Abstract
The structural requirements for DNA/RNA hybrids to be suitable substrates for RNase H1 are well described; however the tolerance level of this enzyme towards modifications that do not alter the duplex conformation is not clearly understood, especially with respect to the sense RNA strand. In order to investigate the molecular requirements of Escherichia coli RNase H1 (termed RNase H1 here) with respect to the sense RNA strand, we synthesized a series of oligonucleotides containing 2′-deoxy-2′-fluoro-β-d-ribose (2′F-RNA) as a substitute for the natural β-d-ribose sugars found in RNA. Our results from a series of RNase H1 binding and cleavage studies indicated that 2′F-RNA/DNA hybrids are not substrates of RNase H1 and ultimately led to the conclusion that the 2′-hydroxyl moiety of the RNA strand in a DNA/RNA hybrid is required for both binding and hydrolysis by RNase H1. Through the synthesis of a series of chimeric sense oligonucleotides of mixed RNA and 2′F-RNA composition, the gap requirements of RNase H1 within the sense strand were examined. Results from these studies showed that RNase H1 requires at least five or six natural RNA residues within the sense RNA strand of a hybrid substrate for both binding and hydrolysis. The RNase H1-mediated degradation patterns of these hybrids agree with previous suggestions on the processivity of RNase H1, mainly that the binding site is located 5′ to the catalytic site with respect to the sense strand. They also suggest, however, that the binding and catalytic domains of RNase H1 might be closer than has been previously suggested. In addition to the above, physicochemical studies have revealed the thermal stabilities and relative conformations of these modified heteroduplexes under physiological conditions. These findings offer further insights into the physical binding and catalytic properties of the RNase H1–substrate interaction, and have been incorporated into a general model summarizing the mechanism of action of this unique enzyme.
INTRODUCTION
The ribonuclease H (RNase H) class of enzymes are endonucleases that cleave the RNA moiety of RNA/DNA hybrids in the presence of divalent cations such as Mg2+ or Mn2+ leaving 5′-phosphate and 3′-hydroxyl oligonucleotide products (1,2). RNase H was first described in 1970 by Hausen and Stein (3) in calf thymus, and since then RNase H proteins have been found in a wide variety of organisms, ranging from bacteria to vertebrates. A significant function of RNase H is its critical action in the life cycle of retroviruses such as the human immunodeficiency virus (HIV) (4) and, as such, RNase H serves as an ideal drug target for anti-HIV therapies. RNase H1 from Escherichia coli is structurally homologous to that of HIV-1 and mammalian RNase H, and is the best characterized member of the family. Although the X-ray crystal structure of E.coli RNase H1 has been determined (5–7), a tertiary structure of the complex between E.coli RNase H1 and its substrate has yet to be resolved. Despite the extensive studies conducted, the biological role of this enzyme is not yet well defined. Escherichia coli RNase H1 has, however, been implicated in DNA replication and repair (8) and in chromosomal DNA replication (9).
The use of antisense oligonucleotides (AONs) as selective inhibitors of gene expression offers a rational approach for the prevention and treatment of some gene-mediated disorders. The three most commonly exploited therapeutic routes of AONs include translation arrest, splicing arrest and activation of RNase H1. The RNase H1-mediated degradation of a target mRNA is considered the most effective mode of action of AONs (10,11) for it is simply more potent than ones which are limited to interfering with the splicing or translational machinery. As such, understanding the mechanisms of catalytic function and substrate recognition for RNase H1 is critical in the design of future potential antisense oligonucleotides, as well as for the design of potential inhibitors of retroviral reverse transcriptases such as HIV.
While many AONs display promising affinities for RNA, only a handful form hybrids with RNA that are recognized and cleaved by RNase H1. These include DNA (12), phosphorothioate DNA (S-DNA) (13,14), cyclohexene nucleic acids (CeNA) (15), β-d-arabinonucleic acids (ANA) (16) and 2′-deoxy-2′-fluoro-β-d-arabinonucleic acids (2′F-ANA) (17). Of these, ANA and 2′F-ANA represent the first and only examples of 2′-modified β-d-furanoside AON that are recognized and cleaved by RNase H1.
The crystal structure of E.coli RNase H1 suggests that the active site is nestled within a cluster of lysines (7). These positively charged amino acids are believed to bind electrostatically to the heteroduplex and thus define the binding surface of the enzyme (18). The interaction between the binding surface of the enzyme and the phosphate groups on the substrate is postulated to take place within the minor groove of the heteroduplex (18,19). For this reason, the minor groove width is believed to be an important structural parameter for DNA/RNA hybrids that enables them to act as substrates of RNase H1 (19). RNase H1 does not bind to double-stranded DNA or single-stranded nucleic acids and although the enzyme is capable of binding to RNA/RNA hybrids, it does not hydrolyze them (20). Studies have shown that the minor groove width of DNA/RNA hybrid duplexes is 8.5 Å, lying between that of A-form RNA/RNA hybrids (11 Å) and B-form DNA/DNA hybrids (7.5 Å) (19). Presumably, it is this difference that allows RNase H1 to discriminate between DNA/RNA and RNA/RNA species. Moreover, it has also been suggested that any heteroduplex which contains protrusions in the minor groove or its proximity (e.g. 2′-O-alkyl RNA/RNA) will interfere with binding to the active site and ultimately preclude the activity of the enzyme (18).
Structural requirements for DNA/RNA hybrids to be suitable substrates for RNase H1 are well described (18–22); however, the tolerance level of this enzyme towards modifications which do not alter the conformation of the duplex is not clearly understood, especially with respect to the sense RNA strand. Not surprisingly, due to their therapeutic applicability, the majority of experiments to date have focused on determining modifications that are tolerated by RNase H1 within the antisense strand (AON). One of the aims of this study was to examine the molecular requirements imposed on RNase H1 by the sense RNA strand.
Our initial objective was to examine whether a fully modified sense oligonucleotide, upon hybridization with a proven RNase H1-inducing DNA target, is capable of being degraded by RNase H1. The most conservative RNA analog with respect to possible changes in chemical structure is probably 2′-deoxy-2′-fluoro-β-d-ribonucleic acid (2′F-RNA), where the 2′-OH group is replaced by a fluorine.
The ability of fluorine to serve as a ‘hydroxyl mimic’ and participate in hydrogen bonding has been extensively described (23). The electronegativities of a hydroxyl group and fluorine are very similar, and the van der Waals radius of fluorine is 1.47 Å, which lies in between that of oxygen (1.57 Å) and hydrogen (1.2 Å) (23). Accordingly, fluorine, just as the natural hydroxyl functionality, drives the sugar of 2′F-RNA to the C3′-endo conformation due to the gauche effect, ultimately leading a fully modified rF-RNA oligonucleotide, like RNA, to adopt the A-form helix conformation (24,25). This is evident from the similar circular dichroic (CD) spectra of 2′F-RNA/RNA duplexes, which indicate that they belong to the A-form conformational family, with the sugars puckering in the C3′-endo conformation (26). The only significant difference between the two moieties is that whereas a hydroxyl group can serve as a strong hydrogen bond donor and acceptor, fluorine is only capable of acting as a weaker acceptor (27).
Oligonucleotides containing 2′-deoxy-2′-fluoro-β-d-ribose were first investigated as potential antisense agents by ISIS Pharmaceuticals in 1993 (26). 2′F-RNA was found to have high binding affinity and selectivity for RNA targets, but unfortunately was not found to be resistant to nucleases and did not elicit RNase H1 activity when bound to RNA. Our objective, however, was not to employ 2′F-RNA for its potential antisense properties, but rather as a modified oligonucleotide to investigate the molecular requirement of RNase H1 within the sense strand.
