Abstract
Based on the analysis of structural features and conserved elements, 27 novel snoRNA genes have been identified from rice. All of them belong to the C/D box-containing snoRNA family except for one that belongs to the H/ACA box type. The newly found genes fall into six clusters that comprise at least three snoRNA genes, and in one case as many as nine genes. Interestingly, four of the six clusters are located within the largest intron of a protein coding gene. The majority of intronic snoRNA gene clusters are simply formed by multiple copies of the same species of snoRNA gene that possess the identical functional elements. This implies a possible mechanism of duplication for the origin of repeating snoRNA coding regions in one intron. However, a few intronic snoRNA gene clusters consisting of different snoRNAs species were also observed. Polycistronic precursors from two independently transcribed clusters were demonstrated by RT–PCR and individual snoRNAs processed from the polycistronic precursors were positively determined by reverse transcription assay. Analyses of the intergenic spacers in the clusters showed that, in addition to a very high AT content, the processing signals in rice snoRNA polycistronic transcripts might be different from those of yeast. Our results demonstrate that, in both plants and mammals, numerous snoRNAs can be produced simultaneously from an mRNA precursor of a host gene despite the different arrangements. The intronic snoRNA gene cluster is a novel gene organization, which is so far unique to plants. The conservation of intronic snoRNA gene clusters in plants was further demonstrated by the study of a similar snoRNA gene organization in the first intron of a Hsp70 gene from wild rice and Zizania caduciflora.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) play an important role in ribosome biogenesis (1). Since the 1990s, a myriad of snoRNA genes have been identified and characterized in a wide range of eukaryotes, from yeast to mammals (2), and recently snoRNA homologs have also been found in Archaea (3–5). Besides RNase MRP, the vast majority of snoRNAs fall into two families, which can be distinguished on the basis of common sequence motifs and structural features (6,7). Many snoRNAs share the conserved 5′-end C box (consensus sequence UGAUGA) and 3′-end D (CUGA) boxes (1,8). The rest possess a common hairpin–hinge–hairpin–tail secondary structure in addition to the H box (AnAnnA) in the hinge region and an ACA motif 3 nt from the 3′-end of the molecule (6,9). As to their diverse functions, several snoRNAs, such as U3, MRP, U14, U8, U22, snR10 and snR30, have been shown to play key roles in rRNA processing, leading to production of mature 18S, 5.8S and 28S rRNAs in both vertebrates and yeast (10–12). The majority of C/D box snoRNAs act as guides for site-specific 2′-O-ribose methylation (7,13,14), while the H/ACA box snoRNAs are responsible for pseudouridylation of rRNA (15,16). Moreover, recent findings have indicated that some snoRNAs target snRNAs or cellular RNAs instead of rRNAs (17–20). In addition to their diverse potential functions, studying snoRNAs is of special interest because of their diverse genomic organizations and the corresponding expression modes, which vary among different organisms. In vertebrates, most snoRNAs are intron-encoded by protein coding genes (1,21). Although a few intronic snoRNAs also exist in yeast, the majority of them are independently transcribed under their own promoters; clustered snoRNA genes driven by a single promoter were reported in a couple of cases (1,22,23). In both vertebrates and yeast, an intron of a host gene encodes, at most, a single copy of a snoRNA. Examples of such an organization, in particular, are vertebrate U22 host genes (UHG), which contain a different snoRNA gene (U22 and U25–U31) in each intron, but whose spliced mRNA lacks an open reading frame (24). The presence of only a single snoRNA coding region per intron is important for both vertebrate and yeast because processing of intronic snoRNAs in these organisms is largely splicing dependent (11,25) and involves exonucleolytic trimming of linearized snoRNA-containing intron lariats (25,26). In contrast to yeast and vertebrates, a distinct feature of snoRNA organization in plants is the prevalence of snoRNA gene clusters (27–29) that require endonucleolytic cleavage in a splicing-independent processing pathway (30). The early study of five clusters in maize (27,28) first revealed this gene organization and its expression mode, which is further demonstrated by recent research on Arabidopsis thaliana (29,31,32). Over 50% of the snoRNAs identified from A.thaliana are organized in clusters and transcribed independently from their own promoters. In contrast, as we have previously reported, a cluster containing six snoRNAs is nested in the first intron of the rice Hsp70 gene (33; L.-H.Qu, L.Zhong and H.Zhou, unpublished results). Clustered snoRNA genes in a single intron have not been reported from yeast and mammals to date. One can therefore ask whether this is a general phenomenon in plants or merely an anomaly encountered by chance. Here, we have taken advantage of our earlier observation that an intronic snoRNA gene cluster was found in rice. With the Rice Genome Project, a large number of rice DNA sequences have now been documented. The relatively large, complex genome of rice is expected to reveal more information than that of A.thaliana. In this study, we report six novel snoRNA gene clusters from rice. Notably, four of them were found located within introns of protein-coding genes. The structural features and conservation of this gene organization in plants was analyzed and is discussed here.
MATERIALS AND METHODS
Search of the database
The Oryza sativa DNA database in GenBank and EMBL was searched for potential C/D box snoRNAs on the basis of structural features and functional elements in addition to comparative analyses with the known snoRNAs in A.thaliana and Zea mays. The flanking sequences of the candidate genes were carefully examined for other possible snoRNA genes. All the newly found rice snoRNA gene candidates were further studied using the PC gene 6.0 package.
