Abstract
The importance of CD8 T cells for the control of cytomegalovirus (CMV) infection has raised interest in the identification of immunogenic viral proteins as candidates for vaccination and cytoimmunotherapy. The final aim is to determine the viral “immunome” for any major histocompatibility complex class I molecule by antigenicity screening of proteome-derived peptides. For human CMV, there is a limitation to this approach: the T cells used as responder cells for peptide screening are usually memory cells that have undergone in vivo selection. On this basis, pUL83 (pp65) and pUL123 (IE1 or pp68 to -72) were classified as immunodominant proteins. It is an open question whether this limited “memory immunome” really reflects the immunogenic potential of the human CMV proteome. Here we document an analogous focus of the memory repertoire on two proteins of murine CMV. Specifically, ca. 80% of all memory CD8 T cells in the spleen as well as in persisting pulmonary infiltrates were found to be specific for the known IE1 peptide 168YPHFMPTNL176 and for the peptide 257AGPPRYSRI265, newly defined here, derived from open reading frame m164. Notably, CD8 T-cell lines of both specificities protected against acute infection upon adoptive transfer. In contrast, the natural immune response to acute infection in draining lymph nodes and in the lungs indicated a somewhat broader specificity repertoire. We conclude that the low number of antigenic peptides identified so far for CMVs reflects a focused memory repertoire, and we predict that more antigenic peptides will be disclosed by analysis of the acute immune response.
CD8 T cells are the principal effector cells that resolve productive cytomegalovirus (CMV) infection (46, 48, 50; for a review, see reference 29). Specifically, control of human CMV (hCMV) infection after bone marrow transplantation (BMT) correlates with the reconstitution of CD8 T cells (51), and clinical trials of a preemptive cytoimmunotherapy of hCMV infection with CD8 T-cell clones gave promising results (63). Likewise, in a murine model of interstitial pneumonia after experimental BMT and infection with murine CMV (mCMV), endogenously reconstituted CD8 T cells infiltrated the lungs, confined the infection to inflammatory foci, and eventually cleared the productive infection (18, 39). The protective capacity of the infiltrating CD8 T cells was demonstrated by two complementary approaches: selective in vivo depletion of CD8 T cells during lymphohematopoietic reconstitution resulted in lethal multiple-organ CMV disease (39, 40), whereas adoptive transfer of pulmonary CD8 T cells (1, 39) and transfer of antiviral CD8 T-cell lines of defined peptide specificities (19, 20) protected against productive infection and disease. In addition, by limiting viral spread during acute infection, antiviral CD8 T cells also limited the latent viral DNA load, resulting in a reduced risk of virus recurrence (58). Furthermore, there is evidence that CD8 T cells are involved in the maintenance of latent mCMV infection (17, 41).
Based on these findings and with the aims of developing a vaccine and optimizing cytoimmunotherapy, there is interest in the antigenic peptides of hCMV that are presented by major histocompatibility complex (MHC) class I molecules. It is a goal of research to identify the complete HLA class I “immunome” of hCMV, which is the list of all antigenic peptides encoded by hCMV and presented by HLA class I molecules. Realistically, a particular HLA molecule and a particular strain of hCMV should be used to start the program. One approach for epitope mapping is to screen peptides selected from the sequence-deduced viral proteome on the basis of computational prediction of MHC class I anchor residues (for a review, see reference 42) in combination with the prediction of proteasomal cleavage fragments (22, 23, 61). This strategy has recently been successfully employed for the identification of nine HLA-B27-restricted peptides of the bacterium Chlamydia trachomatis (30). Another option is a high-throughput analysis of overlapping peptides encompassing the entire proteome. When applied to genes UL83 and UL123 (ie1) of hCMV, new antigenic peptides were identified (24,25).
Cytofluorometric detection of intracellular gamma interferon (IFN-γ) induced in CD8 T cells upon stimulation with the cognate peptide is the method favored by many, because it simultaneously gives the phenotype and the frequency of responding cells (24). However, it should be noted that an enzyme-linked immunospot (ELISPOT) assay (19, 60) performed with purified CD8 T cells gives the same information and consumes a smaller absolute number of cells, which is important for analysis of cells difficult to retrieve in large numbers from extralymphoid and extravascular sites of inflammation. It is also important to recall that relevant antigenic peptides may be missed in cytokine-based screening assays. This was recently documented for an mCMV pM84-derived peptide that was found only by a cytolytic assay after in vitro restimulations (19, 21). Notably, protection-inducing immunogenicity of pM84 had been predicted before by genetic immunization with an M84 expression plasmid (37). Genetic immunization is an unbiased approach to identify the immunome of a given pathogen, but it is realistically applicable only to animal models.
Whatever method one prefers, we have to be aware of what we measure. The antigenicity screening of hCMV peptides by cytokine-based assays depends on the specificity repertoire of the responder cells used in the assays; we can only find what the responder cells can “see.” In the case of hCMV immunomics, the responder cells are blood-derived memory cells from healthy seropositive donors, because it is obviously difficult to do the screenings with CD8 T cells from BMT recipients with primary hCMV infection and almost impossible to recruit volunteers with an asymptomatic primary infection. As a consequence, current knowledge is limited to the memory repertoire. Although hCMV encodes approximately 165 open reading frames (ORFs) (5; Dolan et al., Abstr. 8th Int. CMV Conf., 20 to 25 May 2001, Asilomar, Calif.; and A. J. Davison, personal communication.), HLA class I-presented antigenic peptides have so far been identified only for three proteins, namely pUL55 (gB), pUL83 (pp65), and pUL123 (ie1, exon 4) (for a review, see reference 44), with particularly high frequencies of memory CD8 T cells directed against pUL83 peptides (13, 66).
Here we document that the memory CD8 T-cell response to mCMV in the H-2d haplotype is likewise focused, namely on peptides derived from ORFs m123 (encoding IE1, pp89) and m164, with very minor contributions made by CD8 T cells specific for previously identified peptides from ORFs m04 (20), M83 (19), and M84 (21). Notably, analysis of the acute CD8 T-cell response to mCMV indicates a somewhat broader specificity repertoire and predicts the existence of further antigenic peptides.
MATERIALS AND METHODS
Induction and isolation of antiviral effector and memory CD8 T cells.
Animal experiments were approved by the Ethics Commission (permission no. 177-07/991-35), according to German federal law. An immune response to mCMV was primed by subcutaneous (intraplantar) infection of female, 8- to 10-week-old BALB/c (haplotype H-2d) mice at the left hind footpad with 105 or 106 PFU of cell culture-propagated and sucrose gradient-purified mCMV strain Smith (ATCC VR-194/1981) (32). In a control group, mice were likewise inoculated with UV light (254 nm)-inactivated mCMV (for details of the inactivation and for efficacy control, see reference 47) equivalent to a dose of 106 PFU, referred to as PFUUV.
(i) Acutely sensitized CD8 effector cells from lymph nodes.
Lymphocytes were isolated by standard techniques from pools of 4 to 12 draining popliteal lymph nodes (PLN) at day 8 after intraplantar inoculation of immunocompetent mice with 106 PFU or PFUUV of mCMV, and CD8 T cells were purified to >97% purity by positive immunomagnetic cell sorting (MidiMACS separation unit and kit; Miltenyi Biotec Systems, Bergisch-Gladbach, Germany), as described in greater detail previously (1, 17). The ex vivo-isolated CD8 T cells were directly used for the ELISPOT assay (see below).
(ii) Memory CD8 T cells derived from the spleen.
At 3 to 6 months after intraplantar infection of immunocompetent mice with 105 PFU of mCMV, spleen cells were isolated (from a pool of at least three spleens), and erythrocytes were depleted by standard methods. It should be noted that productive infection of immunocompetent mice is resolved in all organs after 3 months (45). Unseparated splenocytes were used as responder cells for the in vitro generation of cytolytic T lymphocytes (CTLs) and CTL lines (CTLLs). CD8 T cells were purified by positive immunomagnetic cell sorting (described above). For control of priming dependence, CD8 T cells were isolated accordingly from spleens of uninfected, age-matched mice. The ex vivo-isolated CD8 T cells were directly used for the ELISPOT assay (described below).
(iii) CD8 T cells isolated from pulmonary infiltrates.
Interstitial CMV pneumonia (CMV-IP) was induced by intraplantar infection of BMT recipients with 105 PFU of mCMV. For hematoablative and immunosuppressive conditioning, recipients were total-body gamma irradiated with a single dose of 6 Gy delivered by a 137Cs gamma ray source. About 6 h after the irradiation, syngeneic BMT was performed by intravenous application of 5 × 106 femoral and tibial bone marrow cells derived from female BALB/c donors (18); the intraplantar infection followed 2 h later. During acute and latent infection of the lungs, at 1 and at 3 months after BMT, respectively (39), pulmonary infiltrate mononuclear leukocytes (pool derived from 25 lungs) were isolated by collagenase-DNase digestion of lung parenchyma followed by Ficoll density gradient enrichment as described in greater detail previously (18). CD8 T cells were purified by positive immunomagnetic cell sorting (described above) and were directly used in the ELISPOT assay (described below).
IFN-γ-based ELISPOT assays.
The two versions of the ELISPOT assay used herein were described in great detail and with all necessary controls in recent reports (17, 19, 20). The overall frequency of CD8 T cells capable of responding to a stimulus with secretion of IFN-γ was determined by the CD3ɛ-redirected ELISPOT assay (17, 19). In essence, a polyclonal stimulation of CD8 T cells via the signal-transducing CD3ɛ molecule of the T-cell receptor (TCR)-CD3 complex was achieved by using the anti-CD3ɛ monoclonal antibody (MAb)-producing hybridoma 145-2C11 (33) as stimulator cells in the assay. For the detection and quantification of peptide-specific CD8 T cells, the stimulator cells were P815-B7 cells (2) (H-2d mastocytoma cells transfected with human B7-1/CD80 cDNA) that were pulsed for 2 h at 37°C with a saturating dose of synthetic peptide, usually with a concentration of 10−8 M. As we have shown previously, frequencies determined by a peptide-specific ELISPOT assay remain constant over a very wide range of peptide concentrations (19). In a novel application of this method, stimulator cells were pulsed with naturally processed peptides derived from infected cells and separated by high-performance liquid chromatography (HPLC) (30-μl aliquots [described below]). Usually, responder cells were seeded in graded numbers into nylon membrane-backed ELISPOT microwells (three replicate cultures for each test) and were stimulated for ca. 16 h with 105 antigen-presenting stimulator cells. After this short-term cultivation, responder cells and stimulator cells were thoroughly washed off, IFN-γ bound to membrane-fixed anti-IFN-γ MAb was labeled, and brown spots, representing imprints of individual IFN-γ-secreting effector cells, were counted under a zoom stereomicroscope for the cell dilution that resulted in >10 (where appropriate) and <100 spots. It should be noted that the stimulator cells did not secrete IFN-γ. Photodocumentation was made with a digital camera.
