances distance values did not exceed the cutoff limit. The majority of all couples listed in Table 3 (127 of 129) also fell below the cutoff value of the Zambian donor sequences. These data thus indicated that most couples harbored viruses whose sequences were considerably more homologous to one another than to unrelated reference sequences from the database as well as local controls.
TABLE 3.
Genetic distances for linked Zambian transmission pairs
| Sample ID | Sample collection date (mo-day-yr) | Subgenomic regiona | Cutoff valueb | Pairwise distancec | Subtype |
|---|---|---|---|---|---|
| 47 | 6-5-98 | gagA (242) | 2.5 | 0.0 | C |
| 67 | 6-8-98 | gagA | 2.5 | 0.4 | C |
| 82 | 10-19-98 | gagA | 2.5 | 1.7 | C |
| 89 | 6-13-98 | gagA | 2.5 | 0.4 | C |
| 114 | 8-2-98 | gagA | 2.5 | 0.8 | C |
| 119 | 8-9-98 | gagA | 2.5 | 0.0 | C |
| 4 | 11-15-98 | gagB (285) | 4.3 | 1.1 | C |
| 11 | 6-6-98 | gagB | 4.3 | 1.1 | C |
| 23 | 8-7-98 | gagB | 4.3 | 0.4 | C |
| 16 | 2-10-98 | gagB | 4.3 | 0.7 | C |
| 17 | 8-9-96 | gagB | 4.3 | 0.0 | C |
| 25 | 6-12-97 | gagB | 4.3 | 2.5 | C |
| 29 | 6-7-98 | gagB | 4.3 | 4.0 | C |
| 33 | 6-8-98 | gagB | 4.3 | 0.7 | C |
| 42 | 7-22-96 (F) | gagB | 4.3 | 0.0 | C |
| 8-9-96 (M) | |||||
| 49 | 7-9-96 | gagB | 4.3 | 0.4 | C |
| 1 | 10-16-98 | gagC (317) | 4.5 | 2.3 | C |
| 2 | 7-4-98 | gagC | 4.5 | 2.0 | C |
| 5 | 6-15-98 | gagC | 4.5 | 0.6 | C |
| 10 | 2-14-96 | gagC | 4.5 | 1.3 | C |
| 30 | 6-8-98 | gagC | 4.5 | 0.0 | C |
| 71 | 9-5-98 | gagC | 4.5 | 1.7 | C |
| 20 | 10-15-98 | gagD (406) | 3.1 | 1.5 | A |
| 56 | 6-6-98 | gagD | 3.1 | 1.5 | A |
| 142 | 8-13-98 | gagD | 3.1 | 1.0 | A |
| 46 | 6-7-98 | gagD | 4.6 | 0.5 | C |
| 55 | 8-13-98 | gagD | 4.6 | 0.8 | C |
| 62 | 6-6-98 | gagD | 4.6 | 2.2 | C |
| 103 | 7-8-98 | gagD | 4.6 | 1.1 | C |
| 76 | 6-25-98 | gagE (458) | 3.5 | 0.9 | C |
| 101 | 6-20-98 | gagE | 3.5 | 0.4 | C |
| 108 | 6-19-98 (F) | gagE | 3.5 | 1.1 | C |
| 6-20-98 (M) | |||||
| 124 | 8-15-98 | gagE | 3.5 | 0.2 | C |
| 37 | 6-8-98 | gagF (381) | 3.2 | 2.1 | C |
| 51 | 6-6-98 | gagF | 3.2 | 2.4 | C |
| 58 | 6-19-98 | gagF | 3.2 | 0.3 | C |
| 64 | 8-6-98 | gagF | 3.2 | 1.1 | C |
| 70 | 8-27-98 | gagF | 3.2 | 1.1 | C |
| 68 | 8-9-98 | gagG (445) | 3.6 | 2.5 | C |
| 92 | 6-8-98 | gagG | 3.6 | 0.9 | C |
| 97 | 6-6-98 | gagG | 3.6 | 2.9 | C |
| 99 | 6-6-98 | gagG | 3.6 | 0.2 | C |
| 102 | 6-23-98 | gagG | 3.6 | 1.4 | C |
| 105 | 7-21-98 | gagG | 3.6 | 1.4 | C |
| 106 | 6-8-98 | gagG | 3.6 | 1.1 | C |
| 110 | 8-20-98 | gagG | 3.6 | 0.5 | C |
| 22 | 6-7-98 | gagH (373) | 3.7 | 3.1 | C |
| 73 | 6-6-98 | gagH | 3.7 | 0.8 | C |
| 21 | 8-21-98 (F) | gp120A (373) | 5.0 | 2.5 | C |
