Abstract
The Sendai virus P-L polymerase complex binds the NP-encapsidated nucleocapsid (NC) template through a P-NP interaction. To identify P amino acids responsible for binding we performed site-directed mutagenesis on the C-terminal 88 amino acids in the NC binding domain. The mutant P proteins expressed from plasmids were assayed for viral RNA synthesis and for various protein-protein interactions. All the mutants formed P oligomers and bound to L protein. While two mutants, JT3 and JT8, retained all P functions at or near the levels of wild-type (wt) P, three others—JT4, JT6, and JT9—were completely defective for both transcription and genome replication in vitro. Each of the inactive mutants retained significant NC binding but had a different spectrum of other binding interactions and activities, suggesting that the NC binding domain also affects the catalytic function of the polymerase. NC binding was inhibited by combinations of the inactive mutations. The remaining P mutants were active in transcription but defective in various aspects of genome replication. Some P mutants were defective in NP0 binding and abolished the reconstitution of replication from separate P-L and NP0-P complexes. In some of these cases the coexpression of the wt polymerase with the mutant NP0-P complex could rescue the defect in replication, suggesting an interaction between these complexes. For some P mutants replication occurred in vivo, but not in vitro, suggesting that the intact cell is providing an unknown function that cannot be reproduced in extracts of cells. Thus, the C-terminal region of P is complex and possesses multiple functions besides NC binding that can be separated by mutation.
Sendai virus, a paramyxovirus, is an enveloped virus with a single-stranded, negative-sense, nonsegmented RNA genome of about 15 kb (for a review, see reference 24). The genome RNA is completely encapsidated by the nucleocapsid protein, NP, which renders the RNA nuclease resistant. The RNA-dependent RNA polymerases of negative [(−)]-strand RNA viruses are unique in that this helical ribonucleoprotein complex or nucleocapsid (NC), not the RNA alone, serves as the template for all viral RNA synthesis. The viral RNA polymerase is composed of a complex of the phosphoprotein, P (568 amino acids [aa]) and the large protein, L (2228 aa). Transcription by the viral polymerase initiates at the precise 3′ end of the encapsidated genome, yielding first positive [(+)]-strand leader RNA, le+ (55 nucleotides [nt]), which is followed by the sequential synthesis of the six mRNAs in the order NP, P-C-V, M, F, HN, and L. Replication of the genome RNA involves the synthesis of a full-length (+)-sense copy of the genome where its synthesis is coupled to encapsidation with NP. This then serves as the template for the replication and encapsidation of (−)-sense genome RNA. The L protein is believed to contain most of the catalytic activities necessary for viral RNA synthesis and mRNA processing, although presently there are few direct experimental data on this point.
P is a modular protein with discrete regions that have been mapped for the various protein-protein interactions that are required for viral RNA synthesis. First P complexes with L to form the RNA polymerase. The L binding domain within the P protein has been mapped to aa 412 to 478 (13, 33). P appears to be important for the proper folding of the L protein, since L must be coexpressed with P to be stable (19, 20). Binding of the P-L complex to the NC template occurs through the P subunit of the polymerase (21), requiring two noncontiguous nucleocapsid binding sites from aa 345 to 411 and aa 479 to 568 in the C-terminal half of P (29–31). The P protein also binds free NP (NP0) to form a complex required for packaging nascent RNA during genome replication, which requires multiple sites on P at aa 33 to 41, a region between aa 78 and 325, and the region overlapping the NC binding site from aa 479 to 568 (12, 22). The P protein has also been shown to oligomerize, at an oligomerization domain mapped to aa 320 to 411 (11), which overlaps the most N-terminal NC binding domain. Structural studies show that P is a tetramer and the oligomerization domain has a coiled coil structure which extends to aa 433 through part of the L binding domain (34, 35). These data are consistent with a model in which P oligomerization is required for the P-NC interaction. In addition, a supplemental role for P protein in viral RNA synthesis has also been described. Curran (8, 9) showed that while part of the nucleocapsid-associated P protein is found in the P-L complex, the rest is bound to the template independently of L, and this form of P is also essential for mRNA synthesis.
Our goal was to characterize the P-NC interaction with respect to the specific P amino acids which are required for binding assembled NP. We selected clustered charged-to-alanine or hydrophobic-to-alanine mutagenesis of residues that were at least partly conserved in parainfluenza viruses. Charged amino acids may reside on the surface of the protein, and changes to alanine may produce stable mutants capable of exhibiting altered phenotypes (1). This was verified in at least one case for Sendai virus P, where the crystal structure of a portion of P showed clustered charged amino acids on the surface that inactivated the polymerase when changed to alanine (3, 34). The results showed that the C-terminal region of P, initially identified as simply the NP0 and NC binding domains, in fact possesses multiple other functions that can be separated by mutation.
MATERIALS AND METHODS
Cells, viruses, plasmids, and antibodies.
Human lung carcinoma cells (A549 cells [American Type Culture Collection]) were used for all experiments. Recombinant vaccinia virus expressing the phage T7 RNA polymerase (VVT7) (15) was grown on CV1 cells. Sendai virus and the Sendai virus defective interfering particle (DI-H) were propagated on embryonated chicken eggs, and wild-type (wt) polymerase-free Sendai virus and DI-H templates were prepared as described previously (5). Plasmids encoding the Sendai virus genes—pGEM-L, pGEM-NP, and pGEM-Pstop (expressing only the P protein, designated pGEM-P in this work)—were described previously (10). pSPDI-H (21) contains a cDNA copy of the DI-H genome containing only a 3′ portion of the L gene and does not encode any proteins. The genes were all cloned downstream of the phage T7 RNA polymerase promoter. Primary antibodies utilized for immunoblot assays were rabbit polyclonal anti-Sendai virus antibody (α-SV) (5) and rabbit polyclonal antibody specific for the P peptides of aa 274 to 298 and aa 453 to 477 (α-P, provided by K. Gupta, Rush Medical College, Chicago, Ill.) (4).
Mutagenesis.
Clustered charged- or hydrophobic-to-alanine mutagenesis of pGEM-P as shown in Table 1 was carried out using either plasmid-based mutagenesis (mutant oligonucleotide sequences available on request) with the Transformer Site Directed Mutagenesis Kit (Clontech) or by a PCR-based mutagenesis strategy with Vent polymerase (NEB) (18). The second method employs a two-step PCR with complementary mutagenesis primers which also contained a new silent restriction site for screening purposes (sequences available upon request). The first round contained a mutagenic primer and the appropriate upstream (EagI-containing) or downstream (HindIII-containing) outside primer, generating two partially complementary DNAs. The second round used the two DNAs and the two outside primers, generating a full-length mutant PCR product. The mutant plasmid or PCR products were digested with EagI and HindIII and cloned into pGEM-P at those sites. The mutants were sequenced to confirm the correct mutations. To construct P mutants with multiple mutation sites, the JT9 plasmid was digested with NdeI and HindIII, where the mutation sites are contained within the NdeI-HindIII fragment which was then cloned into the JT4 plasmid at those sites. This created the JT4/9 double mutant. Then, the JT4/9 plasmid was used as the target for PCR-based mutagenesis with the primers for JT6 as discussed above to create the triple mutant JT4/6/9. The mutations were confirmed by sequencing.