RNA and DNA preparation
RNA was isolated from rice germs, which were collected and homogenized on ice in the presence of NIB (20 mM Tris–HCl pH 7.4, 10 mM MgCl2, 40% glycerol, 20 mM β-mercaptoethanol, 0.5% Triton X-100, 0.1% bovine serum albumin, 5 vol per 1 g tissue weight). The homogenate was centrifuged at 8000 r.p.m. (Sovall, rotor SL-50T) for 10 min at 4°C. The cell debris was ground into a fine powder in liquid nitrogen. RNA was then extracted and purified as described (34). DNA was extracted from leaf tissue of Zizania caduciflora by the improved potassium acetate method (33). After treatment with RNase A, the DNA was further purified with glass milk or phenol/chloroform.
Detection of polycistronic transcript
Total cellular RNA was treated with DNase I before reverse transcription with primers. About 50 µg RNA in 100 µl of DNase I buffer was incubated (30 min, 37°C) with 10 U RQ1 RNase-free DNase I (Promega) and submitted to phenol/chloroform extraction. The RNA was used for reverse transcription with specific primers. PCR was then carried out with the reverse primer and the corresponding forward primer, using the following program: 30 cycles of denaturation (30 s, 94°C), annealing (30 s, 55°C) and extension (1 min, 72°C), followed by a final extension (10 min, 72°C).
Reverse transcription analyses and mapping of ribose methylation
Reverse transcription was carried out in a 20 µl reaction mixture containing 10 µg RNA, 10 ng 5′-labeled primer and 200 µM dNTPs. After denaturation of the resulting RNA product at 65°C for 5 min, the mixture was cooled to 42°C for 10 min. Then 200 U MMLV reverse transcriptase (Promega) were added and the mixture was further incubated at 42°C for 60 min. The reaction products were examined by electrophoresis on 10% acrylamide–8 M urea gels.
Ribose-methylated nucleotides of rice rRNA were determined by primer extension at low dNTP concentrations as described previously (29). In brief, two reactions of reverse transcription were performed in parallel using 5 µg total RNA and a dNTP concentration of either 4 or 500 µM. The rice 25S rDNA were amplified by PCR with the primer pair 25SF/25SR, then cloned into the SmaI site of plasmid pTZ18. An rDNA sequence ladder was prepared with the same primer used for rRNA methylation mapping and run in parallel with the reverse transcription reaction as a molecular weight marker.
PCR, cloning and sequencing
DNA extracted from Z.caduciflora was amplified with primers Hsp2 and Hsn, which are designed according to the sequence of the rice Hsp70 gene. Hsp2 is located in the boundary region covering 10 nt of exon 1 and 10 nt of intron 1 of the Hsp70 gene while Hsn is complementary to a 20 nt tract of exon 2 near the 3′-end of the intron. The 1972 bp PCR product was cloned into the SmaI site of the pTZ19 plasmid after electrophoretic purification and the recombinant plasmid was named pTZJB. Two fragments were obtained after treating pTZJB with BamHI. The large one, ∼3681 bp in length, is self-ligated and formed pTZJB1. The small fragment, ∼1167 bp in length, was subcloned into the BamHI site of pTZ19. The resulting positive clones identified by enzyme digestion were named pTZJB2. pTZJB1 was then sequenced using the universal sequencing primer and pTZJB2 with both universal sequencing primer and reverse sequencing primer in a 377 sequencer.
Oligonucleotides
Oligonucleotides were synthesized and purified by Sangon Co. (Shanghai, China). The following oligonucleotides were used in reverse transcription of RNA: Pz102, 5′-ATAGAGCTAATACAATTTGAGGCCA-3′; Pz104, 5′-CAAATGCCTCGATTGTCCCCATG-3′; Pz105, 5′-GCATTCAGA ATGAGTAGGAGGA-3′; Pz106, 5′-GAAGTATGAGTGCTTCATTGTAG-3′; Pz107, 5′-TCAGCGGAAAAATCGGCATACAA-3′; Pz109, 5′-GGATTCAGATGCAAAGA TGTGTA-3′. All the above oligonucleotides are complementary to the 3′-end of the corresponding snoRNA genes. The RT–PCR experiments were performed with the follow ing primers: C2101F, 5′-GCAGATGAGGAGGCACAAGATT-3′; C2102R, 5′-ATAGAGCTAATACAATTTGAGGCCA-3′; C2102F, 5′-ATTAGCTCTATCTGATCATCTTCC-3′; C2103R, 5′-CATCATGGCGGCCAATCAAACC-3′; C6107F, 5′-TGGTAATATTCAAGCTCAACAGAC-3′; C6110R, 5′-CAGAAAGAAAAGCCTTCTCATTC-3′; C6110F, 5′-AGGATGAAACCTTTTATAACAATCT-3′; C6113R, 5′-CATCAGGCCCAAACTATCACA-3′. The following oligonucleotides were used for amplification of rice 25S rDNA: 25SF, 5′-TATAGGGGCGAAAGACTAATCG-3′; 25SR, 5′-ATCTCAGTGGATCGTGGCAGCAA-3′. Rice rRNA ribose-methylated nucleotides were assayed by reverse transcription with the following primers: Ri1386R, 5′-GGCTCGCGCCCCGGGTTTTG-3′; Ri2438R, 5′-GGGCTCCCACTTATCCTACA-3′; Ri2948R, 5′-AACTAACCTGTCTCA CGACGGTC-3′. The following oligonucleotides were used for amplification of the first intron of the Hsp70 gene from Z.caduciflora: Hsp2, 5′-ACACCGTCTTCGGTAACTACT-3′; Hsn, 5′-AAGGGCCAGAGCTTAATGTC-3′. The primers used in reverse transcription, RT–PCR and rDNA sequencing were 5′-end labeled with [γ-32P]ATP (Yahui Co.) and submitted to purification according to standard laboratory protocols as described previously (23).
