Abstract
The introduction of cationic 5-(ω-aminoalkyl)-2′-deoxypyrimidines into duplex DNA has been shown to induce DNA bending. In order to understand the energetic and hydration contributions for the incorporation of a cationic side chain in DNA a combination of spectroscopy, calorimetry and density techniques were used. Specifically, the temperature unfolding and isothermal formation was studied for a pair of duplexes with sequence d(CGTAGUCG TGC)/d(GCACGACTACG), where U represents 2′-deoxyuridine (‘control’) or 5-(3-aminopropyl)-2′-deoxyuridine (‘modified’). Continuous variation experiments confirmed 1:1 stoichiometries for each duplex and the circular dichroism spectra show that both duplexes adopted the B conformation. UV and differential scanning calorimetry melting experiments reveal that each duplex unfolds in two-state transitions. In low salt buffer, the ‘modified’ duplex is more stable and unfolds with a lower endothermic heat and lower release of counterion and water. This electrostatic stabilization is entropy driven and disappears at higher salt concentrations. Complete thermodynamic profiles at 15°C show that the favorable formation of each duplex results from the compensation of a favorable exothermic heat with an unfavorable entropy contribution. However, the isothermal profiles yielded a differential enthalpy of 8.8 kcal/mol, which is 4.3 kcal/mol higher than the differential enthalpy observed in the unfolding profiles. This indicates that the presence of the aminopropyl chain induces an increase in base stacking interactions in the modified single strand and a decrease in base stacking interactions in the modified duplex. Furthermore, the formation of the ‘control’ duplex releases water while the ‘modified’ duplex takes up water. Relative to the control duplex, formation of the modified duplex at 15°C yielded a marginal differential ΔG° term, positive ΔΔHITC–Δ(TΔS) compensation, negative ΔΔV and a net release of counterions. The opposite signs of the differential enthalpy–entropy compensation and differential volume change terms show a net uptake of structural water around polar and non-polar groups. This indicates that incorporation of the aminopropyl chain induces a higher exposure of aromatic bases to the solvent, which may be consistent with a small and local bend in the ‘modified’ duplex.
INTRODUCTION
One of the most important physical properties of DNA is its polyelectrolyte behavior; for instance, the repulsive forces between its charges play a role in its conformation (1). Charge repulsion favors extended conformations and as a result DNA is considered a stiff molecule with a persistence length of ∼160 bp that changes if its charge distribution is altered (1). However, this stiff polymer is found wrapped 1.7 times around histone proteins (2). The cationic amino acid residues of histone proteins have been postulated to contribute to DNA bending by neutralizing the phosphate charges via ion pairing (3,4). Furthermore, experimental evidence has shown that phosphate neutralization can also induce DNA bending (3) and theoretical investigations have suggested that an unbalanced electrostatic repulsion between the remaining phosphate charges could lead to a spontaneous collapse of DNA (5). Thus, the incorporation of bases bearing positive charges into DNA may well simulate the electrostatic effects of positively charged protein side chains by neutralizing some of the phosphate negative charges.
Despite numerous reports and current interest in elucidating the mechanisms leading to increased DNA flexibility, the molecular forces involved in DNA bending remain unclear. Some reports have shown that partial replacement of the negatively charged phosphates with neutral methylphosphonates phased with an intrinsically bent A-tract of DNA resulted in significant bending of DNA (3). On the other hand, the neutralization of phosphate charges by tethering ammonium cations to pyrimidine bases resulted in modest bending (6,7). The comparison of these systems suggests that a number of questions remain unanswered. What is the nature of the different behavior in these systems? Does the inclusion of a cationic chain neutralize the local negative phosphate of duplex DNA? If so, what is the role of cations and water in this partial neutralization of charges? Does the overall effect change the DNA conformation by inducing a local bend?
Our current interest is in elucidating the energetic and hydration contributions resulting from the incorporation of tethered ammonium cations in duplex DNA. In this work, we used a variety of physical techniques to determine the unfolding (and formation) energetics and hydration properties of DNA duplexes, shown in Scheme 1, containing a single uridine residue modified with a cationic aminopropyl group. Complete thermodynamic profiles indicate that the favorable formation of each duplex results from the characteristic compensation of a favorable enthalpy with an unfavorable entropy term. The formation of the ‘control’ duplex releases water while the ‘modified’ duplex takes up water and both duplexes yielded uptake of counterions. These profiles allowed us to set up a thermodynamic cycle in which the folding of the ‘control’ duplex is used as the reference reaction. Analysis of this cycle yielded: ΔΔG° = –0.4 kcal/mol, ΔΔHITC = 8.8 kcal/mol, Δ(TΔS) = 9.2 kcal/mol, ΔΔV = –51.4 cm3/mol and ΔΔnNa+ = 0.6 mol Na+/mol duplex. The differential enthalpy–entropy compensation has an opposite sign to the differential volume change. This shows a net uptake of structural water, which is consistent with the small DNA bend induced by the incorporation of the cationic chain in the ‘modified’ duplex.