Peptides. (i) Isolation of endogenously processed peptides from mCMV-infected cells.
Peptides were acid extracted from infected cells by a method originally developed by the group of H.-G. Rammensee (for a review, see reference 42) and adapted by us essentially as described previously (20). In brief, fetal fibroblasts were infected at a multiplicity of infection of 2, which corresponds to 0.1 PFU per cell under conditions of centrifugal enhancement of infectivity (32). Cells were harvested by trypsinization in the late (L) phase of the viral replicative cycle at 22 h postinfection, and acid extraction of peptides was performed with trifluoroacetic acid. Peptides were separated by HPLC with a SuperPac Sephasil C18 5-μm reversed-phase column (Pharmacia). Elution was performed on a linear acetonitrile gradient (20) at a flow rate of 0.8 ml per min. It should be noted that the elution positions of peptides may differ by one or two fractions between different HPLC runs under otherwise identical conditions. Experiments in this report used two HPLC runs: in Fig. 2, data represent HPLC run 1, and in Fig. 4 and 10, data represent HPLC run 2. Aliquots of the collected 0.8-ml fractions were distributed into wells of microwell plates and lyophilized for storage until used for the generation of short-term microculture CTLLs (60-μl aliquots) for pulsing of target cells in the cytolytic assay (60-μl aliquots) and for pulsing of stimulator cells in the ELISPOT assay (30-μl aliquots).
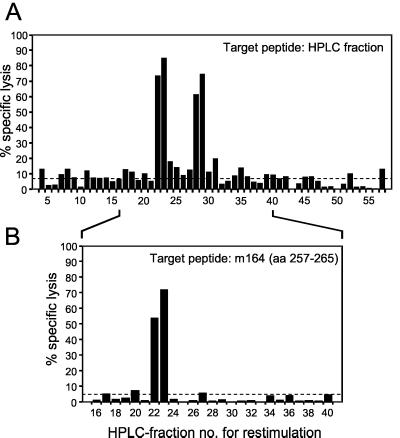
FIG. 2.
HPLC retention of naturally processed ORF m164 peptide. Naturally processed peptides derived from fetal fibroblasts in the L phase of mCMV replication were separated by HPLC (run 1). HPLC fractions were used for repeated restimulation of memory spleen cells to generate short-term microculture CTLLs. (A) Cytolytic assay of HPLC fraction-specific CTLLs with P815 target cells that were pulsed with the naturally processed peptides contained in the corresponding HPLC fractions. (B) Cytolytic assay of HPLC fraction-specific CTLLs with P815 target cells that were pulsed with 10−7 M synthetic m164 peptide aa 257 to 265. Excess of synthetic peptide was washed off before use of the target cells in the cytolytic assay. Bars represent mean values of triplicate CTLL cultures. The dashed line indicates the background lysis of target cells that were not exposed to peptide.
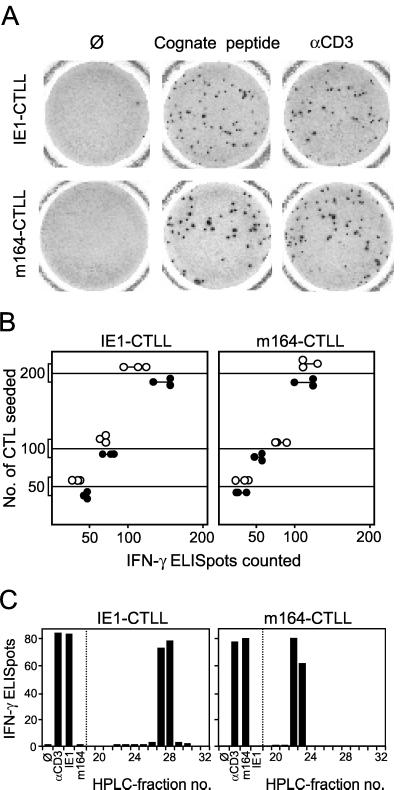
FIG. 4.
Frequencies of IFN-γ-secreting cells in CTLLs determined by ELISPOT assays. Long-term CTLLs were generated by repeated restimulations of memory spleen cells with the respective peptide at a concentration of 10−9 M. (A) Photodocumentation of ELISPOT filters. One representative filter of triplicate assay cultures with 100 cells seeded is shown. Ø, P815-B7 stimulator cells with no peptide added; Cognate peptide, P815-B7 stimulator cells pulsed with the respective peptide at a concentration of 10−8 M; αCD3, stimulation with 145-2C11 hybridoma cells that produce anti-mouse CD3ɛ MAb. (B) Data of triplicate assay cultures for three cell numbers seeded. Solid circles, stimulation with cognate peptide; open circles, stimulation with anti-CD3ɛ hybridoma. (C) HPLC retention of naturally processed peptides detected by IE1-CTLL and m164-CTLL. ELISPOT assays were performed with 100 CTLL cells and with P815-B7 stimulator cells that were pulsed with naturally processed peptides contained in HPLC fractions (run 2). Control stimulations were done as described above. Bars represent mean values of triplicate assay cultures.
(ii) Synthetic peptides.
Custom peptide synthesis in a 1-mg scale and with a purity of >75% was performed by JERINI Bio Tools GmbH (Berlin, Germany). Peptides were dissolved in 30% (vol/vol) acetonitrile in phosphate-buffered saline at a concentration of 10−3 M. Further dilutions were made in culture medium.
Antigenicity tests with CTLs. (i) Generation of short-term microculture CTLLs.
For the screening of peptides contained in HPLC fractions or of synthetic peptides with MHC-binding motifs, short-term CTLLs were generated as described in greater detail previously (20). In brief, 0.2-ml (96-well) microcultures (in triplicates for each HPLC fraction or synthetic peptide) were set up with 2 × 106 unseparated memory spleen cells per culture. The peptides for stimulation were provided either by dissolving the lyophilized HPLC fraction (60-μl aliquot) in 0.1 ml of clone medium or by adjusting the clone medium to the indicated molar concentrations of synthetic peptide. After 1 week of cultivation, half of each culture was restimulated with peptide and 5 × 104 gamma-irradiated (20 Gy) unprimed spleen feeder cells. Cytolytic activity was assayed after the third in vitro stimulation.
(ii) Generation of long-term CTLLs.
The protocol for raising CTLL was essentially that described previously (20), but with some modifications. Specifically, 1.5 × 107 unseparated memory spleen cells per culture were seeded into 24-well cultures in 1.5 ml of clone medium containing antigenic peptide at the molar concentrations indicated (first in vitro restimulation). On day 4, 200 U of recombinant human interleukin-2 (rhIL-2) was added in 0.5 ml of clone medium. On day 8, the 2-ml cultures were split into two, and 1 ml of fresh clone medium was added, containing 200 U of rhIL-2, antigenic peptide, and 8 × 105 P815-B7 stimulator cells gamma irradiated with 90 Gy (second in vitro restimulation). On day 14, CTLs were counted, and 5 × 104 CTLs were seeded per well and restimulated as outlined above (third in vitro restimulation). From here on, further restimulations were accordingly performed every second or third week. It should be noted that the use of P815-B7 stimulator cells greatly improved growth and longevity of the CTLLs.
(iii) Cytolysis assays.
Cytolytic activity of the CTLs was tested in a standard 4-h 51Cr release assay performed with 103 target cells per assay culture. Data represent percentages of specific lysis and are given either for three individual cultures (applies only to Fig. 1A) or as the mean value for triplicate cultures. Throughout, the target cells were P815 mastocytoma cells, expressing the MHC class I molecules Kd, Dd, and Ld. In the case of microculture CTLLs, half of the CTL suspension of a microculture (0.1 ml) was added to a suspension (0.1 ml) of target cells containing synthetic peptide at the molar concentrations indicated or containing a lyophilized HPLC fraction (60-μl aliquot) dissolved in clone medium. In the case of long-term CTLLs, effector cells and peptide-pulsed target cells were incubated at an effector/target cell (E/T) ratio of 15.
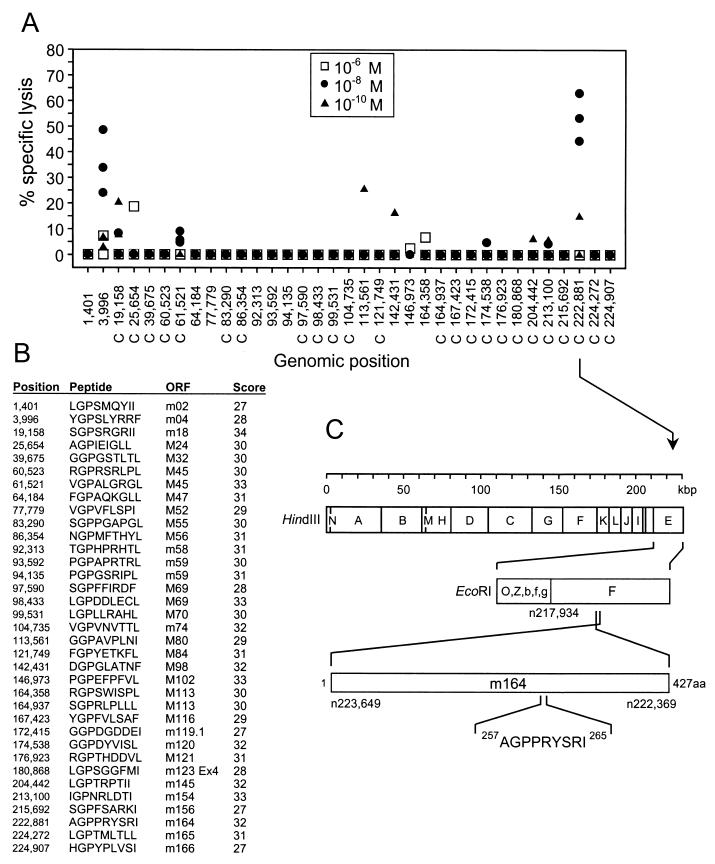
FIG. 1.
