Abstract
The threonyl-tRNA synthetase gene (thrS) is a member of the T-box family of ∼250 genes, found essentially in Gram-positive bacteria, regulated by a tRNA-dependent antitermination mechanism in response to starvation for the cognate amino acid. While interaction between uncharged tRNA and the untranslated leader region of these genes has been firmly established by genetic means, attempts to show this interaction or to reconstitute the antitermination mechanism in vitro using purified tRNAs have so far failed. In addition, a number of conserved sequences have been identified in the T-box leaders, for which no function has yet been assigned. This suggests that factors other than the tRNA are important for this type of control. Here we demonstrate tRNA-mediated antitermination for the first time in vitro, using the regulatory tRNAThr isoacceptor isolated from Bacillus subtilis and a partially purified protein fraction. As predicted by the model, aminoacylation of tRNAThr(GGU) with threonine completely abolishes its ability to act as an effector. The role of the partially purified protein fraction can be functionally substituted by high concentrations of spermidine. However, this polyamine does not play a significant role in the induction of thrS expression in vivo, suggesting that it is specific protein co-factors that promote T-box gene regulation in conjunction with uncharged tRNA.
INTRODUCTION
A large number (∼250) of genes have been identified, primarily in Gram-positive bacteria, whose untranslated leader regions contain a conserved ∼14 nt sequence element known as the T-box, just upstream of a transcription terminator (1–4). Most of these genes encode aminoacyl-tRNA synthetases, amino acid biosynthetic enzymes or enzymes involved in amino acid transport, all of which are likely to be induced upon starvation for their cognate amino acid. The leader regions of these genes are generally ∼300 nt in length and are highly structured. A conserved secondary structure model of T-box leaders has been proposed (2,5) and has essentially been confirmed by experimental data (6). Uncharged cognate tRNA is thought to interact with the leader region in at least two places to stabilise an antiterminator structure at the expense of the terminator (Fig. 1). The specificity of the tRNA:leader interaction is achieved by binding of the anticodon of the tRNA to a ‘specifier codon’ bulged out of the first major RNA structure that forms in the leader, the specifier domain (2,7,8). The antiterminator structure is stabilised by Watson–Crick base pairing between the -NCCA 3′-end of the acceptor arm of all tRNAs and the -UGGN′- sequence in the central region of the T-box, located in a side-bulge of the antiterminator structure (7,9). Steric hindrance by the amino acid is thought to prevent charged tRNA from interacting with this sequence. Although an interaction between the full tRNA and the thrS leader was sought in structure probing experiments both in vivo and in vitro, it was never observed (6). Transfer RNATyr was recently shown, however, to be capable of interacting with a model tyrS antiterminator domain, whose stability was significantly increased by mutation (10).
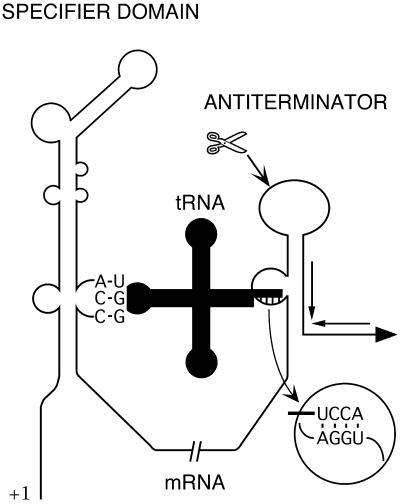
Figure 1.
Model of the interaction of uncharged tRNAThr with the B.subtilis thrS leader RNA. The schematic depicts the genetically identified points of interaction between the specifier codon and the tRNA anticodon and between the 3′-end of the acceptor arm of the tRNA and the side-bulge of the antiterminator structure. The scissors indicate a cleavage site upstream of the terminator believed to occur in many genes regulated by this mechanism. Inverted arrows represent the terminator structure.
In addition to the T-box, a number of other small conserved sequences have been noted in the leaders of genes of this family. These include the so-called GNUG-box and AG-box in the specifier domain and a sequence known as the F-box further downstream (11). Mutations in any of these sequence elements have a dramatic negative effect on antitermination in vivo (12; our unpublished results); however, their function is as yet unknown. Furthermore, only 4 of the 14 nt of the T-box have been assigned a convincing role. Although some of these nucleotides are involved in base pairing interactions with nucleotides which are also used by the terminator, there is no particular reason why other nucleotide pairings would not be tolerated. The existence of the small conserved sequence elements and the fact that we have been unable to reconstitute the tRNA-dependent antitermination mechanism in in vitro transcription assays with the tRNA alone convinced us that protein co-factors are involved in promoting the tRNA:leader interaction.
An initial search for additional factors that might be involved in tRNA-mediated antitermination led to the discovery of an endoribonucleolytic cleavage in the loop of the thrS antiterminator structure (13). Cleavage at this site is RNase E-dependent in Escherichia coli, suggesting Bacillus subtilis has an analogue of this enzyme (14). Bacillus subtilis cleavage is much more efficient under threonine starvation conditions in vivo and results in the production of a shorter transcript, bounded by stable RNA hairpins, that is five times more stable than the full-length mRNA (13). Thus, the effect of antitermination is amplified by cleavage and subsequent stabilisation of the coding portion of the thrS mRNA. The RNAs of several other genes of this family are cleaved in similar positions, suggesting that this phenomenon is conserved among T-box regulated genes.