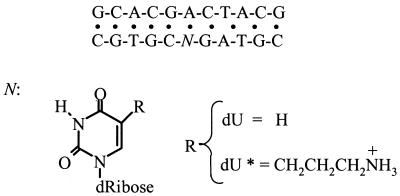
Scheme 1. Sequence of deoxyribonucleotide and structure of base modifications.
MATERIALS AND METHODS
Materials
The zwitterionic phosphoramidite of 5-(3-aminopropyl)-2′-deoxyuridine was synthesized and incorporated into deoxynucleotides as described previously (8,9). The three oligonucleotides were synthesized by the Core Synthetic Facility of the Eppley Research Institute at UNMC, with an ABI synthesizer, and purified by reverse phase HPLC. The solution concentration of oligonucleotides was determined optically at 260 nm and 80°C using the following molar extinction coefficients [per (mM-cm)–1 of strands]: 110 for d(GCACG ACTACG); 108 for d(CGTAGUCGTGC); 130 for d(CGT AGU*CGTGC). These values were calculated by extrapolation of the tabulated values of the dimers and monomer bases (10) at 25°C to high temperatures using procedures reported earlier (11). The uridine contribution was calculated from the RNA values of GU, UG and U, using appropriate comparisons and extrapolations with the corresponding DNA values. To include the chemical modification of uridine, the extinction coefficient of the modified strand has been corrected in continuous variation experiments, as described in the next section. Unless otherwise indicated, all measurements were performed in buffer solutions consisting of 10 mM Na HEPES, adjusted to pH 7.5. Hereafter the single strands are designated as follows: d(GCACGACTACG), ‘common’; d(CGTAGU CGTGC), ‘control’; d(CGTAGU*CGTGC), ‘modified’. The duplex containing the modification is designated the ‘modified’ duplex and that without the modification the ‘control’ duplex.
Duplex stoichiometry
The stoichiometry of each duplex was checked in continuous variation experiments (Job plots) using a Perkin-Elmer Lambda 10 spectrophotometer. In these experiments, the absorbance at 260 and 275 nm of several solutions with similar total strand concentrations (∼10 µM) was measured as a function of the mole fraction of one strand. Plots of the absorbance versus the molar fraction resulted in typical ‘V’ shaped plots, and the intersections of the resulting lines yielded a molar fraction that corresponded to duplex stoichiometry. We also used these curves to confirm the calculated extinction coefficients of all strands, especially of the ‘modified’ strand.
Circular dichroism (CD) spectroscopy
The conformation of each single strand and of the 1:1 duplexes was obtained by simple inspection of their CD spectra. The CD spectra were obtained on a thermoelectrically controlled CD spectrometer Model 202-SF (Aviv, Lakewood, NJ) from 400 to 200 nm at a wavelength step of 1 nm and using a 10 mm quartz cuvette with an absorbance value of 1 at 260 nm. Typical spectra corresponded to the average of at least three scans.
Temperature-dependent UV spectroscopy
Absorbance versus temperature profiles (UV melting curves) for each duplex were measured at 260 nm with a thermoelectrically controlled 14-DS spectrophotometer (Aviv) as a function of strand, salt and osmolyte concentration. The temperature was scanned at a heating rate of ∼0.6°C/min. The analysis of the shape of the melting curves yielded transition temperatures (Tm), which are the midpoint temperatures of the order–disorder transitions, and model-dependent van’t Hoff enthalpies. More accurate van’t Hoff enthalpies were obtained from the slopes of the lines of 1/Tm versus ln(CT/4) plots, according to the equation for non-self-complementary oligonucleotides (12): 1/TM = R/ΔHvH ln(CT/4) + ΔS/ΔHvH. The enthalpy and entropy values are obtained from the slope and intercept of the resulting line, respectively.
UV melting curves were carried out in the range 5–205 mM NaCl to determine the thermodynamic release of counterions (ΔnNa+) in the unfolding of each duplex. The ΔnNa+ term is determined with the following relationship: ΔnNa+ = –1.11(ΔHcal/RTm2)∂Tm/∂ln[Na+] (13), where 1.11 is a proportionality constant for converting ionic activities into concentrations and ∂Tm/∂ln[Na+] is the slope of the lines of the Tm versus ln[Na+] plots. The term in parentheses is a constant measured in differential scanning calorimetric experiments and R is the universal gas constant.