Identification of an antigenic peptide in ORF m164 of mCMV strain Smith. (A) A search for the Dd binding motif xGPxxxxx[L, I, F] was performed for all ORFs of the full-length genomic sequence of mCMV strain Smith (GenBank accession no. MCU68299), and the corresponding synthetic nonapeptides were used at the indicated molar concentrations for the generation of short-term microculture CTLLs. Data represent the cytolytic activity of individual microcultures tested on P815 target cells that were pulsed with the corresponding peptides at the corresponding concentrations. The genomic positions of the nonapeptide-coding sequences are given by the positions of the first nucleotides (n) according to the listing by Rawlinson et al. (43). C, complementary strand. (B) List of nonameric Dd-binding motifs. ORFs of mCMV that are sequence homologs of hCMV ORFs are indicated by capital M. Scores for Dd binding strengths were provided by Stefan Stevanovic (Institute for Cell Biology, Department of Immunology, University of Tuebingen, Tuebingen, Germany). (C) HindIII and EcoRI genetic map locations of ORF m164 peptide C 222881 257AGPPRYSRI265 drawn to scale. Amino acid positions refer to the deduced protein sequence according to Rawlinson et al. (43).
In vivo antiviral function of CTLLs.
The antiviral efficacy of CTLLs in an infected host was tested by adoptive cell transfer as described previously (19, 50). In brief, graded numbers of CTLs were transferred intravenously into gamma-irradiated (6.5 Gy) and infected recipients. The conditions were chosen so as to cause a lethal, multiple-organ CMV disease, unless the transferred cells control the infection. At day 12 after cell transfer, virus titers were determined in tissue homogenates from spleen, lungs, and liver by a plaque assay under conditions of centrifugal enhancement of infectivity. The titers represent the amounts of infectious virus per organ, and are expressed as PFU*, with the asterisk indicating the ca. 20-fold enhancement of infectivity achieved by the centrifugation. For the liver, the extent of tissue infection was visualized by immunohistochemical staining of the intranuclear IE1 protein pp89 of mCMV precisely as described previously (15). In essence, the IE1 protein, which is present in nuclei of infected cells throughout the viral replicative cycle and is concentrated during the L phase in an intranuclear inclusion body, was stained black by the nickel-enhanced avidin-biotin-peroxidase method with MAb CROMA 101 (kindly provided by S. Jonjic, University of Rijeka, Rijeka, Croatia) serving for the specific detection of IE1. A low-intensity counterstaining was performed for 5 s with hematoxylin. Microphotographs were taken with a Zeiss research microscope (Axiophot; Carl Zeiss Jena GmbH, Jena, Germany) with oil-immersion optics (plan-Neofluar; Zeiss) for all magnifications. Diapositives were scanned for computed documentation.
RESULTS
Identification of a novel antigenic peptide in mCMV ORF m164.
The first antigenic peptide identified for mCMV was the IE1 peptide 168YPHFMPTNL176 presented by the MHC class I molecule Ld (47, 49). Since its discovery in 1989, this peptide is generally cited as being the immunodominant antigenic peptide of mCMV in the H-2d haplotype. It protects against lethal mCMV disease (9), it has proven to be useful as a model peptide for studying proteasomal processing (10, 28, 54, 55, 59), and it is the peptide for which immune evasion mechanisms of mCMV, which prevent peptide presentation in the early (E) phase of the viral replication cycle, were originally shown to be operative (8). That its immunodominance is relative rather than absolute became evident from the analysis of the acute CD8 T-cell response to mCMV in pulmonary infiltrates in a model of interstitial pneumonia after BMT (18). While its contribution to the cytolytic activity in the infiltrates was significant, a comparison with the total response predicted the involvement of additional antigenic peptides presented in the E and L phases. Evidence for the existence of antigenic peptides in addition to the IE1 peptide was also given by earlier work of Del Val et al. (8), and protection by CD8 T cells in the Ld gene deletion mutant BALB/c-H-2dm2 had indicated the existence of peptides presented by the MHC class I molecule Kd and/or Dd (1).
Accordingly, we have recently identified additional antigenic peptides, namely E-phase m04/gp34 (26) peptide 243YGPSLYRRF251 presented by Dd (20), early-late M83/pp105 (36) peptide 761YPSKEPFNF769 presented by Ld (19), and E-phase M84/p65 (36) peptide 297AYAGLFTPL305 presented by Kd (21). However, frequency analyses revealed that these three peptides represent subdominant peptides, which even collectively cannot account for the difference between the IE1-specific response and the total response (17, 19). There were two possibilities left: either the total response is made up of many more still unidentified subdominant peptides, or a dominant peptide was missed in all previous studies.
In an earlier mCMV immunome analysis, we had performed an antigenicity screening for naturally processed peptides present in HPLC fractions from lysates of mCMV-infected fetal fibroblasts (20). Antigenicity was detected in a limited number of fractions, with the known IE1 peptide eluting in fractions 27 and 28. A prominent Dd-restricted activity was found for fraction 22, and minor Dd-restricted activities eluted in fractions 23 and 24. A subsequent viral genome-wide screening of synthetic Dd motif xGPxxxxx[L or I or F] (42) peptides led to the identification of the m04 peptide 243YGPSLYRRF251. However, synthetic m04 peptide was then found to elute in fraction 24 instead of in fraction 22. Thus, the fraction 22 peptide was not identified in that screening.
We have here made a second attempt to identify the missing fraction 22 peptide (Fig. 1). The Dd anchor motif peptides (listed in Fig. 1B) were synthesized and used at the indicated molar concentrations to establish short-term microculture CTLLs from memory spleen cells, and cytolytic activity was tested with target cells pulsed with the corresponding peptides (Fig. 1A). This screening detected antigenic activity for the genomic position 3996 (43) (GenBank accession no. MCU68299 [complete genome of mCMV, strain Smith]) peptide YGPSLYRRF, which is the previously identified m04 peptide (20), as well as for the genomic position C 222881 peptide AGPPRYSRI, which is a peptide not identified previously. The coding sequence maps to EcoRI fragment F within HindIII fragment E of mCMV, strain Smith (Fig. 1C), and the peptide represents amino acids (aa) 257 to 265 within the 427 aa of the deduced sequence of the ORF m164 protein (43).
We then raised short-term microculture CTLLs by stimulating memory spleen cells with aliquots of HPLC fractions containing naturally processed peptides derived from mCMV-infected cells (HPLC run 1), and the cytolytic activity of these CTLLs was tested with target cells pulsed with another aliquot of the corresponding HPLC fractions (Fig. 2A). In accordance with the previous HPLC separation that had been performed with a different cell extract (20), strong activities were found in fractions 22 and 23. The same HPLC fraction-specific CTLLs were then tested for cytolytic activity against target cells presenting the synthetic m164 peptide 257AGPPRYSRI265 (Fig. 2B). The only CTLLs that recognized the m164 peptide were those raised with the naturally processed peptides contained in fractions 22 and 23. Thus, the naturally processed m164 peptide eluted in these two HPLC fractions. In conclusion, the fraction 22 peptide is now identified, and fraction 23 contains the same peptide.
Cytoimmunotherapy with an ORF m164 peptide-specific CTLL protects against CMV disease.
For evaluation of the relevance of this new peptide in antiviral protection in comparison to that of the IE1 peptide, long-term CTLLs specific for the IE1 and the m164 peptide were generated in parallel, and are here referred to as IE1-CTLL and m164-CTLL, respectively.
(i) CTLL generation, cytolytic activity, and affinity to the presented cognate peptide.
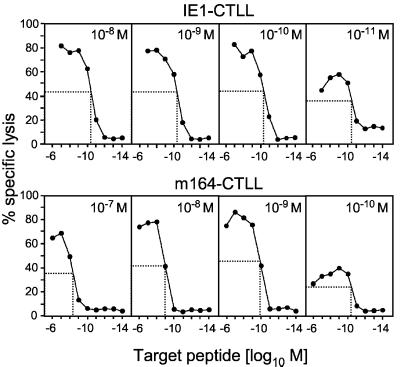
CTLLs were raised from memory spleen cells by restimulations with synthetic peptide at the molar concentrations indicated (Fig. 3). As noted previously for m04-CTLL (20) and for M84-CTLL (21), a range of peptide concentrations must be tested for restimulation, because peptide molarity can be suboptimal as well as supraoptimal for CTLL generation. For IE1-CTLL, 10−11 M was suboptimal, whereas lines of comparable cytolytic activity and affinity could be generated by three restimulations with peptide concentrations ranging from 10−8 M to 10−10 M. Independent of the restimulating peptide molarity, half-maximal lysis of target cells required a peptide concentration of ca. 10−10 M for target cell formation. In the case of m164-CTLL, a peptide concentration of 10−10 M was already suboptimal, and 10−7 M was slightly supraoptimal for the generation of a line. Notably, unlike with IE1-CTLL, the target peptide concentration required for half-maximal lysis shifted to lower values (i.e., to higher affinities) with decreasing concentrations of restimulating peptide. This finding indicates an affinity selection upon restimulation of polyclonal m164 peptide-specific memory cells and suggests a greater affinity heterogeneity of m164 than that of IE1 peptide-specific memory CD8 T cells. Subsequent experiments were performed with CTLLs raised with 10−9 M of the cognate peptide, because these lines are most equivalent with respect to cytolytic activity and affinity. The predicted Dd restriction of m164-CTLL was verified with the L-cell transfectant L-Dd (20) as a target cell (not shown).
FIG. 3.
Generation of long-term CTLLs. Memory spleen cells were restimulated with synthetic peptides IE1 (aa 168 to 176) and m164 (aa 257 to 265) at the indicated molar concentrations for the generation of IE1-CTLL and m164-CTLL, respectively. After three rounds of restimulation, a cytolytic assay was performed at an E/T cell ratio of 15 with P815 target cells that were pulsed with the indicated molar concentrations (abscissa) of the respective peptides. The target peptide concentration for half-maximal lysis (background subtracted) is indicated.
(ii) IFN-γ secretion by CTLLs in response to effector function triggering via TCR-α/β or CD3ɛ.