RESULTS
Identification of six novel snoRNA gene clusters, including four located in introns, from Oryza sativa
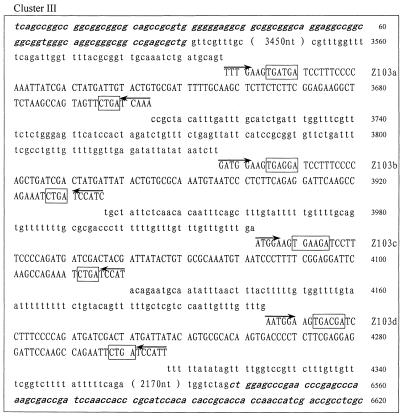
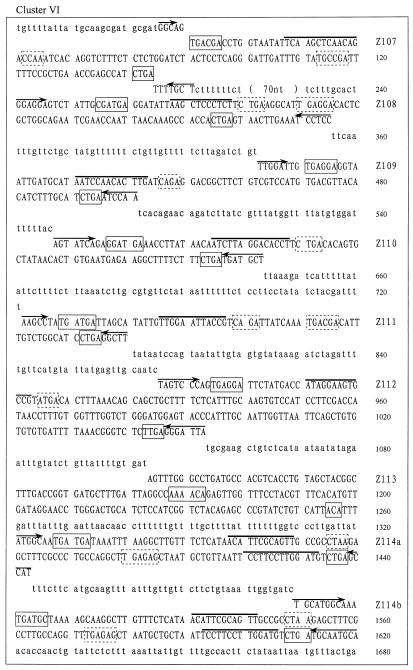
Careful examination of the GenBank and EMBL DNA database allowed us to identify 27 novel snoRNA genes from O.sativa. The snoRNA genes fall into six clusters and were termed rice snoRNA clusters I, II, III, IV, V and VI, respectively. The sequences of each cluster are shown in Figure 1. Interestingly, four of the six clusters, i.e. clusters I, III, IV and V, were found located within the introns of protein coding genes. These host genes encode RPS20, NADH dehydrogenase, RPL30 and an unknown protein, respectively. The host genes possess one to four introns but, in all cases, snoRNA gene clusters were found in only one intron, whose size is usually several-fold larger than the others. The RPL30 gene, for example, is the host gene of cluster IV, which contains four introns of 145, 1872, 369 and 90 bp. Evidently, the second intron was unusually large, and it turned out to contain snoRNA genes. All of the introns in the host genes possess the standard boundary signals, i.e. GU at the 5′-end and AG at the 3′-end of the intron, in spite of considerable variation in their sizes (Fig. 1).
Figure 1.
(Previous page and above) The sequences of the snoRNA gene clusters from rice. snoRNA genes are in capital letters; sequences of exons are in bold and italic; boxes C/D and C′/D′ are boxed with solid and dashed lines, respectively. A bar is drawn over sequences complementary to rRNA and arrows indicate nucleotides involved in the terminal stem.
It appears that most intronic snoRNA gene clusters are simply formed by duplication of the same species of snoRNA gene, which possess identical functional elements. Cluster I consists of four copies of snoRNA Z100. Likewise, clusters III and IV, though scattered over different chromosomes and nested in different protein coding host genes, are both made up of multiple copies of snoRNA Z103, four in cluster III and three in cluster IV. However, cluster V, which contains three different snoRNA species, is far more complex.
As for intronic snoRNAs in mammals, rice intronic clustered snoRNAs were frequently found in introns of genes involved in ribosome biogenesis, i.e. ribosomal proteins or nucleolar proteins. This organization is suggested to have evolved for coordinate expression of functionally related genes. Interestingly, snoRNA cluster V is encoded in an intron of a hypothetical protein-coding gene with a small ORF that is composed of only 25 amino acids. Whether this mRNA accumulates in the cell is not clear, although the evidence from cDNA sequences in the EST database (GenBank accession nos BI797167 and AU065122) supports transcription and splicing of the mRNA precursor from the host gene.