| 5-31-98 (M) | |||||
| 60 | 6-6-98 | gp120A | 5.0 | 0.6 | C |
| 113 | 8-7-98 | gp120A | 5.0 | 1.9 | C |
| 131 | 8-23-98 | gp120A | 5.0 | 0.0 | C |
| 66 | 11-6-98 | gp120B (296) | 3.2 | 0.3 | C |
| 75 | 8-7-98 | gp120B | 3.2 | 1.7 | C |
| 78 | 6-30-98 | gp120B | 3.2 | 0.3 | C |
| 83 | 8-10-98 | gp120B | 3.2 | 1.0 | C |
| 90 | 6-6-98 | gp120B | 3.2 | 0.7 | C |
| 122 | 11-12-98 | gp120B | 3.2 | 1.0 | C |
| 136d | 10-13-99 | gp41 (411) | 2.6 | 2.7e | D |
| 43 | 3-16-97 (F) | gp41 | 3.8 | 1.0 | G |
| 6-24-98 (M) | |||||
| 91 | 6-6-98 | gp41 | 3.8 | 2.3 | G |
| 148 | 3-4-00 | gp41 | 3.8 | 0.5 | G |
| 98d | 8-19-97 (F) | gp41 | n/a | 3.8 | J |
| 10-26-98 (M) | |||||
| 3 | 9-9-99 | gp41 | 5.4 | 0.5 | C |
| 6 | 2-16-00 | gp41 | 5.4 | 1.5 | C |
| 8 | 7-29-96 | gp41 | 5.4 | 2.5 | C |
| 12 | 6-6-00 | gp41 (411) | 5.4 | 1.8 | C |
| 13 | 9-12-99 | gp41 | 5.4 | 0.8 | C |
| 14 | 10-28-99 | gp41 | 5.4 | 0.6 | C |
| 15 | 9-23-99 | gp41 | 5.4 | 1.8 | C |
| 19 | 6-6-98 | gp41 | 5.4 | 2.3 | C |
| 24 | 7-13-96 | gp41 | 5.4 | 1.8 | C |
| 26 | 11-4-99 | gp41 | 5.4 | 0.0 | C |
| 27 | 4-15-97 | gp41 | 5.4 | 1.1 | C |
| 28 | 5-16-96 | gp41 | 5.4 | 0.5 | C |
| 32 | 8-6-99 | gp41 | 5.4 | 0.8 | C |
| 34 | 6-14-96 (F) | gp41 | 5.4 | 1.5 | C |
| 5-14-96 (M) | |||||
| 35 | 12-12-97 (F) | gp41 | 5.4 | 2.0 | C |
| 10-12-96 (M) | |||||
| 36 | 8-25-99 | gp41 | 5.4 | 0.3 | C |
| 38 | 4-8-00 | gp41 | 5.4 | 1.8 | C |
| 39 | 8-27-99 | gp41 | 5.4 | 1.1 | C |
| 40 | 7-25-97 (F) | gp41 | 5.4 | 2.5 | C |
| 10-16-98 (M) | |||||
| 41 | 10-20-99 | gp41 | 5.4 | 2.8 | C |
| 45 | 3-29-00 | gp41 | 5.4 | 2.2 | C |
| 48 | 9-3-99 (F) | gp41 | 5.4 | 1.5 | C |
| 9-19-00 (M) | |||||
| 50 | 9-5-96 | gp41 | 5.4 | 1.5 | C |
| 53 | 2-12-00 | gp41 | 5.4 | 0.8 | C |
| 59 | 9-2-96 | gp41 | 5.4 | 0.6 | C |
| 61 | 9-28-98 | gp41 | 5.4 | 2.4 | C |
| 63 | 4-13-00 | gp41 | 5.4 | 1.8 | C |
| 65 | 11-7-99 | gp41 | 5.4 | 4.8 | C |
| 74 | 8-8-98 | gp41 | 5.4 | 1.7 | C |
| 77 | 4-30-97 | gp41 | 5.4 | 0.5 | C |
| 79 | 9-3-99 (F) | gp41 | 5.4 | 1.3 | C |
| 6-30-98 (M) | |||||
| 80 | 1-14-00 | gp41 | 5.4 | 1.3 | C |
| 81 | 2-11-98 | gp41 | 5.4 | 1.5 | C |
| 84 | 11-7-99 | gp41 | 5.4 | 1.8 | C |
| 85 | 10-8-97 (F) | gp41 | 5.4 | 2.7 | C |
| 5-29-97 (M) | |||||
| 86 | 11-12-98 | gp41 | 5.4 | 0.8 | C |
| 93 | 10-18-99 | gp41 | 5.4 | 0.5 | C |
| 95 | 9-12-00 | gp41 | 5.4 | 2.0 | C |
| 100 | 6-30-00 | gp41 | 5.4 | 0.5 | C |
| 107 | 8-11-00 (F) | gp41 | 5.4 | 1.5 | C |
| 8-16-00 (M) | |||||
| 109 | 3-16-00 | gp41 | 5.4 | 2.2 | C |
| 111 | 9-15-00 | gp41 | 5.4 | 2.0 | C |
| 112 | 7-5-00 | gp41 | 5.4 | 0.5 | C |
| 115 | 1-13-00 | gp41 | 5.4 | 0.8 | C |
| 116 | 6-7-98 | gp41 | 5.4 | 0.3 | C |
