Abstract
The p6 domain of human immunodeficiency virus type 1 (HIV-1) is located at the C terminus of the Gag precursor protein Pr55Gag. Previous studies indicated that p6 plays a critical role in HIV-1 particle budding from virus-expressing HeLa cells. In this study, we performed a detailed mutational analysis of the N terminus of p6 to map the sequences required for efficient virus release. We observed that the highly conserved P-T/S-A-P motif located near the N terminus of p6 is remarkably sensitive to change; even conservative mutations in this sequence imposed profound virus release defects in HeLa cells. In contrast, single and double amino acid substitutions outside the P-T/S-A-P motif had no significant effect on particle release. The introduction of stop codons one or two residues beyond the P-T/S-A-P motif markedly impaired virion release, whereas truncation four residues beyond P-T/S-A-P had no effect on particle production in HeLa cells. By examining the effects of p6 mutation in biological and biochemical analyses and by electron microscopy, we defined the role of p6 in particle release and virus replication in a panel of T-cell and adherent cell lines and in primary lymphocytes and monocyte-derived macrophages. We demonstrated that the effects of p6 mutation on virus replication are markedly cell type dependent. Intriguingly, even in T-cell lines and primary lymphocytes in which p6 mutations block virus replication, these changes had little or no effect on particle release. However, p6-mutant particles produced in T-cell lines and primary lymphocytes exhibited a defect in virion-virion detachment, resulting in the production of tethered chains of virions. Virus release in monocyte-derived macrophages was markedly inhibited by p6 mutation. To examine further the cell type-specific virus release defect in HeLa versus T cells, transient heterokaryons were produced between HeLa cells and the Jurkat T-cell line. These heterokaryons display a T-cell-like phenotype with respect to the requirement for p6 in particle release. The results described here define the role of p6 in virus replication in a wide range of cell types and reveal a strong cell type-dependent requirement for p6 in virus particle budding.
The human immunodeficiency virus type 1 (HIV-1) Gag precursor Pr55Gag, like those of other retroviruses, is necessary and sufficient for the production of noninfectious virus-like particles. Pr55Gag is composed of four major domains, ordered from the N to the C terminus, matrix (MA, p17), capsid (CA, p24), nucleocapsid (NC, p7), and p6 (for reviews, see references 8 and 56). Previous studies have indicated that deletion of p6 markedly inhibits virus particle production from Gag-expressing cells (19, 24). A highly conserved motif, P-T/S-A-P, located near the N terminus of p6 is particularly critical for the p6 virus release function. Examination of Gag-expressing HeLa and COS cells by electron microscopy (EM) indicates that p6 mutation blocks a very late step in virus release; p6- mutant particles fail to bud and remain tethered to the plasma membrane (19, 24).
Domains have been identified in the Gag proteins of other retroviruses that, like HIV-1 p6, appear to be required for virus release. These proteins include p2b of the avian retrovirus Rous sarcoma virus (RSV) (61, 62), p12 of murine leukemia virus (MuLV) (67, 68), p9 of equine infectious anemia virus (47), and pp16 of Mason-Pfizer monkey virus (63). To reflect the function of these sequences late in the virus release process, these domains are collectively referred to as late, or L, domains (43). A similar function appears to be encoded by the matrix protein of the rhabdoviruses and filoviruses (4, 20, 21, 25).
All retroviral and nonretroviral L domains characterized to date contain motifs recognized as being involved in protein-protein interactions among cellular proteins; in each case, this motif has been reported to be critical for L domain function. These motifs include P-X-X-P in HIV-1 p6 (19, 24), Y-X-X-L in equine infectious anemia virus p9 (47), and P-P-P-Y in the L domains of MuLV (68), RSV (62), Mason-Pfizer monkey virus (63), the rhabdoviruses (4, 21), and the filoviruses (20). Among cellular proteins, P-X-X-P, Y-X-X-L, and P-P-P-Y motifs interact with Src homology region 3 (SH3) domains, clathrin-associated adapter protein complexes, and WW domains, respectively (3, 38, 45, 58).
In some cases, interactions or subcellular colocalization between L domain proteins and predicted cellular partners have been demonstrated (18, 20, 21, 48). These observations suggest that L domains may function by interacting with host factors to promote virus release. However, the relevance of these interactions in the L domain virus release function remains unclear. In addition, the HIV-1 L domain can substitute for the L domain of the distantly related RSV (43) and MuLV (67), despite the lack of sequence homology between these L domains.
Our understanding of p6 function is complicated by studies that failed to observe a role for p6 in virus release (22, 23, 27, 32, 33, 46, 49, 52). In some cases, the lack of a requirement for p6 might be explained by differences in Gag expression systems, Gag expression levels, and cell types used in the various studies. The observation that inactivation of the viral protease (PR) mitigated the p6-imposed defect in virus release (24) suggests that PR may also be a factor in differences among studies, since in some cases p6 mutations were evaluated in the context of full-length molecular clones, whereas other studies examined the role of p6 in Gag-only expression systems. However, in another report, PR was not observed to significantly affect p6 L domain function (1). Mutations in p6 have also been reported to reduce levels of pol-encoded enzymes in virions (65) and to affect HIV-1 particle size (16, 17). Finally, a Leu repeat sequence near the C terminus of p6 appears to be necessary for the incorporation of the HIV-1 accessory protein Vpr into virions (30, 46).
To clarify the role of p6 in HIV-1 replication, we performed a detailed mutational analysis of the N terminus of p6 within and surrounding the P-T/S-A-P motif. The effects of the mutations on virion production in HeLa cells and virus replication in the CEM(12D-7) T-cell line were determined. In addition, we evaluated the role of p6 in particle release and virus replication in a panel of T-cell and adherent cell lines and in two primary cell types, peripheral blood lymphocytes (PBLs) and monocyte-derived macrophages (MDM). The results of this study provide a detailed mapping of residues required for the L domain function of p6 and demonstrate intriguing cell type-dependent effects of p6 mutation on virion release and virus replication. To our knowledge, this is the first detailed evaluation of the role that p6 plays in virus production and replication in primary cell types that serve as targets for HIV-1 infection in vivo.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
Cell lines were maintained as described (37). The isolation and culture of human peripheral blood mononuclear cells (PBMC) and primary MDM have been reported in detail elsewhere (9). Primary cells (a kind gift from K. Fields) were purified as previously described (9). PBMC were activated for 48 h with 2 μg of phytohemagglutinin (PHA) (Roche) per ml in the presence of 20 U of interleukin-2 per ml. CD4+ PBLs were purified from PBMC by magnetic cell sorting using CD4 MicroBeads (Miltenyi) according to the manufacturer’s instructions. Elutriated monocytes were plated for 2 h in serum-free medium to facilitate attachment. Human serum was added to 10%, and monocytes were differentiated to macrophages for 8 days at 37°C.
For pseudotyping with the vesicular stomatitis virus G glycoprotein (VSV-G), wild-type (WT) and mutant molecular clones were cotransfected with plasmids encoding VSV-G (pHCMV-G [64]) and the Gag-Pol expression vector pCMVNLGagPolRRE. This latter clone (kindly provided by A. Ono) was constructed by introducing the 2.4-kb BssHII-EcoRV region from pNL4-3 into plasmid pCMVGagPolRRE (53) (kindly provided by D. Rekosh). For pseudotyping of tat-negative molecular clones, in addition to the Gag-Pol- and VSV-G-encoding plasmids, cells were cotransfected with the Tat expression plasmid pSVtat2 (26) (kindly provided by K.-T. Jeang). The gag/pol-deleted molecular clone pEvdl443 was kindly provided by J. Silver. An env-negative version of pEvdl443, pEvd1443/KFS, was constructed by introducing into pEvd1443 the env gene from pNL4-3KFS (14), which contains a frameshift mutation near the 5′ end of the env gene.