Similarly, the thermodynamic uptake (or release) of water (ΔnW) was obtained from the dependence of Tm on water activity according to the equation: ΔnW = – (ΔHcal/RTm2)∂Tm/∂lnaW (14), where ∂Tm/∂lnaW corresponds to the slope of the line of the Tm versus lnaW plots. The activity of water (aW) in these solutions is changed by using different concentrations of ethylene glycol, as this co-solute does not interact specifically with DNA (14). The osmolality of each solution was obtained with a vapor pressure osmometer Model 070 (UIC, Joliet, IL), which was calibrated with standardized solutions of NaCl.
Differential scanning calorimetry (DSC)
Heat capacity functions of the helix coil transition of each duplex were measured with a differential scanning calorimeter Model MC-2 (Microcal, Northampton, MA). Two cells, the sample cell containing 1.6 ml of a duplex solution (∼300 µM in total strands) and the reference cell filled with the same volume of buffer solution, were heated from 10 to 100°C at a heating rate of 0.75°C/min. Analysis of the resulting thermograms, using procedures reported earlier (12) and assuming a negligible heat capacity effect, yielded standard thermodynamic profiles (ΔHcal, ΔScal and ΔG°cal). ΔHcal and ΔScal were measured from the DSC curves according to the following equations: ΔHcal = ∫ΔCpdT and ΔScal = ∫ΔCp/TdT, respectively. ΔG°cal was calculated at 15°C from the Gibbs relationship: ΔG°cal = ΔHcal – TΔScal (12). Shape analysis of the experimental DSC curve allows us to calculate ΔHvH enthalpies according to the equation (12): ΔHvH = 10.14/[(1/T1) – (1/T2)], where 10.14 is a constant for bimolecular transitions, T1 and T2 correspond to the lower and upper temperatures, respectively, at half height of the ΔCp maximum in the ΔCp versus T curve. The ΔHvH/ΔHcal ratio allows us to inspect if duplex unfolding takes place in two-state transitions or through the formation of intermediates. If the ΔHvH/ΔHcal ratio is equal to 1 then the transition takes place in an all or none fashion (12).
Isothermal titration calorimetry (ITC)
The measurement of the heat for mixing a single strand with its complementary strand was carried out with the Omega titration calorimeter from Microcal at 15°C; at this temperature each duplex forms completely. A solution of the common strand was used to titrate each of the complementary strands. A 100 µl syringe was used for the titrant and complete mixing was accomplished by stirring the syringe paddle at 400 r.p.m. The strand concentration in the syringe was generally 25 times higher than the concentration of the complementary strand (∼10 µM) in the reaction cell. These experiments were designed only to obtain the heat evolved upon the folding of each duplex.
Density measurements
We used density techniques to measure the apparent molar volume, for each single strand and duplex, and the volume change for the folding of each duplex at 15°C. The apparent molar volume (ΦV) was calculated using the equation (15): ΦV = M/ρ – [(ρ – ρo)/(ρoC)], where ρo and ρ are the density of the solvent and solution, respectively, and M is the molecular weight of the single strand or duplex. The molar volume change (ΔV) accompanying the folding of a duplex is calculated from the equation (16): ΔV = (m1ρ1 + m2ρ2 – m12ρ12)/(m1ρ1C1), where m and ρ are the mass and density, respectively, of each single strand (subscript 1 or 2) and duplex solutions (subscript 12). All solutions were prepared by weight and their masses were measured with a Mettler microbalance with extreme precautions taken to prevent evaporation of the weighed samples. The density of each single strand, or duplex, solution was measured with a DMA-602 densimeter (Anton Paar, Graz, Austria) with two 0.2 ml cells in a differential set-up. In these batch experiments, one of the strands with a concentration of ∼340 µM was considered as the limiting reagent. Appropriate annealing of duplexes was ensured by heating the 1:1 duplex solutions to 50°C and cooling slowly to the experimental temperature.
The physical meaning of the apparent molar volume (ΦV) is based on the following relationship (17): ΦV = Vm + ΔVh, where Vm is the intrinsic molar volume of the solute molecule and ΔVh is its hydration contribution. The latter contribution corresponds to the change in volume of hydrating water, resulting from the interaction of a solute with water, and the void volume between the solute molecule and that of the surrounding water. The Vm value for simple hydrophilic molecules and oligonucleotides, without significant inner cavities, remains constant in the absence of significant conformational changes (18). The hydration effects that take place in the folding of a non-self-complementary duplex are simply reflected in the changes of ΦV (ΔΦV). Therefore, the ΔΦV (or ΔV) values are simply obtained from their hydration contributions, i.e. ΔV = ΔΔVh. This hydration contribution results from differences in the molar properties of hydrating water; for instance, the molar volume of water around a charge is considered to be ∼14% smaller than bulk water (19).