In this report, we have conducted several binding and RNase H1 induction assays involving a series of modified sense oligoribonucleotides containing 2′-deoxy-2′-fluoro-β-d-ribose acting as a substitute for the natural d-ribose. In the process, we have shown that the 2′-OH moiety of the RNA strand in an RNA/DNA duplex is required for both binding and catalysis by E.coli RNase H1. Furthermore, we have also determined the minimum number of natural RNA residues required within the sense strand to elicit RNase H1 activity. These studies increase our understanding of the specific molecular requirements of RNase H1 with respect to the sense strand and the physical binding properties of the RNase H– substrate interaction.
MATERIALS AND METHODS
Materials
[γ-32P]ATP (>5000 Ci/mmol), E.coli RNase HI and T4 polynucleotide kinase were obtained from Amersham Pharmacia Biotech (Quebec). 5′-O-dimethoxytrityl-3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidites of the various 2′-O-deoxy- and ribonucleosides were purchased from Dalton Chemical Laboratories (Toronto). 1-(5-O-dimethoxytrityl-2-fluoro-3-O-(β-cyanoethyl-N,N-diisopropylphosphoramidite)-β-d-ribofuranosyl)-adenine was obtained from Promega Biosciences (California). All phosphoramidite reagents were stored at –20°C and desiccated under vacuum (over P2O5) for 24 h prior to use.
Oligonucleotide synthesis
Oligonucleotides were synthesized on a PerSeptive Biosystems Expedite 8909 Nucleic Acid Synthesizer using standard phosphoramidite chemistry and 3′-nucleoside–long chain alkylamine controlled pore glass (CPG) (500 Å) derivatized according to the procedure of Damha et al. (28). Nucleoside monomers were dissolved to 0.11 M in anhydrous acetonitrile. Prior to chain assembly, the solid support was treated with the capping reagents acetic anhydride N-methylimidazole 4-(dimethylamino)pyridine. The 5′-terminal trityl group was removed by the synthesizer and the oligomers were then removed from the support and deprotected by treatment of the CPG with a 1 m1 solution containing concentrated ammonium hydroxide/ethanol (3:1 v/v) for 2 days at room temperature. The ammonium hydroxide/ethanol solution was evaporated and the crude product purified by anion exchange HPLC and desalted on a Sephadex G-25 column (29). Oligoribonucleotides and deoxyribonucleotides were assembled on the same instrument using silyl phosphoramidite chemistry as previously described (29).
UV thermal denaturation studies
UV thermal denaturation data were obtained on a Varian Cary 1 UV-vis spectrophotometer equipped with a Peltier temperature controller. Molar extinction coefficients for 2′F-RNA and 2′F-ANA were assumed to be the same as those of normal RNA and DNA strands, respectively. Samples were heated to 80–90°C for 15 min, then cooled slowly to room temperature and stored at 4°C overnight before measurements. Prior to the thermal run, samples were degassed by placing them in a Savant lyophilizer (2 min). Absorbance values were recorded after equilibration as the temperature was increased incrementally by 0.5°C at 1 min intervals (buffer: 140 mM K+, 1 mM Mg2+ and 5 mM Na2HPO4, pH 7.2). To compare relative overall changes in absorbance, normalized ΔA plots were constructed according to the method of Wilson and co-workers (30) by use of the formula (At – Ai)/Af, where Ai is the initial absorbance, Af is the final absorbance and At is the absorbance measured at each time point. Hyperchromicity values (%H) are reported as the percent increase in absorbance at the wavelength of interest with respect to the final absorbance. Tm values were calculated using the baseline method reported by Puglisi and Tinoco (31) and generally have an uncertainty of ±0.5°C.
CD spectra
CD spectra were obtained on a Jasco J-710 spectropolarimeter equipped with a NESLAB RTE-111 circulating bath. Samples were allowed to equilibrate for 5–10 min at the appropriate temperatures (buffer: 140 mM K+, 1 mM Mg2+ and 5 mM Na2HPO4, pH 7.2). Each spectrum was an average of five scans and was collected at a rate of 100 nm/min with a bandwidth of 1 nm and sampling wavelength of 0.2 nm. The CD spectra were recorded from 350 to 200 nm at 5°C and normalized by subtraction of the background scan with buffer. The molar ellipticity was calculated from the equation [Θ] = Θ/Cl, where Θ is the relative ellipticity (millidegrees), C is the molar concentration of oligonucleotides (mol/l) and l is the path length of the cell (cm). The data were processed with Windows-based software supplied by the manufacturer (JASCO, Inc.).
32P labeling of oligonucleotides
The 18mer RNA or RNA analog was 5′ end-labeled with 32P using [γ-32P]ATP and T4 polynucleotide kinase following standard procedures (32). The labeled RNA was purified by 20% denaturing PAGE (33) and desalted using NAP-5 columns containing Sephadex G-25 Medium of DNA grade (APB). The counts per minute for purified labeled samples were quantitated using a Bioscan QC 2000 counter (Washington, DC).
RNase H1 cleavage assays
The ability of oligonucleotides to elicit RNase H1 degradation of the target RNA was determined in assays (50 µl final volume) that comprised 5 pmol test oligonucleotide preannealed with 20 000–50 000 c.p.m. 5′-32P-target RNA, and 10–15 pmol target DNA in 60 mM Tris–HCl pH 7.8 containing 2 mM dithiothreitol, 60 mM KCl and 2.5 mM MgCl2 (unless otherwise indicated). Duplexes were pre-incubated at 90°C for 5 min, followed by slow cooling to room temperature over a 1 h period. Following pre-incubation, reactions were started by the addition of 0.1–1 U E.coli RNase H1 and then allowed to proceed for varying times at 22–24°C. Aliquots of 8 µl were removed at each desired time point and quenched by the addition of 8 µl of loading buffer (98% deionized formamide, 10 mM EDTA, 1 mg/ml bromophenol blue, 1 mg/ml xylene cyanol) and heated at 100°C for 5 min. The reaction products were resolved by electrophoresis using a 16% polyacrylamide sequencing gel containing 7 M urea and visualized by autoradiography.
Competitive binding assays
These experiments measured the ability of various duplexes to inhibit the RNase H-mediated degradation of a 5′-32P-labeled RNA:DNA 18 bp duplex substrate. Each competitor duplex (30 pmol) was pre-annealed prior to addition to an assay comprised of pre-annealed 5′-32P-rA18:dT18 (5 pmol) in 60 mM Tris–HCl pH 7.8, 2 mM dithiothreitol, 60 mM KCl and 2.5 mM MgCl2. Reactions were initiated by the addition of 0.5 U E.coli RNase H1 and then allowed to proceed for varying times at 22°C. Aliquots (8 µl) were quenched and resolved as above.
Quantitation of gel radioactivity
All assays were quantitated in the linear range of the gel. Each film was scanned into a picture file and then digitized using the UN-SCAN-IT® automated gel digitizing software v.4.1 (Silk Scientific Corp.). The percentage of rA18 or I6 remaining over time was calculated by quantitating the amount of the 18mer band in one lane on the gel (representing one time point) as a percentage of the total radioactivity present in that entire lane.
RESULTS AND DISCUSSION
To evaluate the ability of a modified RNA strand to elicit RNase H1 activity, we synthesized oligonucleotides 18 units in length comprised solely of 2′-deoxy-2′-fluoro-β-d-ribonucleotides (2′F-RNA) as well as chimeras containing both 2′F-RNA and RNA residues (Table 1). All oligomers were tested for hybridization affinity (Tm measurements, Table 2) and for the ability to direct cleavage by E.coli RNase H1.