Site-directed mutagenesis and cloning.
The 1.4-kb SphI-SdaI fragment of the full-length infectious molecular clone pNL4-3 (nucleotides [nt] 1443 to 2838; GenBank accession number M19921) was subcloned into M13mp19 between the SphI and PstI sites and mutagenized (Table 1). Oligonucleotide-directed mutagenesis was performed as previously described (31). Whenever possible, we changed multiple nt for a given codon to minimize the rate of reversion in infected cultures back to the original sequence. The entire SphI-PstI region was sequenced, and fragments carrying the correct sequence were recloned back into the pNL4-3 or pNL(AD8) molecular clone. Macrophage-tropic molecular clones were constructed by replacing the 2.7-kb SalI-BamHI fragment in pNL4-3-derived constructs (nt 5785 to 8465) with that of pNL(AD8). env-negative molecular clones were constructed by substituting the same region from the pNL4-3KFS molecular clone (9).
TABLE 1.
Site-directed mutagenesis of the p6 L domaina
Transfections and infections.
Adherent cells were transfected by the calcium phosphate method, and T-cell lines were transfected by the DEAE-dextran procedure as previously described (11). For infection of adherent cell lines, T-cell lines, and PBMC, virus was obtained by transfecting 293T cells with the T-cell line-tropic molecular clone pNL4-3 (2) and p6-mutant derivatives.
For macrophage infectivity analyses, we used the macrophage-tropic molecular clone pNL(AD8) (9, 12). Pseudotyped virus stocks were prepared by cotransfecting 293T cells with the corresponding molecular clone, pHCMV-G, pCMVNLGagPolRRE, or, when specified, pSVtat2. Supernatants from transfected cultures were harvested and filtered. Virus stocks were normalized by reverse transcriptase (RT) activity and used in infections as indicated. Infections of adherent cells, T-cell lines, PBLs, PBMC, and MDM were performed as described (9, 28, 37). Replication kinetics of p6 mutants were monitored for 2 months in T-cell lines and for 1 month in primary cells (PBMC and MDM). RT assays were performed as reported (11).
Analysis of viral revertants.
Genomic DNA was purified from infected cells 2 days before peak virus replication using the QIAamp DNA minikit (Qiagen) under the manufacturer’s instructions. A 1.4-kb fragment of viral DNA (SphI-SdaI; nt 1407 to 2984) was amplified with Elongase PCR mix (Life Technology) in the presence of 2 mM MgSO4, 0.2 mM each of the four deoxynucleoside triphosphates, 0.2 μM primer, 5 μl of genomic DNA, and 1 μl of Elongase enzyme mix. The forward primer was 5′-GGAAGCTGCAGAATGGGATA-3′ (pNL4-3 nt 1407 to 1426), and the reverse primer was 5′-AAAATATGCATCGCCCACAT-3′ (nt 2984 to 2875). Reaction mixes were placed on a thermal cycler preheated to 95°C (hot-start PCR). Reaction conditions were 4.5 min at 95°C for template denaturation, followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, and 90 s at 72°C, and a final extension of 4.5 min at 72°C. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced.
Metabolic labeling and radioimmunoprecipitation.
We routinely used 106 cells for labeling. For infections with VSV-G pseudotypes, we used 107 RT cpm of each virus stock. One day postinfection, cells were labeled in 2 ml of RPMI medium lacking Met and Cys and supplemented with 10% fetal bovine serum. Upon plating of MDM, the medium was supplemented with 10% human serum (Omega). Cells were labeled with 500 μCi of [35S]Met/Cys for 16 h at 37°C.
Preparation of cell lysates, pelleting of labeled virions, and immunoprecipitation of cell- and virion-associated proteins have been described previously (11). Lysates were immunoprecipitated with HIV-1 immunoglobulin (HIV-Ig) or anti-p24 antiserum (National Institutes of Health AIDS Research and Reference Reagent Program). Quantitative analysis of bands visualized by radioimmunoprecipitation was performed on a FujiX BAS2000 Bio-image analyzer. Virus release efficiency was calculated by determining the percentage of virion-associated Gag proteins as a fraction of total (cell plus virion) Gag.
Transmission EM.
The procedures used to prepare and examine virus-expressing cells by EM have been reported previously (15).
Analysis of virus release from transient heterokaryons.
HeLa cells were infected with virus stocks obtained by transfecting 293T cells with pEvd1443 (or pEvd1443/KFS for the negative control), pHCMV-G, and pCMVNLGagPolRRE. Jurkat cells were infected with virus stocks obtained by transfecting 293T cells with a tat-negative pNL4-3 molecular clone (bearing a stop codon at nt 5861 and containing a WT or p6-mutant gag gene), pHCMV-G, pSVtat2, and pCMVNLGagPolRRE. One day postinfection, HeLa and Jurkat cells were washed three times with phosphate-buffered saline and cocultivated for 6 h at 37°C to allow cell fusion. Media were removed, and the cells were labeled as above. Cell and viral lysates were immunoprecipitated with rabbit anti-p24 antiserum.
RESULTS
Role of p6 L domain in virus release in HeLa cells.
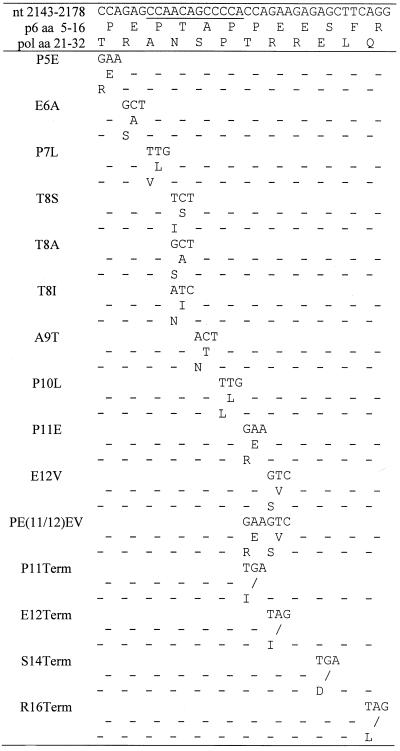
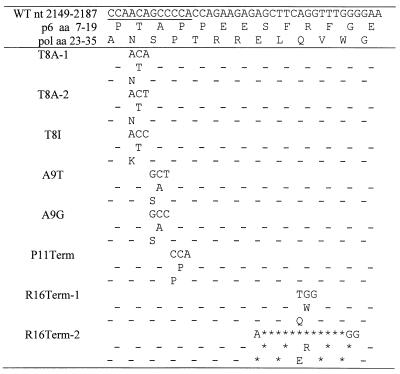
We previously demonstrated that mutations within the highly conserved P-T/S-A-P motif near the N terminus of HIV-1 p6 profoundly disrupted virus production in transfected HeLa cells (24), and a similar defect has been observed in COS cells (19). To map in greater detail the sequence requirements for the p6 virus release function, we introduced a number of additional mutations in and around the P-T/S-A-P motif (Table 1; Fig. 1); these included single and double amino acid substitutions and a series of premature termination codon mutations. Several of the mutations reported in our previous study (24) were reconstructed with a larger number of nucleotide changes to minimize the potential for primary-site reversion in culture. In many cases, the changes in the p6 open reading frame also resulted in substitutions in pol (Table 1).