TABLE 1.
Mutations in the Sendai virus P protein
| Mutant | Amino acid changesa |
|---|---|
| JT1 | R482A, E483A, D484A, E485A |
| JT2 | R487A, D488A, E489A |
| JT3 | E497A, R498A, D499A |
| JT4 | N506A, R509A |
| JT5 | K514A, E515A, K516A |
| JT6 | L524A, V525A, I526A |
| JT7 | R533A, E535A, K536A |
| JT8 | D549A, E551A, K553A |
| JT9 | E560A, E561A, D562A |
Amino acid changes in the 568-aa P protein are listed in the order of the wt amino acid, the position in the P protein, and the amino acid change.
Protein analysis.
For immunoblot analysis cell extracts used for RNA synthesis were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose and then incubated with α-P antibody to detect P protein in transcription assays, with the addition of α-SV antibody to detect P and NP in replication assays. The blots were developed with conjugated goat anti-rabbit secondary antibody using the Enhanced Chemiluminescence (ECL) protein identification system (Amersham Life Science) according to the manufacturer’s protocol. For nucleocapsid binding analysis, 100-mm-diameter dishes of VVT7-infected A549 cells were transfected with the wt or mutant pGEM-P (15 μg) and wt L (15 μg) plasmids and labeled at 4 h posttransfection (h.p.t.) with Tran35S-label (100 μCi/ml) with 0.1× cysteine and methionine. Cell extracts were prepared at 18 h.p.t. using lysolecithin permeabilization as described previously (6, 19) in 300 μl of SV Salts B (0.1 M HEPES [pH 8.5], 50 mM NH4Cl, 7 mM KCl, 1 mM dithiothreitol, 1 mM ATP). Samples (130 μl) were incubated with polymerase-free wt Sendai virus nucleocapsids (1 μg) for 1 h at 30°C. The nucleocapsids were pelleted through a step gradient of 2 ml of 30% (wt/vol) and 2.5 ml of 50% (wt/vol) glycerol in 10 mM HEPES (pH 8.5)-1 mM EDTA at 45,000 rpm for 75 min at 4°C in an SW55 rotor. The nucleocapsid-associated pelleted proteins were analyzed by SDS-7.5% PAGE and visualized by autoradiography and quantitated on the phosphorimager. For glycerol gradient analysis of NP and P complex formation, VVT7-infected cells in 100-mm-diameter dishes were transfected with the wt NP (6 μg) or wt P (15 μg) plasmids alone or the mutant P plasmids together with wt NP plasmid and labeled with Tran35S-label, and cell extracts were made by lysolecithin permeabilization as described above. The samples were fractionated by centrifugation on 5 to 20% (vol/vol) glycerol gradients in 10 mM HEPES (pH 8.5)-1 mM EDTA in the SW41 rotor at 29,000 rpm for 46 h. Gradient fractions (1 ml) were collected and 0.5 ml was immunoprecipitated with α-P and α-SV antibodies and Staphylococcus aureus (Cowen strain) and analyzed by SDS-PAGE and autoradiography.
In vitro transcription.
For normal transcription assays, 60-mm dishes of A549 cells were infected with VVT7 and transfected with the L (0.5 μg) and the wt or mutant P (1.5 μg) plasmids. At 18 h.p.t. cell extracts (110 μl) were prepared by lysolecithin permeabilization in SV Incomplete Reaction Mix (RM) (0.1 M HEPES, pH 8.5, 50 mM NH4Cl, 7 mM KCl, 1 mM dithiothreitol, 1 mM spermidine, 1 mM [each] ATP, GTP, and UTP, 10 μM CTP, and 10% glycerol) and nuclei were pelleted as described previously (6, 19). A portion of extract (10%) was removed for immunoblotting, and the remainder was used for RNA synthesis. The cell extracts were incubated with micrococcal nuclease (MN) (20 μg/ml) plus 1 mM CaCl2 at 30°C for 30 min, followed by 2.2 mM EGTA to inactivate the MN. For mRNA synthesis the nuclease-treated extracts were then supplemented with a 0.1 volume of 10× supplemental mix (45 mM magnesium acetate [MgOAc], RNasin [5 U/μl], actinomycin D [200 μg/ml], creatine phosphokinase [400 U/ml], and creatine phosphate [33 mg/ml]), 1 μg of polymerase-free wt Sendai virus RNA-NP, and 20 μCi of [α-32P]CTP. The samples were incubated for 2 h at 30°C, and the RNA was isolated using Qiagen RNeasy Total RNA kit according to the manufacturer’s protocol and analyzed by 1.5% agarose-6 M urea-citrate gel electrophoresis.
To test the supplemental function of P, VVT7-infected cells (60-mm-diameter dishes) were transfected with no plasmids (mock transfection) or with wt or mutant P plasmids alone (5 μg), and separate dishes were cotransfected in duplicate with the same P plasmids (0.5 μg) and wt L plasmid (3 μg). This ratio of P to L is suboptimal for transcription, since P is limiting. To maintain about the same final amount of viral polymerase used in normal transcription experiments, the amount of L plasmid was doubled from 1.5 to 3 μg, since only 60% of the extract was used for the reaction. Cell extracts (110 μl) were prepared, duplicate dishes were pooled, and nuclei were pelleted as described above. Cytoplasmic extracts with the wt or mutant P-L complexes were then divided into three 60-μl aliquots, and to these were added 30 μl of a cell extract which contained either mock-transfected cells, wt P or mutant P alone. After MN treatment transcription was performed and analyzed as described above.
Leader RNA synthesis was performed as described for in vitro mRNA synthesis with polymerase-free wt template (2.5 μg), except the cell extracts were prepared in SV Incomplete RM with 1 mM CTP without radiolabeled CTP. The incubated samples were then treated with proteinase K and extracted with phenol-chloroform, and the RNA was separated on an 8% polyacrylamide-8 M urea gel and electroblotted onto a Hybond-N nylon membrane (Amersham). le+ RNA was detected by Northern blot analysis using a complementary 32P-end-labeled 55-nt oligonucleotide probe (5′AAATCCTGTA TAACTTCATT ACATATCCCA TACATGTTTT TTCTCTTGTT TGGT3′) as described previously (7).
Viral genome replication in vitro and in vivo.