Table 1. 2′F-RNA oligonucleotide sequences synthesized in this work.
| Oligonucleotidea | Sequenceb |
|---|---|
| rFA | 5′-rF(AAA AAA AAA AAA AAA AAA)-3′ |
| I1 | 5′-rF(AAA AAA AAAOH AAA AAA AAA)-3′ |
| I4 | 5′-rF(AAA AAA AAOHAOH AOHAOHA AAA AAA)-3′ |
| I6 | 5′-rF(AAA AAA AOHAOHAOH AOHAOHAOH AAA AAA)-3′ |
| F1 | 5′-r(AAA AAA AA AF AAA AAA AAA)-3′ |
aPurified by anion exchange HPLC followed by desalting on Sephadex G-25.
brFA and AF refer to 2′-deoxy-2′-fluoro-β-d-riboadenosine nucleotide; rA and AOH refer to the natural 2′-OH-riboadenosine nucleotide.
Table 2. Melting temperatures (Tm) and percent hyperchromicity (%H) values of duplexes of the various rF-RNA modified oligomers with complementary target RNA and DNA strands.
| Sense oligonucleotidea | DNA target | RNA target | ||
|---|---|---|---|---|
| Tmb (°C) | %H | Tmb (°C) | %H | |
| rA18 | 39.0 | 15.4 | 30.4 | 14.0 |
| rF-RNA18 | 48.1 | 20.1 | 39.9 | 20.9 |
| I1 | 44.6 | 17.2 | ND | ND |
| I4 | 35.2 | 15.0 | ND | ND |
aWe were unable to purify sufficient quantities of the I6 sequence to allow for its examination in thermal denaturation or CD studies.
bAqueous solutions: 2.5 × 10–6 M duplex. Buffer: 140 mM KCl, 1 mM MgCl2, 5 mM Na2HPO4, pH 7.2. Uncertainty in Tm values is ±0.5°C. ND indicates that Tm data was not determined.
The melting curves of the modified oligonucleotides hybridized to DNA or RNA exhibited ‘sharp’ monophasic transitions, indicative of formation of a single cooperative complex. As expected, the thermodynamic stability of 2′F-RNA/RNA as well as 2′F-RNA/DNA duplexes are higher than those of corresponding RNA/RNA and RNA/DNA duplexes (26) (Table 2). This is consistent with the higher pre-organization of the 2′F-RNA residues in the C3′-endo conformation which entropically favors double helix formation (24,25). Also worth noting is the fact that all duplexes tested for RNase H1 degradation displayed thermal stability well above 22–24°C (the temperature of the RNase H1 assay), indicating that the strands were fully associated upon incubation with the enzyme.
Interestingly, the Tm of the I4/DNA duplex (35.2°C) is lower than that of rA:dT (39.0°C). This is puzzling considering that the 14 pre-organized 2′F-RNA residues present in I4/DNA should in theory enhance the thermodynamic stability of this duplex relative to the native RNA/DNA duplex. The role of the 2′-OH groups in hydration of the duplex may be responsible for the observed decrease in Tm. This proposal is consistent with the suggestion that RNA/RNA duplexes are generally more stable than DNA/DNA duplexes because the 2′-OH groups in RNA duplexes propagate conserved networks of water molecules that stabilize the duplex (34). Despite the suggestion that fluorine atoms are also hydrated (35), the stabilization provided by fluorine as compared to hydroxyl groups is likely not as significant. It is hence clear that 2′F-RNA residues can provide a stabilizing entropic effect at the expense of a stabilizing hydration effect provided by 2′-OH RNA residues. Therefore, the drop in Tm for the F1:dT and F4:dT systems would suggest that the destabilization generated by introducing RNA units is more significant than the gain in stability provided by hydration. There will be a point, however, where these effects will ‘balance out’ such that one would start seeing an overall gain in stability after introducing even more RNA units (e.g. I6 or I10).
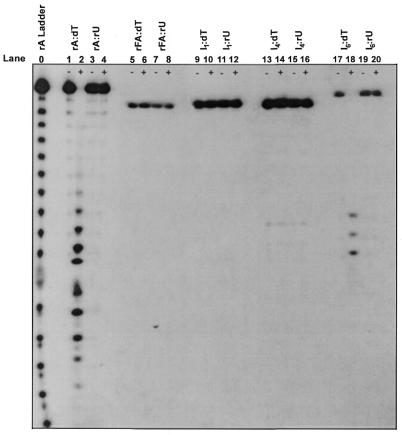
Figure 1 summarizes the results of the RNase H1 assays conducted in this study. The lanes indicated by a – sign represent the controls of each assay prior to the addition of RNase H, and those with a + sign represent reactions of duplexes after a 30 min incubation time with RNase H. Our initial objective was to examine whether a modified sense oligonucleotide, upon binding to a DNA target, is capable of being degraded by RNase H1. Hence, we first examined the susceptibility of fully modified 32P-labeled rFA18 (rFA) bound to dT18 (dT) to degradation by E.coli RNase H1 (Fig. 1, lanes 5 and 6). This was done in conjunction with three controls: an RNA:DNA control, rA:dT (Fig. 2, lanes 1 and 2) and two RNA:RNA controls, rA:rU (Fig. 1, lanes 3 and 4) and rFA:rU (Fig. 1, lanes 7 and 8). The degradation ladder found in Figure 1 (lane 2), which corresponds to the natural RNA/DNA substrate, confirms the viability of the assay. Furthermore, as expected, neither RNA/RNA (lane 4) nor 2′F-RNA/RNA (lane 8) served as substrates of RNase H1. With respect to the test in question (rFA:dT), the absence of a degradation ladder in lane 6 demonstrates that the fully modified 32P-labeled rF-RNA sense strand bound to its complementary DNA antisense strand is not a substrate of RNase H1.
Figure 1.
Electrophoresis gel showing RNase H-mediated cleavage of duplexes. dT18 or rU18 (15 pmol) was pre-annealed with 20 000 c.p.m. of 5′-32P-target RNA in 60 mM Tris–HCl pH 7.8 containing 2 mM dithiothreitol, 60 mM KCl, 2.5 mM MgCl2. Reactions were started by the addition of 0.5 U E.coli RNase H at 22°C and were quenched after 30 min reaction time. The base hydrolysis ladder was prepared by incubation of 5′ end- labeled RNA at 90°C for 15 min in 10 µl of 100 mM sodium carbonate pH 9.0. –, an aliquot from the reaction mixture prior to the addition of enzyme; +, an aliquot from the reaction mixture after a 30 min incubation with enzyme. rFA, rFA18; I1, rFA8(rA)1rFA9; I4, rFA7(rA)4rFA7; I6, rFA6(rA)6rFA6. Note that the low intensity bands of degradation observed in the – lanes are indicative of non-enzymatic background degradation of labeled oligonucleotides.
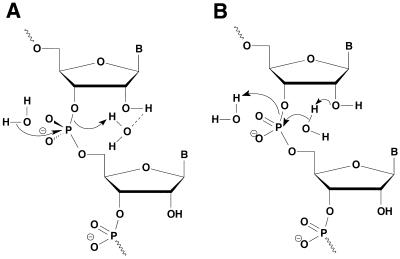
Figure 2.
Hydrolysis of RNA strands in RNA/DNA hybrids by E.coli RNase H1. (A) The 2′-OH group acting as a general acid to increase the electrophilicity of the phosphorus center. (B) The 2′-OH group acting as a general base to increase the nucleophilicity of the attacking water molecule. In both mechanisms, the products are the observed fragments containing 3′-hydroxyl and 5′-phosphate termini.