Solvent-accessible surfaces were modeled with MOLCAD, which is a subprogram in SYBYL (Tripos Associates, St Louis, MO).
RESULTS
Duplex stoichiometry
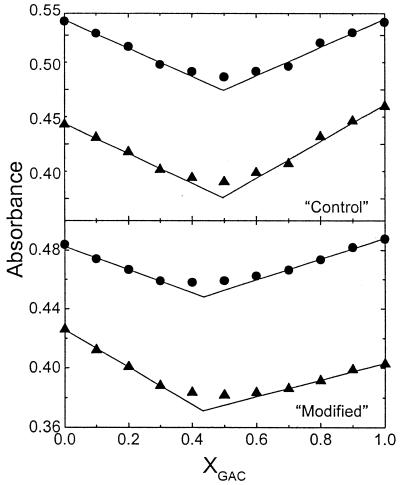
Continuous variation experiments were used to determine duplex stoichiometries, as shown in Figure 1. At these two wavelengths the lines intercepted at mole fractions of 0.492 (‘control’) and 0.440 (‘modified’), which corresponded to duplex stoichiometries of 1:1 and 1:1.25, respectively. The deviation from 1:1 stoichiometry of the modified duplex clearly indicates that the presence of the chemically modified uridine has affected its molar extinction coefficient. The resulting stoichiometry of the unmodified duplex was used to correct this value by recalculating its extinction coefficient to yield a 1:1 stoichiometry. This correction changed the extinction coefficient from 108 to 130 (mM-cm)–1.
Figure 1.
Continuous variation experiments (Job plot) for the determination of duplex stoichiometry in 10 mM HEPES buffer, pH 7.5, at 19°C. The wavelengths used are 260 (circles) and 275 nm (triangles).
CD spectroscopy
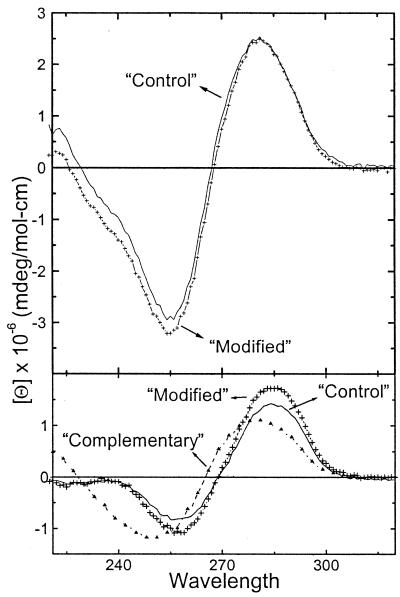
The CD spectra of each single strand and duplex at 5°C are shown in Figure 2. The 1:1 duplexes exhibit CD spectra characteristic of right-handed helices in the B conformation. The spectra similarity indicates that the incorporation of the cationic aminopropyl chain does not alter the overall DNA conformation. In addition, the sum of the single strands spectra at high temperature superimposes with the spectra of the corresponding unfolded duplex at high temperature (data not shown). These results confirm the formation of 1:1 duplexes and the accuracy of the calculated molar extinction coefficients.
Figure 2.
CD spectra of duplexes (top) and single strands (bottom) in 10 mM HEPES buffer, pH 7.5, at 5°C.
Overall hydration properties of single strands and duplexes at 15°C
Density techniques were used to measure the apparent molar volume of each strand and duplex at 15°C. These ΦV values (in cm3/nt) are as follows: ‘common’ strand, 94.7; ‘control’ strand, 101.5; ‘modified’ strand, 119.3; ‘control’ duplex, 99.9; ‘modified’ duplex, 106.3. Since ΦV includes contributions from the van der Waals volume and the hydration volume, the resulting positive values are consistent with the size of their average molar masses, which are proportional to their intrinsic molar volume. The overall hydration effects are small, however, the magnitude of the ΦV values suggests that the modified strand is more hydrated than the other two strands and that the modified duplex is slightly more hydrated than the control duplex. The contribution of water in the hydration shell depends on the type of hydrating water: structural around organic moieties and non-polar atomic groups or electrostricted around charged and polar atomic groups (20–22). In the case of nucleic acids, the single-stranded bases are more exposed to the solvent; therefore, on average, the single strands have a slightly higher structural hydration while the duplexes have a higher electrostricted hydration. In the formation of a duplex from mixing its complementary strands, these hydration contributions tend to compensate each other.