FIG. 1.
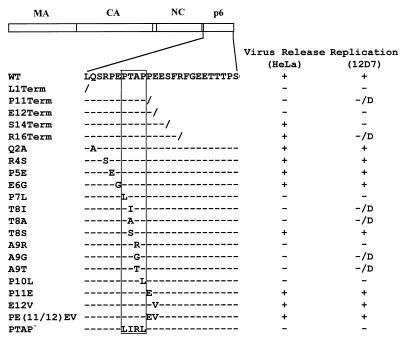
Schematic representation of p6 mutants analyzed in this study. The arrangement of the HIV-1 Gag domains is depicted at the top. The amino acid sequence of the N-terminal region of p6 encompassing the L domain is shown below. The P-T/S-A-P motif is boxed. Backslashes represent stop codons; dashes denote amino acid sequence identity with the WT. The columns at the right summarize the phenotypes of the mutants in HeLa and CEM(12D-7) cells: +, virus release or replication kinetics similar to WT; −, severe defect in virus release or a block in virus replication; D, delayed virus replication. The L1Term, Q2A, R4S, E6G, A9R, and PTAP− mutants were constructed previously (24) and were analyzed in more detail in this study. P7L, T8I, and P10L (24) were reconstructed here to increase the number of nt substitutions. (Table 1).
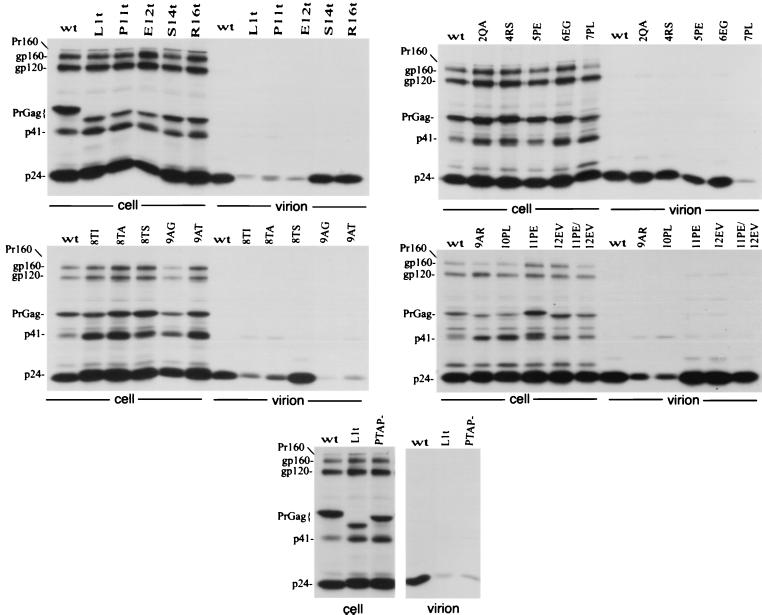
The changes were cloned into the full-length infectious molecular clone pNL4-3, and the effects on virus particle production in transfected HeLa cells were determined (Fig. 2). Single and double amino acid substitutions N- and C-terminal to the P-T/S-A-P motif had no significant effect on virus particle production, as monitored by the levels of virion-associated protein released into the medium. A Thr→Ser substitution at residue 8 (T8S) had no effect on the efficiency of virus release. In contrast, all other mutations that fell within the P-T/S-A-P motif markedly reduced virion production. PhosphorImager analysis of the gels shown in Fig. 2 indicated that the single amino acid changes in the P-T/S-A-P motif reduced the efficiency of Gag release five- to eightfold (Fig. 2 and data not shown). The premature termination codon mutations varied in the degree to which they affected the release of virion-associated Gag; the P11Term and E12Term mutations markedly disrupted particle production, whereas S14Term and R16Term had no effect on the efficiency of virus assembly and release (Fig. 2).
FIG. 2.
Immunoprecipitation of cell- and virion-associated proteins produced in HeLa cells. Transfected cells were metabolically labeled overnight with [35S]Met/Cys; cell and virion lysates were immunoprecipitated with anti-HIV-Ig. For the mutants containing termination codons, Term is abbreviated t. At the left side of each panel are indicated the positions of the Gag-Pol precursor Pr160GagPol (Pr160); the Env glycoproteins gp160 and gp120; the Gag precursor (PrGag); the Gag processing intermediate p41; and the CA protein (p24).
Role of p6 L domain in virus replication in CEM(12D-7) T-cell line.
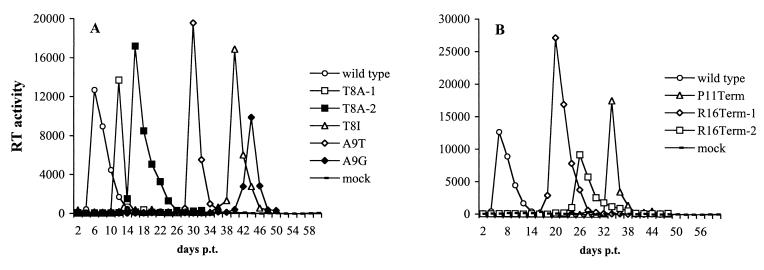
The p6 mutants described above were analyzed for their ability to establish a productive spreading infection in the CEM(12D-7) T-cell line (Fig. 3). Cells were transfected in parallel with WT or mutant molecular clones, and RT activity was monitored over time. All transfections were performed in duplicate or triplicate. The results confirmed our previous observation (24) that clones bearing mutations within the P-T/S-A-P motif failed to replicate efficiently in CEM(12D-7) cells. Only the T8S mutant replicated with WT kinetics (data not shown). In several cases, virus replication was observed with a significant delay relative to the WT (Fig. 3A and data not shown).
FIG. 3.
Replication kinetics of p6 mutants in the CEM(12D-7) T-cell line. CEM(12D-7) cells were transfected in parallel with the indicated molecular clones. Cells were split every 2 days, and RT activity was determined at each time point. Only those mutants that replicated with a delay relative to the WT (i.e., that reverted in culture) are shown. (A) Point mutants within P-T/S-A-P; (B) truncation mutants. p.t., posttransfection.
Mutations outside the P-T/S-A-P sequence were also evaluated for their effect on virus replication in the CEM(12D-7) T-cell line. These transfections were performed in triplicate. The single and double amino acid substitution mutants replicated with WT kinetics (data not shown). P11Term and E12Term, which as indicated above markedly reduced virus release in HeLa cells, failed to produce any detectable RT activity or replicated with a significant delay relative to the WT (Fig. 3B and data not shown). Interestingly, the S14Term mutant, which released WT levels of virus in transfected HeLa cells, completely failed to replicate (data not shown). The R16Term mutant, which also showed no impairment in virus production in HeLa cells, consistently replicated with a significant delay relative to the WT (Fig. 3B). When virus from the RT peak was repassaged, it replicated with WT kinetics (data not shown), suggesting the emergence of viral revertants.
To identify the location of genetic changes present in the putative revertant virus populations, we PCR amplified and sequenced viral DNA obtained from infected cultures near the peak of RT production. In every case, reversion events were evident; for the P-T/S-A-P motif mutants, the revertants contained changes back to the original or synonymous codon, so that the WT Gag amino acid was restored (Table 2). The P11Term revertants encoded a Pro at position 11. In one R16Term-infected culture (R16Term-1), a revertant emerged that contained a Trp codon at residue 16; in the other culture (R16Term-2), a 12-nt deletion appeared that removed the codon 16 stop, thereby reopening the reading frame (Table 2).