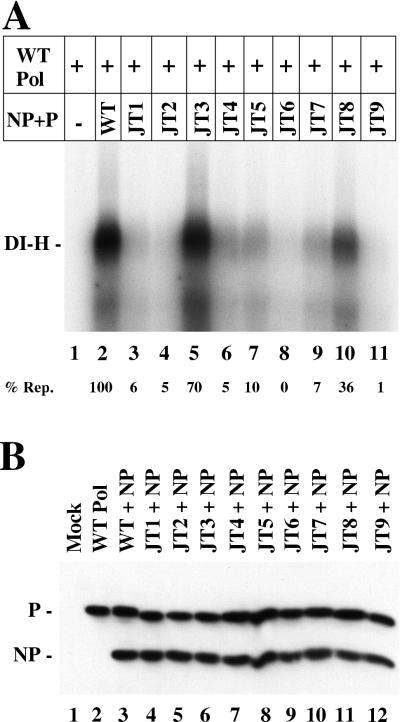
For in vitro replication 60-mm-diameter dishes of A549 cells were infected with VVT7 and transfected with the L (0.5 μg), NP (2 μg), and the wt or mutant P (5 μg) plasmids. Extracts were prepared by lysolecithin permeabilization in SV Incomplete RM supplemented with 4.5 mM MgOAc and RNasin (0.5 U/μl). Actinomycin D (20 μg/ml), 50 μCi of [α-32P]CTP, and 2 μg of polymerase-free DI-H RNA-NP were added to the samples and incubated for 2 h at 30°C. The samples were then MN treated, and nuclease-resistant RNA was isolated using the Qiagen RNeasy Total RNA kit and analyzed by electrophoresis on a 1.5% agarose-6 M urea-citrate gel. Alternatively for the reconstitution of replication, one set of cells was transfected with wt P (1 μg) and wt L (1 μg) plasmids to synthesize the wt polymerase (wt Pol) and another set was transfected with the wt NP (8 μg) and wt or mutant P (8 μg) plasmids. After preparation of the cell extracts, aliquots (45 μl each) of the wt Pol and NP0-P extracts were mixed, and replication was assayed as described above. The replication in the reconstituted wt reaction was similar to that obtained when the NP, P, and L plasmids were coexpressed (data not shown).
For in vivo replication, infected cells were transfected as for in vitro replication with all the L, NP, and P plasmids with the addition of plasmid pSPDI-H (2.5 μg) to all samples with pGEM4 (7.5 μg) for the mock sample. Extracts were prepared in SV Salts B and treated with MN, and the nuclease-resistant nucleocapsid-associated RNA was isolated using the Qiagen RNeasy Total RNA kit. The RNA was separated on a 1.5% agarose-6 M urea-citrate gel and transferred to Hybond-N nylon membrane. The unlabeled DI-H replication products (− sense) were detected by Northern blot analysis with 32P-labeled T7 DI-H transcript (+ sense) generated from XbaI-linearized pSPDI-H plasmid. Hybridization was carried out overnight at 57°C, and the blot was washed in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])−0.1% SDS at room temperature for 15 min, once in the same solution at 57°C for 30 min, and twice in 2× SSPE-0.1% SDS at 57°C for 30 min. All RNA products were visualized by autoradiography and quantitated on the phosphorimager (Molecular Dynamics).
RESULTS
Mutant P proteins defective in transcription in vitro.
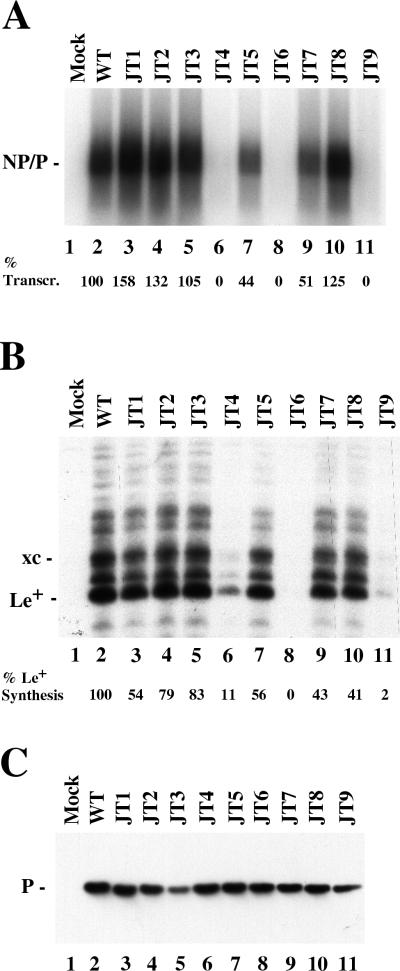
Eight charged-to-alanine mutants and one hydrophobic-to-alanine mutant were constructed in the NC binding domain of the P protein as shown in Table 1. To test for the transcription activity of the mutant P proteins, VVT7-infected A549 cells were cotransfected with L plasmid together with the wt or mutant P plasmid and incubated overnight. Cytoplasmic cell extracts were prepared and incubated with polymerase-free Sendai virus RNA-NP template and radiolabeled nucleotide. Total RNA was isolated and analyzed as described in Materials and Methods. The wt P and L proteins were active in the synthesis of full-length NP and P mRNA products which comigrate, whereas no synthesis of mRNAs was detected in the absence of viral proteins (Fig. 1A, lanes 2 and 1, respectively). The mutants JT1, JT2, JT3, and JT8 gave wt or better levels of mRNA synthesis (Fig. 1A, lanes 3 to 5 and 10), while JT5 and JT7 gave about half the activity (Fig. 1A, lanes 7 and 9). In contrast, the mutants JT4, JT6, and JT9 were completely inactive (Fig. 1A, lanes 6, 8, and 11). We also tested if the P mutants could synthesize the first transcription product, the 55-nt le+ RNA, from the wt RNA-NP template. The infected, transfected extracts were used for the synthesis of unlabeled viral RNAs which were separated by polyacrylamide-urea gel electrophoresis and analyzed by Northern blotting with a le+ specific probe. The wt P-L complex synthesized le+ RNA as well as some longer products due to read-through of the leader-NP gene boundary as reported earlier (6, 36), while no le+ RNA was detected in the mock sample (Fig. 1B, lanes 2 and 1). JT1, JT2, JT3, and JT8, which showed no reduction of mRNA synthesis, synthesized le+ RNA but at reduced levels (41 to 83%) compared to wt P (Fig. 1B, lanes 3 to 5 and 10). Mutants JT5 and JT7 were reduced in their ability to synthesize le+ RNA (Fig. 1B, lanes 7 and 9) by amounts similar to their reduction in mRNA synthesis. JT6 gave no le+ RNA synthesis, while JT4 and JT9 synthesized small amounts (2 to 11%) (Fig. 1B, lanes 8, 6, and 11). Thus, JT6 is defective in all transcription, while JT4 and JT9 can synthesize only le+ RNA. Immunoblot analysis of samples of the cell extracts showed that the mutant P proteins were nearly equally expressed and as stable as wt P after overnight incubation (Fig. 1C), so the differences were in the activities of the proteins. JT3 was somewhat reduced here, but not in duplicate experiments (not shown). Thus, mutagenesis of P amino acids throughout the NC binding domain produced a spectrum of transcription phenotypes.