UV unfolding of duplexes
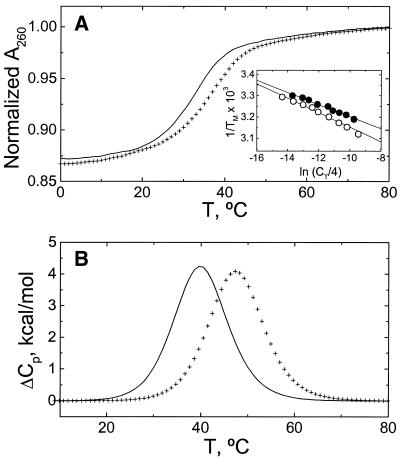
The UV melting curves of the 1:1 duplexes at 260 nm and their Tm dependences on strand concentration are shown in Figure 3A. Both melting curves follow the characteristic sigmoidal behavior for the unfolding of a nucleic acid duplex. The molecules are in the duplex state up to 18°C, which allows us to choose 15°C as the experimental temperature for volumetric and isothermal calorimetric measurements. At low salt concentration and at a total strand concentration of 300 µM, the Tm dependence on strand concentration, as shown in the inset of Figure 3A, yielded Tm values of 39.7°C (‘control’ duplex) and 45.7°C (‘modified’ duplex). The slopes of these plots yielded ΔHVH values of 71 (± 10) kcal/mol (control) and 59 (± 9) kcal/mol (modified), which are in good agreement with the van’t Hoff enhalpies obtained from the shape of the melting curves (data not shown).
Figure 3.
Thermodynamic characterization of the helix–coil transition of duplexes in 10 mM HEPES buffer, pH 7.5. Line, ‘control’ duplex; crosses, ‘modified’ duplex. (A) Typical UV melting curves at a strand concentration of 25 µM. (Inset) Tm dependence on total strand concentration. Filled circles, ‘control’; open circles, ‘modified’. (B) DSC curves at oligonucleotide strand concentrations of ∼300 µM.
Calorimetric unfolding of duplexes
Typical DSC melting curves for each duplex at similar total strand concentrations of 300 µM and low salt concentration are shown in Figure 3B and the resulting unfolding thermodynamic profiles are summarized in Table 1. Both DSC curves are broad and monophasic. The Tm value of the modified duplex is higher by 6°C, which indicates that the incorporation of a single aminopropyl chain increases the thermal stability of the duplex at this salt concentration. Another observation is that we do not measure heat capacity effects between the initial and final states, which is consistent with our assumption in calculating unfolding enthalpies and entropies. However, if apparent heat capacity effects are present, then these are buried in the experimental curves, which have a noise level of 150 cal/°C mol duplex. Inspection of Table 1 shows that the melting of each duplex is accompanied by unfavorable free energy terms resulting from the characteristic compensation of unfavorable enthalpy and favorable entropy terms. The ΔHvH values obtained from the shape of the DSC curves are in good agreement with the enthalpies obtained from UV melting curves. Furthermore, we obtained ΔHvH/ΔHcal ratios of 1.02 (‘control’) and 1.04 (‘modified’) that confirm that each duplex is unfolding in a two-state transition.
Table 1. Thermodyamic profiles for the unfolding of duplexes.
| |
Differential scanning calorimetry |
UV melting curves |
||||||
|---|---|---|---|---|---|---|---|---|
| Tm (°C) | ΔG°cal (kcal/mol) | ΔHcal (kcal/mol) | ΔHvH (kcal/mol) | TΔScal (kcal/mol) | ΔHvH (kcal/mol) | ΔnNa+ (per duplex) | ΔnW (per duplex) | |
| ‘Control’ duplex | 39.7 | 5.4 | 68.6 | 70 | 63.2 | 71 | 2.4 | 24.4 |
| ‘Modified’ duplex | 45.7 | 6.2 | 64.5 | 67 | 58.3 | 59 | 1.9 | 21.5 |
All experiments were done in 10 mM HEPES buffer, pH 7.5. The Tm (± 0.5°C) corresponds to a total strand concentration of 300 µM. Experimental errors for each parameter are as follows: ΔHcal, 3%; ΔG°cal and TΔScal, 5%; ΔHvH, 10%; ΔnNa+, 6%; ΔnW, 8%.