In this study, we describe the reconstitution of the tRNA-mediated antitermination mechanism in in vitro transcription assays, using uncharged tRNAThr and a partially purified protein extract. As predicted by the model, aminoacylation of this tRNA abolishes its ability to promote antitermination. We show that while high concentrations of spermidine can substitute for the protein fraction in vitro, a B.subtilis strain unable to produce spermidine in vivo has unaltered thrS expression following threonine starvation. These results provide strong evidence that specific protein co-factors are required for tRNA-mediated antitermination.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The B.subtilis strains used in this study are the prototrophic strain 168 (BGSC 1A2) and derivatives thereof. Strain SSB275 (amyE::thrS′-lacZ) contains a thrS-lacZ transcriptional fusion (plasmid pHMS69) integrated at the amyE locus. Strain SSB315 (amyE::thrS′-lacZ, ΔspeA::spc) was constructed by transformation of strain SSB275 with chromosomal DNA isolated from strain BSIP7010 (15) and selection for spectinomycin resistance. The correct deletion of the speA gene was verified by Southern blot. Strain SSB1031 (amyE::thrS′-lacZ, pnpA::spc) was obtained by transform ing strain SSB275 with chromosomal DNA from strain BD2411 (16).
Strain HP24 (amyE::thrS′-lacZ, pHMS11) contains the thrS overproducing plasmid pHMS3 (1). For the purification of the ThrS protein this strain was grown in LB medium supplemented with 0.5% glucose. For starvation experiments, cells were grown in SMS minimal salt medium (17) with 0.5% glucose. Threonine starvation was achieved by the addition of 650 µg/ml dl-threonine hydroxamate at A600 = 0.4. Cells were harvested 2 h after the addition of threonine hydroxamate. Antibiotics for selection of chromosomal plasmid integrants were added at 4 µg/ml for chloramphenicol, 100 µg/ml for spectinomycin and for the selection of the replicative plasmid pHMS3 at 20 µg/ml for tetracycline.
Plasmid constructs
pHMS17. A 0.5 kb DraI–ClaI fragment of the thrS gene containing the promoter, the entire leader sequence and part of the thrS structural gene was inserted between the SmaI and ClaI sites of the plasmid Bluescript KS+.
pHMS69. The 0.44 kb DraI–EcoRV fragment of the thrS gene containing the promoter, the entire leader sequence and part of the thrS structural gene was isolated as an EcoRI–BamHI fragment and inserted between the EcoRI and BamHI sites of the lacZ fusion plasmid pHM2 (18).
pHMS71. Plasmid pHMS71 is derived from plasmid pHMS17. The 65 nt between the EcoRV and ClaI sites of the thrS structural gene were replaced with the third leader region transcription terminator of the thrZ gene (1). The terminator-containing fragment was constructed by hybridising two complementary oligonucleotides carrying an EcoRV and a ClaI site, respectively, at either end.
Preparation of tRNAs
A 110 g frozen cell pellet harvested from an exponentially growing culture of strain BGSC 1A2 was added to 500 ml of a solution preheated to 50°C containing 250 ml of buffer I (10 mM Na–acetate pH 5, 10 mM MgCl2, 0.5% SDS, 0.5% bentonite) and 250 ml phenol, pH 6. The contents were incubated at 50°C on a rotary shaker at 300 r.p.m. for 30 min. The phenol phase was separated by centrifugation for 15 min at 6000 g at 4°C. The aqueous phase was ethanol precipitated, dissolved in 30 ml water and phenolised once more (phenol pH 8). After ethanol precipitation 166 mg of RNA was obtained, consisting of >90% tRNA.
The whole RNA preparation was loaded on a MonoQ HR10/10 (Pharmacia) anion exchange column equilibrated with buffer A (20 mM K-PO4, pH 7). After an initial wash step with 350 mM NaCl the tRNAs were fractionated with a 350–750 mM NaCl gradient. Fractions containing threonine accepting tRNA species (identified by aminoacylation assays using purified threonyl-tRNA synthetase) were pooled and (NH4)2SO4 was added to a final concentration of 1.8 M. The tRNA preparation was then loaded on a Phenylsuperose HR10/10 (Pharmacia) column equilibrated with buffer B [1.8 M (NH4)2SO4 in buffer A]. The two threonine isoaccepting tRNA species were resolved by a 1.8–0.4 M (NH4)2SO4 gradient. The separation of the tRNAThr(GGU) and tRNAThr(UGU) isoacceptors was confirmed by dot-blot analysis (Fig. 2) using labelled oligonucleotides specific for tRNAThr(UGU) (5′-CCCCCAACCTACTGATTACAAGTCAGT-3′) and tRNAThr(GGU) (5′-CGCTGACCTCTTCCTTACCATGGA-3′). For each isoacceptor several fractions were pooled and the concentration of the specific tRNA was determined in aminoacylation assays with purified threonyl-tRNA synthetase. The tRNAThr(UGU) pool used here contained 1.6 pmol of the isoacceptor/µg tRNA; the concentration of tRNAThr(GGU) was 2.2 pmol/µg tRNA.