| 117 | 10-2-97 | gp41 | 5.4 | 2.6 | C |
| 118 | 10-31-99 | gp41 | 5.4 | 1.8 | C |
| 120 | 11-5-99 | gp41 | 5.4 | 1.0 | C |
| 125 | 4-14-00 | gp41 | 5.4 | 0.8 | C |
| 127 | 10-31-99 | gp41 | 5.4 | 1.8 | C |
| 128 | 9-25-98 | gp41 | 5.4 | 2.2 | C |
| 129 | 8-13-99 | gp41 | 5.4 | 2.3 | C |
| 130 | 6-16-00 (F) | gp41 | 5.4 | 1.0 | C |
| 9-14-00 (M) | |||||
| 132 | 8-11-00 | gp41 | 5.4 | 3.5 | C |
| 134 | 1-12-00 | gp41 | 5.4 | 3.8 | C |
| 135 | 6-28-98 | gp41 | 5.4 | 2.5 | C |
| 137 | 10-10-99 | gp41 | 5.4 | 2.5 | C |
| 138 | 10-2-98 | gp41 | 5.4 | 3.3 | C |
| 139 | 8-2-00 | gp41 | 5.4 | 1.3 | C |
| 140 | 11-11-99 | gp41 | 5.4 | 1.3 | C |
| 141 | 8-13-99 | gp41 | 5.4 | 1.5 | C |
| 143 | 8-26-99 | gp41 | 5.4 | 0.8 | C |
| 145 | 8-18-99 | gp41 | 5.4 | 0.8 | C |
| 146 | 8-23-00 | gp41 | 5.4 | 0.8 | C |
| 147 | 10-23-99 | gp41 | 5.4 | 0.5 | C |
| 149 | 9-30-00 | gp41 | 5.4 | 3.8 | C |
The subgenomic region was analyzed as shown in Fig. 1. The subgenomic region size (in base pairs) is given in parentheses. F, female partner; M, male partner.
The cutoff value is 2 SD below the mean (see Table 2).
Pairwise distances are the percent sequence differences in the analyzed genomic region.
Determined as epidemiologically linked by phylogenetic tree analysis.
Genetic distance is primarily due to G-to-A hypermutation.
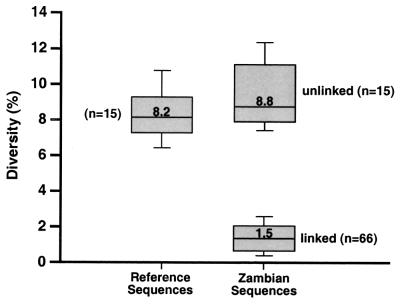
Distance calculations also identified 20 couples harboring HIV-1 strains whose uncorrected pairwise distances exceeded the corresponding cutoff values, and this was confirmed by sequencing two independent genomic regions (Table 4). The great majority of pairwise distances from these transmission pairs fell well above the cutoff values of both sets of reference sequences (compare with Table 2), thus indicating a clearly discernible difference between linked and unlinked transmission pairs (in the LTR region, Zambian donor sequences served as the sole reference set). This is best illustrated in Fig. 2, where the pairwise distances of 15 subtype C reference sequences in the gp41 region are contrasted to the corresponding gp41 distances from 66 linked and 15 unlinked (subtype C) cohort transmission pairs. The median sequence distance of the viral group tentatively classified as linked was significantly different from the median distance of the viral group tentatively classified as unlinked as well as the median distance of the reference sequence group (P < 0.0001) by using a one-sided Mann-Whitney test (17). In contrast, the median sequence distance of the unlinked viral group was not statistically different from that of the reference sequences (P > 0.05, Mann-Whitney test).
TABLE 4.