Previous studies have elucidated the importance of the 2′-OH groups in the general catalytic mechanism of RNase H1, mainly that the divalent metal cation interacts with the 2′-OH group of the nucleotide 5′ to the scissile linkage by forming an outer sphere metal hydrate complex (36). Furthermore, other studies have suggested that the 2′-OH group of the nucleoside 3′ to the scissile linkage acts as both a proton donor and an acceptor (37). In other words, the 2′-OH functionality may act as a general base to facilitate the nucleophilicity of an attacking water molecule or as a general acid to increase the electrophilicity of the phosphorus center (Fig. 2). By replacing a hydroxyl group with a fluorine, not only have the general acid capabilities of the hydroxyl group been abolished, but the basic capabilities of the 2′ moiety have also been reduced because fluorine generally acts as a weak base. All other factors being equal, a modified sense strand containing a 2′ functionality with diminished general acid and base properties loses the power to elicit RNase H1 activity. These findings could explain why a fully modified sense strand (rFA18) that lacks the critical 2′-OH moiety (rFA18) is not cleaved by the enzyme.
We next examined the susceptibility of the modified sequence I1 (with one 2′-OH substitution within a 2′F-RNA18 strand) to RNase H1 degradation in anticipation of possible cleavage at the site of hydroxyl substitution. As with the fully 2′F-modified sense strand, I1:dT was also not a substrate of RNase H1, as can be seen from the absence of a degradation ladder in Figure 1 (lane 10). This implies that RNase H1 requires a minimum number of hydroxyl groups within the backbone of the sense strand for recognition and/or hydrolysis of the substrate. We hence set out to determine this number by treating oligomer I4 (with four hydroxyl substitutions) bound to dT18 with RNase H1 (Fig. 1, lane 13). Interestingly, four hydroxyl insertions (I4:dT) were not sufficient to elicit cleavage of the hybrid. This is somewhat different to the findings of Hogrefe et al. (38), who determined that a DNA/DNA duplex incorporating four contiguous RNA units is a substrate for RNase H1. The discrepancies likely result from the different structural features of duplex DNA (Hogrefe system) as compared to a DNA/RNA-like hybrid (I4/DNA). By using a system that more closely mimics the natural DNA/RNA substrate, we feel we were able to more precisely ascertain the sense strand gap requirements of RNase H1.
Next we prepared the six hydroxyl substitution sequence (I6) and tested its ability to activate RNase H1. Indeed, the data shows that the I6:dT hybrid duplex served as a substrate of RNase H1 (Fig. 1, lane 18). Unfortunately, since 2′F-RNA monomers are no longer commercially available, we were unable to synthesize and test the I5 sequence (with five hydroxyl substitutions), and hence cannot state at this time that RNase H1 requires a minimum of six 2′-OH residues within the sense strand to hydrolyze its substrate. Nevertheless, it is safe to conclude that in order for RNase H1 to bind and cleave a DNA/RNA substrate, the enzyme requires more than four contiguous 2′-OH groups within the sense strand.
The majority of studies to date have shown that the minimum number of consecutive natural DNA residues required within the antisense strand to elicit E.coli RNase H1 is also only four (20,39). In theory, one would expect the minimum gap requirement for both the antisense and sense strands to be similar, considering that RNase H1 is binding to both strands of the duplex. This theory may still hold, however, for one cannot ignore the possible discrepancies with respect to this ‘minimum’ DNA antisense gap required to elicit the enzyme, with some research groups reporting five as the minimum gap requirement (40). These discrepancies may be attributed to the different sources of E.coli RNase H1 and/or differences based on the location of the gap within the antisense strand. Nevertheless, the majority of studies have shown that E.coli RNase H1 requires only four natural DNA residues within the antisense strand for recognition and hydrolysis of a duplex substrate.
In a similar argument to that presented above, the difference in gap requirement within the sense and antisense strands may also be due to structural differences amongst the two hybrid systems studied. Hybrids used to study the gap requirements within the antisense strand have been mainly RNA/RNA-like (A-form), whereas hybrids used in this study to examine the gap requirements within the sense strand are more DNA/RNA-like (‘A-like form’). Thus, helical structure, which has been shown to play a key role in RNase H1 recognition (19), could have accounted for the deviating gap requirements observed.
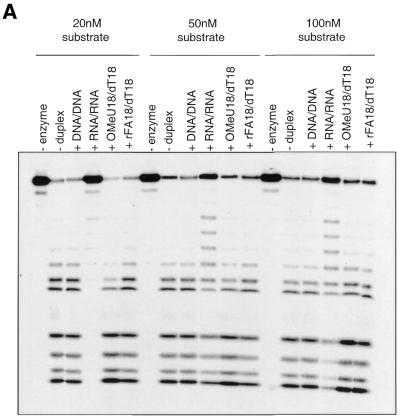
Next, we conducted a series of competitive binding studies in order to determine whether RNase H1 is capable of binding to a DNA/RNA-like substrate lacking 2′-OH moieties within the sense strand. To do this, we tested the binding affinity of both 2′F-RNA:DNA and 2′OMe-RNA:DNA via experiments that measured the ability of these duplexes to inhibit the RNase H1-mediated degradation of a wild-type 5′-32P-labeled RNA:DNA duplex substrate (32P-rA18:dT18) (Fig. 3A). In this experiment, natural RNA/RNA and DNA/DNA substrates were used as competitive controls. RNA/RNA duplexes have the proper A-form conformation required for binding to RNase H1 (20), but are not hydrolyzed by the enzyme due to a non-optimal minor groove width (41) and/or the presence of protruding 2′-OH groups in the minor groove of the helix which block the catalytic site of the enzyme upon binding (18). Accordingly, one can see from Figure 3A that the competitor RNA/RNA duplex is clearly binding to the enzyme at all three concentrations of substrate as it is retarding the rate of degradation of the wild-type substrate. A DNA/DNA duplex, however, is not recognized by RNase H1 and, accordingly, does not retard the rate of degradation of the wild-type substrate in Figure 3A. Clearly then, the lack of inhibition displayed by either the 2′OMe- or 2′F-RNA/DNA duplexes, even at a 10-fold concentration of competitor duplex (20 nM concentration of substrate), indicates that these substrates bind weakly or not at all to the substrate. This is in line with the results obtained by Lima and Crooke (20), who tested the binding affinity of RNase H1 to 2′OMe-RNA/DNA, a hybrid that mimics the conformation of a normal RNA/DNA hybrid. Their data showed that the association constant between RNase H1 and the 2′-O-methylated substrate was one-thirtieth of that between enzyme and the natural substrate. They suggested that very weak affinity consequent to 2′-O-methylation may be due to steric hindrance by the methyl groups and/or loss of the hydrogen bonds. Choosing a modification such as fluorine rather than a methoxy group allowed us to bypass the steric hindrance argument imposed by the methyl group. Thus, these findings strongly suggest that the 2′-OH group within the sense strand plays a key role in the binding step of the substrate to RNase H1.
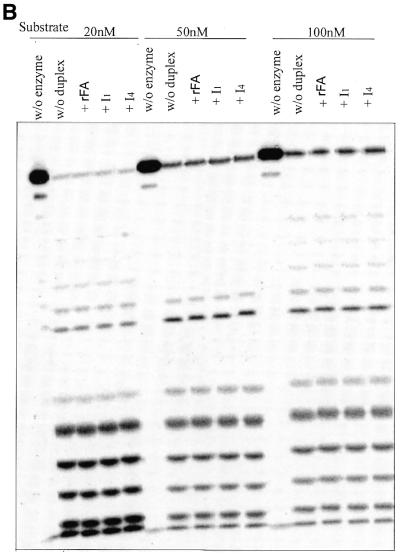
Figure 3.
(A and B) RNase H1 competitive binding study. Competing substrates were added at a final concentration of 200 nM (corresponding to a 10-, 4- and 2-fold increase in concentration when the substrate concentration is 20, 50 and 100 nM, respectively). Note that in all cases, 18 bp oligonucleotide sequences were used with adenine as the RNA base and thymine as the DNA base.