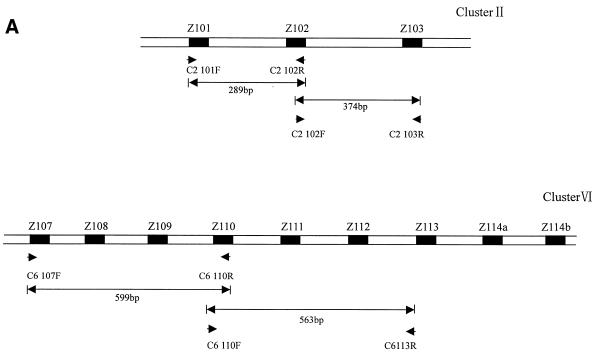
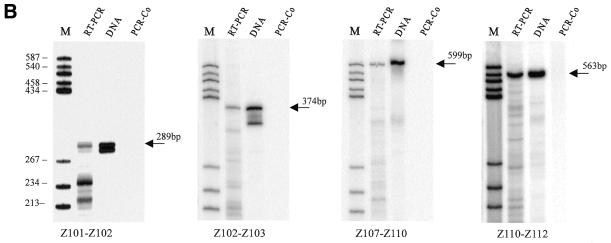
Clusters II and VI lie in a spacer between two protein coding genes and appear to be independently transcribed. Cluster II consists of genes encoding three different species of snoRNA, including Z103, which is also found in two other intronic clusters (clusters III and IV), as stated above. Cluster VI is particularly large and contains as many as nine genes corresponding to eight different snoRNA species. The clusters may be transcribed from an upstream promoter and multiple snoRNAs are released by processing from polycistronic precursors. Transcription of the clusters as polycistronic precursors was determined by RT–PCR with specific primer pairs, as shown in Figure 2.
Figure 2.
Detection of snoRNA polycistronic precursors by RT–PCR. (A) Schematic representations of clusters II and IV. The coding regions of snoRNAs are shown as black boxes. Arrowheads represent primer pairs used for the detection of the precursors by RT–PCR. The lengths of the expected products are given. (B) Detection of polycistronic precursors. Lane DNA, positive control PCR performed on rice total DNA; lane RT–PCR, PCR amplification after reverse transcription of rice total RNA; lane PCR-Co, control PCR performed on rice total RNA without reverse transcription; lane M, molecular weight marker (pBR322 digested with HaeIII and 5′-end labeled with [γ-32P]ATP). PCR amplification products exhibiting the expected size are indicated by arrows.
Predicted structure and function of the novel snoRNA genes in each cluster
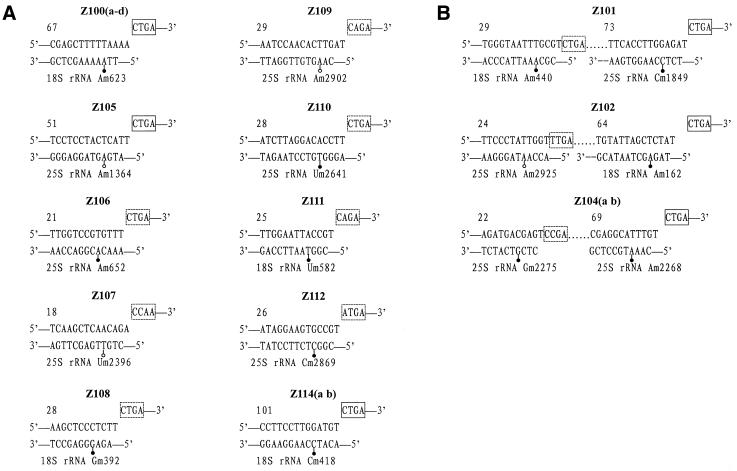
Among the 27 snoRNA genes in the clusters, 26 encode C/D box-containing snoRNAs. They clearly exhibit the hallmark structures of C/D box antisense snoRNAs, which include the presence of two conserved motifs, the 5′-end C box (5′-UGAUGA-3′) and the 3′-end D box (5′-CUGA-3′), immediately flanked by a 4–10 bp inverse repeat. These form the 5′–3′ terminal stem structure in which the C and D boxes are in close proximity, an important feature for the stability and accumulation of snoRNAs as well as for snoRNA binding to proteins (26,35,36). Two other conserved motifs, the C′ and D′ boxes, can be found in the central region of most snoRNA genes. In particular, 18 snoRNA genes have one or two regions of complementarity to rRNAs, 10–14 nt long, which have been shown to function in guiding 2′-O-ribose methylation of rRNA (37). However, the remaining eight snoRNA genes of Z103, which are distributed over different chromosomes in both intronic and non-intronic clusters, do not show any complementarity to rRNAs, implying that the function of Z103 may differ from those of other snoRNAs. For example, it may interact with RNAs other than rRNAs. Based on the known relationships between antisense snoRNA structure and function (7,13,14,38,39), proposed 2′-O-ribose methylation sites in rice rRNAs, complementary to the new guide RNAs, are shown in Figure 3.
Figure 3.
Predicted methylation guide duplexes between snoRNAs and rRNAs. 2′-O-ribose methylation sites homologous to those of yeast or vertebrates are shown by filled circles and novel methylation sites predicted from the present work are depicted by open circles. Boxes D and/or D′ are indicated. (A) snoRNAs with one complementarity to rRNAs; (B) snoRNAs with two complementarities to rRNAs.