Genetic distances for unlinked Zambian transmission pairs
| Sample ID | Sample collection date (mo-day-yr) | Subgenomic regiona | Cutoff valueb | Pairwise distancec | Subtype |
|---|---|---|---|---|---|
| 7 | 9-23-98 | gagB | 4.3 | 5.7 | C |
| 9-23-98 | gp120C | 8.3 | 11.1 | C | |
| 9 | 8-16-00 | gp41 | 5.4 | 11.2 | C |
| 8-16-00 | gagF | 3.2 | 5.9 | C | |
| 18 | 10-19-98 | gagB | 4.3 | 7.5 | C |
| 10-19-98 | gp120C | 8.3 | 12.1 | C | |
| 31 | 2-29-00 | gp41 | 5.4 | 8.5 | C |
| 2-29-00 | gagE | 3.5 | 7.5 | C | |
| 44 | 11-11-98 (F) | gp120C | 8.3 | 16.9 | C |
| 8-11-98 (M) | gp41 | 5.4 | 7.7 | C | |
| 52 | 6-7-98 | gagH | 3.7 | 6.8 | C |
| 6-7-98 | gp120B | 3.2 | 6.6 | C | |
| 54 | 11-17-99 | gp41 | 5.4 | 7.8 | C |
| 11-17-99 | gagF | 3.2 | 4.0 | C | |
| 57 | 5-29-97 | gp41 | 5.4 | 11.0 | C |
| 5-29-97 | LTR | 2.4d | 3.8 | C | |
| 69 | 6-7-98 | gagD | 4.6 | 11.0 | C |
| 6-7-98 | gp120B | 3.2 | 3.7 | C | |
| 72 | 6-12-98 | gp120B | 3.2 | 5.8 | C |
| 6-12-98 | gp41 | 5.4 | 9.0 | C | |
| 87 | 6-6-00 | gp41 | 5.4 | 7.5 | C |
| 6-6-00 | gagE | 3.5 | 6.8 | C | |
| 88 | 8-22-99 (F) | gp41 | 5.4 | 8.5 | C |
| 9-19-00 (M) | gagC | 4.5 | 9.8 | C | |
| 94 | 10-25-98 | gagD | 4.6 | 7.6 | C |
| 10-25-98 | gp41 | 5.4 | 12.4 | C | |
| 96 | 6-12-98 (F) | gp41 | 5.4 | 13.1 | C |
| 2-3-99 (M) | LTR | 2.4d | 3.1 | C | |
| 104 | 6-26-98 | gp41 | 5.4 | 9.5 | C |
| 6-26-98 | LTR | 2.4d | 3.8 | C | |
| 121 | 4-10-98 | gagH | 3.7 | 10.4 | C |
| 4-10-98 | gp120B | 3.2 | 5.2 | C | |
| 123 | 6-26-00 | gagG | 3.6 | 7.0 | C |
| 6-26-00 | gp41 | 5.4 | 6.8 | C | |
| 126 | 10-27-98 | gp41 | 5.4 | 8.4 | C |
| 10-27-98 | gp120B | 3.2 | 4.1 | C | |
| 133 | 3-16-00 | gp41 | 5.4 | 12.2 | C |
| 3-16-00 | gagE | 3.5 | 5.6 | C | |
| 144 | 7-29-00 | gp41 | 5.4 | 8.8 | C |
| 7-29-00 | gagF | 3.2 | 3.7 | C |
Values are as defined in Table 3, footnote a. The subgenomic region was analyzed as shown in Fig. 1. F, female partner; M, male partner.
The cutoff value was 2 SD below the mean of the reference sequence set (see Table 2).
Pairwise distances are the percent sequence differences in the analyzed genomic region.
The cutoff value for LTR sequences was derived from Zambian donor sequences.
FIG. 2.
Within-group diversity among linked and unlinked Zambian transmission pairs and corresponding reference sequences. Subtype C reference sequences (n = 15) from the Los Alamos HIV/SIV Sequence Database (Table 1) were subjected to pairwise sequence comparisons in the region corresponding to the PCR-amplified gp41 fragment shown in Fig. 1. Pairwise sequence distances were also calculated for 66 subtype C transmission pairs classified as linked and 15 subtype C transmission pairs classified as unlinked in the same genomic region. The distribution of distance values for these three different groups is depicted as boxes, with the lower and upper limits of the box delineating the 25th and 75th percentiles and the bars indicating the 10th and 90th percentiles, respectively. The median distance of the linked viral group (median = 1.5) was significantly different from that of both the unlinked viral group (median distance = 8.8) and the reference sequence group (median distance = 8.2) (P < 0.0001, one-sided Mann-Whitney test [17]). In contrast, the median sequence distance of the unlinked viral group was not statistically different from that of the reference sequence group (P > 0.05, Mann-Whitney test).