Thermodynamic release of counterions
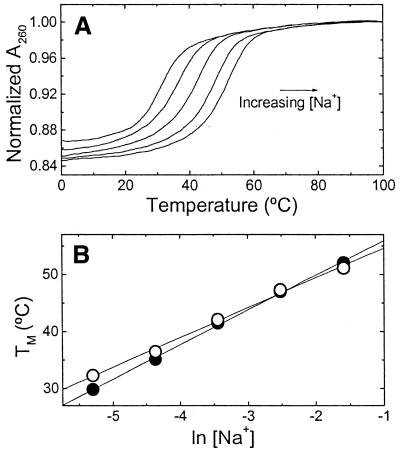
UV melting curves at several salt concentrations are shown in Figure 4A, the increase in the salt concentration resulted in a shift of the curves to higher temperatures. This is the characteristic effect of salt stabilizing the duplex state with a higher negative charge density parameter. The Tm dependence on salt concentration is shown in Figure 4B for each duplex. The lines cross at a salt concentration of 80 mM, which indicates that below this salt concentration the unmodified duplex is more thermally stable, while at higher salt concentrations the control duplex is more stable. The slopes of these lines are 6.1°C (‘control’) and 5.2°C (‘modified’). Over this range of salt concentration we obtained counterion releases of 2.38 (‘control’) and 1.86 mol Na+/mol duplex (‘modified’). The overall effect is that the presence of the cationic aminopropyl chain significantly decreases the overall counterion release by 0.52 mol Na+/mol duplex.
Figure 4.
UV melting curves of oligonucleotides, at a constant strand concentration of 5 µM, as a function of salt concentration in 10 mM HEPES buffer, pH 7.5. (A) Typical melting curves over the NaCl concentration range 5–205 mM. (B) Dependence of Tm on salt concentration. Filled circles, ‘control’; open circles, ‘modified’.
Thermodynamic release of water
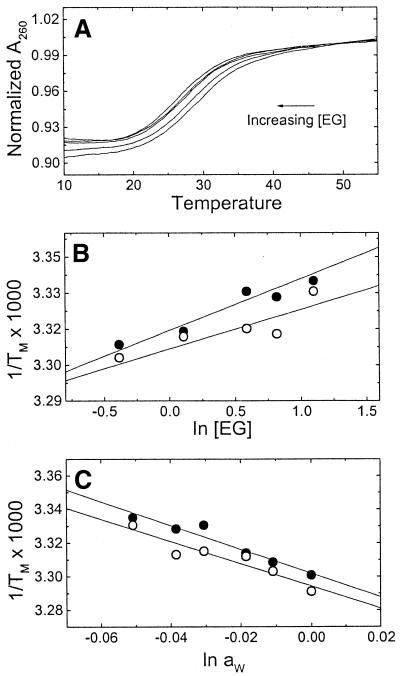
UV melting curves at several concentrations of ethylene glycol in 10 mM HEPES buffer are shown in Figure 5A. The increase in osmolyte concentration shifts the melting curves to lower temperatures. The 1/Tm dependence on osmolyte concentration is shown in Figure 5B for each duplex. The osmolarity values were converted to water activity using a calibration curve determined with a vapor pressure osmometer and the resulting curves for the dependence of 1/Tm on water activity are shown in Figure 5C. The slopes of these lines are 7.1 × 10–4 (‘control’ duplex) and 6.6 × 10–4°C (‘modified’ duplex), yielding water releases of 24.4 (‘control’ duplex) and 21.5 mol H2O/mol duplex (‘modified’ duplex). The increase in osmolyte concentration lowers the activity of water, which in turn shifts the helix–coil equilibrium towards the least hydrated state; therefore, the incorporation of the cationic aminopropyl chain slightly decreases the release of water molecules.
Figure 5.
UV melting curves of oligonucleotides, at a constant strand concentration of 3 µM, as a function of osmolyte concentration in 10 mM HEPES buffer, pH 7.5. (A) Typical melting curves of the ‘control’ duplex over the ethylene glycol molal concentration range 0.6–3 mol/kg of solvent. (B) Dependence of 1/Tm on ethylene glycol concentration. Filled circles, ‘control’; open circles, ‘modified’. (C) Dependence of 1/Tm on water activity. Symbols as in the previous plot.
Thermodynamic profiles for the formation of duplexes
In order to obtain complete thermodynamic profiles for the formation of each duplex at 15°C, titration and batch experiments were performed to obtain directly the ΔHITC and ΔV parameters, respectively. The resulting parameters are shown in Table 2. We measured exothermic enthalpies for the formation of each duplex; the ‘control’ duplex yielded a higher heat by 8.8 kcal/mol duplex. However, these enthalpies are lower in magnitude than the corresponding unfolding enthalpies, by 2.9 (‘control’) and 7.6 kcal/mol (‘modified’). The reason for this discrepancy is that at low temperatures the contributions of base–base stacking interactions of the single strands need to be considered, as has been shown earlier with other duplexes (22–25). The overall effect indicates that at 15°C the ‘modified’ strand has a larger endothermic contribution than the ‘control’ strand and means that base stacking interactions in the ‘modified’ strand are more pronounced or simply this particular strand is more ordered.