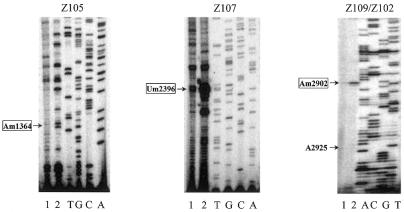
Based on the complementarity to rRNA and the corresponding target sites of methylation, a comparison with known snoRNA genes from various organisms revealed that 15 of the 26 C/D box snoRNA genes showed structural similarity to yeast or human snoRNAs (Table 1). This may imply that their functions have been conserved during the course of evolution. However, some distinct structural features of plant snoRNAs were also observed. For example, Z112 and its maize counterpart show high homology to each other, but both seem hardly related to their human counterpart other than the complementarity to rRNA. Rice Z112, with extra plant-specific regions, is more than twice the length of its vertebrate counterpart. The four novel snoRNAs, Z101, Z102, Z104a and Z104b, with dual antisense elements, display a mosaic structure as compared with their counterparts in yeast and mammals. The two methylation sites predicted by each of these snoRNAs are targeted by two different individual snoRNAs in yeast and mammals (Table 1). Based on complementarity to Z102, we predict a novel site of methylation in rice rRNAs, although this snoRNA shares another antisense element with human U44. The putative target sites of Z108 and Z110 are conserved between plants and vertebrates, but not yeast, and only the cognate of Z108 has been identified from mouse (GenBank accession no. AJ278763). Three snoRNA genes, Z105, Z107 and Z109, can form a 13–14 bp long duplex with rice rRNAs at sites that do not match any known ribose-methylated nucleotide of rRNAs in yeast and vertebrates. Thus, it is possible that there may be plant-specific methylation of rRNA. Recently, the counterparts of these three snoRNAs have also been reported in A.thaliana, but the methylation site predicted by the snoRNAs was not experimentally mapped (32). Using primer extension at low dNTP concentrations, we determined three new methylated nucleotides in rice 25S rRNA (Fig. 4), i.e. Am1364, Um2396 and Am2902, which were targeted by Z105, Z107 and Z109, respectively. However, A2925 in 25S rRNA, a methylation site predicted by Z102, was found to be unmethylated in our experiment (Fig. 4).
Table 1. Localization and constitution of the six snoRNA gene clusters.
| Cluster | Chromosome | snoRNA | Modification (homology) | Location | Acccesion no. |
|---|---|---|---|---|---|
| I | 6 | Z100a | SSU Am623 (yeast snR47) | 2nd intron of rpS20 gene | AJ320255 |
| Z100b | AJ320256 | ||||
| Z100c | AJ307912 | ||||
| Z100d | AJ307913 | ||||
| II | 1 | Z101 | SSU Am440 (human U16) | Spacer between two protein coding genes | AJ307914 |
| LSU Cm1849 (human U39) | |||||
| Z102 | LSU Am2925 (at snoR18) | ||||
| SSU Am162 (human U44) | AJ307915 | ||||
| Z103h | No complementarity (at snoR28) | AJ307916 | |||
| III | 3 | Z103a | 2nd intron of NADH dehydrogenase gene | AJ307917 | |
| Z103b | AJ307918 | ||||
| Z103c | AJ307919 | ||||
| Z103d | AJ307920 | ||||
| IV | 1 | Z103e | 2nd intron of rpL30 gene | AJ307921 | |
| Z103f | AJ307922 | ||||
| Z103g | AJ307923 | ||||
| V | 3 | Z104a | LSU Gm2275 (yeast snR75) | Intron of an unknown protein | AJ307924 |
| Z104b | LSU Am2268 (human U15) | AJ307925 | |||
| Z105 | LSU Am1364 (at snoR7) | AJ307926 | |||
| Z106 | LSU Am625 (human U18) | AJ307927 | |||
| VI | 2 | Z107 | LSU Um2396 (at R87) | Spacer between two protein coding genes | AJ307928 |
| Z108 | SSU Gm392 (mouse Z51) | AJ307929 | |||
| Z109 | LSU Am2902 (at snoR31) | AJ307930 | |||
| Z110 | LSU Um2641 (at Z27) | AJ307931 | |||
| Z111 | SSU Um582 (yeast snR77) | AJ307932 | |||
| Z112 | LSU Cm2869 (human U49) | AJ320263 | |||
| Z113 | (maize snoR2) | AJ320264 | |||
| Z114a | SSU Cm418 (human U14) | AJ315478 | |||
| Z114b | AJ315479 |
Figure 4.
Determination of rRNA methylation sites predicted by the novel snoRNAs. Lane 1, control reaction at 500 mM dNTP; lane 2, primer extension at 4 µM dNTP; lanes A, C, G and T, the rDNA sequence ladder. The sites of ribose methylation in rRNA were revealed by RT pauses at low dNTP concentrations. Arrows indicate potential methylation sites predicted by the novel snoRNAs.
Z113 is the only H/ACA box snoRNA found in the clusters. It appears homologous to maize snoR2 by comparative analysis. In maize, snoR2 is accompanied by two other C/D box snoRNAs, U49 and U14, and this clustering has been found several times (28). Intriguingly, rice Z113 is positioned in a similar genomic context, with Z112 (the cognate gene of U49) upstream and Z114 (the homolog of U14) downstream, but there are also other different genes within the cluster.