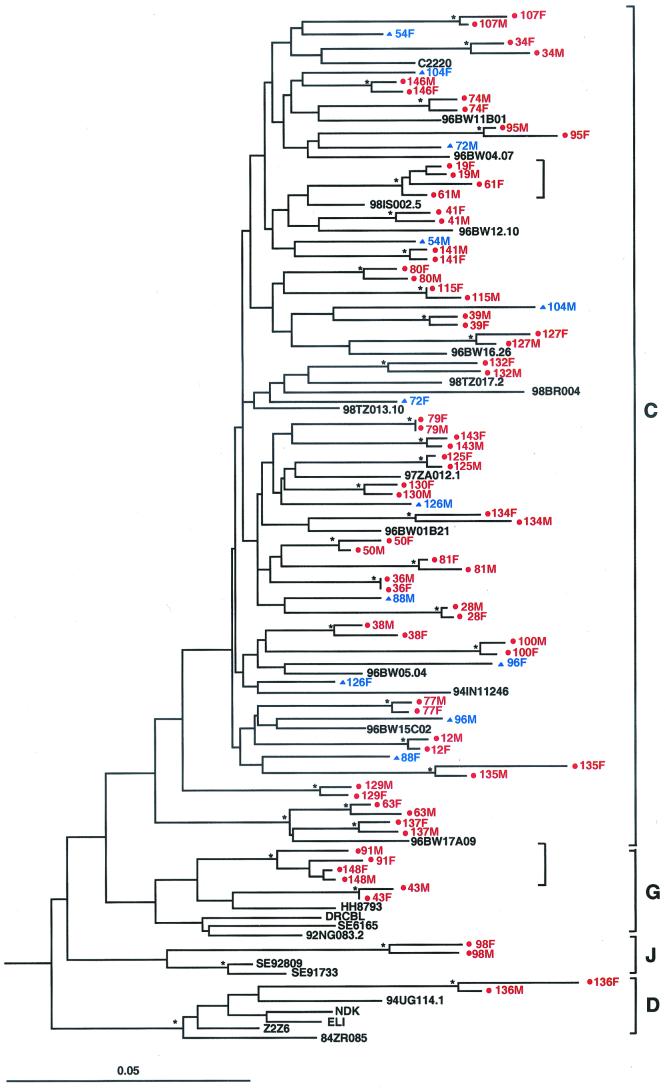
In a final set of experiments, epidemiological linkage was assessed by phylogenetic tree analysis. PCR-derived viral sequences from both partners were added to an existing master alignment (obtained from the Los Alamos HIV/SIV Sequence Database [http://hiv-web.lanl.gov/HTML/alignments.html]) that contained all reference sequences listed in Table 1. Sequences were aligned by using CLUSTAL W (19) and adjusted manually by using MASE (10). Sites with a gap in any of the sequences or sites that were ambiguous due to the population sequence approach were excluded from further analyses. Evolutionary distances were corrected for superimposed hits by using Kimura’s two-parameter method (22). Phylogenetic trees were constructed by using the neighbor-joining method (34), and the reliability of topologies was estimated by using the bootstrap approach (11). Bootstrap values of ≥80% were considered significant (4, 20, 29, 36, 44). An example of a phylogenetic tree constructed from gp41 sequences of 42 transmission pairs and 26 reference sequences is shown in Fig. 3. All transmission pairs initially classified as linked by pairwise distance analysis (depicted in red) also clustered together in phylogenetic trees with significant bootstrap values (indicated by asterisks). Similarly, all transmission pairs initially classified as unlinked (depicted in blue) were not significantly related in phylogenetic trees. The latter was true for the two independent genomic regions analyzed (not shown). Finally, viral sequences derived from 98M and 98F (Fig. 3), which clustered with subtype J viruses, were significantly related to each other and thus classified as epidemiologically linked.
FIG. 3.
Molecular linkage analysis for a subset of putative HIV-1 transmission pairs. A phylogenetic tree was constructed from partial gp41 sequences (consensus length, 276 bp) by using the neighbor-joining method (34) and the Kimura two-parameter model (22). Horizontal branch lengths are drawn to scale (the scale bar represents 0.05 nucleotide substitutions per site); vertical separation is for clarity only. Asterisks indicate bootstrap values in which the cluster to the right is supported in >80% replicates (out of 1,000). Newly derived sequences from 42 transmission pairs (84 individuals) are shown, along with 26 reference sequences from the Los Alamos Sequence Database (http://hiv-web.lanl.gov/HTML/alignments.html). Viruses from 36 couples are closely related to one another and cluster together with significant bootstrap values, indicating that they are epidemiologically linked (highlighted in red and denoted by dots). Viruses from six couples do not cluster together and exhibit a range of within-couple diversity that is similar to that of the reference sequences (highlighted in blue and denoted by triangles), indicating that they are epidemiologically unlinked. Two small brackets denote viral subclusters, each involving viruses from two sets of couples (see text for details). Brackets on the far right indicate major group M sequence subtypes.
Phylogenetic tree analysis also yielded a subtype designation for each of the viruses infecting the 149 transmission pairs (data not shown). As shown in Fig. 3, the overwhelming majority (141 of 149; 95%) of enrolled couples were infected with subtype C viruses. Three couples harbored subtype G viruses, three couples harbored subtype A viruses, one couple harbored subtype D viruses, and one couple harbored subtype J viruses, all representing linked transmissions. To determine whether non-subtype C viruses were introduced more recently, patient records were examined for the first occurrence of non-subtype C viruses (not shown). The results revealed no particular association between the date of enrollment and the appearance of non-subtype C strains within the ZUHRP cohort: couples infected with subtype A viruses were enrolled in 1996 and 1999; couples infected with subtype G were enrolled in 1995, 1996, and 1998; couples infected with subtype D were enrolled in 1998; and couples infected with subtype J were enrolled in 1997. If we assume no recombination in the remainder of the genome, these results indicate that subtype C predominates within the ZUHRP cohort.