TABLE 2.
Sequence analysis of viral revertantsa
Role of p6 in virus replication in T-cell lines, PBMC, and MDM.
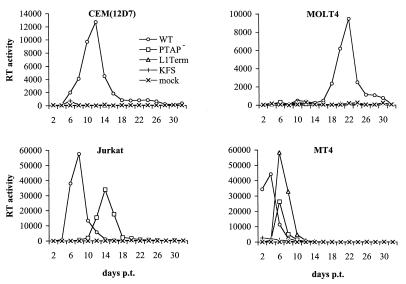
To extend the analysis of replication kinetics to other T-cell lines, we examined in parallel the effect of the L1Term and PTAP− mutations on virus replication in the CEM(12D-7), Jurkat, MOLT-4, and MT-4 T-cell lines. Cells were transfected with WT or mutant molecular clones, and RT activity was measured over time (Fig. 4). In addition to CEM(12D-7) cells, MOLT-4 cells were also restrictive for p6-mutant virus replication. In MT-4 cells, both p6 mutants replicated, although with a delay relative to the WT. Jurkat cells were partially permissive; the PTAP− clone replicated with a delay of 6 days relative to the WT, while L1Term failed to produce detectable RT activity. The inability of L1Term to replicate in Jurkat has been noted previously (19).
FIG. 4.
Replication kinetics of the L1Term and PTAP− mutants in T-cell lines. The indicated T-cell lines were transfected in parallel with WT or mutant molecular clones. Cells were split every 2 days, and RT activity was determined at each time point. Untransfected cells (mock) and cells transfected with the env-negative molecular clone pNL4-3KFS (KFS) served as negative controls. p.t., posttransfection.
When virus stocks were obtained from the delayed virus peak and repassaged in parallel with the WT, the mutants again replicated with similarly delayed kinetics (data not shown). This observation suggested that the delayed peaks observed in Fig. 4 were not due to the emergence of viral revertants or recombinants. This was confirmed by PCR amplifying and sequencing the viral DNA from the mutant-infected cultures; in each case, the original changes remained intact.
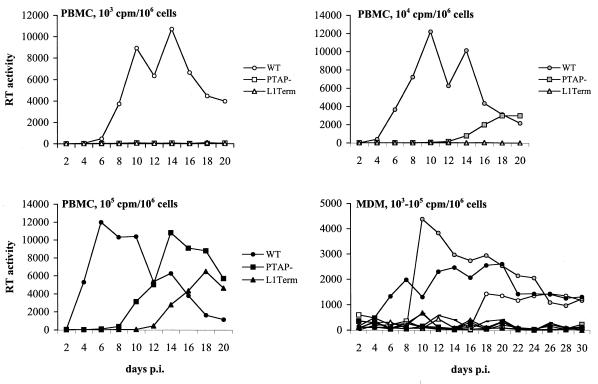
We also examined the phenotype of the L1Term and PTAP− mutations on virus replication in two primary cell types that serve as natural targets for HIV-1 infection, PBMC and MDM. Because primary cell types generally cannot be transfected with high efficiency and p6-mutant virus stocks cannot be generated readily by transfection because of their virus release defect, we developed the following approach to monitor p6-mutant replication kinetics. 293T cells were cotansfected with WT or p6-mutant molecular clones, a cytomegalovirus (CMV) promoter-driven Gag-Pol expression vector, and a plasmid expressing VSV-G (Materials and Methods). Virus stocks were harvested and used to infect PBMC and MDM. For the MDM infections, the WT and p6-mutant molecular clones were modified by the introduction of the M-tropic NL(AD8) env gene (9, 12).
Infections of PBMC and MDM were performed over a three-log range of virus input (Fig. 5). In PBMC, WT virus replication was observed at each input level; peak RT production was observed between day 6 (for the 105 RT cpm input) and day 14 (for the 103 input) postinfection (Fig. 5). Virus replication in PBMC cultures infected with the PTAP− p6 mutant peaked on day 14 (for the 105 RT cpm input) or day 20 (for the 104 input) postinfection. No virus replication was observed when PTAP− infections were performed with a 103 RT cpm input. Low but readily detectable virus replication was observed in PBMC cultures infected with L1Term at the highest input; however, no replication was observed for this mutant at the 104 and 103 inputs. While we cannot exclude the possibility that the virus replication observed in PBMC might have been the result of reversion or recombination, when this possibility was directly investigated in the T-cell line experiments, the original mutations were found to be intact (see above). In primary MDM, virus replication was observed in all cultures infected with WT virus (Fig. 5). In contrast, even at the highest inputs, the p6 mutations completely abolished virus replication.
FIG. 5.
Replication kinetics of the L1Term and PTAP− mutants in primary cell types. In each case, 106 cells (PBMC or MDM) were infected at the indicated RT input with virus stocks prepared by triple transfection of 293T cells with WT or mutant molecular clones, pHCMV-G, and pCMVNLGagPolRRE (Materials and Methods). pNL4-3 and pNL(AD8) were used as the parental molecular clones in PBMC and MDM infections, respectively. Supernatants from infected cells were collected every 2 days postinfection (p.i.) for measurement of RT activity. Virus inputs (in RT cpm) per 106 cells are indicated as follows: white symbols, 103; gray symbols, 104; and black symbols, 105. WT, PTAP−, and L1Term are indicated by circles, squares, and triangles, respectively.
The VSV-G pseudotyping system used here to analyze replication kinetics in primary cell types is very efficient. We observed that in CEM(12D-7) cells transfected with PTAP− or L1Term, no virus replication could be detected (Fig. 4); however, when this cell line was infected with a high input of VSV-G-pseudotyped virus stocks (>104 cpm/106 cells), virus replication was observed (albeit with delayed kinetics relative to the WT) (data not shown).
Biochemical analysis of role of p6 in virus production in diverse cell types.
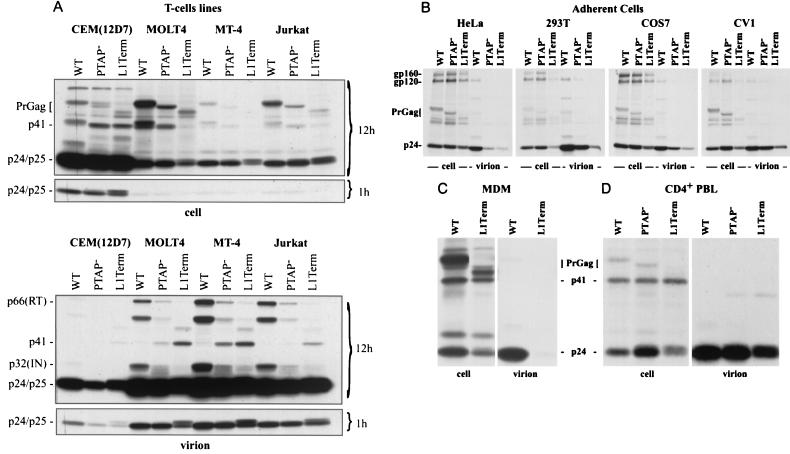
We have shown here (Fig. 2) and previously (24) that p6 is critical for efficient virus release in HeLa cells. We have also demonstrated that virus replication in most T-cell lines and in PBMC and MDM is profoundly impaired by mutations in p6 (Fig. 3 to 5). We next sought to determine what step in the virus replication cycle is disrupted by p6 mutations in T-cell lines and primary cell types. In particular, we wished to assess the role of p6 in virus production. To perform this analysis, we again used virus stocks obtained by triple transfection of 293T cells with WT and mutant pNL4-3 molecular clones, a Gag-Pol expression vector, and a VSV-G expression plasmid (Materials and Methods). T-cell lines were infected at a high input and metabolically labeled overnight with [35S]Met/Cys. Cell- and virion-associated proteins were immunoprecipitated with HIV-Ig.