Figure 2.
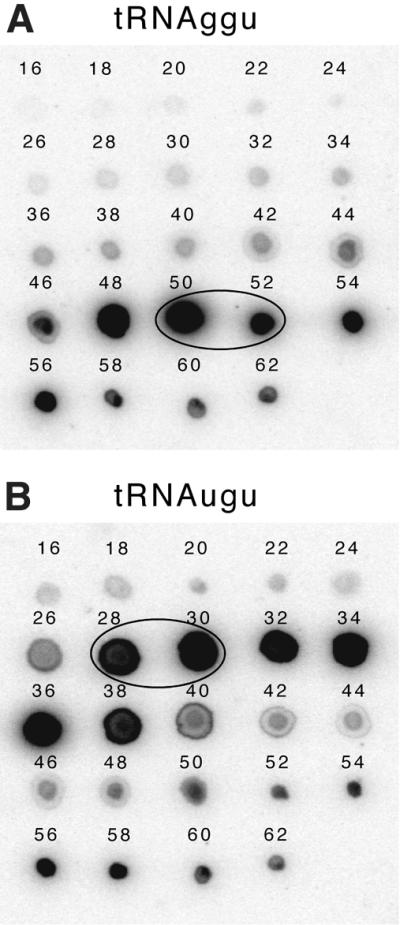
Dot-blot analysis of tRNAThr isoacceptors following fractionation on a phenylsuperose column. (A) Aliquots of tRNA fractions were spotted on a membrane and hybridised to an oligonucleotide probe specific for tRNAThr(GGU). (B) The same fractions as in (A) were hybridised to an oligonucleotide probe specific for tRNAThr(UGU). Numbers correspond to the fractions as they eluted from the column. Encircled spots indicate the fractions that were combined to create the pools of tRNAThr(GGU) and tRNAThr(UGU) used in the in vitro transcription assays.
Preparation of protein fractions
The cell pellet from a 100 ml culture of threonine-starved strain SSB1031 was sonicated in 2 ml buffer C (20 mM Tris–HCl pH 7.5, 4 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol) supplemented with 2 mM PMSF. An S30 supernatant containing 15 mg of total protein was loaded on a Mono Q HR5/5 (Pharmacia) anion exchange column equilibrated with buffer C. Proteins were fractionated by a 0–1 M KCl gradient. Samples of the fractions were combined in four separate pools and then tested in all possible combinations for their capacity to stimulate tRNA-dependent antitermination in vitro. The major activity was found in fractions eluting at 500 mM KCl. One of these fractions was used in the in vitro transcription assays described in the text.
Purification of the B.subtilis threonyl-tRNA synthetase ThrS
A 24 g cell pellet of strain HP24 overproducing the ThrS enzyme 10-fold was washed in 100 ml of buffer A (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 20% sucrose), centrifuged (10 000 g × 30 min) and resuspended in 50 ml buffer A. The suspension was heated to 37°C before addition of 6 mg of lysozyme and incubated for 20 min with gentle agitation. All subsequent steps were carried out at 4°C. The protoplasts were centrifuged (12 500 g × 30 min) and resuspended in 50 ml of buffer B (10 mM Tris–HCl pH 8.0, 10 mM MgCl2, 10% glycerol, 20 mM β-mercaptoethanol, 1 mM PMSF). After passage through a French Press (20 000 psi) the lysate was centrifuged (18 000 g × 45 min) and an S100 supernatant was obtained by ultracentrifugation (100 000 g × 4 h). Proteins were precipitated with 80% (NH4)2SO4, the pellet dissolved in a minimum of buffer C (10 mM Tris–HCl pH 7.8, 10 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, 0.1 mM PMSF) and dialysed against buffer C. Half of the preparation was loaded on a MonoS HR10/10 (Pharmacia) column and fractionated by a 0–200 mM KCl gradient in buffer C. ThrS eluted at 115 mM KCl. ThrS-containing fractions were directly loaded on a DEAE AP-1 8HR (Waters) column and fractionated by a 0–300 mM KCl gradient in buffer C. ThrS eluted at 200 mM KCl and was >95% pure. The pH of all buffers was adjusted at room temperature.
In vitro transcription assays
RNA polymerase was isolated from the B.subtilis wild-type strain BGSC 1A2 as described by Moran (19). The enzyme was stored at –20°C in storage buffer (10 mM Tris–HCl pH 8.0, 10 mM MgCl2, 0.1 mM EDTA, 100 mM KCl, 50% glycerol).
The template plasmids pHMS71 and pHMS17 were purified using Nucleobond AX columns (Macherey-Nagel), as described by the manufacturer. They contain the thrS promoter, leader and leader region terminator, and the beginning of the structural gene. Plasmid pHMS71 also contains the third terminator of the thrZ leader (1) cloned immediately upstream of the ClaI cleavage site used to linearise the template DNA.