FIG. 1.
The effect of the P mutants on viral transcription. (A) In vitro mRNA synthesis. VVT7-infected cells were transfected with no plasmids (Mock) or with L and the indicated wt or mutant P plasmids. Cytoplasmic extracts were prepared and incubated with wt RNA-NP and [α-32P]CTP, and the RNA transcripts were isolated and analyzed by agarose-urea-citrate gel electrophoresis. The position of the NP and P transcripts is indicated. The level of transcription (% Transcr.), as an average of three experiments, is shown below the gel relative to wt P as 100%, with a variation of <10%. (B) le+ RNA synthesis. VVT7-infected cells were transfected as described for panel A, extracts were incubated with wt RNA-NP, and le+ RNA synthesis was analyzed by Northern blotting with a complementary end-labeled probe as described in Materials and Methods. The positions of le+ RNA and xylene cyanol (xc) dye are indicated. The level of le+ synthesis, as an average of two experiments relative to wt P as 100%, is shown below the gel. (C) Immunoblot analysis of samples in panel A with α-P antibody. The position of the P protein is indicated.
The inactive P mutants are differentially active in supplemental P transcription function.
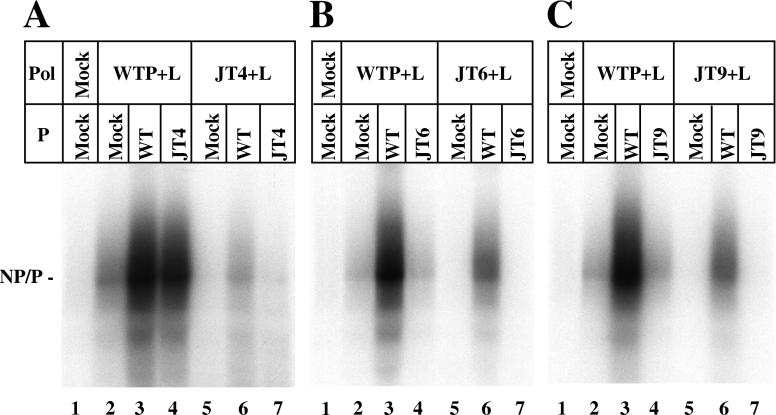
A required supplemental role for the P tetramer alone, in addition to its activity as part of the P-L polymerase complex, has been described (3, 8, 9), although the mechanism for this additional function of P has yet to be delineated. To test if the defect in the transcription activities of the JT4, JT6, and JT9 proteins are independent of or dependent on L, we designed experiments to ask if mutant P proteins had supplemental activity with the wt P-L complex or if wt P protein could rescue the defective polymerase complexes, respectively, as described in Materials and Methods. The wt P+L or mutant (JT+L) polymerase complexes lacking supplemental P function were expressed in one set of cells by utilizing a suboptimal ratio of P and L plasmids. The wt P+L gave very reduced (<5%) transcription with the addition of a mock-transfected extract as expected (Fig. 2, lane 2), since P was limiting. When a wt P extract, in which P protein was expressed alone, was added to the suboptimal wt P+L complex, transcription activity was significantly stimulated (Fig. 2, lane 3), up to the level observed when P and L were coexpressed at the optimum ratio of plasmids (data not shown). The addition of the JT4 protein to wt P-L complex, was able to rescue 71% of activity (Fig. 2A, lane 4); however, neither JT6 nor JT9 P was able to restore any activity (Fig. 2B and C, lanes 4). Furthermore, none of the JT+L mutant polymerases could be rescued to full activity by the addition of either wt P or mutant P (Fig. 2, lanes 6 and 7). JT4+L polymerase could not be activated at all by wt P, while wt P, but not the homologous mutant P, stimulated the activity of the JT6+L and JT9+L complexes about 35%. Thus, these P mutants are differentially defective in supplemental P function and polymerase activity.
FIG. 2.
Supplemental function of mutant P proteins. VVT7-infected A549 cells were transfected with no plasmids (Mock) or the wt or mutant P plasmids alone, and separate dishes were cotransfected with the same P plasmids and wt L plasmid at a suboptimal ratio. Cell extracts containing the viral polymerases were then divided into three aliquots, and to these were added a cell extract containing either mock-transfected cells, wt P, or mutant P protein, where the P proteins were expressed alone as described in Materials and Methods. The samples were incubated with wt RNA-NP in the presence of [α-32P]CTP. The RNA was purified and analyzed by agarose-urea-citrate gel electrophoresis. The position of the NP and P transcripts is indicated.
The mutant P proteins bind the nucleocapsid template.
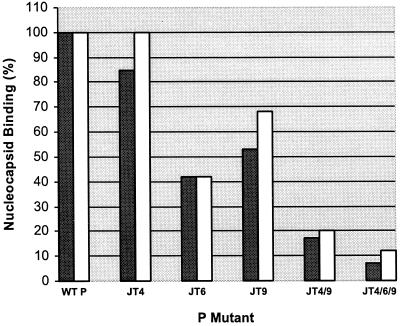
Since these P mutations reside in the NC binding domain, perhaps JT4, JT6 and JT9 were inactive in RNA synthesis due to impaired binding to the template. To test NC binding VVT7 infected cells were transfected with wt or mutant P plasmids together with L plasmid and labeled with Tran35S-label overnight. Cytoplasmic extracts were incubated in the presence of NC and pelleted through glycerol as described in Materials and Methods. Quantitation of the amount of P associated with NC showed JT4 retained significant binding to NC, while JT6 and JT9 were reduced 60% and 50%, respectively, compared to wt P (Fig. 3). The amount of L protein sedimenting with NC by virtue of its binding to P was directly proportional to the level of P (Fig. 3), where the ratio of P to L varied from 1 to 1.3. Thus, the single sets of mutations did not abolish NC binding and the degree of binding did not correlate with the complete lack of transcription activity of these mutants. All of the P mutants that could transcribe did bind to NC as expected (data not shown). Since there was some reduction in binding for two of the mutants, we constructed P mutants, JT4/6 and JT4/6/9, that contained multiple sets of mutations as described in Materials and Methods. Each of these mutants was synthesized like wt P and was completely inactive in all RNA synthesis (data not shown). Analysis of their binding to NC showed a progressive and proportional loss of binding of both P and L with the double and triple mutants (Fig. 3). These data show that the defect in RNA synthesis with some of the individual P mutants is not proportional to the degree of polymerase binding to NC.