Similar studies were conducted with sequences I1 and I4 as well (Fig. 3B). As can be seen from the figure, neither rFA:dT, I1:dT nor I4:dT seem to be acting as competitive inhibitors of the enzyme at a substrate concentration of 20, 50 or 100 nM. Thus, one can conclude that the lack of sufficient 2′-OH groups within the sense strand disrupts the binding affinity of RNase H1 for these helices. In fact, these results reinforce our notion that the 2′-OH groups of the sense strand are involved in both the binding and catalytic steps of E.coli RNase H1. The possible role of the 2′-OH groups in hydration of the duplex (34) and in providing essential water molecules that can act as anchors for the enzyme might be an explanation as to why these molecules do not bind to the enzyme.
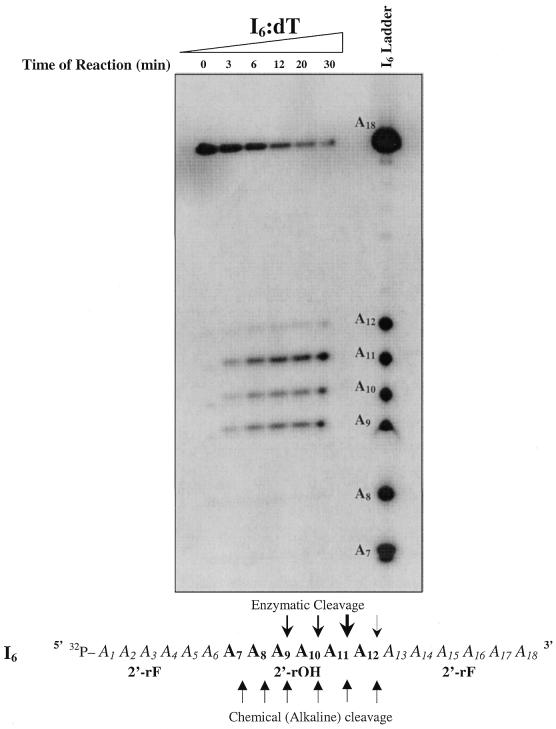
Next, the exact locations of the RNase H1-mediated cleavage sites in the I6:dT sequence were determined by examining the degradation products adjacent to an I6 alkaline cleavage ladder (Fig. 4). Upon treatment with base, sugars containing 2′-OH groups are susceptible to internucleotide cleavage which occurs via an in-line attack on the internucleotide phosphorus center, followed by hydrolysis of the 2′,3′-cyclic phosphate intermediate to form 2′/3′-monophosphate and 5′-hydroxyl products (42). 2′F-RNA sugars are less susceptible to base hydrolysis due to the lack of the 2′-OH moiety within the sugar backbone. Accordingly, when I6 alone is exposed to an alkaline solution (pH 9.0, 90°C), six products are formed, consistent with hydrolysis occurring only at each of the six ribose (2′-OH) units present within oligomer I6 (Fig. 4). It is important to note, however, that there exists a slight shift in mobility between enzymatically hydrolyzed 5′-32P-labeled fragments (I6/dT) and chemically hydrolyzed 5′-32P-labeled fragments (I6 alkaline ladder). This shift originates from the fact that RNase H1 hydrolyzes an RNA strand leaving 5′-phosphate and 3′-hydroxyl oligonucleotide products (1,2) (refer to the mechanism in Fig. 2), whereas chemical hydrolysis results in 5′-hydroxyl and 2′/3′-phosphate products. This same reasoning can be used to explain the presence of two distinct adjacent degradation bands in some RNase H1 assays where background chemical cleavage (induced by the reaction medium prior to the addition of enzyme) was present.
Figure 4.
RNase H1-mediated degradation of I6/dT adjacent to an I6 non-enzymatic alkaline cleavage ladder. AON (dT18) (15 pmol) was pre-annealed with 15 000 c.p.m. of 5′-32P-target I6 [rFA6(rOH)6rFA6] in 60 mM Tris–HCl pH 7.8 containing 2 mM dithiothreitol, 60 mM KCl, 2.5 mM MgCl2. The reaction was started by the addition of 0.2 U E.coli RNase H at 22°C and aliquots of the reaction were quenched after 0, 3, 6, 12, 20 and 30 min by the addition of denaturing buffer. Note that time 0 represents an aliquot from the reaction mixture taken prior to the addition of enzyme and hence represents a negative control for the reaction. The non-enzymatic alkaline cleavage ladder was prepared by incubation of 5′ end-labeled I6 at 90°C for 15 min in 10 µl of 100 mM sodium carbonate pH 9.0.
By examining the pattern of cleavage of I6 by RNase H1 in Figure 4, it is clear that cleavage only occurred at positions within the modified RNA strand containing ribose sugars (2′-OH), implying that the 2′-OH moiety is indeed involved in the hydrolytic step of the enzyme as well. Furthermore, we observe three dominant sites of cleavage corresponding to positions A9, A10 and A11, and a slower rate of cleavage at position A12. Very little cleavage, if any, was observed at positions A8 and A7, which correspond to the ribose (2′-OH) residues closest to the 5′ end. Previous structural studies have suggested that the enzyme exhibits a selective binding directionality with respect to the RNA strand of the heteroduplex substrate such that the binding region of the enzyme is positioned several residues 5′ to the catalytic region (6,18). Furthermore, it was also suggested that RNase H1 initiates an endonucleolytic cleavage near the 3′ end of the RNA and then exonucleolytically degrades the RNA in the 3′→5′ direction with respect to the RNA sense strand (43). Our degradation patterns conform with these findings.
We have shown that RNase H1 is not capable of binding a fully modified 2′F-RNA strand bound to its DNA complement and that RNase H1 requires a minimum number of hydroxyl groups within the sense strand to bind to its substrate. This, along with the suggested catalytic importance of the 2′-OH moiety, suggests that the RNase H1 binding domain cannot bind to regions of the duplex lacking 2′-OH groups within the sugar backbone of the sense strand. Extending this to our I6:dT substrate, we can hence assume that RNase H1 does not bind between regions A1–A6 and A13–A18 because they are 2′F-RNA domains lacking a 2′-OH group. The only possible region where binding could occur is between sugars A7 and A12. Considering the cleavage pattern observed, and assuming that the binding domain exists 5′ to the catalytic domain, we propose that RNase H1 binds close to sugars A7 and A8 within the duplex in a way that masks those sites from cleavage. The catalytic domain, located 3′ to the binding domain, is then able to cleave the sense strand at positions A9, A10, A11 and A12. Also worth noting is the fact that the intensity in none of the degradation bands decreased over time, indicating that RNase H1 cuts each I6 strand at only one position and does not further process the degraded substrate.
Since the I4:dT duplex (with four hydroxyl substitutions) did not bind to the enzyme, we can conclude that the binding domain requires a DNA/RNA hybrid segment of at least 5 or 6 bp (region A6–A12 in I6/dT). According to this model, and to the observed cleavage patterns, the binding and catalytic domains must somehow overlap because cleavage in I6:dT is only observed between positions A8 and A12, which constitutes a segment where RNase H1 is likely binding due to the presence of the essential 2′-OH groups. This contrasts with the proposal by Lima and Crooke (44) that there exist at least five residues between the binding and catalytic sites of E.coli RNase H1. The only way that the binding and catalytic domain could be at least five residues apart in our system is if the RNase H1 binding domain is capable of binding to the region of the helix lacking 2′-OH moieties (A1–A6). But if this is possible, one cannot explain why cleavage at position A8 did not occur. We propose instead that the catalytic and binding domains of E.coli RNase H1 are closer in proximity than has been previously suggested and might not be completely separate entities in themselves.