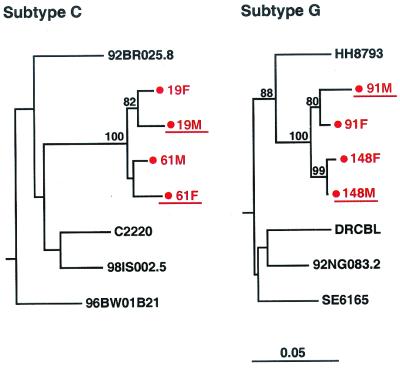
Finally, phylogenetic analysis allowed us to examine the evolutionary history of the cohort viruses compared to other HIV-1 strains from the same subtype. In particular, we were interested in determining whether ZUHRP couples were participating in transmission networks involving closely related viruses. Inspection of the phylogenetic tree in Fig. 3 revealed only two significant subclusters (indicated by brackets), each involving viruses from two sets of couples, which are shown in greater detail in Fig. 4. One subcluster involved subtype C viruses infecting couples 19 and 61, while the other involved subtype G viruses infecting couples 91 and 148. Given the short genomic region analyzed and the nonsignificant or borderline significant bootstrap values for three of the four couples (couples 19, 61, and 91), we could not determine with confidence that transmission had occurred between the partners of the same rather than different couples. The exact sequence of transmission events involving couples 19 and 61 and couples 91 and 148, respectively, thus remains to be determined. Rapid viral passage from a donor through one or more unidentified intermediaries to his or her putative recipient remains a theoretical possibility for all transmission pairs classified as epidemiologically linked in this study. However, since no other viral subclusters were identified in the data set, the existence of extensive transmission networks within the ZUHRP cohort is highly unlikely.
FIG. 4.
Transmission networks within the ZUHRP cohort. Phylogenetic trees were constructed from gp41 sequences of viruses infecting four different transmission pairs putatively classified as linked by pairwise sequence analysis. Couple identifiers are indicated in red (F, female partner; M, male partner). Horizontal branch lengths are drawn to scale (the scale bar represents 0.05 nucleotide substitutions per site); vertical separation is for clarity only. Values at nodes indicate the percentage of bootstraps in which the cluster to the right was found; only values of ≥80% are shown. Representative subtype C (left) and subtype G (right) reference sequences are included in each tree. Donor partners are underlined.
In summary, this report describes the first comprehensive molecular epidemiological analysis of heterosexual transmission events occurring among discordant couples in an African urban setting. Our analysis allowed us to (i) determine the proportion of linked and unlinked infections with a high level of certainty, (ii) identify the sequence subtype for all transmitted viruses in the genomic regions analyzed, and (iii) examine the cohort for evidence of transmission networks. The results show that of 149 cohabitating couples assumed to have transmitted to each other, 129 (87%) were molecularly confirmed as epidemiologically linked. Nevertheless, approximately 1 in every 10 transmission events involved an individual outside of the partnership. Assumptions concerning transmission linkage based on patient self-reporting alone are thus unlikely to be accurate, and this needs to be factored into the interpretation of transmission data from cohorts in which linkage has not been independently verified. For example, we found a stronger association between plasma viral load and transmission for female-to-male than for male-to-female transmissions in the ZUHRP cohort (12), while such a gender-based difference was not observed in a discordant couple cohort studied in Rakai, Uganda (32). Because transmission linkage was not confirmed at the molecular level, it is possible that some of the putative transmitters in this Ugandan cohort were misclassified. The proportion of unlinked transmissions is likely to vary considerably depending on the demographic, ethnic, and behavioral circumstances characterizing a cohort (45) but will undoubtedly be >0%. Thus, for investigations that require accurate assessment of HIV-1 transmission, such as studies aimed at identifying host and viral transmission correlates or determining the effectiveness of certain prevention strategies, the molecular characterization of viruses from both partners is essential.
Acknowledgments
We thank the staff, participants, and project management group of the ZUHRP cohort and Maria Salazar for expert technical assistance.
This work was supported by grants N01 AI-85338, R01 AI-40951, and U01 AI-41530 from the National Institutes of Health. DNA sequencing was performed in the DNA Sequence Analysis Core of the UAB Center for AIDS Research, supported by grant P30 A1-27767.
REFERENCES
- 1.Albert, J., J. Wahlberg, T. Leitner, D. Escanilla, and M. Uhlen. 1994. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J. Virol. 68:5918–5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizon, M., S. Wain-Hobson, L. Montagnier, and P. Sonigo. 1986. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell 46:63–74. [DOI] [PubMed] [Google Scholar]
- 3.Allen, S., K. E. N’Gandu, and A. Tichacek. 1998. The evolution of voluntary testing and counseling as an HIV prevention strategy: preventing HIV in developing countries: biomedical and behavioral approaches. Platinum Press, New York, N.Y.