Transcription reactions (50 µl) contained 20 mM HEPES–KOH pH 8.0, 4 mM MgCl2, 1 mM DTT, 40 mM KCl, 2 µg bovine serum albumin, 12 U RNasin (Promega), CTP, GTP and UTP at 10 µM, ATP at 400 µM, [α-32P]UTP (0.5 µCi/reaction), template DNA at 4 nM and RNA polymerase at 0.5 nM. Pools of tRNA containing either tRNAThr(UGU) or tRNAThr(GGU) were added to 4 µM; this corresponds to 160 nM of tRNAThr(UGU) and 220 nM of tRNAThr(GGU). Charged tRNA was prepared by incubating the uncharged tRNA for 5 min at 30°C in a 20 µl reaction containing 20 mM HEPES–KOH pH 8.0, 4 mM MgCl2, 1 mM DTT, 2 mM ATP, 1 mM threonine and 0.3 µg of purified B.subtilis threonyl-tRNA synthetase ThrS. Threonine was replaced by 1 mM valine as a negative control. The entire aminoacylation reaction was then added to the in vitro transcription reaction. Where indicated, 10 µg of a protein fraction from a total B.subtilis extract prepared as described above were added. Spermidine was added as indicated at various concentrations between 0.5 and 4 mM.
In vitro transcription reactions were incubated at 30°C for 20 min, and were then were stopped by addition of 5 µl of 3 M Na–acetate pH 5, phenolised and precipitated with 3 vol ethanol. The pellet was dissolved in 0.3× gel loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), heated to 95°C for 2 min and loaded on a 5% sequencing gel.
β-Galactosidase assay
The β-galactosidase activity of lacZ fusions was measured as described previously (1). Each experiment was performed three times.
RESULTS
The experimental system
In order to study tRNA-mediated transcription antitermination in vitro, we were careful to use a system which reproduced conditions in vivo as closely as possible, using native RNA polymerase and tRNAs purified from B.subtilis. As template, we used the leader region of the B.subtilis threonyl-tRNA synthetase gene (thrS), cloned on a plasmid and linearised at a ClaI site downstream of the leader transcription terminator. ThrS is a member of the T-box family of genes whose regulation has been extensively studied in vivo (1,7). The secondary structure of the entire thrS leader RNA has also been experimentally determined both in vitro and in vivo (6). Transcription of the thrS leader with B.subtilis RNA polymerase under classical in vitro transcription conditions, i.e. 100–200 µM NTPs, results in quite efficient read-through of the terminator (50% and more), in the absence of additional factors. This significantly reduces the sensitivity of the in vitro assay, as the maximum increase in terminator read-through that can be measured under these conditions is 2-fold. By lowering the NTP concentration in the assay to 10 µM NTP, only 5–10% of all transcripts extend beyond the terminator to produce a run-off (read-through) band at the site of template linearisation. The low NTP concentration is thought to reduce the elongation rate of RNA polymerase, thereby increasing the efficiency of the terminator (20). Under these conditions, 10–20-fold effects on antitermination can be measured.
The effector of the T-box regulatory system is the cognate uncharged tRNA. Bacillus subtilis has two tRNAThr isoacceptors, with UGU and GGU anticodons. We have shown previously that only tRNAThr(GGU) is able to interact productively with the thrS leader in vivo (7). To study the effect of tRNAThr on thrS transcription antitermination in vitro, while protecting against the possibility that modified bases were necessary for the tRNA:leader interactions, we decided to use tRNAs purified from standard B.subtilis cultures, rather than in vitro synthesised tRNAs or tRNAs overproduced in vivo. Total tRNA was fractionated using HPLC columns creating two tRNA pools containing either the tRNAThr(UGU) or the tRNAThr(GGU) isoacceptor. Each was present in very similar quantities as measured by hybridisation assays with probes specific for each tRNA and by aminoacylation assays using purified threonyl-tRNA synthetase. The complete separation of the two isoacceptors is illustrated by dot-blot analysis of phenylsuperose column fractions (Fig. 2). We studied the effect of addition of tRNAThr(GGU) on thrS transcription antitermination in vitro, using the tRNAThr(UGU)-containing fraction as a negative control.
tRNA-dependent antitermination in vitro
The addition of the tRNAThr(GGU) isoacceptor to our in vitro assay was not sufficient to promote antitermination (see below). Thus, either our in vitro reaction conditions were not optimal or other factors, present in vivo, are required for tRNA-dependent terminator read-through in vitro. We decided to search for proteins capable of promoting antitermination in concert with, and dependent on, uncharged tRNAThr(GGU). A pnp mutant strain was used for the preparation of B.subtilis cell extracts to help limit RNA degradation of the read-through transcript in particular during in vitro transcription. For these assays, we also used a template (pHMS71) where the 3′-end of the run-off transcript could fold back into a stable RNA hairpin in an attempt to further protect it from RNA degradation. A complete extract (S30) from a threonine-starved culture (thrS inducing conditions) was fractionated by anion exchange chromatography (see Materials and Methods) and fractions were tested for their ability to promote antitermination. Several fractions eluting at 500 mM salt clearly increased antitermination in the presence of uncharged tRNAThr(GGU) but not tRNAThr(UGU) isoacceptor. The effect of the addition of one of these fractions is shown in Figure 3. Addition of the tRNAThr(GGU) isoacceptor alone to the in vitro transcription assay does not increase antitermination compared with tRNAThr(UGU) (Fig. 3, lanes 1 and 2). In contrast, further addition of the partially purified protein fraction specifically stimulated read-through of the thrS terminator 6-fold in the presence of tRNAThr(GGU) relative to that seen with the tRNAThr(UGU) isoacceptor (Fig. 3, lanes 3 and 4). It is noteworthy that E.coli RNA polymerase can quite effectively replace the B.subtilis enzyme under these conditions (data not shown).