Surprisingly, the L1Term and PTAP− mutations had no significant effect on virus particle production from MOLT-4, MT-4, and Jurkat cells, as determined by the ratios of cell-associated to virion-associated Gag (Fig. 6A). In CEM(12D-7), these p6 mutations caused a modest (approximately twofold, as determined by PhosphorImager analysis) reduction in the levels of virion-associated Gag proteins. Two other interesting observations could be made from this analysis (Fig. 6A): (i) the p6 mutations, in particular L1Term, caused defects in Gag processing, as indicated by the increased levels of the Gag processing intermediates p41 and p25 (35) in virus particles, and (ii) levels of the pol-encoded enzymes p66 (RT) and p32 (IN) were reduced in p6-mutant virions. Again, this phenotype was most dramatic for L1Term.
FIG. 6.
Immunoprecipitation of cell- and virion-associated proteins produced in diverse cell types. In each case, 106 cells were infected with 107 cpm of virus stocks prepared by triple transfection of 293T cells with WT or mutant pNL4-3 molecular clones, pHCMV-G, and pCMVNLGagPolRRE (Materials and Methods). At 24 h postinfection, cells were labeled overnight with [35S]Met-Cys; cell and virion lysates were immunoprecipitated with anti-HIV-Ig. (A) T-cell lines; (B) adherent cell lines; (C) MDM; (D) CD4+ PBLs. In panel A, 12-h and 1-h exposures are shown. The positions of the following viral proteins are indicated: the Env glycoproteins gp160 and gp120; the Gag precursor (PrGag); the Gag processing intermediates p41 and p25; the CA protein (p24); and (in virions) p32 (IN) and p66 (RT).
The results presented above indicate that while p6 plays a crucial role in virus release in HeLa cells, it is not required for efficient particle production from T-cell lines. To examine further the cell type-dependent nature of the p6 L domain function, we expanded our analysis of virus release to include several additional adherent cell types commonly used in virus assembly studies: COS-7, CV-1, and 293T. For these experiments, we again used VSV-G-pseudotyped virus stocks, prepared as described above. Since the possibility existed that the differences we observed between HeLa cells and T-cell lines were not due to differences in cell type but rather depended on the expression system used (i.e., transfection versus VSV-G pseudotype infection), we also retested HeLa cells in parallel with COS-7, 293T, and CV-1.
Consistent with the results obtained by transfection, the p6 mutants showed a severe defect (more than 10-fold reduction, as determined by PhosphorImager analysis) in virus production in HeLa cells (Fig. 6B). A reduced efficiency of virus production was also observed in 293T cells; this defect was also evident in transfection experiments (data not shown). PhosphorImager analysis of the gel shown in Fig. 6B indicated that in 293T cells the PTAP− and L1Term mutations reduced virus production by approximately two- and sixfold, respectively. Reduced virus release was also observed in COS-7 and CV-1 cells; we measured a three- to fourfold reduction in virus release efficiency in these cell lines.
Finally, it was of interest to determine the effect of p6 mutations on the efficiency of virus particle production in primary cell types. To this end, we infected primary MDM with VSV-G-pseudotyped virus stocks and analyzed virus release early postinfection. The results indicated that mutation of p6 markedly impaired (by approximately fivefold) virus particle release in primary MDM (Fig 6C). To examine the effect of p6 mutations on virus release in primary CD4+ lymphocytes, we purified CD4+ T cells from activated PBMC (see Materials and Methods) and infected them with VSV-G-pseudotyped virus stocks. As was observed in the majority of T-cell lines tested, the L1Term and PTAP− mutations had only a minor effect on virus particle production (Fig. 6D).
From the data presented above, it appears that, in general, adherent cell lines and cell types are more sensitive to virus release inhibition by p6 mutation than are nonadherent cells. To address the possibility that the differences we observed in the virus release phenotype were dependent on the state of adherence of each particular cell type, we tested the effect of p6 mutations on virus release in an adherent derivative of the CEM T-cell line (CEM-A) (57). As we observed in nonadherent T-cell lines, the L1Term and PTAP− mutations did not induce a virus release defect in CEM-A (data not shown). We also labeled HeLa cells in suspension and observed a potent virus release defect with L1Term and PTAP− (data not shown). Thus, the virus release phenotype of p6 mutants varied between cell types regardless of whether the cells were adherent or in suspension. Together, these results clearly demonstrate that the requirement for p6 in virus particle release is strongly cell type dependent.
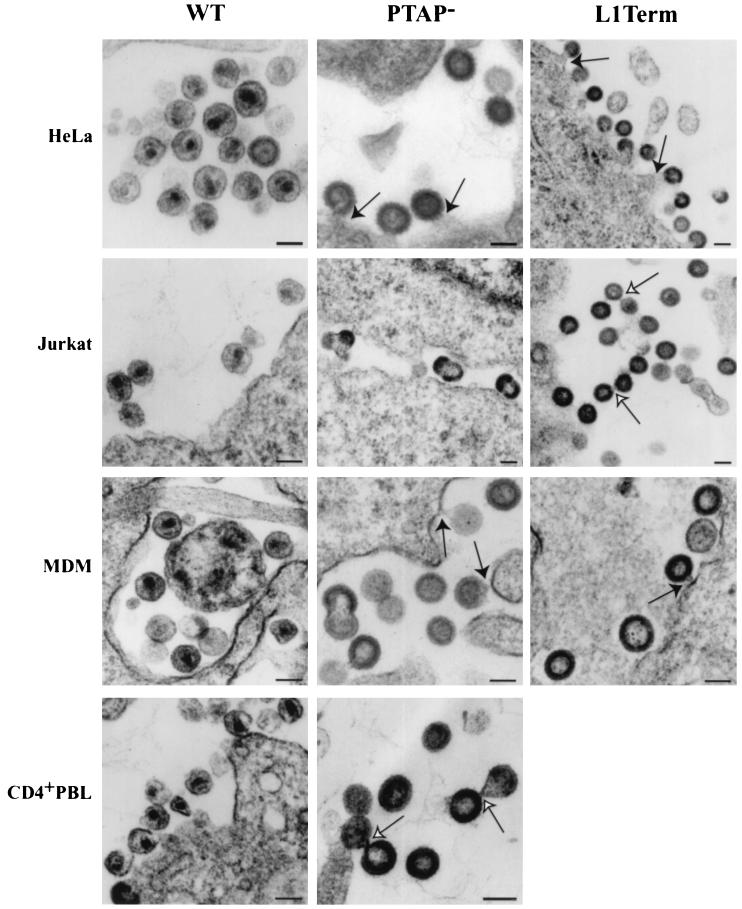
Examination of virus release in HeLa cells, T-cell lines, MDM, and primary CD4+ lymphocytes by EM.