FIG. 3.
Binding of the mutant P proteins to nucleocapsids. VVT7-infected A549 cells were cotransfected with wt L and the wt or mutant P plasmids. The cells were labeled with Tran35S-label, and cytoplasmic cell extracts were prepared, incubated in the presence of polymerase-free wt RNA-NP, and pelleted through glycerol as described in Materials and Methods. The pellets were analyzed by SDS-PAGE, and the amount of P (dark boxes) and L (open boxes) associated with NC was quantitated on the phosphorimager and plotted relative to the binding of wt P or L, respectively, as 100%.
P mutants inactive in replication.
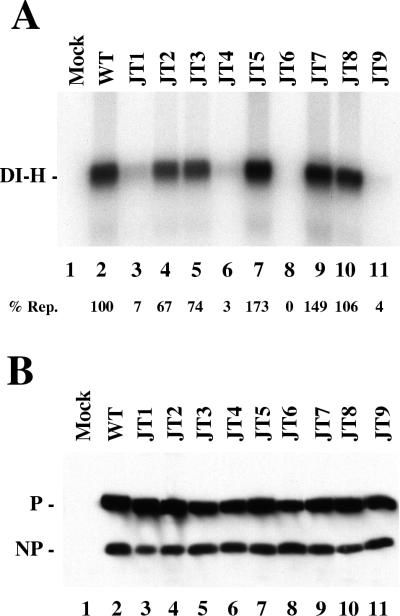
The P mutants were tested for their ability to function in DI-H viral replication under two different conditions, in vitro with added template and in vivo with a DI-H clone. For in vitro replication there is a large molar excess of input DI-H RNA-NP template compared with newly synthesized DI-H to serve as template for replication. Therefore, this assay measures the ability of the viral polymerase to carry out a single round of replication from preassembled DI-H nucleocapsids. VVT7-infected cells were transfected with the L, the NP, and the wt or mutant P plasmids. Cytoplasmic extracts were prepared and incubated with polymerase-free DI-H nucleocapsids and [α-32P]CTP. Nuclease-resistant, nucleocapsid-associated RNA was isolated and analyzed by agarose-urea-citrate gel electrophoresis. Replication of full-length DI-H genome RNA occurred in the presence, but not in the absence, of the wt viral proteins (Fig. 4A, lanes 1 and 2). Mutants JT4, JT6, and JT9, which were inactive in transcription (Fig. 2), were also virtually inactive in replication in vitro (Fig. 4A, lanes 6, 8, and 11). JT5, JT7, and JT8 each replicated at wt or better levels (lanes 7, 9, and 10), while JT2 and JT3 were reduced by 26 to 33% (lanes 4 and 5). Transcription for these five mutants was also significant (Fig. 1); however, the levels did not precisely correlate with those for replication. For JT2, JT3, and JT8 transcription was higher than replication, but the reverse was true for JT5 and JT7. In contrast, JT1 was nearly inactive in replication (7% [Fig. 4A, lane 3]), although it transcribed better than wt P (Fig. 1). Thus, for JT1 P transcription and replication are uncoupled. Immunoblot analysis on a portion of each extract showed similar expression of the P mutants in each sample (Fig. 4B).
FIG. 4.
In vitro replication with the P mutants. VVT7-infected cells were either mock transfected or cotransfected with the L, the NP, and the indicated wt or mutant P plasmids. (A) Cytoplasmic extracts were prepared and incubated with polymerase-free DI-H nucleocapsids and [α-32P]CTP at 30°C. Nuclease-resistant radiolabeled RNA was isolated and separated by agarose-urea-citrate gel electrophoresis as described in Materials and Methods. The position of the DI-H RNA is indicated. The level of replication (% Rep.), as an average of three experiments relative to wt P as 100%, is shown below the gel with a difference of <10%. (B) Immunoblot analysis of the extracts with α-P and α-SV antibodies. The positions of the P and NP proteins are indicated.
For replication P must form P-L polymerase and NP0-P complexes, which are both made when NP, P, and L are cosynthesized as they were in the experiment described in the legend to Fig. 4. While we know the polymerase is defective in JT4, JT6, and JT9 (Fig. 1), it is unknown if the NP0-P complex of each mutant is also defective. To examine just the NP0-P function of the mutants, we conducted replication reconstitution experiments as described in Materials and Methods. wt P-L (Pol) was synthesized in one set of dishes, an extract of these cells was mixed with an extract of cells expressing just the wt or mutant NP0-P complex, and DI-H replication from added template was measured. Genome replication with wt Pol was dependent on the addition of the wt NP0-P complex (Fig. 5A, lanes 1 and 2). JT1 gave minimal activity (6 to 7%) in both the reconstitution of replication (lane 3) as well as normal in vitro DI-H replication, although it could transcribe well (Fig. 1). JT4, JT6, and JT9 in complex with NP were nearly or completely inactive in the reconstitution of replication (Fig. 5A, lanes 6, 8, and 11), showing that both the NP0-P complexes, as well as the polymerases (Fig. 4), of these P mutants were defective. Interestingly, only the NP0-P complexes of JT3 and JT8 retained partial activity, while surprisingly the remainder of the mutant complexes were virtually or completely inactive (Fig. 1A). We had expected JT2, JT5, and JT7 to have significant activity as they had when the P, NP, and L proteins were all expressed together (Fig. 4). Apparently coexpression of the three viral proteins rescues the defect in NP0-P complexes formed by expressing just NP and P. Immunoblot analysis on a portion of each extract showed equivalent NP and P expression in each sample (Fig. 5B).
FIG. 5.
Reconstitution of replication in vitro from separate P-L and NP0-P complexes. In separate dishes VVT7-infected cells were mock transfected (−) and cotransfected with wt L and wt P plasmids (WT Pol) at levels optimal for transcription or with NP and the indicated wt or mutant P plasmids (NP+P). (A) Cytoplasmic extracts were prepared, and an extract with WT Pol was mixed with an extract with wt or mutant NP+P and the samples were incubated with polymerase-free DI-H nucleocapsids and [α-32P]CTP as described in Materials and Methods. Nuclease-resistant radiolabeled RNA was isolated and separated by agarose-urea-citrate gel electrophoresis. The position of the DI-H RNA is indicated. The level of replication (% Rep.), as an average of three experiments relative to wt P as 100%, is shown below the gel. (B) Immunoblot analysis of the WT Pol and NP+P extracts with α-P and α-SV antibodies. The positions of the P and NP proteins are indicated.