Cytokine secretion (here IFN-γ) is another relevant effector function of CTLs. In previous work (17, 19), we have already noted that not all cells of a CTLL are in a metabolic state enabling them to respond to a TCR-CD3 complex-mediated stimulus with IFN-γ secretion. For the example of M84-CTLL, it was found that CTLs may be deficient in responding to the cognate MHC-peptide complex, while being capable of responding to a direct stimulation via the signal-transducing CD3ɛ molecule (19).
For IE1-CTLL and m164-CTLL, the proportions of cells responding in IFN-γ-based ELISPOT assays (Fig. 4) were comparable, and there was no significant difference between the CD3ɛ-mediated and peptide-specific responses. Representative ELISPOT membranes for 100 cells seeded are documented in Fig. 4A, and triplicate data for all three cell numbers seeded are plotted in Fig. 4B.
The two CTLLs were used as effector cells in an ELISPOT assay performed with stimulator cells pulsed with HPLC fractions from infected cell lysate (HPLC run 2) in order to localize the elution of the respective naturally processed peptides (Fig. 4C). The control panel using synthetic peptides for stimulation verified that each CTLL recognized only the cognate peptide and not the heterologous peptide. IE1-CTLL identified the naturally processed IE1 peptide in fractions 27 and 28. In accordance with Fig. 2B, m164-CTLL identified the naturally processed m164 peptide in fractions 22 and 23. To make the difference between the two approaches very clear, Fig. 2B shows that a CTLL raised with naturally processed peptide contained in fractions 22 and 23 recognized the synthetic m164 peptide, whereas here it is shown that a CTLL raised with the synthetic m164 peptide recognized naturally processed peptide present in fractions 22 and 23. Altogether, the fractions containing the IE1 and m164 peptides are now unequivocally identified as fractions 27/28 and 22/23, respectively.
(iii) Antiviral efficacy of CTLLs upon adoptive transfer.
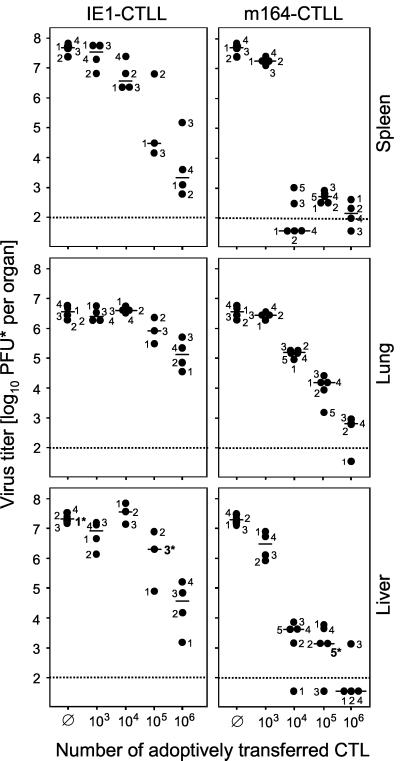
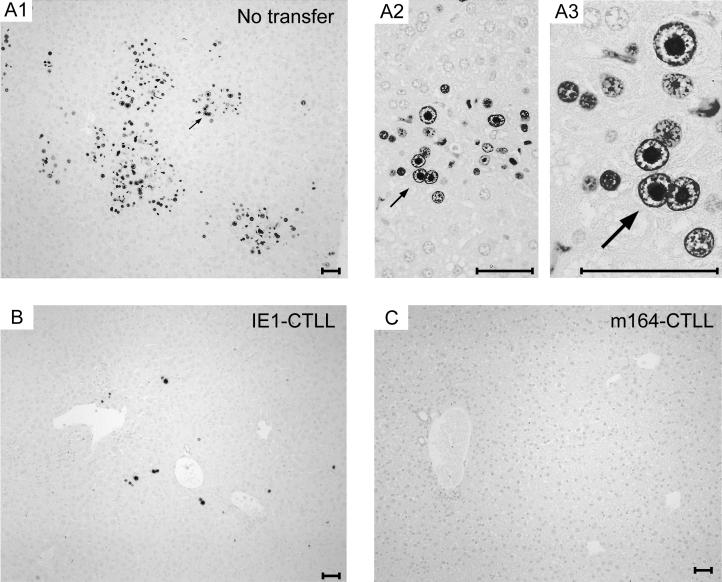
According to all in vitro parameters tested (see above), IE1-CTLL and m164-CTLL were functionally comparable, which was a good basis for a comparison of antiviral function in vivo. Here we have used again the long-established in vivo protection assay (46, 50, 58), which is based on the transfer of immune cells into immunodeficient recipients under conditions that cause a lethal multiple-organ CMV disease, unless the transferred cells are efficient in controlling the infection. Both IE1-CTLL and m164-CTLL were capable of controlling mCMV infection in spleen, lung, and liver (Fig. 5), with m164 CTLs being more efficient than IE1 CTLs in reducing virus titers. The histopathological correlate of virus replication in the absence of protective cells is documented for the liver by immunohistological staining of IE1 protein in nuclei of infected hepatocytes (Fig. 6). Figure 6A1 (corresponds to mouse 1 in Fig. 5) gives an overview showing many foci of infection in liver parenchyma. Resolved to greater detail, Fig. 6A2 and A3 show IE1 protein in intranuclear inclusion bodies that are characteristic of the L phase of the viral productive cycle. Transfer of 105 IE1 CTLs (Fig. 6B; median mouse 3 in Fig. 5) significantly reduced and transfer of 105 m164 CTLs (Fig. 6C, median mouse 5 in Fig. 5) largely prevented virus replication as well as histopathology.
FIG. 5.
In vivo antiviral function of CTLLs. Graded numbers of CTLs were transferred intravenously into BALB/c recipients under lethal conditions of infection (6.5 Gy of total-body gamma irradiation followed by intraplantar infection with 105 PFU of mCMV). Ø, no adoptive cell transfer. Virus titers in homogenates of spleen, lung, and liver were determined on day 12 after infection. The virus plaque assay was performed under conditions of centrifugal enhancement of infectivity. Accordingly, titers of infectious virus are expressed as PFU*. Dots represent virus titers in numbered individual mice. Asterisks at the numerals mark individual mice for which the liver histopathology is documented in Fig. 6. The median values are marked by horizontal bars. The dotted line indicates the detection limit of the plaque assay.
FIG. 6.
Clearance of infection in the liver by cytoimmunotherapy with CTLLs. Immunohistochemical analysis specific for the intranuclear IE1 protein pp89 of mCMV (black staining) was performed on liver sections from mice for which virus titers are documented in Fig. 5 (see numerals marked by an asterisk). (A1 to A3) Infection of liver parenchyma in the absence of protective CTLs. Panel A1 gives an overview at low magnification. The arrow points to a site that is resolved to greater detail in panels A2 and A3, highlighting intranuclear inclusion bodies in infected hepatocytes. These so-called “owl’s eyes” are characteristic of the L phase of the productive cycle. (B and C) Protection mediated by adoptive transfer of 105 cells of IE1-CTLL and m164-CTLL, respectively. Light counterstaining was performed with hematoxylin. Bars, 50 μm.
ORF m164 and m123 peptides codominate the memory CD8 T-cell response.
So far, the data have documented an antiviral in vivo function of m164 peptide-specific CTL effector cells. The prominence of m164 peptide-specific CD8 T cells in the natural immune response to mCMV depends on the efficacy of in vivo priming, and this needed to be tested next.
The antigenicity screening shown in Fig. 2A had indicated the existence of just two dominant specificities, namely the already known IE1 peptide and the newly identified m164 peptide eluting in fractions 22 and 23. However, this limited repertoire could result from differential efficacies in the in vitro restimulation during CTLL generation, which might have caused a loss of clones. We therefore used an ex vivo IFN-γ-based ELISPOT assay to determine the frequencies of IE1 and m164 peptide-specific cells among CD8 T cells derived from the spleen (Fig. 7). To serve as a reference for evaluating the quantitative contribution of particular peptides to the memory repertoire, the total frequency of CD8 T cells capable of responding in the assay was determined by polyclonal triggering via CD3ɛ (see appendix in reference 19), a method that identifies presensitized cells.
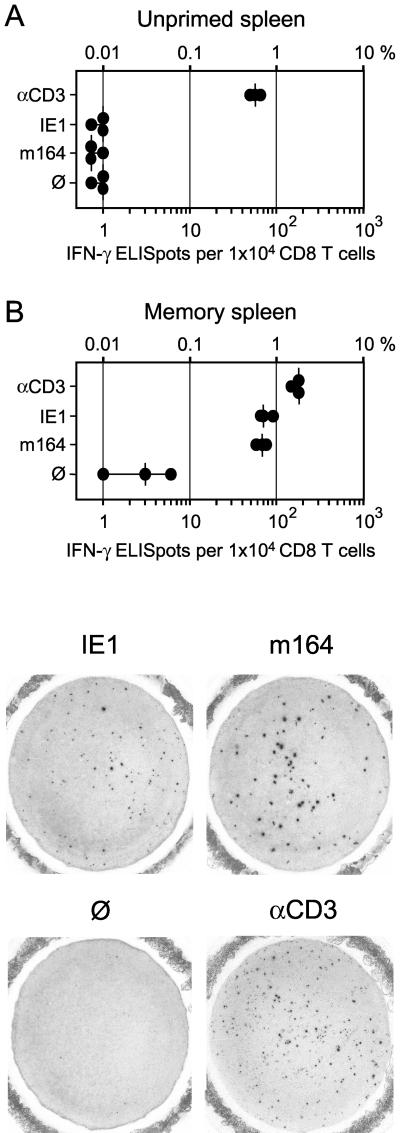
FIG. 7.
Frequencies of memory CD8 T cells determined by IFN-γ-based ELISPOT assays. (A) Responder cells were CD8 T cells isolated from the spleens of unprimed BALB/c mice at 5 months of age. (B) Responder cells were CD8 T cells isolated from age-matched BALB/c mice at 3 months after intraplantar infection with 105 PFU of mCMV. For peptide-specific stimulation of responder cells, P815-B7 cells were pulsed with a 10−8 M concentration of antigenic peptides IE1 (aa 168 to 176) and m164 (aa 257 to 265). αCD3, polyclonal stimulation with 145-2C11 hybridoma cells that produce MAb anti-mouse CD3ɛ; Ø, P815-B7 cells with no peptide added. Dots represent data from triplicate assay cultures. The median values are marked by vertical bars. For 104 CD8 T cells seeded, one representative filter of each of the triplicates is documented as a photograph.