The data presented in the previous section reveal some striking cell type-dependent effects of p6 mutation on virus particle production. Most notably, we were intrigued by the lack of a requirement for p6 in virion release from T-cell lines. However, both our biological and biochemical data in T-cell lines indicated a variety of p6-imposed defects. To address these issues in more detail, we examined cells expressing WT and p6-mutant molecular clones by EM. In agreement with our previous observations (24), p6-mutant virions in HeLa cells remained largely tethered to the plasma membrane, consistent with a defect late in the budding process (Fig. 7).
FIG. 7.
Visualization of virus assembly and budding in diverse cell types by EM. The WT and mutant molecular clones were introduced into Jurkat cells, MDM, and PBLs as indicated in Fig. 6; clones were introduced into HeLa cells by transfection. Solid arrows indicate particles tethered to the plasma membrane or (in MDM) intracellular membranes. Open arrows indicate particles tethered to each other, occasionally forming long chains (e.g., L1Term in Jurkat). Bar, 100 nm.
To examine virus assembly and release in T-cell lines and in primary lymphocytes, we infected Jurkat and purified primary CD4+ T cells with VSV-G pseudotypes under conditions similar to those used in the radioimmunoprecipitation assays presented above. In both T-cell lines and in the primary T cells, considerable amounts of released virion-associated material were observed (Fig. 7), in agreement with the radioimmunoprecipitation data. Remarkably, however, many of these virions were tethered to each other in long chains. Doublet particles were also seen frequently. The morphology of the released particles was largely immature.
Consistent with a previous report (40), WT HIV-1 assembly in primary MDM occurred predominantly within intracytoplasmic vesicles (Fig. 7). Interestingly, in p6-mutant-expressing MDM, we observed a marked increase in the number of particles that remained tethered to the luminal surface of these intracytoplasmic vesicles, consistent with the virus release defect observed in our immunoprecipitation analyses (Fig. 6C). These observations confirm the cell type-dependent nature of the p6-mutant phenotypes and reveal a novel p6-mutant defect in T-cell lines and PBLs.
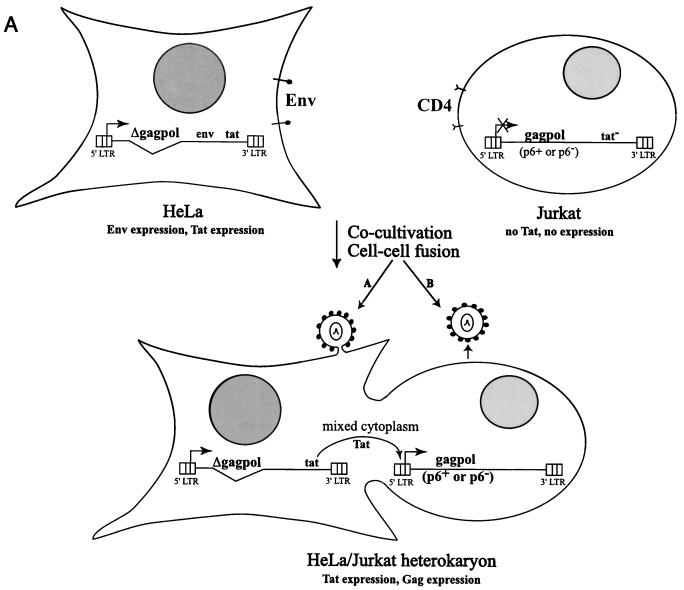
Requirement for p6 in virus release from transient HeLa/T-cell heterokaryons.
The data presented above indicate that p6 is required for budding from HeLa cells but that in T-cell lines virus release is not significantly impaired by p6 mutation. To explore further the basis for this cell type-specific difference, we analyzed the effects of p6 mutation on virus production in transient heterokaryons formed between HeLa and Jurkat cells. We developed a system in which Gag expression is dependent on cell-cell fusion (Materials and Methods) (Fig. 8A). HeLa cells were infected with virus stocks obtained by cotransfecting the NL4-3-derived gag/pol deletion construct pEvd1443 (7), a CMV-Gag-Pol expression vector, and pHCMV-G. Jurkat cells were infected with virus stocks obtained by cotransfecting a tat-negative molecular clone containing either a WT or p6-mutant gag gene, the CMV-Gag-Pol expression vector, the VSV-G expression vector, and pSVtat2.
FIG. 8.
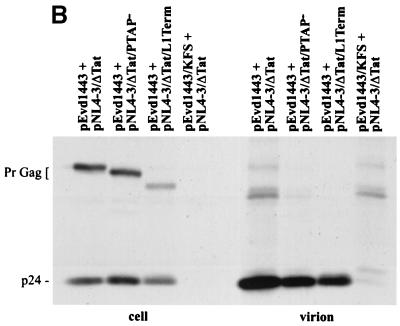
Immunoprecipitation of cell- and virion-associated proteins produced in transient HeLa/Jurkat heterokaryons. (A) Strategy for generating and expressing WT and p6-mutant Gag in heterokaryons. Details are provided in the text (Materials and Methods and Results). (B) Immunoprecipitation analysis. Cocultivated cells were metabolically labeled for 16 h with [35S]Met/Cys. Cell- and virion-associated proteins were immunoprecipitated with rabbit anti-p24. The positions of the Gag precursor (PrGag) and CA (p24) are shown.
Prior to cocultivation, the infected HeLa cells expressed high levels of Env glycoproteins, Rev, and Tat but no Gag; the Jurkat cells did not express HIV-1 proteins because they harbor tat-negative clones (data not shown). Upon cocultivation of the HeLa and Jurkat cells, cell-cell fusion was induced by Env-CD4 interactions, and Gag expression was transactivated by Tat present in the HeLa partner. As a negative control, an env-negative version of pEvd1443 was constructed by introducing the env gene from pNL4-3KFS into pEVdl443.
Cocultivated cells were metabolically labeled, and cell and virion proteins were immunoprecipitated with anti-p24 antiserum. In the absence of Env expression, Gag proteins were not synthesized (Fig. 7B, pEvd1443/KFS + pNL4-3Δtat lanes). When the HeLa partner expressed Env, cocultivation with Jurkat cells harboring the tat-negative molecular clone resulted in the expression of the appropriate (WT or p6-mutant) Gag proteins. In this context, the L1Term and PTAP− mutations reduced virus production by only 20 to 30% relative to the WT. Thus, in HeLa/T-cell line heterokaryons, the T-cell line phenotype is dominant, and p6 is not required for efficient virus release.
DISCUSSION
As a first step towards revealing the mechanism of p6 L domain-mediated virus release, we performed extensive site-directed mutagenesis of the region comprising and surrounding the P-T/S-A-P motif (Table 1, Fig. 1). The mutant molecular clones were used to transfect HeLa cells, in which we previously observed a marked virus release defect with p6 mutants (24). Our initial observation of a strong requirement for the Pro-rich domain was confirmed in this study. While the T8S mutant was released with WT efficiency, all other mutations within P-T/S-A-P markedly impaired virus release. Sequences flanking the Pro-rich domain were dispensable. Although we did not identify any particular sequence requirement for the residues flanking P-T/S-A-P, we found that to retain function, this motif must be followed by at least four amino acid residues.
The Pro-rich motif in the L domain of HIV-1 p6 matches the consensus sequence (Pro-X-X-Pro) found in proteins interacting with SH3 domains (51). Currently, these Pro-rich ligands are divided into two general classes, I and II, based on the position of a basic residue N- or C-terminal to the core Pro-X-X-Pro motif (34). As the P-T/S-A-P motif of p6 is preceded by an Arg residue (Arg4) (Fig. 1), one might assume that the Pro-rich L domain of p6 functions as a class I SH3-binding motif (consensus Arg-X-X-Pro-X-X-Pro [34]). However, we observed previously (24) and now (Fig. 2) that mutation of Arg4 does not affect virus release, suggesting that the L domain of p6 may represent a separate class of ligands for interaction with SH3 domains (5) or that this motif does not function as an SH3 binding domain.