Modeling studies based on the crystal structure of the enzyme alone suggest that RNase H1 interacts with 9–10 bp of RNA/DNA hybrids (41,45). According to this, the enzyme is likely interacting with I6:dT at both the modified (2′F-RNA) and natural (RNA) portions of the I6 sense strand. Although the enzyme might contact 9–10 bp within the I6:dT duplex, our conclusions suggest that the actual binding interaction likely constitutes a segment consisting of only about 5 or 6 bp. Confirming such findings would require a co-crystal structure of RNase H1 with its substrate.
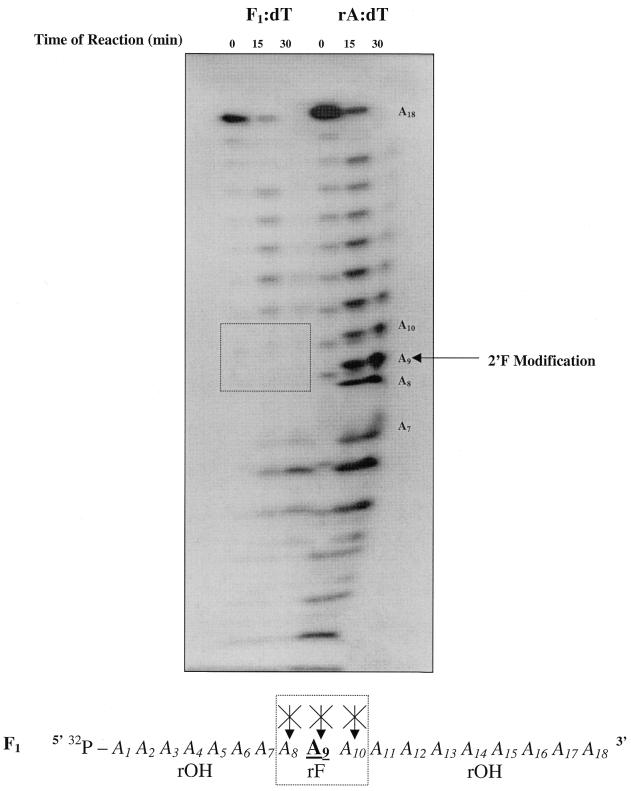
Another modified sense strand that was tested in our RNase H1 assays is F1 (Table 1). This sequence represents an oligoribonucleotide containing a single 2′F-RNA sugar substitution in the center. As expected, the F1:dT duplex was a substrate of RNase H1, but to study more precisely the effects of a single 2′-OH → 2′F-RNA substitution on processivity of this modified sense strand, we compared the RNase H1-mediated degradation products of an F1/dT duplex adjacent to those of an RNA/DNA substrate (rA/dT) (Fig. 5). Note that these assays revealed the presence of two distinctly close degradation bands that resulted from the presence of both background chemical cleavage products (represented in the lane 0 min) and enzymatic cleavage products (represented by the lanes 15 and 30 min).
Figure 5.
RNase H-mediated degradation of F1/dT adjacent to rA/dT. AON (dT18) (15 pmol) was pre-annealed with 15 000 c.p.m. of 5′-32P-target F1 [5′-rA8(rFA)1rA9-3′] and 30 000 c.p.m. of rA18 in 60 mM Tris–HCl pH 7.8 containing 2 mM dithiothreitol, 60 mM KCl, 2.5 mM MgCl2. The reactions were started by the addition of 0.2 U E.coli RNase H at 22°C and aliquots of the reaction were quenched after 0, 15 and 30 min by the addition of denaturing buffer. Note that time 0 represents an aliquot from the reaction mixture taken prior to the addition of enzyme. Also note that a ‘smiling effect’ occurred at the lower end of the gel due to a prolonged electrophoretic exposure time.
As can be seen from Figure 5, the ‘interference fingerprint’ (denoted by the box) indicates that the single 2′F-RNA modification at position A9 interfered with the ability of RNase H1 to cleave at that site, as well as at the two adjacent sites (A8 and A10) around it. The absence of degradation at position A9 was expected and confirms the notion that RNase H1 requires the 2′-OH moiety for cleavage. Furthermore, the lack of degradation at positions A8 and A10 agrees with the propositions recently made by Iwai et al. (37). They proposed that the 2′-OH group of the nucleoside on the 3′ side of the scissile linkage acts as both a proton donor and an acceptor and that the 2′-OH group of the second nucleoside 5′ to the scissile linkage acts as a proton acceptor. Translating this to our system, cleavage cannot occur at position A10 due to the absence of the essential 2′-OH group on the nucleoside (A9) 5′ to the scissile linkage. Moreover, cleavage cannot occur at position A8 due to the lack of the same 2′-OH group on the nucleoside (A9) now 3′ to this scissile linkage.
Circular dichroism studies
If by modifying the 2′-OH group into a 2′-F within the sense strand in turn changes the overall conformation of the resulting helix, a negative result from a RNase H1 assay cannot be used to directly implicate the 2′-OH group in the mechanism of cleavage of RNase H. For this reason, CD spectral analysis was used to establish whether incorporation of 2′F-RNA units within RNA has a fundamental influence on the conformation of the hybrid duplex structure.
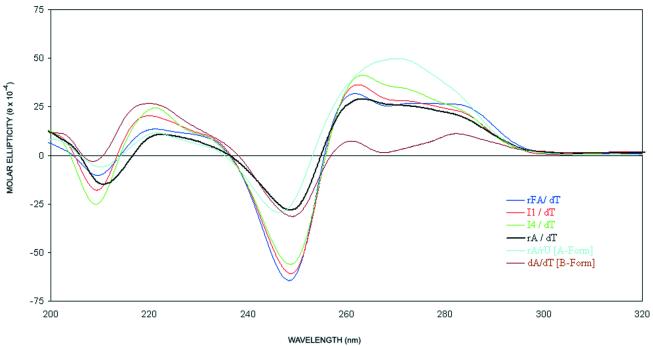
The CD spectra of the duplexes studied are shown in Figure 6. The rA18/dT18 duplex, which is intermediate between the A-form and B-form, has a large positive peak from 255 to 290 nm, a negative peak around 247 nm and a positive peak at 220 nm. The CD spectra of the riboF-A18/dT18, I1:dT and I4:dT duplexes are similar to the rA18/dT18 spectrum, suggesting that all duplexes share similar conformations in solution.
Figure 6.
CD spectra of oligonucleotides hybridized to dT18 at 5°C (2.3 µM, in 140 mM KCl, 1 mM MgCl2 and 5 mM Na2HPO4, pH 7.2).
General model reviewing the molecular requirements and mechanism of action of RNase H
In conclusion, our findings have been incorporated into a general model reviewing the mechanism of action of E.coli RNase H1. In the first stage, it has been suggested that RNase H1 assists in the formation of the mRNA/AON complex (46). This is followed by binding of RNase H1 to its substrate. For binding to occur, the following prerequisites must be met: (i) an A-form helix, which includes DNA/RNA and RNA/RNA type duplexes (19,20,47); (ii) at least five or six ribonucleotide (2′-OH) residues in the sense strand of the substrate (this work); (iii) the presence of a divalent metal cation (1) such as Mn2+ or Mg2+; (iv) 9–10 bp within a DNA/RNA hybrid (45,46), even though the actual binding interaction likely only requires 5–6 bp (this work); (v) the key amino acid residues required for binding (Cys13, Asn44, Asn16 and Asn45), which have been shown to contact both the RNA and DNA strands (19).