As one could have predicted, spleens from unprimed 5-month-old mice, kept specific pathogen free but not germ free, contained a memory population of CD8 T cells responsive to stimulation via CD3ɛ (namely, 0.6% of all CD8 T cells), but no CD8 T cells specific for mCMV peptides IE1 and m164 (Fig. 7A). In contrast, in spleens of age-matched mice that had been infected with mCMV at 2 months of age, the frequency of CD3ɛ-responsive cells was elevated to ca. 2%, and ca. 0.7% of the cells (median values) were found to be specific for either of the two mCMV peptides IE1 and m164 (Fig. 7B, top). For each group, one of the triplicate ELISPOT filters was photographed to document a characteristic difference between the IE1 and the m164 peptide-specific responses (Fig. 7B, bottom). While polyclonal CD8 T-cell populations always comprise a mixture of cells differing in IFN-γ secretion, as reflected by different spot sizes, it was our consistent observation from many experiments that cells with high secretion of IFN-γ are more frequent in the m164 peptide-specific population. This may indicate a higher proportion of clones with high-affinity TCRs specific for the Dd-presented m164 peptide as compared to a higher proportion of clones with low-affinity TCRs specific for the Ld-presented IE1 peptide. The more pronounced heterogeneity within the m164 peptide-specific population may also explain the observation of affinity selection by restimulations during the generation of m164-CTLL (Fig. 3).
In conclusion, the frequency analysis based on IFN-γ secretion of ex vivo-responding CD8 T cells and the screening of naturally processed peptides based on cytolytic function of short-term in vitro-propagated CTLLs gave concordant results. The memory repertoire specific for mCMV is dominated in a quantitative sense by just two antigenic peptides, and the newly identified m164 peptide is as immunodominant as the long-known IE1 peptide.
Peptide specificity of CD8 T cells constituting the acute immune response in draining lymph nodes.
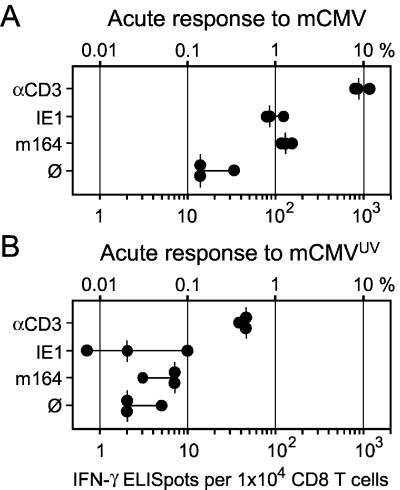
There are examples in the literature of a difference between the specificity repertoires in the acute and memory CD8 T-cell responses to viral infections (3; for a review, see reference 67). We therefore tested the contribution of the newly defined m164 peptide to the acute immune response in the PLN that drains the site of intraplantar local infection. An IFN-γ-based ELISPOT assay was performed with purified CD8 T cells isolated on day 8 after infection with 106 PFU of mCMV (Fig. 8A). In good accordance with a likewise performed recent analysis (19), the frequencies (median values) of CD3ɛ-responsive cells and of IE1 peptide-specific cells were ca. 8 and 0.8%, respectively. The new information is that the frequency of m164 peptide-specific cells was within the range measured for IE1. Thus, both peptides contributed equally to the acute CD8 T-cell response. Notably, however, unlike during memory in the spleen (Fig. 7B), the two frequencies did not add up to the frequency of CD8 T cells responding to signaling via CD3ɛ. In confirmation of previous data (19), the overall response was significantly diminished when viral replication was abolished by UV inactivation of the virions (Fig. 8B), and IE1 peptide-specific CD8 T cells were then not activated. CD8 T cells specific for the m164 peptide were not activated either, which indicated that priming with m164 peptide too depends on effective viral replication at the site of infection.
FIG. 8.
Frequencies of acutely sensitized CD8 T cells present in the draining PLNs during the primary immune response. Stimulation conditions for the IFN-γ-based ELISPOT assays were as described for Fig. 7. (A) Primary immune response in the ipsilateral PLN at day 8 after intraplantar infection with 106 PFU of mCMV. (B) Primary immune response in the ipsilateral PLN at day 8 after intraplantar inoculation with 106 PFUUV of inactivated mCMV virions.
Peptide specificity of CD8 T cells recruited to an extralymphoid site of viral pathogenesis.
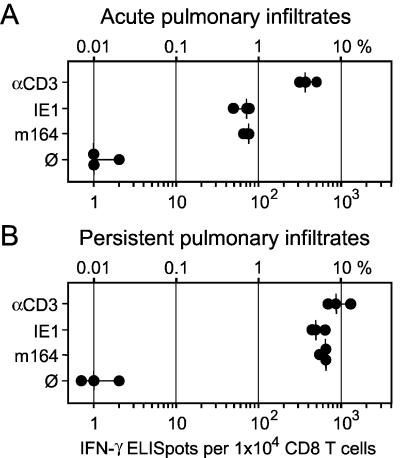
In a murine model of CMV-IP after experimental BMT, CD8 T cells are specifically recruited to the infected lungs and confine tissue infection to inflammatory foci (1, 18, 39). After resolution of the productive infection, memory CD8 T cells persist in the interstitium during the latent state of the infection (39). A previous study comparing the peptide specificities of CD8 T cells in acute and persisting pulmonary infiltrates has shown that IE1-specific CD8 T cells are enriched during latency and belong to a CD44+ CD62Llo population, whereas the frequency of M83 peptide-specific CD8 T cells remains unchanged between acute and latent infection (17). It was proposed that selective expression of IE1 in latently infected lungs (31) may account for the selective expansion of the IE1 peptide-specific memory cell pool (17). It was therefore of interest to test whether the m164 peptide behaves like IE1 peptide or like M83 peptide in this respect (Fig. 9). With frequencies (median values) of 4 and 0.7% for CD3ɛ-responsive and IE1 peptide-specific CD8 T cells, respectively, the response in pulmonary infiltrates during acute infection of the lungs at 4 weeks after BMT (Fig. 9A) was in good accordance with previous results (17). The new information is that m164 and IE1 peptide-specific CD8 T cells contributed equally to the response. Notably, these two specificities codominated the response by the interstitial CD8 T cells that persisted in the lungs during latent infection at 3 months after BMT (Fig. 9B). Thus, unexpectedly, the m164 peptide-specific cells are also enriched in the pulmonary memory cell pool. In essence, qualitatively at least, the response in acute and persisting pulmonary infiltrates strikingly resembled the acute response in draining PLN and the memory response in the spleen, respectively. Thus, apparently, there appears to be no general difference between lymphoid and extralymphoid sites with regard to CD8 T-cell specificities participating in the response.
FIG. 9.
Frequencies of CD8 T cells present in acute and persisting pulmonary infiltrates. Stimulation conditions for the IFN-γ-based ELISPOT assays were as described for Fig. 7. (A) Pulmonary CD8 T cells isolated from infected lungs at 1 month after BMT and intraplantar infection with 105 PFU of mCMV. (B) Interstitial CD8 T cells isolated accordingly from the lungs of mice at 3 months after BMT and intraplantar infection, a time point at which productive infection of the lungs was resolved and latent infection was established.
Antigenicity spectra of mCMV during acute and memory CD8 T-cell responses.
Operationally, one can define the CD8 T-cell immunome of a pathogen as the sum of the antigenic peptides that constitute the total CD8 T-cell response. However, as we have seen from the comparison between acute and memory responses (described above), the relative contribution of a particular antigenic peptide to the CD3ɛ-defined total response was lower in the acute immune response, suggesting that a broader-specificity repertoire is involved in the acute immune response. Another possibility is that the acute immune response recruits cells with many unrelated specificities by polyclonal “bystander” activation (62). This would lead to an elevated CD3ɛ-defined response, but should remain invisible in the scanning of naturally processed peptides from lysates of infected cells.
With this rationale in mind, we tested naturally processed peptides of one HPLC separation (HPLC run 2), first with CD8 T cells isolated from memory spleens and then (a corresponding set of HPLC aliquots) with CD8 T cells derived from draining PLNs (Fig. 10). As a new combination of methods, we used the IFN-γ-based ELISPOT assay for the scanning, because this test directly reveals the frequency of CD8 T cells that respond to naturally processed peptides contained in a particular HPLC fraction.
FIG. 10.
Empirical immunome analysis. The frequencies of CD8 T cells responding to known antigenic peptides or to antigenic peptides processed naturally in fetal fibroblasts during the L phase of mCMV infection were determined by IFN-γ-based ELISPOT assays. P815-B7 stimulator cells were pulsed either with the respective synthetic peptides or with HPLC fractions (run 2). (A) Responder cells for the first scan were CD8 T cells isolated from memory spleens at 5 months after intraplantar infection with 105 PFU of mCMV. (B) Responder cells for the second scan were CD8 T cells isolated from draining PLNs on day 8 after intraplantar infection with 106 PFU of mCMV. Controls (left panels) include stimulations at a saturating peptide concentration of 10−8 M with all currently known antigenic, H-2d-restricted peptides of mCMV, namely IE1 (aa 168 to 176) presented by Ld, m164 (aa 257 to 265) presented by Dd, M83 (aa 761 to 769) presented by Ld, M84 (aa 297 to 305) presented by Kd, and m04 (aa 243 to 251) presented by Dd. Ø, P815-B7 cells with no peptide added; αCD3, polyclonal stimulation with 145-2C11 hybridoma cells that produce a MAb directed against mouse CD3ɛ. Throughout, bars represent mean values of triplicate assay cultures.
(i) Antigenicity spectrum representing the memory immunome.