Figure 3.
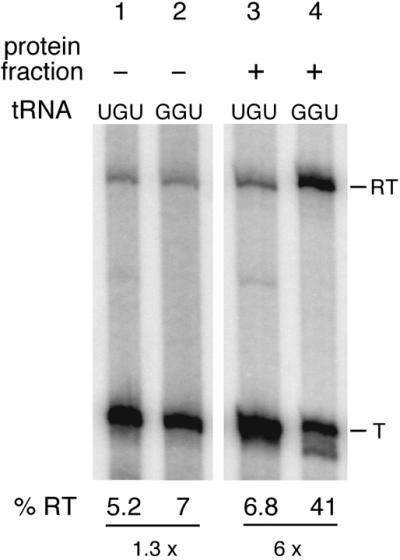
In vitro transcription of the thrS leader region. All reactions contained linearised pHMS71 plasmid DNA as template (4 nM) and B.subtilis RNA polymerase (0.5 nM). Pools of uncharged tRNA containing either tRNAThr(GGU) (220 nM) or tRNAThr(UGU) (160 nM) were added as indicated. Lanes 3 and 4, addition of 10 µg of a protein fraction from a total extract eluting at 500 mM KCl on a MonoQ (Pharmacia) column (see Materials and Methods). T and RT indicate products terminated at the thrS leader region terminator and read-through transcripts, respectively. % RT represents the percentage of read-through transcripts; the terminated and read-through transcripts taken together as 100%. The fold increase in antitermination is shown at the bottom of the figure.
The T-box regulation model predicts that only uncharged tRNA can interact productively with the leader mRNA to promote antitermination. It is assumed that base pairing between the side-bulge of the antiterminator (-UGGN′-) and the -NCCA 3′-end of the cognate tRNA, which stabilises the antiterminator, is sterically hindered if the tRNA is aminoacylated. A prediction of this hypothesis is that charging of the tRNA should abolish tRNAThr(GGU)-dependent antitermination in our in vitro transcription assay. This is exactly what we observe (Fig. 4). In the presence of the active protein fraction, stimulation of antitermination is tRNAThr(GGU)-dependent as above (Fig. 4, lanes 1 and 2). When the tRNAThr(GGU) was aminoacylated with its cognate amino acid (threonine) by purified threonyl-tRNA synthetase prior to its addition to the in vitro system it was completely unable to stimulate antitermination (Fig. 4, lane 4). In a key control experiment, where threonine was replaced by valine in the aminoacylation reaction, tRNAThr(GGU) was still able to promote antitermination, as it remains uncharged (Fig. 4, lane 3). Our data clearly indicate that B.subtilis contains proteins which are capable of promoting tRNA-mediated antitermination in vitro and presumably necessary for this type of control in vivo. The protein fraction used here still contains a relatively large number of proteins (data not shown) and attempts to identify the individual proteins involved in T-box gene regulation are underway.
Figure 4.
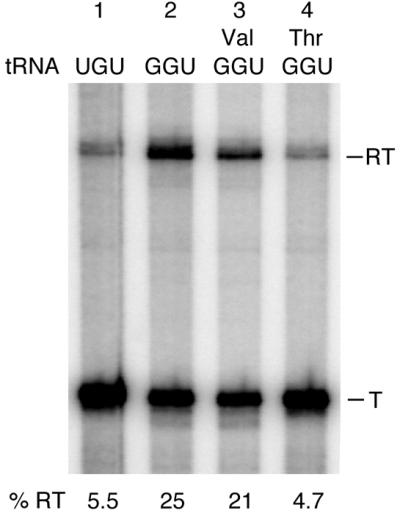
Effect of tRNA aminoacylation on protein-mediated thrS antitermination in vitro. Reaction conditions are as described in the legend to Figure 3. Val and Thr indicate that the tRNAThr(GGU) preparation was incubated in the presence of valine or threonine, respectively, and purified threonyl-tRNA synthetase, prior to addition to the in vitro transcription assay. T and RT indicate products terminated at the thrS leader region terminator and read-through transcripts, respectively. % RT represents the percentage of read-through transcripts; the terminated and read-through transcripts taken together as 100%.
tRNA-dependent antitermination in the absence of added proteins
Previous studies from our laboratory aimed at the determination of the structure of the B.subtilis thrS leader revealed that the specifier domain (Fig. 1) is significantly more stable in vivo than in vitro (6). The high degree of thermodynamic instability of this domain probably explains why the tRNA:leader interaction could not be demonstrated in vitro (6), as under these circumstances it is unlikely that the specifier codon is correctly bulged out of the secondary structure and available for interaction with the anticodon of the tRNA. We believe that one of the roles of the protein fraction we have identified above is to stabilise the specifier domain and promote the tRNA:leader interaction necessary for this type of control.