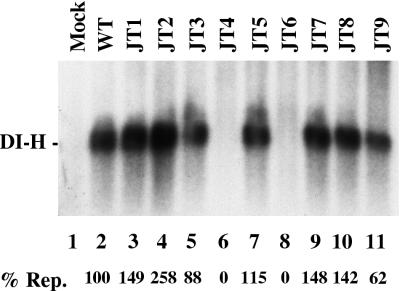
The activity of the mutant P proteins was also assessed for DI-H replication in vivo. This assay differs from its in vitro counterpart in that following VVT7 infection, plasmids encoding NP, L, and the indicated P proteins and a plasmid containing a cDNA copy of the DI-H genome are all transfected simultaneously. T7 transcription from the DI-H plasmid yields (+)-sense DI-H genome RNA, which is then nonspecifically encapsidated by the NP protein encoded by pGEM-NP. The encapsidated (+)-sense DI-H is then replicated in vivo by the viral polymerase with multiple rounds of amplification required to detect product. Cell extracts were prepared and digested with MN to eliminate any nonencapsidated RNA. The nuclease-resistant replication products were then isolated, separated by gel electrophoresis, and detected by Northern blot analysis with a probe specific for (−)-sense DI-H genome RNA as described in Materials and Methods. In the absence of viral proteins no DI-H RNA was replicated, while full-length DI-H RNA was detected in the presence of the wt P, NP, and L proteins (Fig. 6, lanes 1 and 2). JT4 and JT6 were also inactive in replication in vivo, as they were in transcription and replication in vitro, however, for the first time JT9 also gave activity (Fig. 6, lanes 6, 8 and 11). Each of the mutants (JT2, JT3, JT5, JT7, and JT8) that could function in replication in vitro where the NP, L, and P proteins were cosynthesized (Fig. 4) also did so in vivo (Fig. 6). JT1 did synthesize DI-H RNA in vivo (Fig. 6, lane 3), but not in vitro (Fig. 4 and 5). Thus, for two mutants, JT1 and JT9, the intact cell somehow rescued a replication defect of P mutants observed in extracts of these cells.
FIG. 6.
In vivo replication with the P mutants. VVT7 infected cells were transfected with pSPDI-H only (Mock) or with pSPDI-H, L, NP, and the indicated wt or mutant P plasmids. After overnight incubation cytoplasmic extracts were prepared and directly treated with MN. The nuclease-resistant RNA was then isolated, separated by agarose-urea-citrate gel electrophoresis, and analyzed by Northern blotting with a DI-H-specific (+)-sense 32P-labeled riboprobe as detailed in Materials and Methods. The position of the DI-H RNA is indicated. The level of replication (% Rep.), as an average of two experiments relative to wt P as 100%, is shown below the gel.
Correlation of the formation of the mutant NP0-P complex with viral replication.
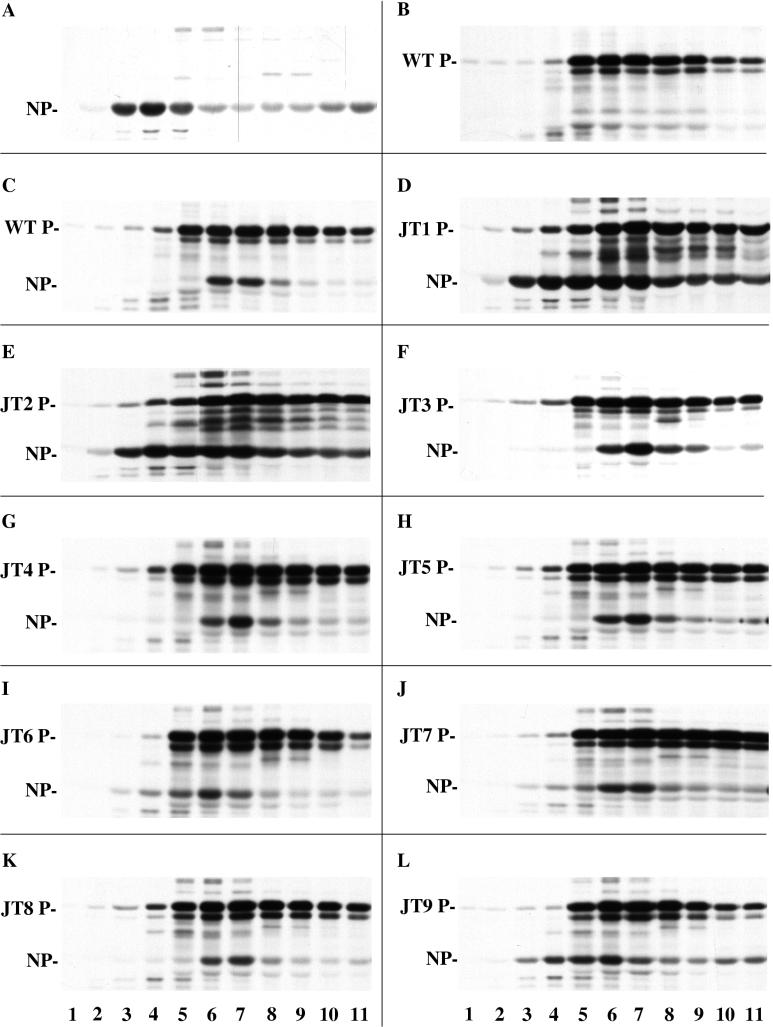
Replication defects in the P protein could be due to a defect in its function in the polymerase complex and/or to a defect in NP0-P complex formation or function. By sedimentation analysis of coexpressed 35S-labeled NP and mutant P proteins as described in Materials and Methods, one can monitor NP0-P complex formation by a shift of NP from a soluble form to a form in complex with P. Part of NP protein expressed alone sedimented near the top of the gradient in fractions 3 to 5 (Fig. 7A) which we designate NP0, while the majority pelleted as assembled nucleocapsid-like structures (data not shown). P protein expressed alone sedimented as a tetramer (34, 35) in fractions 5 to 9, but with some faster-sedimenting species (Fig. 7B). The sedimentation profile of each of the P mutants was the same as that of wt P (Fig. 7), suggesting that none of the changes affected the oligomerization of P, in agreement with direct gstP-P binding studies (data not shown). When wt P was coexpressed with NP in amounts optimal for replication, the sedimentation profile of P remained the same, while NP now cosedimented with P in fractions 6 and 7 (Fig. 7C) rather than as NP0. The P mutants, JT3 and JT8, also formed an NP0-P complex since NP0 shifted to sediment with the mutant P (Fig. 7F and K), consistent with their wt activity in all replication in vitro and in vivo (Fig. 8). JT4 and JT5 also cosedimented with NP (Fig. 7G and H); however, in these two cases this apparent complex formation did not correlate with their ability to function in replication (Fig. 8). The NP0-JT4 complex was inactive in all the replication assays, so apparently the complex was completely defective, and while NP0-JT5 was also inactive in the reconstitution of replication, it was active in normal replication in vitro and in vivo. When coexpressed with JT1, JT2, JT6, JT7, or JT9, NP sedimented in a different pattern from fractions 3 to 7 (Fig. 7D, E, I, J, and L), suggesting that complex formation might be impaired. Indeed each of these P mutants was inactive in the reconstitution of replication; however, their activities in the other assays for replication were not the same. JT2 and JT7 did give DI-H replication with the coexpression of all the plasmids both in vitro and in vivo, while JT1 and JT9 only showed activity in vivo (Fig. 8). JT6, on the other hand, was never active under any replication conditions. Thus, the relationship between the NP0-P complex formation and replication activity is very complicated and dependent upon how the proteins are expressed.