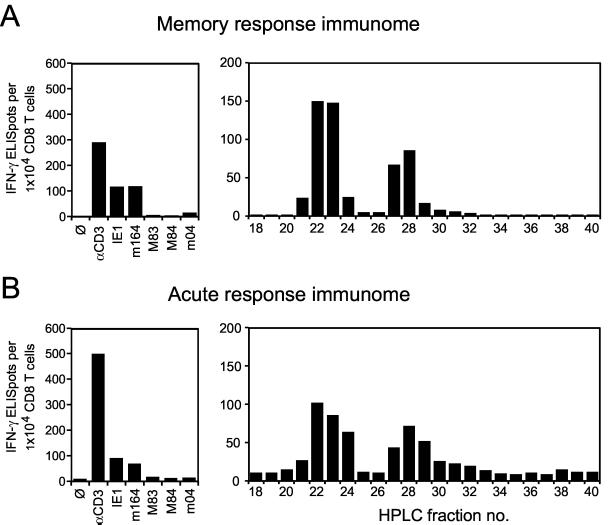
The HPLC fraction scan in Fig. 10A (right panel) should involve all antigenic peptides that are recognized by memory CD8 T cells, which of course includes peptides encoded by mCMV. Possibly included as well are peptides processed from self proteins, in particular from those induced by the infection. Notably, even though it is likely that numerous virally encoded and self peptides were present in the fractions, significant frequencies of responding memory cells were limited to two peaks, namely to fractions 22/23 and fractions 27/28 known to contain mCMV-encoded peptides m164 and IE1, respectively (recall data in Fig. 4C for the same HPLC run 2). Minor proportions of CD8 T cells responded to fraction 21; to fraction 24, which contains the m04 peptide (20); and to fraction 29. In addition, there were several fractions that were recognized by only very few CD8 T cells. One may argue that the prominent signals could represent more than one peptide and that relevant peptides may be missed because of an insufficient peptide concentration in the respective HPLC fractions. It was therefore important to determine the frequencies for the already defined peptides in the same experiment with the very same memory cell population (Fig. 10A, left panel) under conditions of optimal peptide presentation. Peptides IE1 and m164 accounted for ca. 80% of the CD3ɛ-defined total response, and minor contributions were made by known peptides M83, M84, and m04. If one checks carefully, the frequency for HPLC fractions 22 and 23 was somewhat higher than for optimally presented peptide m164. Indeed, we have evidence for a peptide derived from mCMV ORF m18 coeluting with the m164 peptide (R.H., unpublished data).
(ii) Antigenicity spectrum representing the acute response immunome.
The HPLC scan in Fig. 10B (right panel) should include all antigenic peptides recognized by acutely sensitized CD8 T cells involved in the primary immune response to mCMV in the draining PLN. As compared to memory, the spectrum is not essentially different, but it is somewhat less focused and indicates a higher contribution of peptides contained in fractions 24 and 29. Again, the frequency observed for fraction 22 is not explained by peptide m164 alone. Apparently, there is a significant gap between the CD3ɛ-defined response and the response to the currently known antigenic peptides (Fig. 10B, left panel).
Discussion.
Identification of the immunogenic potential of the proteome of a pathogen (i.e., of its immunome) is a current goal for rational vaccine design. For the identification of all antigenic peptides presented to CD8 or CD4 T cells by MHC class I or II molecules (here referred to as the MHC class I or II immunome of a pathogen), two approaches exist. One approach is to perform a high-throughput screening for antigenicity, for instance by cytofluorometric detection of induced intracellular IFN-γ, by using libraries of overlapping synthetic peptides that encompass the entire proteome. This strategy has been exemplified for two ORFs of hCMV, namely UL83 and UL123 (ie1), and has led to the mapping of several new antigenic peptides (24, 25). This empirical approach is independent of the precise optimal length of an antigenic peptide and is applicable in one step to all allomorphs of MHC class I and II molecules represented in the responder cell population used for the screening. A second approach to determine the MHC class I immunome is based on a computational prediction of antigenic sequences by combining the MHC binding motifs identified by H.-G. Rammensee and colleagues (11, 12; for an overview, see reference 42) with the prediction of proteasomal cleavage fragments (22, 23, 61). This approach has recently been employed to define the immunome of the bacterium C. trachomatis for a selected MHC class I molecule, namely HLA-B27, and has identified 9 antigenic peptides out of 199 that had been predicted for the bacterial proteome (30).
There is a lot of evidence in the literature to suggest that the natural immune response to a pathogen in any given MHC haplotype involves only a very limited set of target proteins and antigenic peptides derived thereof (for a review, see reference 67). While originally noted for viruses with low protein-coding capacity, the principle of a highly focused immune response appears to hold true as well for viruses, such as hCMV and mCMV, which have a substantial protein-coding capacity (5, 43). Many parameters are in discussion for explaining immunodominance. Besides the TCR repertoire of the host, these parameters include the amount and turnover of the respective proteins in the infected antigen-presenting cell, as well as the efficacy of antigenic peptide liberation by cleavage in the proteasome (54; for reviews, see references 27 and 68). Cleavage efficacy and the repertoire of generated peptides depend on the type of proteasome, constitutive proteasome or immunoproteasome (6, 61), as well as on proteasome regulator PA28 (10, 55, 59). Additional parameters are the efficacy of peptide transport into the endoplasmic reticulum (28), and, most importantly, peptide affinity for MHC class I molecules (reviewed in references 67 and 68). From a probability point of view, the number of peptides that optimally meet all of these criteria should be proportional to the coding capacity. Accordingly, pathogens with high coding capacity should have a more complex immunome. However, high coding capacity can also code for more immunosubversive functions. Specifically, CMVs have put additional hurdles in the pathway of antigen processing and presentation by coding for immune evasion proteins (for reviews, see references 16 and 65), which a “successful” peptide must be able to overcome or bypass. Thus, algorithms based on proteasomal cleavage preferences and on MHC binding affinities of peptides with anchor motifs predict more antigenic peptides than are eventually presented at the cell surface.
A limitation of both approaches discussed above is the fact that the readout system depends on responder cells usually derived from donors primed in the course of a natural immune response. As already pointed out in the introduction, identification of antigenic peptides specified by hCMV has so far been based mainly on the specificity repertoire of the memory T-cell pool in healthy, seropositive donors. The list of antigenic MHC class I-restricted peptides of hCMV (for a review, see reference 44) therefore reflects our current knowledge of the memory MHC class I immunome of hCMV. The memory repertoire in CMV infections may be shaped by a long period of in vivo selection, possibly by antigen presentation during intermittent reactivations (13) and during latency-associated gene expression (17, 31), and may thus not reflect the original immunogenic potential of the infection. Specifically, for human CD8 T-cell clones recognizing hCMV pUL83 (pp65) peptides, Weekes et al. (64) documented extensive expansion in vivo. For influenza virus, Belz et al. (3) demonstrated a difference between primary and recall response in that an antigenic peptide that was prominent in the primary response was less apparent in the memory pool. Of utmost importance in this respect is recent work from the group of R. M. Welsh (56) demonstrating that the memory pool of T cells specific to a previously encountered virus can be quantitatively deleted or qualitatively altered by subsequent unrelated infections. Furthermore, a prior virus infection was shown to alter the immunodominance hierarchy of antigenic peptides involved in the response to a second virus. As shown for lymphocytic choriomeningitis virus infection, the T-cell repertoire can be qualitatively conserved between acute response and memory under defined experimental conditions (4, 34, 57). Yet, the highly variable infection and immunization history of human donors may unpredictably modulate memory to the virus under investigation. All in all, memory repertoires are subject to change and are not necessarily representative of the immunogenic potential of a pathogen.
Approaches to identify the immunome of mCMV.
A much more direct and unbiased approach to identify an immunome was started for the example of mCMV by the group of D. H. Spector (14, 37). An ORF-by-ORF screening performed by using genetic immunization with mCMV ORF expression plasmids confirmed the previously known protective function of ORF m123 (encoding IE1, pp89) (14) and identified ORF M84 as a new immunogenic ORF (37). Based on this prediction, the M84 peptide (aa 297 to 305) was identified (21). Notably, this peptide was invisible in IFN-γ-based screening assays, such as the ELISPOT assay (21) and the intracellular cytokine cytofluorometry (19). It was disclosed only in a cytolysis assay performed with CTLLs generated by repeated restimulations of memory spleen cells. While the frequency of M84 peptide-specific CD8 T cells was below the detection limit during acute and memory immune responses to mCMV infection (19), an M84 peptide-specific CTLL was protective in cytoimmunotherapy (19), and a significant number of CD8 T cells specific for the M84 peptide were elicited by genetic immunization (M. Ye et al., Abstr. 8th Int. CMV Workshop). Likewise, in a work on antigenic peptides of lymphocytic choriomeningitis virus, Rodriguez et al. (52) demonstrated the power of genetic immunization in the identification of subdominant epitopes. These examples show that currently favored methods for antigenicity screening (24) bear a risk of failing to identify all relevant immunogenic peptides. In turn, we identified antigenic peptide m04 (aa 243 to 251) (20), and its protective potential was later confirmed by genetic immunization (D. H. Spector, CDC Workshop on Cytomegalovirus Vaccine Development, Atlanta, Ga., 25 to 27 October 2000). The genetic immunization approach indicated that not every ORF mediates protection. Specifically, a screening of murine homologs of hCMV ORFs that encode virion proteins gave negative or only inconsistently positive results (37). Reproducible protection against a low-dose challenge infection with mCMV was achieved only if these expression plasmids were administered as a pool (C. S. Morello, M. Ye, and D. H. Spector, 26th Int. Herpesvirus Workshop, 28 July to 3 August 2001; Regensburg, Germany, abstract no. 11.41).
We took an approach here that was different from all of the strategies discussed above. With the rationale in mind that all naturally processed and MHC-protected peptides encoded or induced by mCMV should be present in infected cells, we tested HPLC-fractionated cell extract for the presence of peptides that are recognized by ex vivo (i.e., not in vitro selected) CD8 T cells derived from immune mice. An HPLC fraction scanning with an IFN-γ-based ELISPOT assay directly revealed the frequency of the responding CD8 T cells. For memory cells, the scan showed just two high-frequency peaks, namely for HPLC fractions 22/23 and 27/28, and a few low-frequency signals. This already suggested a very focused memory repertoire. Peptides constituting the dominant signals were identified as ORF m164 aa 257 to 265 and ORF m123 (ie1) aa 168 to 176, respectively. Because an HPLC fraction may contain more than one antigenic peptide, the frequency of responding CD8 T cells was then also determined by stimulation with the two corresponding synthetic peptides. In essence, the results of the HPLC scan were confirmed, and a comparison with polyclonal stimulation via CD3ɛ showed that these two peptides accounted for ca. 80% of the response.