Previously, we found that p6-mutant clones that failed to bud efficiently from HeLa cells were unable to replicate in the CEM(12D-7) T-cell line (24). We reasoned that L domain mutants that are replication defective might occasionally give rise to revertants, the characterization of which could provide interesting clues to p6 function. In this study, we isolated and characterized a series of p6 revertants. These revertants arose either by regenerating the original P-T/S-A-P sequence (with either the same or a synonymous codon) or by acquiring point mutations or in-frame deletions that removed stop codons (Table 2). The absence of second-site changes distal to the site of the original mutation, which we have observed frequently with mutations in MA and Env (10, 11, 36, 39), highlights the importance of the P-T/S-A-P motif in HIV-1 replication and is consistent with the hypothesis that the p6 L domain functions by interacting with a host factor (see below).
The cell type-specific differences in replication kinetics observed with the L1Term and PTAP− mutants (Fig. 4) prompted us to examine the efficiency of virus release from four different T-cell lines and from primary PBLs and MDM. Interestingly, we found that the L1Term and PTAP− mutations had no effect on virus release in three of four T-cell lines tested and reduced virus production by only twofold in the fourth T-cell line [CEM(12D-7)]. In primary lymphocytes, the L1Term and PTAP− mutations caused only a slight reduction in virus production. The lack of a strong virus release defect induced by these p6 mutations in T cells was confirmed by EM (Fig. 7).
In contrast to the phenotype observed in HeLa cells, in T cells the p6-mutant particles were released abundantly into the medium. The mutant particles displayed a predominantly immature morphology and frequently formed clusters of two or more particles that failed to detach from each other. Interestingly, a similar tethered phenotype has also been reported for MuLV L domain mutants (67). Thus, while p6 is not strictly required for virus release from T cells, it nevertheless promotes proper virion-virion detachment and core maturation in this context.
Although WT and mutant virus particles were released with similar efficiency in most T-cell lines and in primary lymphocytes, we observed differences in the amounts of both RT and IN in the mutant particles (Fig. 6A), a defect that was particularly striking with L1Term. A similar effect of p6 mutation on the levels of pol products in virions has been observed previously (6, 65, 66). It is not clear how p6 L domain mutations cause a reduction in pol-encoded enzymes in virions. This phenotype cannot be explained by a defect in the synthesis of Pr160GagPol, since the levels of this precursor protein are not diminished by the p6 mutations (Fig. 2). Also, in the absence of PR, levels of Pr160GagPol incorporated into virions are not affected by the PTAP− mutation (65). Although levels of released virions were not significantly reduced in T-cell lines in steady-state assays, it remains possible that the kinetics of virus production may be slightly delayed even in these cell lines. Such a delay in the kinetics of virus pinching off from the cell surface could result in PR-mediated cleavage of Pr160GagPol before the connection between the host cell cytoplasm and the virion has been broken. Cleavage of the Gag-Pol precursor while the particles are still tethered to the plasma membrane could result in the “leakage” of mature pol products back into the cytoplasm of the virus-producing cell. We should note in this context that the levels of RT and IN in virions were similarly decreased by p6 mutation in all four T-cell lines tested (Fig. 6A and longer exposures thereof). This phenomenon is therefore unlikely to explain the cell type-specific differences observed in virus replication in these cell lines (Fig. 4). Interestingly, longer exposures of the gels presented in Fig. 2 indicated that while release of virion-associated S14Term and R16Term Gag occurred with WT efficiency, the released virions contained reduced levels of both RT and IN (unpublished data). This reduction in virion-associated pol products might well contribute to the replication defect observed with S14Term and R16Term in CEM(12D-7) cells.
It has been suggested, based on the observation that p6- mutant virions appeared very large in rate zonal sucrose gradients, that p6 is a determinant of HIV-1 particle size (16, 17). In light of the EM observations presented here, it is possible that the behavior of p6-mutant particles in velocity gradients reflects their failure to detach from each other, since tethered particles would be indistinguishable from large particles in velocity sedimentation analyses.
HIV-1 p6 contains near its C terminus a Leu-rich motif implicated in the incorporation of the accessory protein Vpr into virus particles (30, 46). A variety of roles have been proposed for Vpr, including the stimulation of nuclear import in nondividing cells (e.g., MDM) (13). Since the L1Term mutant would be predicted to disrupt Vpr incorporation, this defect may contribute to the replication block observed with L1Term in MDM. However, the Vpr defect in MDM is relatively modest (a severalfold reduction in peak RT production [9]) and, in addition, the PTAP− mutant, which is also replication incompetent in MDM, would not be predicted to possess a Vpr incorporation defect. Thus, it appears likely that the inability of the p6 mutants to replicate in MDM is primarily caused by the p6 mutation itself.
We should note that a number of the p6 mutations introduced in this study also result in changes in the pol open reading frame in a region upstream of PR (referred to as p6*) (Table 1). Several observations suggest that these pol changes have little or no effect on the phenotype of the p6 mutants analyzed here. (i) Similar L domain defects were observed whether or not pol was altered. In fact, the PTAP− mutation is silent in pol. (ii) Several of the mutants that we analyzed previously (e.g., P7L, T8I, and P10L [24]) were reconstructed in this study to increase the number of nucleotides changed. The virus release phenotypes of the original mutants (which were silent in pol) and the reconstructed mutants (which contained pol changes) were essentially identical. (iii) In our previous study, the pol changes that resulted from mutation of p6 were introduced and analyzed in the absence of the gag changes. Invariably, the pol mutants replicated with WT kinetics (24). (iv) The revertants that arose during passage of several of the p6 mutants showed a restoration of the WT p6 coding region, whereas in the case of the T8I revertant, a pol change remained. (Table 2). Finally, (v) both the T8S mutant and the R16Term-2 revertant contained pol changes, yet replicated with WT kinetics. Together, these results highlight the importance of the gag mutations and suggest a minimal contribution of the pol changes to the phenotypes observed in this study.
To explore further the intriguing cell type-specific differences in the effect of p6 mutation on virus release, we constructed transient heterokaryons between HeLa cells (which displayed a strong requirement for p6 in virus release) and Jurkat cells (which released high levels of p6-mutant virions). The strategy used (Fig. 8A) ensured that Gag would be expressed only in cells that had undergone fusion between HeLa and Jurkat partners. Interestingly, the Jurkat phenotype was dominant in this context, as only a slight reduction in particle production was observed with PTAP− and L1Term (Fig. 8B). Although at this time we cannot formally exclude the possibility that the permissive phenotype is in some way a consequence of heterokaryon formation, these results suggest that T cells may harbor a factor that overcomes the requirement for p6 in virus release from the plasma membrane. Alternatively, the T-cell membrane itself may be more permissive for the extrusion of p6-mutant particles, and this property may dominate in the heterokaryons. In any case, further analysis of HeLa/Jurkat heterokaryons as well as analogous heterokaryons formed between different cellular partners will be useful in future studies of p6 function.