Table 2. Thermodynamic profiles for the folding of duplexes at 15°C.
| ΔHITC (kcal/mol) | ΔV (cm3/mol) | ΔG° (kcal/mol) | TΔS (kcal/mol) | ΔnNa+ (per duplex) | |
|---|---|---|---|---|---|
| ‘Control’ duplex | –65.7 | 34.7 | –5.2 | –60.5 | –2.3 |
| ‘Modified’ duplex | –56.9 | –16.7 | –5.5 | –51.4 | –1.7 |
All experiments were done in 10 mM HEPES buffer, pH 7.5. The ΔG° term was corrected by the ΔHITC/ΔHcal factor while the TΔS term was calculated from the Gibbs relationship. Experimental errors for each parameter are as follows: ΔHITC, 3%; ΔV, 5%; ΔG° and TΔS, 5%; ΔnNa+, 6%.
Furthermore, the formation of the control duplex yielded a volume expansion (positive ΔV), while the modified duplex yielded a volume contraction (negative ΔV). The ΔV term at constant temperature and pressure results from the net change in the molar volume of water and is essentially the net change in compression of the water dipoles in response to intermolecular solute–solute interactions. Thus, the equilibrium ΔV values are indicative of a net release of water molecules (‘control’) and uptake of water (‘modified’).
In order to discuss the heat and hydration effects appropriately and to correlate them with standard thermodynamic profiles, the ΔG°cal, TΔScal and ΔnNa+ unfolding parameters were extrapolated to 15°C by using the enthalpy factor, ΔHITC/ΔHcal (25,26). This correction factor takes into account the contribution of single-strand stacking interactions. For instance, extrapolated parameters were obtained with the relationships: ΔG° = ΔG°cal(ΔHITC/ΔHcal) and TΔS = ΔHITC – ΔG°. Complete profiles for the formation of duplexes at 15°C are shown in Table 2. The favorable folding of each duplex results from the characteristic compensation of a favorable enthalpy with an unfavorable entropy term. The favorable enthalpy term corresponds to exothermic contributions of base pairing and base pair stacking interactions and endothermic contributions of the release of electrostricted water molecules and/or uptake of structural water. The unfavorable entropy contributions are the result of the conformational rearrangement of the bimolecular association of two strands and the uptake of counterions and uptake of water molecules in the case of the ‘modified’ duplex.
DISCUSSION
Duplex design and stoichiometry
In this work, the sequences of the oligonucleotide strands were chosen to form non-self-complementary duplexes exclusively. UV melting curves of the individual single strands confirmed that these strands do not form intramolecular hairpin structures (data not shown); however, their hyperchromicities below 20°C are above the characteristic 3% changes of a random coil. This indicates some degree of base stacking interactions of the single strands at low temperatures.
The ‘modified’ duplex contained an aminopropyl chain at the C5 position of a pyrimidine base, which is a convenient position for the attachment of chemical groups of different sizes without adversely affecting the formation of a duplex (27). The modified uridine was placed in the middle of the duplex to ensure that interactions of this substituent with DNA atomic groups take place in either the 5′ or 3′ direction. However, earlier electrostatic footprinting (28) and molecular modeling calculations have indicated that this aminopropyl chain faces the major groove in the 3′ direction of the modified strand (29,30). This observation has been confirmed recently in NMR investigations of the solution structure of a DNA dodecamer duplex containing this base modification (31).
We have checked the molar extinction coefficients of the single strands in order to obtain accurate solution concentration of the strands and duplexes, which in turn would yield accurate thermodynamic parameters. The presence of the aminopropyl chain affected the extinction coefficient of the modified strand, yielding deviations from a 1:1 stoichiometry. The overall effect is due to the electron withdrawing properties of the aminopropyl chain that is affecting the electron delocalization of the uridine aromatic ring. This molar extinction coefficient was corrected and doubled checked in the CD experiments. We obtained a superimposition of the CD spectrum of the melted 1:1 duplex, at 70°C, with the CD spectrum of the sum of the individual strands.
Thermodynamic unfolding profiles
In the temperature unfolding of a nucleic acid duplex, it is important to take into account all the physical and chemical contributions from each of the participating species. These contributions include: base pairing and base pair stacking of the duplex at low temperatures, base–base stacking of the single strands at high temperatures and the differences in the counterion and water binding between the duplex and single stranded states. Complete thermodynamic profiles for the unfolding of each duplex are shown in Table 1. The unfolding of each duplex is accompanied by unfavorable free energy terms, a compensation of an unfavorable enthalpy with a favorable entropy contribution and the release of counterions and water molecules. The observed endothermic enthalpies result primarily from the endothermic heats for disrupting base pairs and base pair stacks of the duplex, relative to the remaining base–base stacking of the single strands at 80°C, if any. The release of electrostricted water by the duplex and uptake of structural water by the single strands may also contribute with endothermic heats (32,33). However, the release of counterions from the duplex is normally considered to have a negligible heat contribution (34). The favorable entropy contributions in the unfolding of a duplex correspond to the bimolecular dissociation of a duplex into two strands and the net release of counterions and water molecules.