Positive identification of specific snoRNAs from the clusters
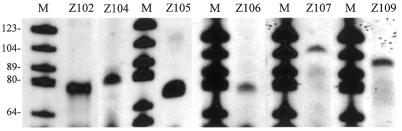
To confirm that the expected snoRNAs were transcribed from the gene clusters and processed from a polycistronic precursor, reverse transcription analyses were carried out with RNA isolated from partially purified nuclei from O.sativa cells. These experiments employed six oligonucleotide primers, designed and synthesized to pair with the coding regions of six novel snoRNA genes, Z102, Z104, Z105, Z106, Z107 and Z109, which were selected from clusters II, V and VI, respectively. As shown in Figure 5, a major cDNA product consistent with the expected size was observed in each case. The snoRNAs yielded signals of different intensities under the same experimental conditions, possibly reflecting different cellular abundances. These appear to correlate with gene number. For example, in cluster V, two nearly identical copies of Z104 are present, while only one copy of Z106 is present, and the band representing the cDNA product of Z104 is stronger than that representing Z106. Intriguingly, the signal due to Z105 does not accumulate at the same level as that due to Z106, and is even stronger than that due to Z104, suggesting that other copies of Z105 might exist in the rice genome. In fact, we have found another copy of the Z105 snoRNA gene on rice chromosome I by searching the growing rice DNA database (our unpublished result). Another possible explanation for the different signal levels of snoRNAs is differences among snoRNA gene promoters. However, at present little is known concerning plant snoRNA gene promoters and differential expression.
Figure 5.
Reverse transcription analyses of snoRNAs. The experiments were carried out with 10 µg rice RNA and 5′-end labeled primers specific to candidate snoRNAs, as described in Materials and Methods. Lane M, molecular weight markers (pBR322 digested with HaeIII and 5′-end labeled with [γ-32P]ATP).
Analyses of the intergenic sequences in the gene clusters
All of the snoRNA genes in the clusters are arranged in a head-to-tail fashion and are closely linked. The sizes of the intergenic regions range from 46 to 456 bp. Sequence analyses have shown that all of them are rich in uridine, with >40% and even as high as 61% in one case (the spacer between Z108 and Z109 in cluster VI). This phenomenon is similar to the one observation for the snoRNA gene clusters from A.thaliana (29). However, in addition to being U-rich, the sequences of the intergenic spacers of each cluster are very different from each other. Although hairpin-like structures can be deduced for the spacers with long sequences with the RNA folding program, some small spacers in the cluster (for example the spacer between Z109 and Z110) do not tend to form any stable secondary structure. Moreover, no conserved motif was found among the folded spacers. Thus, unlike the hairpin secondary structure with a tetranucleotide loop that is recognized by RNase III in yeast (23), the spacers in rice snoRNA gene clusters may adopt other structures that contribute to the signal for processing of the polycistronic transcripts. In addition, since the polycistronic transcript of plant snoRNAs can be efficiently processed from both intronic and non-intronic contexts, the intergenic regions of the clusters in both contexts may share a common processing mechanism that remains to be elucidated.
An intronic snoRNA gene cluster is conserved in Gramineae
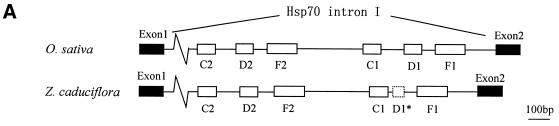
Five intronic snoRNA gene clusters have been identified from rice so far, together with our previous report of a snoRNA gene cluster in the first intron of the rice Hsp70 gene, which contains four C/D box and two H/ACA box snoRNAs (33; our unpublished results). To demonstrate the conservation of intronic snoRNA gene clusters in plants, we have systematically studied the first intron of the Hsp70 gene, a well conserved protein coding gene in plants, from two subspecies of rice, i.e. O.sativa ssp. indica and O.sativa ssp. japonica, and the wild rice (Oryza rufipogan). The results showed that both the sequences and the order of the six snoRNA genes in the cluster are highly conserved (data not shown). To further study the divergence and distribution of the cluster, the first intron of the Hsp70 gene from Z.caduciflora (Gramineae) was cloned and sequenced (Fig. 6). The sequence similarity of the first introns of the Hsp70 genes between O.sativa and Z.caduciflora is 66%. Five snoRNA genes can be easily spotted from the sequence of the first intron of the Z.caduciflora Hsp70 gene. All of them exhibit extensive homology to their counterparts in O.sativa and are organized in a similar order in spite of extensive divergence of the intergenic sequences. However, in the region where D1 was expected, only the 3′-half of the snoRNA gene is found, while the 5′-half, including one of the two key motifs, the C box, and the 5′ terminal repeat, is completely missing. Therefore, the partial sequence of D1 in Z.caduciflora may be a pseudogene. This result demonstrates that the intronic snoRNA gene cluster in the first intron of the Hsp70 gene is conserved between rice and Z.caduciflora and may extend to all plants of the Gramineae family.
Figure 6.
Sequence comparison of the first intron of the Hsp70 genes from O.sativa and Z.caduciflora. (A) Schematic diagrams of gene organization. The relative positions of the genes are drawn to scale with the exception of the region between exon 1 and C2, which is shown by a wavy line. Exons are shown as black boxes and snoRNA genes as open boxes with solid lines. The open box with dashed lines indicates the D1 pseudogene. (B) Alignment of the two sequences. snoRNA genes are in capital letters and boxed and the name of each gene is shown. Sequences of exons are in bold capital letters. Nucleotide identities are indicated by hyphens and those absent in either sequence by asterisks. The size of each intron is given.