Once bound, the following criteria must be met by the substrate for efficient RNase H mediated hydrolysis to occur: (i) the AON/RNA duplex should have an intermediate minor groove width similar to that of DNA/RNA-like hybrids, which is wide enough to accommodate the chemical reaction that will take place at the cleavage site (19,48); (ii) the presence of a 2′-OH moiety in the sugars 5′ and 3′ to the scissile linkage (37; supported by this work); (iii) no protrusions in the minor groove that might interfere with the 2′-OH metal hydrate complex (36); (iv) the key amino acids required for catalysis are Asp10, Asp70, His124, Asp34 and Glu48 (6,49–51).
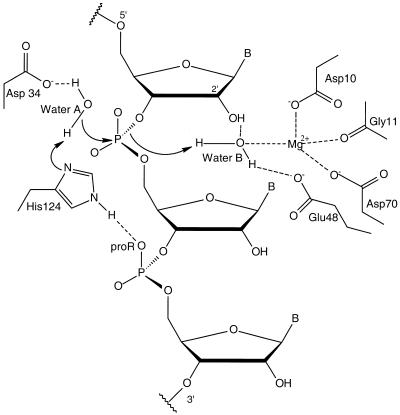
With respect to the molecular mechanism of action of RNase H1 at the catalytic site, the most recent general acid–base mechanism (49), outlined schematically in Figure 7, suggests that Asp10 and Asp70 are responsible for the binding of the Mg2+ ion to the correct position in the active site of the enzyme; His124 accepts a proton from the attacking water molecule A, which acts as a general base; Asp34 holds this water molecule; finally, Glu48 anchors another water molecule B that acts as a general acid. Furthermore, it has also been proposed that the pro-Rp-oxygen 3′ to the scissile linkage contributes to orient His124 to the best position for the catalytic function through the formation of a hydrogen bond (52). This entire process occurs in the minor groove of the DNA/RNA substrate (19).
Figure 7.
Proposed general acid–base catalytic mechanism of E.coli RNase H1 [adapted from Haruki et al. (52)].
Once all criteria for binding and cleavage have been met, RNase H1, with its binding site located slightly 5′ to its catalytic site with respect to the RNA strand, induces an endonucleolytic cleavage near the 3′-terminus of the RNA strand, and then processively and exonucleolytically degrades the RNA in a 3′→5′ direction (with respect to the RNA strand) (6,18,43; supported by this work). Upon degradation of the RNA strand, or upon reaching a segment of the duplex that does not conform to the binding and/or catalytic requirements of RNase H1, the enzyme dissociates from the complex and is thus available to process another substrate.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Robert H. E. Hudson and Maria Mangos for discussion and help in preparation of this manuscript. This work was supported by a grant from the Canadian Institute of Health Research (CIHR). D.R.Y. thanks FCAR for a post-graduate scholarship.
REFERENCES
- 1.Berkower L., Leis,J. and Hurwitz,J. (1973) Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid.deoxyribonucleic acid hybrid structures. J. Biol. Chem., 248, 5914–5921. [PubMed] [Google Scholar]
- 2.Crouch R.J., and Dirksen,M.L. (1982) Ribonuclease H. In Linn,S.M. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 211–241.
- 3.Hausen P., and Stein,H. (1970) Ribonuclease H, enzyme degrading the RNA moiety of DNA/RNA hybrids. Eur. J. Biochem., 14, 278–283. [DOI] [PubMed] [Google Scholar]
- 4.Hostomsky Z., Hostomska,Z. and Matthews,D. (1993) Ribonuclease H. In Linn,S.M. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 341–376.
- 5.Yang W., Hendrickson,W., Crouch,R.J. and Satow,Y. (1990) Structure of ribonuclease H phased at 2Å: resolution by MAD analysis of the selenomethionyl protein. Science, 249, 1398–1405. [DOI] [PubMed] [Google Scholar]
- 6.Katayanagi K., Miyagawa,M., Matsushima,M., Ishikawa,M., Kanaya,S., Ikehara,M., Matsuzaki,T. and Morikawa,K. (1990) Three-dimensional structure of ribonuclease H from E. coli. Nature, 347, 306–309. [DOI] [PubMed] [Google Scholar]
- 7.Katayanagi K., Miyagawa,M., Matsushima,M., Ishikawa,M., Kanaya,S., Nakamura,H., Ikehara,M., Matsuzaki,T. and Morikawa,K. (1992) Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution. J. Mol. Biol., 223, 1029–1052. [DOI] [PubMed] [Google Scholar]
- 8.Casregola S., Khidhir,M. and Holland,I.B. (1987) Effects of modulation on RNase H production on the recovery of DNA synthesis following UV-irradiation in Escherichia coli. Mol. Gen. Genet., 209, 494–498. [DOI] [PubMed] [Google Scholar]
- 9.Hong X., and Kogoma,T.J. (1993) Absence of a direct role for RNase H1 in initiation of DNA replication of oriC site on the Escherichia coli chromosome. J. Bacteriol., 175, 6731–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minshull J., and Hunt,T. (1986) The use of single-stranded DNA and RNase H to promote quantitative ‘hybrid arrest of translation’ of mRNA/DNA hybrids in reticulocyte lysate cell-free translations. Nucleic Acids Res., 14, 6433–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crooke S.T., (1999) Molecular mechanism of action of antisense drugs. Biochim. Biophys. Acta, 1489, 31–44. [DOI] [PubMed] [Google Scholar]
- 12.Walder R.T., and Walder,J.A. (1988) Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein C., and Cheng,Y. (1993) Antisense oligonucleotides as therapeutic agents—is the bullet really magical? Science, 261, 1004–1012. [DOI] [PubMed] [Google Scholar]
- 14.Cazenave C., Stein,C., Loreau,N., Thuong,N.T., Neckers,L., Subasinghe,C., Helene,C., Cohen,J. and Toulm,J. (1989) Comparative inhibition of rabbit globin mRNA translation by modified antisense oligodeoxynucleotides. Nucleic Acids Res., 17, 4255–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wengel J., Verbeure,B., Luyten,I., Lescrinier,R., Froeyen,M. and Hendrix,C. (2000) Cyclohexene nucleic acids (CeNA): serum stable oligonucleotides that activate RNase H and increase duplex stability. J. Am. Chem. Soc., 122, 8595–8602. [Google Scholar]
- 16.Noronha A.M., Wilds,C.J., Lok,C.N., Viazovkina,K., Arion,D., Parniak,M.A. and Damha,M.J. (2000) Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA·RNA hybrid duplexes. Biochemistry, 24, 7050–7062. [DOI] [PubMed] [Google Scholar]
- 17.Damha M.J., Wilds,C.J., Noronha,A., Brukner,I., Borkow,G., Arion,D. and Parniak,M. (1998) Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soc., 120, 12976–12977. [Google Scholar]
- 18.Nakamura H., Oda,Y., Iwai,S., Inoue,H., Ohtsuka,E., Kanaya,S., Kimura,S., Katsuda,C., Katayanagi,K., Morika-wa,K., Miyashiro,H. and Ikehara,M. (1991) How does RNase H recognize a DNA/RNA hybrid? Proc. Natl Acad. Sci. USA, 88, 11535–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedoroff O.Y., Ge,T. and Reid,B.R. (1993) Structure of a DNA/RNA hybrid duplex. Why RNase H does not cleave pure RNA. J. Mol. Biol., 233, 509–523. [DOI] [PubMed] [Google Scholar]
- 20.Lima W.F., and Crooke,S.T. (1997) Binding affinity and specificity of E. coli ribonuclease H1: impact on the kinetic catalysis of antisense oligonucleotide-RNA hybrids. Biochemistry, 36, 390–396. [DOI] [PubMed] [Google Scholar]
- 21.Ratmeyer L., Vinayak,R., Zhong,Y., Zon,G. and Wilson,W. (1994) Sequence specific thermodynamic and structural properties for DNA:RNA duplexes. Biochemistry, 33, 5298–5304. [DOI] [PubMed] [Google Scholar]
- 22.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Solution structure of DNA/RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- 23.Smart B.E., (1994) Characteristics of CF compounds. In Banks,R.E (ed.), Organofluorine Chemistry: Principles and Commercial Applications. Plenum Press, New York, NY, pp. 72–93.