- 4.Belec, L., A. Si Mohamed, M. C. Müller-Trutwin, J. Gilquin, L. Gutmann, M. Safar, F. Barre-Sinoussi, and M. D. Kazatchkine. 1998. Genetically related human immunodeficiency virus type 1 in three adults of a family with no identified risk factor for intrafamilial transmission. J. Virol. 72:5831–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, A., S. Ferris, S. Chamaret, D. Guetard, and L. Montagnier. 1998. Molecular evidence for nosocomial transmission of human immunodeficiency virus from a surgeon to one of his patients. J. Virol. 72:4537–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burger, H., B. Weiser, K. Flaherty, J. Gulla, P. N. Nguyen, and R. A. Gibbs. 1991. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc. Natl. Acad. Sci. USA 88:11236–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, J. K., T. Laukkanen, M. O. Salminen, J. Albert, A. Alaeus, B. Kim, E. Sanders-Buell, D. L. Birx, and F. E. McCutchan. 1999. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS 13:1819–1826. [DOI] [PubMed] [Google Scholar]
- 8.Carr, J. K., M. O. Salminen, J. Albert, E. Sanders-Buell, D. Gotte, D. L. Birx, and F. E. McCutchan. 1998. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology 247:22–31. [DOI] [PubMed] [Google Scholar]
- 9.Cassol, S. A., S. Read, B. G. Weniger, P. Gomez, N. Lapointe, C. Y. Ou, and P. G. Babu. 1996. Dried blood spots collected on filter paper: an international resource for the diagnosis and genetic characterization of human immunodeficiency virus type-1. Mem. Inst. Oswaldo Cruz 91:351–358. [DOI] [PubMed] [Google Scholar]
- 10.Faulkner, D. V., and J. Jurka. 1988. Multiple aligned sequence editor (MASE). Trends Biochem. Sci. 13:321–322. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1992. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet. Res. 60:209–220. [DOI] [PubMed] [Google Scholar]
- 12.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frenkel, L. M., J. I. Mullins, G. H. Learn, L. Manns-Arcuino, B. L. Herring, M. L. Kalish, R. W. Steketee, D. M. Thea, J. E. Nichols, S. L. Liu, A. Harmache, X. He, D. Muthui, A. Madan, L. Hood, A. T. Haase, M. Zupancic, K. Staskus, S. Wolinsky, P. Krogstad, J. Zhao, I. Chen, R. Koup, D. Ho, B. Korber, R. J. Apple, R. W. Coombs, S. Pahwa, and N. J. Roberts, Jr. 1998. Genetic evaluation of suspected cases of transient HIV-1 infection of infants. Science 280:1073–1077. [DOI] [PubMed] [Google Scholar]
- 14.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, B. H. Hahn, and the W.H.O. Network for HIV Isolation and Characterization. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res. Hum. Retrovir. 10:1359–1368. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons, J. D. 1997. Inferences concerning location based on two or more samples, p. 169–223. In Nonparametric methods for quantitative analysis, 3rd ed. American Sciences Press, Columbus, Ohio.
- 18.Goujon, C. P., V. M. Schneider, J. Grofti, J. Montigny, V. Jeantils, P. Astagneau, W. Rozenbaum, F. Lot, C. Frocrain-Herchkovitch, N. Delphin, F. Le Gal, J. C. Nicolas, M. C. Milinkovitch, and P. Deny. 2000. Phylogenetic analyses indicate an atypical nurse-to-patient transmission of human immunodeficiency virus type 1. J. Virol. 74:2525–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383–402. [DOI] [PubMed] [Google Scholar]
- 20.Hillis, D. M., and J. J. Bull. 1991. Of genes and genomes. Science 254:528. [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson, S. J., S. M. Gore, D. J. Goldberg, D. L. Yirrell, J. McGregor, A. G. Bird, and A. J. Leigh-Brown. 1999. Method used to identify previously undiagnosed infections in the HIV outbreak at Glenochil prison. Epidemiol. Infect. 123:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. [DOI] [PubMed] [Google Scholar]
- 23.Laukkanen, T., J. Albert, K. Liitsola, S. D. Green, J. K. Carr, T. Leitner, F. E. McCutchan, and M. O. Salminen. 1999. Virtually full-length sequences of HIV type 1 subtype J reference strains. AIDS Res. Hum. Retrovir. 15:293–297. [DOI] [PubMed] [Google Scholar]
- 24.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna, S. L., G. K. Muyinda, D. Roth, M. Mwali, N. Ng’andu, A. Myrick, C. Luo, F. H. Priddy, V. M. Hall, A. A. von Lieven, J. R. Sabatino, K. Mark, and S. A. Allen. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS 11:S103–S110. [PubMed] [Google Scholar]