FIG. 7.
Sedimentation analysis of NP0-P complex formation. VVT7-infected A549 cells were transfected with the indicated NP and/or wt or mutant P plasmids. The cells were labeled with Tran35S-label, and cytoplasmic cell extracts were prepared and fractionated on a glycerol gradient as described in Materials and Methods. Samples of the gradient fractions were immunoprecipitated with α-P and α-SV antibodies and analyzed by SDS-PAGE. Sedimentation is from left to right. The positions of the proteins are indicated.
FIG. 8.
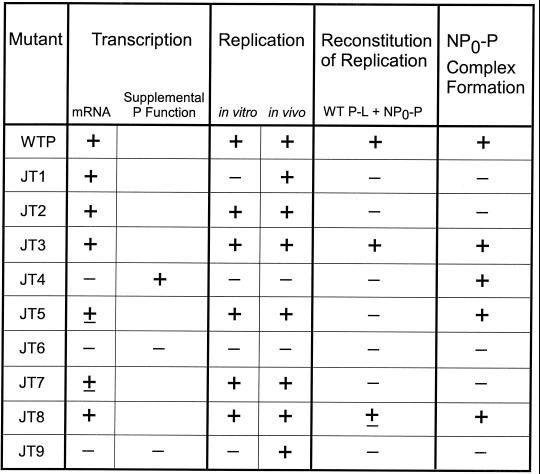
Summary of the protein-protein interactions and activities of the Sendai virus P mutants. The interactions and activities of each mutant were quantitated in FIG. 1, 2, and 4 to 7 but are summarized here by the following symbols: +, significant activity (greater than 60% of wt P); ±, intermediate activity (between 10 and 60% of wt P); and −, little or no activity (less than 10% of wt P). For supplemental P function only mutants JT4, JT6, and JT9 were tested.
DISCUSSION
The initial deletion mapping of protein-protein interaction sites on the Sendai virus P protein suggested that the binding domains for L, P, NP0, and NC resided in very discrete nonoverlapping regions in the C-terminal half of the protein. We designed a mutagenesis strategy to identify amino acids that were important specifically for NC and NP0 binding, functions which had been mapped to the C-terminal 481 to 568 aa of P (29, 30). All the mutants formed P oligomers (not shown) and bound to L protein (Fig. 3), which was expected since these domains were previously mapped as N-terminal to aa 481 (11, 33). Two mutants, JT3 and JT8, retained all P functions at or near the levels of wt P, so these amino acids are nonessential (summarized in Fig. 8). Analysis of RNA synthesis showed that six of the P mutants had significant or better than wt P activity in transcription, while three others—JT4, JT6, and JT9—were completely defective for both transcription and genome replication in vitro (Fig. 8). JT4, however, had normal supplemental P function and normal NC and NP0 binding, so its RNA synthesis defect resided only in the P-L catalytic activity. JT6 and JT9 abolished both P supplemental and P-L function, as well as NP0 binding, although they bound NC at a level of about 50% of wt P. This reduction, however, did not account for the complete inactivity of these mutants in transcription, so in these cases as well there appears to be an actual defect in the polymerase itself. Thus, mutation in the NC binding domain can affect and even uncouple the supplemental P and P-L catalytic activities which were previously thought to reside only in the L binding domain, since deletion (9) or specific changes in amino acids (3) in this region abolished activity. We postulate that the mutant P proteins bind to, but cause an improper folding of, L that leads to the loss of catalytic function.
NC binding was, in fact, inhibited by combinations of the JT4, JT6, and JT9 mutations (Fig. 3), suggesting that multiple, nonlinear regions of P within this domain are needed for binding to the template. Comparison of the sequence of Sendai virus P with the P proteins of other parainfluenza viruses shows that the amino acids changed in JT4 and JT9 are conserved, and one of three is conserved in JT6, while there is less conservation at the positions of the other mutants (25). It is interesting to speculate that where there is conservation these amino acids would also be essential for similar activities with the P proteins of other viruses.
Three mutants—JT1, JT2, and JT7—gave some transcription but were defective in NP0-P complex formation and the function of this complex in the reconstitution of genome replication as assayed by the addition of wt polymerase to the mutant NP0-P complex (FIG. 8). Surprisingly, the coexpression of the NP, L, and mutant P proteins, as opposed to reconstitution with separately expressed complexes, rescued the defect in replication in vitro and in vivo for JT2 and JT7, but only in vivo for JT1 as well as JT9. We analyzed NP0-P formation for JT2 and JT7 when the P, NP, and L proteins were all coexpressed but still saw a defective sedimentation pattern identical to that of the expression of just NP and mutant P (data not shown). Nonetheless, the mutants were active in replication under these conditions. We have observed a similar phenotype with some Sendai virus L mutants which can replicate in vivo but not in vitro (7), so the effect occurs with mutants in both subunits of the polymerase. Expression of all the viral proteins and an intact cell thus overcame defects in some, but not all, of the mutant proteins.
The data suggest that the intact cell is providing a function(s) to rescue a replication defect for some, but not all, of the defective P proteins; however, this function cannot be reproduced in extracts of the cells. The mechanism for this rescue is unknown; however, the same effect was observed in another cell line. Mutants JT1 and JT9 also replicated in vivo, but not in vitro, in BHK cells (data not shown), so these results were not cell type specific. Perhaps it is the architecture of the intact cell that somehow facilitates the proper folding of the mutant proteins. The data do show that the use of a single assay for determining the function of a mutant viral protein might be misleading and emphasize also that in vitro assays may not measure all the possible interactions and functions of a given protein. Nonetheless, the C-terminal region of P possesses multiple functions besides NC binding that can be separated by mutation.