Nevertheless, we were curious to see whether it was possible to get the T-box system to function in vitro without proteins. In this respect, we turned our attention to polyamines, which, although dispensable under normal laboratory growth conditions, play an important role in the cell, in particular modulation of RNA structure and translational accuracy (21). Spermidine is the predominant polyamine (90–95%) in B.subtilis (22) and its capacity to mould RNA structure is becoming increasingly appreciated (21). To analyse a potential effect of spermidine on tRNA-dependent antitermination, we set up the same in vitro transcription assays used for testing protein fractions but where the proteins were replaced by varying concentrations of spermidine. In these assays, as no protection from RNA degradation is necessary, we used a template (pHMS17) where the 3′-end of the run-off transcript consists of native unstructured thrS sequence. This template results in two read-through bands, the reason for which is unclear. As shown in Figure 5, tRNAThr(GGU) was unable to stimulate terminator read-through compared with the tRNAThr(UGU) isoacceptor in in vitro transcription reactions performed in the absence of spermidine (Fig. 5, lanes 1 and 2). Addition of spermidine to concentrations of 0.5 mM or more specifically increased the relative abundance of read-through transcript in the presence of the tRNAThr(GGU) isoacceptor compared with that of tRNAThr(UGU). The stimulatory effect of spermidine was dose-dependent, ranging from 1.3-fold at 0.5 mM to almost 6-fold at 4 mM (Fig. 5, lanes 3–8 and Fig. 6, lanes 1 and 2). Moreover, just as in the case of the partially purified protein fraction, spermidine-stimulated antitermination was strictly dependent on the uncharged state of tRNAThr(GGU). Prior charging of the tRNAThr(GGU) isoacceptor with threonine completely abolished the spermidine effect, whereas the substitution of threonine with valine in the aminoacylation reaction left the tRNA competent for antitermination (Fig. 6, lanes 3 and 4).
Figure 5.
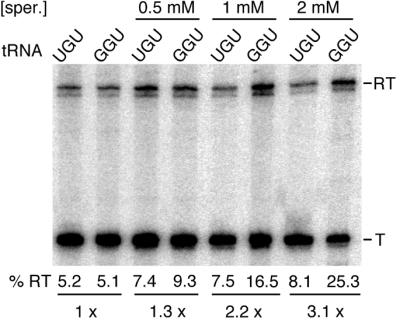
Effect of spermidine on thrS antitermination in vitro. Reaction conditions are as described in the legend to Figure 3, except that template pHMS17 was used. Spermidine [sper.] was added to the reactions at the indicated concentrations. T and RT indicate products terminated at the thrS leader region terminator and read-through (run-off) transcripts, respectively. % RT represents the percentage of read-through transcripts; the terminated and read-through transcripts taken together as 100%. The fold-increase in antitermination is shown at the bottom of the figure.
Figure 6.
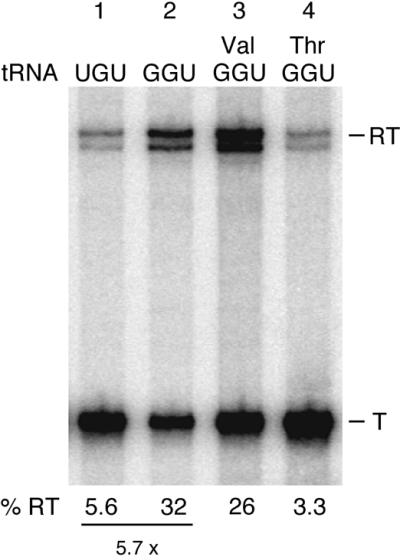
Effect of tRNA aminoacylation on spermidine-mediated thrS antitermination in vitro. Reaction conditions are as described in the legend to Figure 3, except that template pHMS17 was used. Val and Thr indicate that the tRNAThr(GGU) preparation was incubated in the presence of valine or threonine, respectively, and purified threonyl-tRNA synthetase, prior to addition to the in vitro transcription assay. T and RT indicate products terminated at the thrS leader region terminator and read-through (run-off) transcripts, respectively. % RT represents the percentage of read-through transcripts; the terminated and read-through transcripts taken together as 100%.
Effect of a speA mutation on tRNA-mediated antitermination in vivo
Bacillus subtilis cells contain millimolar quantities of spermidine, most of it complexed with RNA (23). The concentration of free spermidine is only ∼10 µM (24). The question thus arose as to what extent the rather high spermidine concentrations, required to observe tRNA-mediated antitermination in vitro, reflect the physiological context. Unlike in E.coli, spermidine is produced by a single pathway from arginine in B.subtilis, catalysed by the speA, B and E gene products (15). Danchin and co-workers (15) have thus shown that in a speA deletion strain no trace of spermidine or its precursor, putrescine, can be detected. To see whether spermidine plays a role in the regulation of thrS expression in vivo, we transferred this speA deletion construct (BSIP7010) into a strain containing a thrS leader-lacZ transcriptional fusion and compared β-galactosidase production from this strain to that of the wild-type. Under conditions of threonine starvation, both wild-type and speA strains showed very similar levels of thrS induction (Table 1). Thus, it appears that while spermidine may have an effect on tRNA-mediated antitermination in vitro, this role is played by proteins rather than spermidine in vivo.