- 26.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung’u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oelrichs, R. B., A. M. Vandamme, K. Van Laethem, Z. Debyser, F. E. McCutchan, and N. J. Deacon. 1999. Full-length genomic sequence of an HIV type 1 subtype G from Kinshasa. AIDS Res. Hum. Retrovir. 15:585–589. [DOI] [PubMed] [Google Scholar]
- 28.Oram, J. D., R. G. Downing, M. Roff, J. C. Clegg, D. Serwadda, and J. W. Carswell. 1990. Nucleotide sequence of a Ugandan HIV-1 provirus reveals genetic diversity from other HIV-1 isolates. AIDS Res. Hum. Retrovir. 6:1073–1078. [DOI] [PubMed] [Google Scholar]
- 29.Ou, C. Y., C. A. Ciesielski, G. Myers, C. I. Bandea, C. C. Luo, B. T. Korber, J. I. Mullins, G. Schochetman, R. L. Berkelman, A. N. Economou, J. J. Witte, L. J. Furman, G. A. Satten, K. A. MacInnes, J. W. Curran, H. W. Jaffe, J. Moore, Y. Villamarzo, C. Schable, E. G. Shaper, T. Liberti, S. Lieb, R. Scott, J. Howell, R. Dumbaugh, A. Lasch, B. Kroesen, L. Ryan, K. Bell, V. Munn, D. Marianos, and B. Gooch. 1992. Molecular epidemiology of HIV transmission in a dental practice. Science 256:1165–1171. [DOI] [PubMed] [Google Scholar]
- 30.Piot, P., M. Bartos, P. D. Ghys, N. Walker, and B. Schwartlander. 2001. The global impact of HIV/AIDS. Nature 410:968–973. [DOI] [PubMed] [Google Scholar]
- 31.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, and R. H. Gray. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. N. Engl. J. Med. 342:921–929. [DOI] [PubMed] [Google Scholar]
- 33.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161–168. [DOI] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- 35.Salminen, M. O., B. Johansson, A. Sonnerborg, S. Ayehunie, D. Gotte, P. Leinikki, D. S. Burke, and F. E. McCutchan. 1996. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res. Hum. Retrovir. 12:1329–1339. [DOI] [PubMed] [Google Scholar]
- 36.Song, J. Z., B. Wang, Y. C. Ge, D. E. Dwyer, A. L. Cunningham, and N. K. Saksena. 1999. Significance of plasma and peripheral blood mononuclear cell derived HIV-1 sequences in establishing epidemiologic linkage between two individuals multiply exposed to HIV-1. Microb. Pathog. 26:287–298. [DOI] [PubMed] [Google Scholar]
- 37.Spire, B., J. Sire, V. Zachar, F. Rey, F. Barre-Sinoussi, F. Galibert, A. Hampe, and J. C. Chermann. 1989. Nucleotide sequence of HIV1-NDK: a highly cytopathic strain of the human immunodeficiency virus. Gene 81:275–284. [DOI] [PubMed] [Google Scholar]
- 38.Srinivasan, A., R. Anand, D. York, P. Ranganathan, P. Feorino, G. Schochetman, J. Curran, V. S. Kalyanaraman, P. A. Luciw, and R. Sanchez-Pescador. 1987. Molecular characterization of human immunodeficiency virus from Zaire: nucleotide sequence analysis identifies conserved and variable domains in the envelope gene. Gene 52:71–82. [DOI] [PubMed] [Google Scholar]
- 39.UNAIDS-W.H.O. 2000. AIDS epidemic update: December 2000. Joint United Nations Programme on HIV/AIDS and World Health Organization. World Health Organization, Geneva, Switzerland.
- 40.UNAIDS-W.H.O. 2000. AIDS in Africa, country by country. Joint United Nations Programme on HIV/AIDS and World Health Organization. World Health Organization, Geneva, Switzerland.
- 41.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103–110. [DOI] [PubMed] [Google Scholar]
- 43.Yang, C., D. Pieniazek, S. M. Owen, C. Fridlund, J. Nkengasong, T. D. Mastro, M. A. Rayfield, R. Downing, B. Biryawaho, A. Tanuri, L. Zekeng, G. van der Groen, F. Gao, and R. B. Lal. 1999. Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers. J. Clin. Microbiol. 37:2581–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yirrell, D. L., S. J. Hutchinson, M. Griffin, S. M. Gore, A. J. Leigh-Brown, and D. J. Goldberg. 1999. Completing the molecular investigation into the HIV outbreak at Glenochil prison. Epidemiol. Infect. 123:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yirrell, D. L., H. Pickering, G. Palmarini, L. Hamilton, A. Rutemberwa, B. Biryahwaho, J. Whitworth, and A. J. Leigh Brown. 1998. Molecular epidemiological analysis of HIV in sexual networks in Uganda. AIDS 12:285–290. [DOI] [PubMed] [Google Scholar]