To various degrees the protein-protein interactions mediated by the P proteins of several paramyxoviruses have been studied, mainly by deletion mapping. In the most complete experiments, the C terminus of the P protein of human parainfluenza virus type 2 (hPIV2), has been shown to form NP0-P, P-P, P-L, and P-NC complexes, and the mapping of the domains showed a modular structure like that of Sendai virus P (26–28). In contrast, both the N terminus and C terminus of the P proteins of measles virus (17), respiratory syncytial virus (16, 23), hPIV3 (14), and rinderpest virus (32) have been shown to be required for the NP-P interaction, although whether the interaction was with NP0 or NC was not always distinguished. In Sendai virus P, regions of the N terminus are required only for binding to N0 (12, 22). The data show that each virus has similarities as well as differences in the details of the binding sites from that of Sendai virus P. Analogous to the experiments presented here in which a functional polymerase required both the P-L binding domain and the NC binding site on the P protein, Bousse et al. (2) showed with chimeric hPIV1 and Sendai virus P proteins that both the L binding domain and the C terminus were required to interact with the hPIV1 L protein. Whether this involves specifically the polymerase or supplemental function of P remains to be determined. Thus, the details of all the protein-protein interactions and functions of the P proteins of other paramyxoviruses will require further analysis of structure and function by more detailed site-directed mutagenesis.
Acknowledgments
This work was funded by NIH grant AI14594 (to S.A.M.).
REFERENCES
- 1.Bass, S. H., M. G. Mulkerrin, and J. A. Wells. 1991. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc. Natl. Acad. Sci. USA 88: 4498–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousse, T., T. Takimoto, T. Matrosovich, and A. Portner. 2001. Two regions of the P protein are required to be active with the L protein for human parainfluenza virus type 1 RNA polymerase activity. Virology 283: 306–314. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73: 6474–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrappa, S., S. Hendricks, Y.-B. Pan, J. M. Seyer, and K. G. Gupta. 1995. Intracellular phosphorylation of the Sendai virus P protein. Virology 208: 408–413. [DOI] [PubMed] [Google Scholar]
- 5.Carlsen, S. R., R. W. Peluso, and S. A. Moyer. 1985. In vitro replication of Sendai virus wild-type and defective interfering particle genome RNAs. J. Virol. 54: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrika, R., S. M. Horikami, S. Smallwood, and S. A. Moyer. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213: 352–363. [DOI] [PubMed] [Google Scholar]
- 7.Cortese, C. K., J. A. Feller, and S. A. Moyer. 2000. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology 277: 387–396. [DOI] [PubMed] [Google Scholar]
- 8.Curran, J. 1996. Reexamination of the Sendai virus P protein domains required for RNA synthesis: a possible supplemental role for the P protein. Virology 221: 130–140. [DOI] [PubMed] [Google Scholar]
- 9.Curran, J. 1998. A role for the Sendai virus P protein trimer in RNA synthesis. J. Virol. 72: 4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10: 3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran, J., R. Boeck, N. Lin-Marq, A. Lupas, and D. Kolakofsky. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214: 139–149. [DOI] [PubMed] [Google Scholar]
- 12.Curran, J., J.-B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran, J., T. Pelet, and D. Kolakofsky. 1994. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology 202: 875–884. [DOI] [PubMed] [Google Scholar]
- 14.De, B. P., M. A. Hoffman, S. Choudhary, C. C. Huntley, and A. K. Banerjee. 2000. Role of NH2- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J. Virol. 74: 5886–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83: 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76: 2863–2867. [DOI] [PubMed] [Google Scholar]
- 18.Higuichi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16: 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66: 4901–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikami, S. M., R. E. Hector, S. Smallwood, and S. A. Moyer. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235: 261–270. [DOI] [PubMed] [Google Scholar]
- 21.Horikami, S. M., and S. A. Moyer. 1995. Alternative amino acids at a single site in the Sendai virus L protein produce multiple defects in RNA synthesis in vitro. Virology 211: 577–582. [DOI] [PubMed] [Google Scholar]
- 22.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology 222: 383–390. [DOI] [PubMed] [Google Scholar]
- 23.Khattar, S. K., A. S. Yunus, P. L. Collins, and S. K. Samal. 2001. Deletion and substitution analysis defines regions and residues within the phosphoprotein of bovine respiratory syncytial virus that affect transcription, RNA replication, and interaction with the nucleoprotein. Virology 285: 253–269. [DOI] [PubMed] [Google Scholar]
- 24.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 577–604. In B. N. Fields, D. M. Knipe, and A. Kato (ed.), Fundamental virology. Lippincott-Raven, New York, N.Y..
- 25.Matsuoka, Y., J. Curran, T. Pelet, D. Kolakofsky, R. Ray, and R. W. Compans. 1991. The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J. Virol. 65: 3406–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishio, M., M. Tsurudome, M. Ito, and Y. Ito. 2000. Mapping of domains on the human parainfluenza type 2 virus P and NP proteins that are involved in the interaction with the L protein. Virology 273: 241–247. [DOI] [PubMed] [Google Scholar]
- 27.Nishio, M., M. Tsurudome, M. Ito, N. Watanabe, M. Kawano, H. Komada, and Y. Ito. 1997. Human parainfluenza virus type 2 phosphoprotein: mapping of monoclonal antibody epitopes and location of the multimerization domain. J. Gen. Virol. 78: 1303–1308. [DOI] [PubMed] [Google Scholar]
- 28.Nishio, M., M. Tsurudome, M. Kawano, N. Watanabe, S. Ohgimoto, M. Ito, H. Komada, and Y. Ito. 1996. Interaction between nucleocapsid protein (NP) and phosphoprotein (P) of human parainfluenza virus type 2: one of the two NP binding sites on P is essential for granule formation. J. Gen. Virol. 77: 2457–2463. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, K. W., and D. W. Kingsbury. 1988. Carboxyl-terminal region of Sendai virus P protein is required for binding to viral nucleocapsids. Virology 167: 106–112. [Erratum, 169:489,1989.] [DOI] [PubMed] [Google Scholar]
- 30.Ryan, K. W., E. M. Morgan, and A. Portner. 1991. Two noncontiguous regions of Sendai virus P protein combine to form a single nucleocapsid binding domain. Virology 180: 126–134. [DOI] [PubMed] [Google Scholar]
- 31.Ryan, K. W., and A. Portner. 1990. Separate domains of Sendai virus P protein are required for binding to viral nucleocapsids. Virology 174: 515–521. [DOI] [PubMed] [Google Scholar]
- 32.Shaji, D., and M. S. Shaila. 1999. Domains of rinderpest virus phosphoprotein involved in interaction with itself and the nucleocapsid protein. Virology 258: 415–424. [DOI] [PubMed] [Google Scholar]
- 33.Smallwood, S., K. W. Ryan, and S. A. Moyer. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202: 154–163. [DOI] [PubMed] [Google Scholar]
- 34.Tarbouriech, N., J. Curran, R. W. H. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7: 777–781. [DOI] [PubMed] [Google Scholar]
- 35.Tarbouriech, N., J. Curran, C. Ebel, R. W. H. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266: 99–109. [DOI] [PubMed] [Google Scholar]
- 36.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63: 1951–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]