What fills the gap? First of all, we have already identified three minor antigenic peptides, namely ORF M83 aa 761 to 769 (19), ORF M84 aa 297 to 305 (21), and ORF m04 aa 243 to 251 (20). In addition, we know of a peptide derived from ORF m18 that coelutes with the ORF m164 peptide (R.H., unpublished). An unidentified peptide maps to HindIII fragment E and EcoRI fragment F, which is actually like the ORF m164 peptide. Yet, it must be a different one, because it is presented by Ld instead of by Dd (8). Still missing is the peptide (or even more peptides) that is derived from virion protein(s) and processed after exogenous loading of the antigen presentation pathway (19). It should be noted that the ORF m164 peptide is not recognized by the m164-CTLL on target cells that selectively present virion-derived peptides (not shown).
An unknown factor in this puzzle is the frequency of CD8 T cells with memory for peptides unrelated to mCMV infection. The frequency that we have determined in age-matched unprimed mice (Fig. 7A) would already fill the gap. However, as the work by Selin et al. (56) has impressively taught us, memory frequencies are not additive. Accordingly, the memory response to mCMV may have deleted preceding memory.
Altogether, we are very close to the MHC class I memory immunome of mCMV in the BALB/c strain. For sure, additional antigenic peptides of mCMV will be disclosed in future studies, but we predict that none of those will make a quantitatively dominant contribution to the memory response.
The situation is clearly different for the acute immune response. In essence, the shape of the response was already formed on day 8. Specifically, the ORF m123 and m164 peptides already dominated the response at this very early stage. Thus, apparently, immunodominance of these two peptides is an intrinsic quality and not the result of frequent restimulations in the course of reactivation of latent mCMV or by latency-associated gene expression. This does not exclude the possibility of clonal expansion during the memory state by selective restimulations that sharpen the repertoire (17). Elevated frequencies for HPLC fractions 24 and 29 suggest that CD8 T cells recognizing naturally processed peptides contained in these fractions get lost during the selection of the memory repertoire.
The most obvious difference is reflected by the significant gap between the CD3ɛ-defined acute response and the contributions of the already defined antigenic peptides (Fig. 10B). We currently see three possible explanations, which are not mutually exclusive. First, myriad unidentified minor antigenic peptides of mCMV may collectively account for this response, while any single peptide remains invisible in the HPLC fractions, because the frequency of the responding CD8 T cells is below the detection limit. Actually, this is the case for the already known minor antigenic peptides from ORFs M83 and M84. Second, we might have missed one or more immunodominant peptides in the analysis of HPLC fractions, because antigen processing in fetal fibroblasts may fail to generate all relevant peptides. In fact, recent work by Toes et al. (61) has shown that discrete cleavage motifs of constitutive and immunoproteasomes are responsible for qualitative differences in the generation of antigenic peptides. Likewise, different cell types involved in mCMV infection in vivo might process and present different sets of peptides. There is a third aspect to be considered: even though the CD3ɛ-defined response is clearly induced by mCMV infection (recall the mCMVUV control in Fig. 8), the responding cells are not necessarily all specific for peptides encoded by mCMV or by cellular genes induced by mCMV. An unknown factor is the “bystander activation” of CD8 T cells with unrelated specificities (62); cells that participate in the acute immune response but subsequently undergo apoptosis and do not contribute to memory (35). Although the magnitude of bystander activation was revisited based on results obtained with MHC-peptide tetramer staining of peptide-specific CD8 T cells (38), some contribution of bystander cells to the CD3ɛ-defined total response is likely. All in all, the question of the specificity repertoire involved in the acute immune response to mCMV is not yet settled.
ORF m164 protein, a novel player in mCMV immunology.
The role of pORFm164 in the immune response is currently the only known function of this protein. With the exception of the ORF m123 peptide, all antigenic peptides of mCMV were identified by the reverse immunology approach based on Rammensee’s MHC binding motifs. So far, the corresponding viral proteins had all been known before: gp34 of ORF m04 (26), pp105 of ORF M83 (36), and p65 of ORF M84 (36). The ORF m164 peptide is the first for which the corresponding protein was only predicted from the genomic sequence. Based on Rawlinson et al. (43), ORF m164 encodes a putative glycoprotein of 427 aa with a sequence-deduced molecular mass of 46.6 kDa. The computer program PredictProtein PHDhtm version 1.96 (53; Columbia University Bioinformatics Center, http://cubic.bioc.columbia.edu) predicts membrane-spanning α-helices for amino acid segments 92 to 109 and 360 to 379, thus suggesting that pORFm164 may be an integral membrane protein. A concordant prediction is given by the Dense Alignment Surface (DAS) method (7; DAS Transmembrane Prediction Server; Stockholm Bioinformatics Center, http://www.sbc.su.se/∼miklos/DAS/). Experiments are in progress to determine the kinetics of m164 gene expression, as well as the kinetics of peptide processing and presentation during the viral replication cycle in infected cells. Current evidence strongly suggests that pORFm164 is an E-phase protein.
Conclusion.
The MHC class I memory immunome of mCMV in BALB/c mice (H-2d haplotype) is dominated in a quantitative sense by just two antigenic peptides: the long-known peptide ORF m123 (encoding IE1, pp89) aa 168 to 176 presented by Ld and the peptide newly identified here, ORF m164 aa 257 to 265, presented by Dd. While the memory CD8 T-cell repertoire is highly focused on these two peptides, a somewhat broader repertoire constitutes the acute immune response. Our data from the mCMV model predict a likewise focused CD8 T-cell repertoire for hCMV, in particular for immunological memory in the responder cell donors that are included in programs of proteome-spanning peptide library screening. While these programs may therefore not identify the complete immunome, they will certainly reveal the immunodominant peptides for different HLA allomorphs.
Acknowledgments
We thank Aysel Rojan for technical assistance with the immunohistological analysis and Stefan Stevanovic (Institute for Cell Biology, Department of Immunology, University of Tübingen, Tübingen, Germany) for predicting scores for MHC binding motifs.
This work was supported by the Deutsche Forschungsgemeinschaft, project RE 712/3-2 (R.H. and M.J.R); Sonderforschungsbereich 490, individual project B1 (M.J.R.); and Sonderforschungsbereich 432, individual project A10 (J.P., A.R., and M.J.R.).
REFERENCES
- 1.Alterio de Goss, M., R. Holtappels, H.-P. Steffens, J. Podlech, P. Angele, L. Dreher, D. Thomas, and M. J. Reddehase. 1998. Control of cytomegalovirus in bone marrow transplantation chimeras lacking the prevailing antigen-presenting molecule in recipient tissues rests primarily on recipient-derived CD8 T cells. J. Virol. 72:7733–7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azuma, M., M. Cayabyab, D. Buck, J. H. Philipps, and L. L. Lanier. 1992. CD28 interaction with B7 costimulates primary allogeneic proliferative responses and cytotoxicity mediated by small, resting T lymphocytes. J. Exp. Med. 175:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belz, G. T., W. Xie, J. D. Altman, and P. C. Doherty. 2000. A previously unrecognized H-2Db-restricted peptide prominent in the primary influenza A virus-specific CD8+ T-cell response is much less apparent following secondary challenge. J. Virol. 74:3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattmann, J. N., D. J. Sourdive, K. Murali-Krishna, R. Ahmed, and J. D. Altman. 2000. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J. Immunol. 165:6081–6090. [DOI] [PubMed] [Google Scholar]
- 5.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., C. C. Norbury, Y. Cho, J. W. Yewdell, and J. R. Bennink. 2001. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 193:1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10:673–676. [DOI] [PubMed] [Google Scholar]
- 8.Del Val, M., K. Münch, M. J. Reddehase, and U. H. Koszinowski. 1989. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell 58:305–315. [DOI] [PubMed] [Google Scholar]
- 9.Del Val, M., H.-J. Schlicht, H. Volkmer, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 1991. Protection against lethal cytomegalovirus infection by a recombinant vaccine containing a single nonameric T-cell epitope. J. Virol. 65:3641–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick, T. P., T. Ruppert, M.Groettrup, P. M. Kloetzel, L. Kuehn, U. H. Koszinowski, S. Stevanovic, H. Schild, and H.-G. Rammensee. 1996. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell 86:253–262. [DOI] [PubMed] [Google Scholar]
- 11.Falk, K., O. Rötzschke, K. Deres, J. Metzger, G. Jung, and H.-G. Rammensee. 1991. Identification of naturally processed viral nonapeptides allows their quantification in infected cells and suggests an allele-specific T cell epitope forecast. J. Exp. Med. 174:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk, K., O. Rötzschke, S. Stevanovic, G. Jung, and H.-G. Rammensee. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature (London) 351:290–296. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie, G. M. A., M. R. Wills, V. Appay, C. O’Callaghan, M. Murphy, N. Smith, P. Sissons, S. Rowland-Jones, J. I. Bell, and P. A. H. Moss. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. J. Virol. 74:8140–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzáles Armas, J. C., C. S. Morello, L. D. Cranmer, and D. H. Spector. 1996. DNA immunization confers protection against murine cytomegalovirus infection. J. Virol. 70:7921–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzimek, N. K. A., J. Podlech, H.-P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 6:190–197. [DOI] [PubMed] [Google Scholar]
- 17.Holtappels, R., M.-F. Pahl-Seibert, D. Thomas, and M. J. Reddehase. 2000. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62Llo memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 74:11495–11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtappels, R., J. Podlech, G. Geginat, H.-P. Steffens, D. Thomas, and M. J. Reddehase. 1998. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J. Virol. 72:7201–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtappels, R., J. Podlech, N. K. A. Grzimek, D. Thomas, M.-F. Pahl-Seibert, and M. J. Reddehase. 2001. Experimental preemptive immunotherapy of murine cytomegalovirus disease with CD8 T-cell lines specific for ppM83 and pM84, the two homologs of human cytomegalovirus tegument protein ppUL83 (pp65). J. Virol. 75:6584–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtappels, R., D. Thomas, J. Podlech, G. Geginat, H.-P. Steffens, and M. J. Reddehase. 2000. The putative natural killer decoy early gene m04 (gp34) of murine cytomegalovirus encodes an antigenic peptide recognized by protective antiviral CD8 T cells. J. Virol. 74:1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtappels, R., D. Thomas, and M. J. Reddehase. 2000. Identification of a Kd-restricted antigenic peptide encoded by murine cytomegalovirus early gene M84. J. Gen. Virol. 81:3037–3042. [DOI] [PubMed] [Google Scholar]
- 22.Holzhütter, H.-G., C. Frömmel, and P.-M. Kloetzel. 1999. A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20S proteasome. J. Mol. Biol. 286:1251–1265. [DOI] [PubMed] [Google Scholar]
- 23.Holzhütter, H.-G., and P.-M. Kloetzel. 2000. A kinetic model of vertebrate 20S proteasome accounting for the generation of major proteolytic fragments from oligomeric peptide substrates. Biophys. J. 79:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern, F., I. P. Surel, C. Brock, B. Freistedt, H. Radtke, A. Scheffold, R. Blasczyk, P. Reinke, J. Schneider-Mergener, A. Radbruch, P. Walden, and H. D. Volk. 1998. T-cell epitope mapping by flow cytometry. Nat. Med. 4:975–978. [DOI] [PubMed] [Google Scholar]
- 25.Kern, F., I. P. Surel, N. Faulhaber, C. Frömmel, J. Schneider-Mergener, C. Schönemann, P. Reinke, and H.-D. Volk. 1999. Target structures of the CD8+ T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleijnen, M. F., J. B. Huppa, P. Lucin, S. Mukherjee, H. Farrell, A. E. Campbell, U. H. Koszinowski, A. B. Hill, and H. L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloetzel, P.-M. 2001. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2:179–187. [DOI] [PubMed] [Google Scholar]
- 28.Knuehl, C., P. Spee, T. Ruppert, U. Kuckelkorn, P. Henklein, J. Neefjes, and P.-M. Kloetzel. 2001. The murine cytomegalovirus pp89 immunodominant H-2 Ld epitope is generated and translocated into the endoplasmic reticulum as an 11-mer precursor peptide. J. Immunol. 167:1515–1521. [DOI] [PubMed] [Google Scholar]
- 29.Koszinowski, U. H., M. J. Reddehase, and S. Jonjic. 1993. The role of T-lymphocyte subsets in the control of cytomegalovirus infection, p. 429–445. In D. B. Thomas (ed.), Viruses and the cellular immune response. Marcel Dekker, Inc., New York, N.Y.