Although the physiological ligand for p6 has not been definitively identified, a variety of observations suggest that retroviral L domains function by interacting with the host ubiquitination machinery (reviewed in reference 60): (i) The L domain-containing proteins of MuLV (p12) and HIV (p6) are ubiquitinated (42), (ii) depletion of free ubiquitin in virus-producing cells with proteasome inhibitors impairs retrovirus budding (44, 50), (iii) the presence of L domains in HIV-1 minimal Gag constructs induces ubiquitination of the minimal Gag proteins (54), (iv) the ubiquitin ligase Nedd4 interacts with the RSV L domain (29), and (v) the cellular protein TSG101, which contains at its N terminus a ubiquitin-conjugating enzyme-like sequence, interacts with HIV-1 Gag in a p6-dependent fashion (59).
Interestingly, we have observed that overexpression of the ubiquitin-conjugating enzyme-like domain of TSG101 inhibits HIV-1 budding in a manner dependent on a functional p6 L domain (D. G. Demirov, A. Ono, and E. O. Freed, Cold Spring Harbor Retroviruses Conference, abstr. 7, 2001; D. G. Demirov, A. Ono, J. M. Orenstein, and E. O. Freed, submitted for publication). We should note that S14Term and R16Term, which lack the Lys residues that serve as targets for p6 ubiquitination, are released with WT efficiency in HeLa cells. This observation is consistent with the finding of Ott et al. (41) that p6 ubiquitination is not required for HIV-1 budding.
Although it is currently not clear how the interaction of retroviral L domains with proteins of the host ubiquitin pathway would promote virus budding, it is intriguing to speculate that by recruiting a ubiquitin-conjugating enzyme or ubiquitin ligase to the site of budding, the L domain could stimulate the ubiquitination and internalization of a host plasma membrane protein(s) (55), thereby modifying the protein composition at the site of budding. Future efforts will be aimed at further understanding the relationship between host proteins and retrovirus budding. The panel of mutants and the cell type-specific phenotypes described here will be useful is this regard.
Acknowledgments
We thank A. Ono and T. Murakami for helpful discussion and critical review of the manuscript. We acknowledge C. Dye and P. Sicurello for assistance with EM. We thank J. Silver, D. Rekosh, A. Ono, S. Venkatesan, K.-T. Jeang, R. Willey, and J. Burns for plasmids. The following reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program: HIV-1 Ig (from NABI), anti-p24 (from Chiron Corp.), and CEM-A (from M. Wainberg and J. McMahon).
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bork, P., and M. Sudol. 1994. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 19:531–533. [DOI] [PubMed] [Google Scholar]
- 4.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalgarno, D. C., M. C. Botfield, and R. J. Rickles. 1997. SH3 domains and drug design: ligands, structure, and biological function. Biopolymers 43:383–400. [DOI] [PubMed] [Google Scholar]
- 6.Dettenhofer, M., and X. F. Yu. 1999. Proline residues in human immunodeficiency virus type 1 p6gag exert a cell type-dependent effect on viral replication and virion incorporation of Pol proteins. J. Virol. 73:4696–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felser, J. M., T. Klimkait, and J. Silver. 1989. A syncytia assay for human immunodeficiency virus type I (HIV-I) envelope protein and its use in studying HIV-I mutations. Virology 170:566–570. [DOI] [PubMed] [Google Scholar]
- 8.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1–15. [DOI] [PubMed] [Google Scholar]
- 9.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O., and M. A. Martin. 1994. HIV-1 infection of non-dividing cells. Nature 369:107–108. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 2001. HIVs and their replication, p. 1971–2041. In D. M. Knipe and P. M. Howley (ed.). Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 14.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, L., L. J. Parent, B. Rovinski, S. X. Cao, and J. W. Wills. 1999. Identification of retroviral late domains as determinants of particle size. J. Virol. 73:2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnier, L., L. Ratner, B. Rovinski, S. X. Cao, and J. W. Wills. 1998. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J. Virol. 72:4667–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381:744–745. [DOI] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871–13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen Virol. 75:2985–2997. [DOI] [PubMed] [Google Scholar]
- 23.Hoshikawa, N., A. Kojima, A. Yasuda, E. Takayashiki, S. Masuko, J. Chiba, T. Sata, and T. Kurata. 1991. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J. Gen Virol. 72:2509–2517. [DOI] [PubMed] [Google Scholar]
- 24.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 69:6810–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818–98127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeang, K. T., P. R. Shank, A. B. Rabson, and A. Kumar. 1988. Synthesis of functional human immunodeficiency virus tat protein in baculovirus as determined by a cell-cell fusion assay. J. Virol. 62:3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079–3086. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. A. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo, E., F. Mamma no, E. A. Cohen, and H. G. Gottlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69:2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, L., Y. Li, S. Dales, and C. Y. Kang. 1994. Mapping of functional domains for HIV-2 gag assembly into virus-like particles. Virology. 205:496–502. [DOI] [PubMed] [Google Scholar]
- 34.Mayer, B. J., and M. J. Eck. 1995. SH3 domains: minding your p’s and q’s. Curr. Biol. 5:364–367. [DOI] [PubMed] [Google Scholar]
- 35.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative Gag precursors. J. Virol. 62:3993–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269:1872–1875. [DOI] [PubMed] [Google Scholar]
- 39.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orenstein, J. M., M. S. Meltzer, T. Phipps, and H. E. Gendelman. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 62:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111–121. [DOI] [PubMed] [Google Scholar]
- 42.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, 2nd, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12gag protein of Moloney murine leukemia virus. J. Virol. 72:2962–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawson, T., and G. D. Gish. 1992. SH2 and SH3 domains: from structure to function. Cell 71:359–362. [DOI] [PubMed] [Google Scholar]
- 46.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of Gag and mutational analysis. J Virol. 67:7229–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218–10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royer, M., M. Cerutti, B. Gay, S. S. Hong, G. Devauchelle, and P. Boulanger. 1991. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology 184:417–422. [DOI] [PubMed] [Google Scholar]
- 50.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sparks, A. B., J. E. Rider, N. G. Hoffman, D. M. Fowlkes, L. A. Quillam, and B. K. Kay. 1996. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCγ, Crk, and Grb2. Proc. Natl. Acad. Sci. USA 93:1540–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasakumar, N., N. Chazal, C. Helga-Maria, S. Prasad, M. L. Hammarskjold, and D. Rekosh. 1997. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 71:5841–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strous, G. J., and R. Govers. 1999. The ubiquitin-proteasome system and endocytosis. J. Cell Sci. 112:1417–1423. [DOI] [PubMed] [Google Scholar]
- 56.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263–334. In J. M. Coffin, S. H. Huges, and H. E. Varmus (ed.) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 57.Tremblay, M., A. K. Sullivan, R. Rooke, R. Geleziunas, C. Tsoukas, G. Shematek, N. Gilmore, and M. A. Wainberg. 1989. New CD4+ cell line susceptible to infection by HIV-1. J. Med. Virol. 28:243–249. [DOI] [PubMed] [Google Scholar]
- 58.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9:129–161. [DOI] [PubMed] [Google Scholar]
- 59.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55gag. Proc. Natl. Acad. Sci. USA 98:7724–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99–112. [DOI] [PubMed] [Google Scholar]
- 65.Yu, X. F., L. Dawson, C. J. Tian, C. Flexner, and M. Dettenhofer. 1998. Mutations of the human immunodeficiency virus type 1 p6gag domain result in reduced retention of Pol proteins during virus assembly. J. Virol. 72:3412–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, X. F., Z. Matsuda, Q. C. Yu, T. H. Lee, and M. Essex. 1995. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J. Gen. Virol. 76:3171–3179. [DOI] [PubMed] [Google Scholar]
- 67.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]