- 24.Guschilbauer W., Blandin,M., Drocourt,J.L. and Thang,M.N. (1977) Poly-2′-deoxy-2′-fluoro-cytidynic acid: enzymatic synthesis, spectroscopic characterization and interaction with poly-inosinic acid. Nucleic Acids Res., 4, 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakiuchi N., Marek,C., Rousseau,N., Leng,M., De Clereq,E. and Guschilbauer,W. (1982) Polynucleotide helix geometry and stability. Spectroscopic, antigenic and interferon-inducing properties of deoxyribose-, ribose-, or 2′-deoxy-2′-fluororibose-containing duplexes of poly(inosinic acid).poly(cytidylic acid). J. Biol. Chem., 257, 1924–1928. [PubMed] [Google Scholar]
- 26.Kawasaki A.M., Casper,M.D., Freiser,S.M., Lesnik,E.A., Zounes,M.C., Cummins,L.L., Gonzalez,C. and Cook,P.D. (1993) Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem., 36, 831–841. [DOI] [PubMed] [Google Scholar]
- 27.Howard J.K., Hoy,V.J., O’Hagan,D. and Smith,G.T. (1996) How good is fluorine as a hydrogen bond acceptor? Tetrahedron, 52, 12613–12619. [Google Scholar]
- 28.Damha M.J., Giannaris,P.A. and Zabarylo,S.V. (1990) An improved procedure for derivitization of controlled-pore glass beads for solid-phase oligonucleotide synthesis. Nucleic Acids Res., 18, 3813–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damha M.J., and Ogilvie,K.K. (1993) Oligoribonucleotide synthesis. In Agrawal,S. (ed.), Methods in Molecular Biology, Protocols for Oligonucleotides and Analogues: Synthesis and Properties. Humana Press, Totowa, NJ, Vol. 20, pp. 81–114. [DOI] [PubMed]
- 30.Kibler-Herzog L., Zon,G., Whittier,G., Shaikh,M. and Wilson,W.D. (1993) Stabilities of duplexes and triplexes of dA18 and dT19 with alternating methylphosphonate and phosphodiester linkages. Anticancer Drug Des., 8, 63–68. [PubMed] [Google Scholar]
- 31.Puglisi J.D., and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 32.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. John Wiley, New York, NY.
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Egli M., Portmann,S. and Usman,N. (1996) RNA hydration: a detailed look. Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 35.Berger I., Tereshko,V., Ikeda,H., Marquez,V.E. and Egli,M. (1998) Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res., 26, 2473–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiyama Y., Miura,Y., Inoue,H., Ohtsuka,E., Ueno,Y., Ikehara,M. and Iwai,S. (1994) Studies of the interactions between Escherichia coli ribonuclease H1 and its substrate. J. Mol. Biol., 243, 782–791. [DOI] [PubMed] [Google Scholar]
- 37.Iwai S., Kataoka,S., Wakasa,M., Ohtsuka,E. and Nakamura,H. (1995) Recognition of 2′-hydroxyl groups by Escherichia coli ribonuclease H1. FEBS Lett., 368, 315–320. [DOI] [PubMed] [Google Scholar]
- 38.Hogrefe H.H., Hogrefe,R.I., Walder,R.Y. and Walder,J.A. (1990) Kinetic analysis of E. coli RNase H using DNA-RNA-DNA/DNA substrates. J. Biol. Chem., 265, 5561–5566. [PubMed] [Google Scholar]
- 39.Crooke S.T., Lemonidis,K.M., Neilson,L., Griffey,R., Lesnik,E.A. and Monia,B.P. (1995) Kinetic characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide-RNA duplexes. Biochem. J., 312, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen L.X., Kindimalla,E.R. and Agrawal,S. (1998) Impact of mixed-backbone oligonucleotides on target binding affinity and target cleavage specificity and selectivity by Escherichia coli RNAse H. Bioorg. Med. Chem., 6, 1695–1705. [DOI] [PubMed] [Google Scholar]
- 41.Kanaya E., and Kanaya,S. (1995) Kinetic analysis of Escherichia coli ribonuclease H1 using oligomeric DNA/RNA substrates suggests an alternative mechanism for the interaction between the enzyme and the substrate. Eur. J. Biochem., 231, 557–562. [DOI] [PubMed] [Google Scholar]
- 42.Roberts G.C., Dennis E.A., Meadows,D.H., Cohen,J.S. and Jardetzky,O. (1969) The mechanism of action of ribonuclease. Proc. Natl Acad. Sci. USA, 62, 1151–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agrawal S., Maynard,S.H., Zamecnik,P.C. and Pederson,T. (1990) Site specific excision from RNA by RNase H and mixed-phosphate-backbone oligonucleotides. Proc. Natl Acad. Sci. USA, 87, 1401–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima W.F., and Crooke,S.T. (1997) Cleavage of single strand RNA adjacent to RNA-DNA duplex regions by Escherichia coli RNase H1. J. Biol. Chem., 272, 27513–27516. [DOI] [PubMed] [Google Scholar]
- 45.Iwai S., Wakasa,M., Ohtsuka,E., Kanaya,S., Kidera,A. and Nakamura,H. (1996) Interaction of the basic protrusion of Escherichia coli ribonuclease H1 with its substrate. J. Mol. Biol., 263, 699–706. [DOI] [PubMed] [Google Scholar]
- 46.Li J., and Wartell,R.M. (1998) RNase H1 can catalyze RNA/DNA formation and cleavage with stable hairpin or duplex DNA oligomers. Biochemistry, 37, 5154–5161. [DOI] [PubMed] [Google Scholar]
- 47.Lane A.N., Ebel,S. and Brown,T. (1993) NMR assignments and solution conformation of a DNA-RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur. J. Biochem., 215, 297–306. [DOI] [PubMed] [Google Scholar]
- 48.Salazar M., Fedoroff,O.Y., Miller,J.M., Ribeiro,N.S. and Reid,B.R. (1993) The DNA strand in DNA/RNA hybrid duplex is neither B-form nor A-form in solution. Biochemistry, 32, 4207–4215. [DOI] [PubMed] [Google Scholar]
- 49.Kanaya S., Oobatake,M. and Liu,Y.Y. (1996) Thermal stability of Escherichia coli ribonuclease H1 and its active site mutants in the presence and absence of the Mg2+ ion. Proposal of a novel catalytic role for glu-48. J. Biol. Chem., 271, 32729–32736. [DOI] [PubMed] [Google Scholar]
- 50.Oda Y., Yoshida,M. and Kanaya,S. (1993) Role of His124 in the catalytic function of ribonuclease H from Escherichia coli. J. Biol. Chem., 268, 88–92. [PubMed] [Google Scholar]
- 51.Kanaya S., Kohara,A., Miura,Y., Sekiguchi,A., Iwai,S., Inoue,H., Ohtsuka,E. and Ikehara,M. (1990) Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J. Biol. Chem., 265, 4615–4621. [PubMed] [Google Scholar]
- 52.Haruki M., Tsunaka,Y., Morikawa,M., Iwai,S. and Kanaya,S. (2000) Catalysis by Escherichia coli ribonuclease H1 is facilitated by a phosphate group of the substrate. Biochemistry, 39, 13939–13944. [DOI] [PubMed] [Google Scholar]