DISCUSSION
Intronic snoRNA gene clusters most likely predominate in the genome of rice
Rice, after A.thaliana, is the second plant whose genome has been chosen to be fully sequenced. The rapid progress of the rice genome project will certainly provide a solid base for further studies of rice and other crop grasses. In addition to the sequence itself, rice has already become a good source for the identification of new genes. In this study, we took advantage of the growing rice DNA database in an effort to shed new light on snoRNA gene organization. So far, seven clusters including 33 snoRNA genes have been identified, and a novel gene organization, i.e. intronic snoRNA gene clusters, was first revealed in rice. Indeed, among the seven clusters, five are located within introns of protein-coding genes. This relatively high proportion of intronic snoRNA gene clusters suggests that this novel gene organization may be prevalent in the rice genome. Our results also reveal that the intronic snoRNA gene cluster in the Hsp70 gene is conserved among rice, wild rice and Z.caduciflora, and probably in many other plants of the Gramineae family. On the other hand, we have identified 35 snoRNA gene clusters from the complete genome of A.thaliana, among which three are intronic snoRNA gene clusters (40; our unpublished results). To our knowledge, no intronic snoRNA gene cluster has been found in yeast or animals so far. Therefore, this gene organization may be unique to plants. The relative scarcity of intronic snoRNA gene clusters in the dicotyledon A.thaliana may reflect a genome that is particularly small and compact. Most introns in A.thaliana are of small size (∼170 bp on average) (41), which is merely long enough to accommodate a single snoRNA gene at most. Conversely, the rice genome, estimated to be 430 Mb, is more than three times larger than that of A.thaliana (∼120 Mb). Thus, it is not surprising that a large number of intronic snoRNAs in clusters are found in rice and other crops with large genome sizes.
Some features of the intronic snoRNA gene clusters
There are usually multiple copies of each snoRNA gene in plants (29,31,32). Although snoRNA genes can be found both in independently transcribed and intronic clusters, the sequences from different copies of a snoRNA gene are very conserved in spite of their different locations. However, compared with the richness of components in non-intronic clusters, the constitution of most rice intronic snoRNA clusters is relatively monotonous. All three intronic snoRNA gene clusters in A.thaliana are also made up of multiple copies of a single species of snoRNA gene (our unpublished results). This implies that frequent duplication has occurred within introns during the evolution of the plant genome. However, rice snoRNA cluster V contains two additional species of snoRNA genes besides isoforms of one species, and the snoRNA cluster located in the first intron of the Hsp70 gene consists of six snoRNAs, which belong not only to different species but also to different types, C/D box and H/ACA box. This shows that the structure of intronic snoRNA gene clusters can be more complex than initially thought. Leader et al. (28) have successfully expressed an independently transcribed cluster of maize from both non-intronic and intronic contexts in tobacco protoplasts. This result demonstrates that the processing of snoRNA polycistronic transcripts is independent of splicing (28,30). However, the processing signals in polycistronic transcripts that are recognized by endonucleases and exonucleases in plants have not yet been elucidated. The discovery of different kinds of intronic snoRNA gene clusters provides impetus for further studies of the expression and processing of intronic snoRNA in plant.
The introns that contain a snoRNA gene cluster are exceptionally large as compared with other introns in the same host gene, even if the sequence of the snoRNA gene is not considered. It therefore seems that it is not simply the internal snoRNA cluster that makes an intron large. It remains to be elucidated whether there is a length threshold for an intron to contain a snoRNA gene cluster or whether introns without snoRNA gene clusters have become smaller during evolution.
Diversity of snoRNA gene organization
One main goal of studying snoRNAs is to elucidate their diverse modes of gene organization in various organisms. Earlier studies focused mainly on yeast and vertebrates and revealed two distinct genomic organizations (1). In yeast, a majority of snoRNA genes are dispersed as independently transcribed singlets, while in vertebrates, most snoRNA genes are located within introns. The latter organization was later also found in yeast, but in only a few cases (1,22). The discovery of clustered snoRNAs, which are transcribed under an upstream promoter, in yeast and plants adds a third mode, i.e. snoRNA gene clusters, to the diversity of snoRNA gene organization. This gene organization implies the transcription of snoRNAs as polycistronic precursors and a splicing-independent processing pathway involving endonucleolytic cleavage as well as exonucleolytic trimming to release individual snoRNAs (23,30,42). In this study we report intronic snoRNA gene clusters as a novel polycistronic organization. In contrast to independently transcribed clusters, the biosynthesis of multiple snoRNAs from an intronic polycistron depends absolutely on transcription from the promoter of a protein-coding gene and processing of the mRNA precursor.
snoRNA gene clusters, either non-intronic or intronic, permit the coordinated expression of multiple or different snoRNAs at high efficiency. The latter further permits coordinated regulation of the expression of snoRNAs and the host gene. Interestingly, in both plants and mammals, numerous snoRNAs can be produced simultaneously from a mRNA precursor of a host gene despite the different arrangements. We notice that in plants, usually only one intron of a host gene encodes multiple snoRNAs, whereas in mammals a host gene may contain numerous snoRNA sequences, but never more than one snoRNA per intron (1,42). The adoption of two different modes of snoRNA gene organization may reflect intrinsic differences in gene expression and evolution between plants and vertebrates.