Table 1. Effect of a speA deletion on the induction of thrS-lacZ expression.
| Strain | Genotype | Threonine hydroxamate | β-Galactosidase specific activity (U/mg) | Factor of induction |
|---|---|---|---|---|
| SSB275 | amyE::(thrS′-lacZ) | – | 276 | |
| 6.6× | ||||
| + | 1818 | |||
| SSB315 | amyE::(thrS′-lacZ) ΔspeA::spc | – | 208 | |
| 6.7× | ||||
| + | 1409 |
DISCUSSION
In recent years several important aspects of the tRNA-mediated antitermination mechanism have been revealed, mostly through in vivo experiments. Two points of interaction between the cognate uncharged tRNA and the leader mRNA have been clearly defined by mutational studies on several genes of the T-box family: the specifier codon:anticodon interaction, which confers the specificity of induction, and the base pairing between 3′-end of the uncharged tRNA and the central region of the T-box, believed to transiently stabilise the antiterminator (reviewed in 4,25,26). The experimentally determined structure of the thrS leader is in good agreement with this model (6), but no specific interaction between the leader transcript and the tRNA could be observed in these experiments. Nevertheless, on a smaller scale mini-helices corresponding to the acceptor arm of tRNATyr can interact with the bulge of a model antiterminator RNA in the absence of additional factors (10). For some time now, we and others (3) have attempted to reconstitute the T-box regulatory system in vitro by addition of uncharged cognate tRNA, but without success. This strongly suggested that additional factors present in vivo must be required for the tRNA:mRNA interaction to occur in a productive fashion. This is also supported by the fact that, besides the T-box, several conserved sequence elements have been identified within the leaders of genes of this family for which no function has yet been assigned.
At the outset of this study, we optimised our in vitro transcription assay to improve the chances of detecting antitermination activity in protein extracts. This was done by first lowering the NTP concentration in order to obtain termination levels >90%, a condition sine qua non if one wants to achieve the sensitivity required to detect small changes in termination efficiency. Secondly, we used native non-overproduced tRNAs rather than in vitro synthesised tRNA transcripts to circumvent the possibility that modified tRNA nucleotides play an important role in leader recognition. Thirdly, we took precautions against degradation of the read-through transcript in particular, upon addition of complex protein mixtures, by preparing our extracts from a pnp mutant strain and by cloning a stable hairpin at the 3′-end of the run-off transcript. Lastly, we used an all-inclusive S30 cell extract (rather than an S100 supernatant) from a threonine-starved culture as a starting point to look for potential antitermination factors. While we are not sure what contribution any of these individual precautions provided to the identification of a partially purified protein fraction capable of tRNA-dependent antitermination in vitro, undoubtedly their combination has led to success, where other attempts have failed.
The lack of thermodynamic stability of the specifier domain in vitro prompted us to examine the role of potential stabilisers of RNA structure in the tRNA-mediated antitermination mechanism. At high concentrations, the polyamine spermidine could essentially substitute for the partially purified protein fraction in promoting antitermination by uncharged tRNAThr(GGU). The effect of spermidine on the in vitro antitermination assay was intellectually unsatisfying from two points of view. First, the concentrations of spermidine required for the antitermination effect are much higher than its free concentration in vivo. Secondly, the conservation of small sequence elements in the T-box leaders could not be explained by a relatively non-specific co-effector such as spermidine. The fact that induction of a thrS-lacZ fusion following threonine starvation was not affected by deletion of the speA gene provided evidence that this intuition was correct. Spermidine is by far the dominant polyamine in B.subtilis but no traces of it nor of putrescine, the sole precursor of polyamine synthesis, are detected in a speA mutant (15). As spermidine is itself the precursor for another well known polyamine, spermine, it seems unlikely that other polyamines can replace spermidine function in vivo. Thus, while the effect of spermidine on tRNA-mediated antitermination in vitro may provide an interesting clue as to how the protein antitermination co-factors function, we do not believe spermidine is involved in the T-box control mechanism in vivo.
The threonine-starved S30 extract, still containing ribosomes, was fractionated on an anion exchange column, with the proteins eluting at 500 mM KCl containing the tRNA-dependent antitermination activity. However, the elution profile does not necessarily reflect the binding characteristics of the individual proteins to the column matrix. As ribosomes bind very strongly to this type of column, we cannot exclude the possibility that the antitermination activity was eluted from the ribosome rather than directly from the column matrix. Indeed, preliminary experiments suggest that an essential component of the antitermination activity is actually associated with the ribosome (data not shown). Further fractionation steps lead to complete loss of antitermination activity, suggesting that several proteins are involved. This would also explain the difficulty we have encountered in selecting mutants defective in tRNA-mediated control by genetic means. Although laborious, we believe the pooling of inactive fractions from subsequent chromatography steps, followed by the purification of the individual components one by one, provides the best chance of identifying the proteins involved in the control of T-box regulated genes.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to O. Pellegrini for the purification of RNA polymerase, to A. Sekowska and A. Danchin for the strain BSIP7010 and to D. Bechhofer for the chromosomal DNA of strain BD2411. We thank M. Springer and P. Stragier for helpful discussions. R.B. was supported by the ERASMUS program. This work was supported by funds from the CNRS (UPR 9073), MRE (Contract 92C0315), Université Paris VII (Contract DRED) and PRFMMIP from the Ministère de l’Education Nationale.