- 30.Kuon, W., H.-G. Holzhütter, H. Appel, M. Grolms, S. Kollnberger, A. Traeder, P. Henklein, E. Weiss, A. Thiel, R. Lauster, P. Bowness, A. Radbruch, P.-M. Kloetzel, and J. Sieper. 2001. Identification of HLA-B27-restricted peptides from the Chlamydia trachomatis proteome with possible relevance to HLA-B27-associated diseases. J. Immunol. 167:4738–4746. [DOI] [PubMed] [Google Scholar]
- 31.Kurz, S. K., M. Rapp, H.-P. Steffens, N. K. A. Grzimek, S. Schmalz, and M. J. Reddehase. 1999. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurz, S. K., H.-P. Steffens, A. Mayer, J. R. Harris, and M. J. Reddehase. 1997. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J. Virol. 71:2980–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leo, O., M. Foo, D. H. Sachs, L. E. Samelson, and J. A. Bluestone. 1987. Identification of a monoclonal antibody specific for a murine T3 polypeptide. Proc. Natl. Acad. Sci. USA 84:1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, M. Y., and R. M. Welsh. 1998. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection in mice. J. Exp. Med. 188:1993–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally, J. M., C. C. Zarozinski, M. Y. Lin, M. A. Brehm, H. D. Chen, and R. M. Welsh. 2001. Attrition of bystander CD8 T cells during virus-induced T-cell and interferon responses. J. Virol. 75:5965–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morello, C. S., L. D. Cranmer, and D. H. Spector. 2000. Suppression of murine cytomegalovirus (MCMV) replication with a DNA vaccine encoding MCMV M84 (a homolog of human cytomegalovirus pp65). J. Virol. 74:3696–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murali-Krishna, K., J. D. Altman, M. Suresh, D. J. D. Sourdive, A. J. Zajac, J. D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177–187. [DOI] [PubMed] [Google Scholar]
- 39.Podlech, J., R. Holtappels, M.-F. Pahl-Seibert, H.-P. Steffens, and M. J. Reddehase. 2000. Murine model of interstitial cytomegalovirus pneumonia in syngeneic bone marrow transplantation: persistence of protective pulmonary CD8-T-cell infiltrates after clearance of acute infection. J. Virol. 74:7496–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podlech, J., R. Holtappels, N. Wirtz, H.-P. Steffens, and M. J. Reddehase. 1998. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J. Gen. Virol. 79:2099–2104. [DOI] [PubMed] [Google Scholar]
- 41.Polic, B., H. Hengel, A. Krmpotic, J. Trgovcich, I. Pavic, P. Lucin, S. Jonjic, and U. H. Koszinowski. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rammensee, H.-G., J. Bachmann, and S. Stevanovic. 1997. MHC ligands and peptide motifs. Molecular Biology Intelligence Unit, Landes Bioscience, Austin, Tex.
- 43.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddehase, M. J. 2000. The immunogenicity of human and murine cytomegaloviruses. Curr. Opin. Immunol. 12:390–396 and 738. [DOI] [PubMed] [Google Scholar]
- 45.Reddehase, M. J., M. Balthesen, M. Rapp, S. Jonjic, I. Pavic, and U. H. Koszinowski. 1994. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J. Exp. Med. 179:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddehase, M. J., S. Jonjić, F. Weiland, W. Mutter, and U. H. Koszinowski. 1988. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J. Virol. 62:1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddehase, M. J., and U. H. Koszinowski. 1984. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature (London) 312:369–371. [DOI] [PubMed] [Google Scholar]
- 48.Reddehase, M. J., W. Mutter, and U. H. Koszinowski. 1987. In vivo application of recombinant interleukin 2 in the immunotherapy of established cytomegalovirus infection. J. Exp. Med. 165:650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddehase, M. J., J. B. Rothbard, and U. H. Koszinowski. 1989. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature (London) 337:651–653. [DOI] [PubMed] [Google Scholar]
- 50.Reddehase, M. J., F. Weiland, K. Münch, S. Jonjic, A. Lüske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reusser, P., S. R. Riddell, J. D. Meyers, and P. D. Greenberg. 1991. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood 78:1373–1380. [PubMed] [Google Scholar]
- 52.Rodriguez, F., M. K. Slifka, S. Harkins, and J. L. Whitton. 2001. Two overlapping subdominant epitopes identified by DNA immunization induce protective CD8+ T-cell populations with differing cytolytic activities. J. Virol. 75:7399–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidtke, G., M. Eggers, T. Ruppert, M. Groettrup, U. H. Koszinowski, and P.-M. Kloetzel. 1998. Inactivation of a defined active site in the mouse 20S proteasome complex enhances major histocompatibility complex class I antigen presentation of a murine cytomegalovirus protein. J. Exp. Med. 187:1641–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz, K., M. Eggers, A. Soza, U. H. Koszinowski, P.-M. Kloetzel, and M. Groettrup. 2000. The proteasome regulator PA28 alpha/beta can enhance antigen presentation without affecting 20S proteasome subunit composition. Eur. J. Immunol. 30:3672–3679. [DOI] [PubMed] [Google Scholar]
- 56.Selin, L. K., M. Y. Yin, K. A. Kraemer, D. M. Pardoll, J. P. Schneck, S. M. Varga, P. A. Santolucito, A. K. Pinto, and R. M. Welsh. 1999. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity 11:733–742. [DOI] [PubMed] [Google Scholar]
- 57.Sourdive, D. J., K. Murali-Krishna, J. D. Altman, A. J. Zajac, J. K. Whitmire, C. Pannetier, P. Kourilsky, B. Evavold, A. Sette, and R. Ahmed. 1998. Conserved T cell repertoire in primary and memory CD8 T cell responses to an acute viral infection. J. Exp. Med. 188:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steffens, H.-P., S. Kurz, R. Holtappels, and M. J. Reddehase. 1998. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J. Virol. 72:1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stohwasser, R., A. Soza, M. Eggers, U. H. Koszinowski, and P. M. Kloetzel. 2000. PA28α/β double and PA28β single transfectant mouse B8 cell lines reveal enhanced presentation of a mouse cytomegalovirus (MCMV) pp89 MHC class I epitope. Mol. Immunol. 37:13–19. [DOI] [PubMed] [Google Scholar]
- 60.Taguchi, T., J. R. McGhee, R. L. Coffman, K. W. Beagley, J. H. Eldridge, K. Takatsu, and H. Kyono. 1990. Detection of individual mouse splenic T cells producing IFN-γ and IL-5 using the enzyme-linked immunospot (ELISPOT) assay. J. Immunol. Methods 128:65–73. [DOI] [PubMed] [Google Scholar]
- 61.Toes, R. E., A. K. Nussbaum, S. Degermann, M. Schirle, N. P. Emmerich, M. Kraft, C. Laplace, A. Zwinderman, T. P. Dick, J. Müller, B. Schönfisch, C. Schmid, H. J. Fehling, S. Stevanovic, H. G. Rammensee, and H. Schild. 2001. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tough, D. F., P. Borrow, and J. Sprent. 1996. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science 272:1947–1950. [DOI] [PubMed] [Google Scholar]
- 63.Walter, E. A., P. D. Greenberg, M. J. Gilbert, R. J. Finch, K. S. Watanabe, E. D. Thomas, and S. R. Riddell. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 333:1038–1044. [DOI] [PubMed] [Google Scholar]
- 64.Weekes, M. P., M. R. Wills, K. Mynard, A. J. Carmichael, and J. G. P. Sissons. 1999. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J. Virol. 73:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiertz, E., A. Hill, D. Tortorella, and H. Ploegh. 1997. Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol. Lett. 57:213–216. [DOI] [PubMed] [Google Scholar]
- 66.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and J. G. P. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51–88. [DOI] [PubMed] [Google Scholar]
- 68.Yewdell, J. W., and J. R. Bennink. 2001. Cut and trim: generating MHC class I peptide ligands. Curr. Opin. Immunol. 13:13–18. [DOI] [PubMed] [Google Scholar]