The finding of snoRNA gene clusters in various organisms suggests an ancient origin of this gene organization (29). It is not yet clear whether this is also true of intronic snoRNA gene clusters, which are so far unique to plants. More detailed analyses of the rice genome and comparative studies using data from other organisms may provide answers to this important question.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the technical assistance of Xiao-Hong Chen in rice culture. We also thank Dr Henri Grosjean for revising the text of the manuscript. This research was supported by the National Natural Science Foundation of China (key projects 39730300, 39525007 and 39970171), by the Fund for Distinguished Young Scholars from the Ministry of Education of China and the Fund for Rice Resource and Gene Engineering (J00-A-009) from the Ministry of Sciences and Technology of China.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +86 20 84112399; Fax: +86 20 84036551; Email: AJ320255, AJ320256, AJ307912–AJ307932, AJ320263, AJ320264, AJ315478, AJ315479
REFERENCES
- 1.Maxwell E.S., and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 35, 897–934. [DOI] [PubMed] [Google Scholar]
- 2.Smith C.M., and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 3.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 4.Gaspin C., Cavaille,G., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 5.Dennis P.P., Omer,A. and Lowe,T. (2001) A guided tour: small RNA function in Archaea. Mol. Microbiol., 40, 509–519. [DOI] [PubMed] [Google Scholar]
- 6.Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small nucleolar RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 7.Tollervey D., and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 3, 337–342. [DOI] [PubMed] [Google Scholar]
- 8.Tyc K., and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily defined secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- 10.Eichler D.C., and Craig,N. (1995) Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 49, 197–239. [DOI] [PubMed] [Google Scholar]
- 11.Lafontaine D., and Tollervey,D. (1995) Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem. Cell Biol., 73, 803–812. [DOI] [PubMed] [Google Scholar]
- 12.Venema J., and Tollervey,D. (1995) Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast, 11, 1629–1650. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-Laszlo A., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 14.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation rRNA. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 15.Ni J., Tie,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 16.Ganot P.H., Bortolin,M.L. and Kiss,T. (1997) Site-specific pseudouridine formation in eukaryotic pre-rRNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 17.Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 splicesomal RNA is guided by another small RNA. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 18.Ganot P.H., Jady,B.E., Bortolin,M., Darzcq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 splicesomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hüttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.-P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jady B., and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstein L.B., and Steitz,J.A. (1999) Guided tours: from precursors snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 22.Lowe T.M., and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 23.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-Ferrer,M. and Bachellerie,J.P. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tycowski K.T., Shu,M.-D. and Steitz,J.A. (1996) A mammalian gene with introns instead of exons generating stable RNA products. Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- 25.Kiss T., and Filipowicz,W. (1995) Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev., 9, 1411–1424. [DOI] [PubMed] [Google Scholar]
- 26.Cavaillé J., and Bachellerie,J.-P. (1996) Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie, 78, 443–456. [DOI] [PubMed] [Google Scholar]
- 27.Leader D.J., Sanders,J.F., Waugh,R., Shaw,P.J. and Brown,J.W.S. (1994) Molecular characterization of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as a polycistronic U14 transcript. Nucleic Acids Res., 22, 5196–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W. (1997) Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNA. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu L.H., Meng,Q., Zhou,H. and Chen,Y.Q. (2001) Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res., 29, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1999) Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol. Biol., 39, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 31.Barneche F., Gaspin,C., Guyot,R. and Echeverría,M. (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- 32.Brown J.W., Clark G.P., Leader D.J.,.Simpson C.G. and Lowe,T. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 33.Qu L.H., Zhong,L., Shi,S.H., Lu,Y.J., Fang,R. and Wang,Q. (1997) Two snoRNAs are encoded in the first intron of the rice hsp70 gene. Prog. Nat. Sci., 7, 371–377. [Google Scholar]
- 34.Chomczynski P., and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 732–735. [DOI] [PubMed] [Google Scholar]
- 35.Caffarelli E., Fatica,A., Prisley,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 36.Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3743–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bachellerie J.P., and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- 38.Tycowski K.T., Smith,C.M., Shu,M.D. and Steitz,J.A. (1996) A small nucleolar RNA required for site-specific ribose methylation of rRNA in Xenopus.Proc. Natl Acad. Sci. USA, 93, 14480–14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollervey D., (1996) Small nucleolar RNAs guide ribosomal RNA methylation. Science, 273, 1056–1057. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H., Meng,Q. and Qu,L.H. (2000) Identification of Z2 snoRNA gene cluster from Arabidopsis thaliana genome. Sci. China Ser. C, 43, 449–453. [Google Scholar]
- 41.The, Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 42.Bachellerie J.-P., Cavaille,J. and Qu,L.-H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNA guides revisited. In Carrett,R.A. (ed.), The Ribosome: Structure, Function, Antibiotics and Cellular Interaction. ASM Press, Washington, DC, pp. 191–203.