REFERENCES
- 1.Putzer H., Gendron,N. and Grunberg-Manago,M. (1992) Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J., 11, 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundy F.J., and Henkin,T.M. (1993) Transfer RNA as a positive regulator of transcription antitermination in B. subtilis. Cell, 74, 475–482. [DOI] [PubMed] [Google Scholar]
- 3.Grundy F.J., Moir,T.R., Haldeman,M.T. and Henkin,T.M. (2002) Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res., 30, 1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putzer H., and Laalami,S. (2002) Regulation of the expression of aminoacyl-tRNA synthetases and translational factors. In Lapointe,J. and Brakier-Gingras,L. (eds), Translational Mechanisms. Landes Bioscience, Georgetown, TX, http://www.eurekah.com/chapter.php?chapid=678&bookid=59&catid=54.
- 5.Grundy F.J., and Henkin,T.M. (1994) Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in Gram-positive bacteria. J. Mol. Biol., 235, 798–804. [DOI] [PubMed] [Google Scholar]
- 6.Luo D., Condon,C., Grunberg-Manago,M. and Putzer,H. (1998) In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis. Nucleic Acids Res., 26, 5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putzer H., Laalami,S., Brakhage,A.A., Condon,C. and Grunberg-Manago,M. (1995) Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: induction, repression and growth-rate regulation. Mol. Microbiol., 16, 709–718. [DOI] [PubMed] [Google Scholar]
- 8.Luo D., Leautey,J., Grunberg-Manago,M. and Putzer,H. (1997) Structure and regulation of expression of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol., 179, 2472–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundy F.J., Rollins,S.M. and Henkin,T.M. (1994) Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol., 176, 4518–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdeman M.S., Henkin,T.M. and Hines,J.V. (2002) In vitro structure–function studies of the Bacillus subtilis tyrS mRNA antiterminator: evidence for factor-independent tRNA acceptor stem binding specificity. Nucleic Acids Res., 30, 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henkin T.M., (1994) tRNA-directed transcription antitermination. Mol. Microbiol., 13, 381–387. [DOI] [PubMed] [Google Scholar]
- 12.Rollins S.M., Grundy,F.J. and Henkin,T.M. (1997) Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol., 25, 411–421. [DOI] [PubMed] [Google Scholar]
- 13.Condon C., Putzer,H. and Grunberg-Manago,M. (1996) Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 93, 6992–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condon C., Putzer,H., Luo,D. and Grunberg-Manago,M. (1997) Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol., 268, 235–242. [DOI] [PubMed] [Google Scholar]
- 15.Sekowska A., Bertin,P. and Danchin,A. (1998) Characterization of polyamine synthesis pathway in Bacillus subtilis 168. Mol. Microbiol., 29, 851–858. [DOI] [PubMed] [Google Scholar]
- 16.Luttinger A., Hahn,J. and Dubnau,D. (1996) Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol., 19, 343–356. [DOI] [PubMed] [Google Scholar]
- 17.Anagnostopoulos C., and Spizizen,J. (1961) Requirements for transformation in Bacillus subtilis. J. Bacteriol., 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendron N., Putzer,H. and Grunberg-Manago,M. (1994) Expression of both Bacillus subtilis threonyl-tRNA synthetase genes is autogenously regulated. J. Bacteriol., 176, 486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran C.P., (1990) Measuring gene expression in Bacillus. In Harwood,C.R. and Cutting,S.M. (eds), Molecular Biological Methods for Bacillus. John Wiley & Sons Ltd, Chichester, pp. 267–293.
- 20.McDowell J.C., Roberts,J.W., Jin,D.J. and Gross,C. (1994) Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science, 266, 822–825. [DOI] [PubMed] [Google Scholar]
- 21.Cohen S., (1998) A Guide to Polyamines. Oxford University Press, Oxford, UK.
- 22.Ishii I., Takada,H., Terao,K., Kakegawa,T., Igarashi,K. and Hirose,S. (1994) Decrease in spermidine content during logarithmic phase of cell growth delays spore formation of Bacillus subtilis. Cell. Mol. Biol., 40, 925–931. [PubMed] [Google Scholar]
- 23.Watanabe S., Kusama-Eguchi,K., Kobayashi,H. and Igarashi,K. (1991) Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem., 266, 20803–20809. [PubMed] [Google Scholar]
- 24.Woolridge D.P., Vazquez-Laslop,N., Markham,P.N., Chevalier,M.S., Gerner,E.W. and Neyfakh,A.A. (1997) Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J. Biol. Chem., 272, 8864–8866. [DOI] [PubMed] [Google Scholar]
- 25.Henkin T.M., (1996) Control of transcription termination in prokaryotes. Annu. Rev. Genet., 30, 35–57. [DOI] [PubMed] [Google Scholar]
- 26.Pelchat M., and Lapointe,J. (1999) Aminoacyl-tRNA synthetase genes of Bacillus subtilis: organization and regulation. Biochem. Cell Biol., 77, 343–347. [PubMed] [Google Scholar]