To discuss the physical effects for the incorporation of a cationic chain in DNA, we initially used the thermodynamic unfolding profiles, which allowed us to set up a thermodynamic cycle. In this Hess cycle, the contribution of the ‘common’ strand cancels out exactly; the species with an asterisk represent the ‘modified’ strand or duplex while the species without an asterisk represent ‘control’ strand or duplex. The resulting reaction is shown below:
duplex(aNa+, vH2O, 15°C) + strand*(bNa+, xH2O, 80°C) → duplex*(cNa+, yH2O, 15°C) + strand(dNa+, zH2O, 80°C) + ΔΔnNa+Na+ + ΔΔnWH2O
In this reaction, each duplex or single strand contains both associated counterions and water molecules at the indicated temperatures. The ΔΔnNa+ and ΔΔnW terms correspond to the differential ion and water releases, respectively. The first row of Table 3 shows the resulting differential thermodynamic profile pertaining to the above reaction. In 10 mM HEPES buffer (or 5 mM Na+), the higher Tm value of the modified duplex, by 6°C, corresponds to a small favorable ΔΔG°cal term. The unfavorable ΔΔHcal term is also small and corresponds to differences in base pairing and base pair stacking contributions among these two duplexes. Therefore, the stabilizing effect for the incorporation of a single aminopropyl chain in DNA is primarily driven by a favorable entropic contribution. Since the bimolecular dissociation of each duplex is contributing equally, this entropic effect is due to the lower release of counterions and water by the unfolding of the modified duplex. Furthermore, this stabilization is electrostatic in nature, because it gradually disappears with increasing salt concentration, as shown by the crossover lines of Figure 4B.
Table 3. Differential thermodynamic profiles for the incorporation of a cationic chain in DNA.
| ΔTM (°C) | ΔΔGcal (kcal/mol) | ΔΔHcal (kcal/mol) | Δ(TΔS) (kcal/mol) | ΔΔnNa+ (per duplex) | ΔΔnW (per duplex) | |
|---|---|---|---|---|---|---|
| Duplexes | 6.0 | –0.8 | 4.1 | 4.9 | 0.52 | 2.9 |
| Hairpins | 0.6 | 0.1 | 0.0 | –0.1 | 0.11 | 1.2 |
The sign of the relatively small differential enthalpy– entropy compensation term is similar to the sign of the unfolding volume change (positive ΔΔnW), which indicates the empirical involvement of electrostricted water (19). This means that the incorporation of the aminopropyl chain is accompanied by a release of nearly 3 mol electrostricted H2O/mol duplex. However, if the enthalpy term (4.1 kcal/mol) is fully assigned to hydration changes, then we estimate a higher removal of water (∼14 ± 7 mol) from this heat exchange, based on a molar heat of 0.3 kcal/mol H2O (32). The discrepancy in the number of water molecules released may be explained in terms of hydration differences among the single strands and duplexes and/or actual differences in stacking interactions, as will be explained in the following paragraph.
Relative to the duplex state, the single strands at 80°C may be considered as true random coils with a higher degree of structural hydration that is due to the exposure of aromatic rings to the solvent. The presence of the aminopropyl group in the modified strand would render it slightly more hydrated if this group is completely exposed to the solvent. However, the heat contribution to this hydration is the result of the compensation of an endothermic contribution, by the structural hydration of the propyl group, and an exothermic contribution, by the electrostricted hydration of the amino charged group. On the other hand, the differential enthalpy term may be explained in terms of a small decrease in stacking interactions by the modified duplex, resulting from conformational changes, such as the induction of a small and local bend. Therefore, the expectation is that the decrease in stacking interactions results from a partial exposure of DNA bases to the solvent that immobilizes structural water, which would be accompanied by endothermic heat. The overall enthalpy effect in this case is that the loss of stacking interactions is enhanced by the uptake of structural water by the modified duplex and release of electrostricted water by the control duplex. Furthermore, the uptake of structural water by the modified duplex at 15°C may disappear with an increase in temperature, and this may well be the reason that this additional structural hydration is not detected by the osmotic stress technique.
For the interpretation of the lower ion release by the modified duplex, we suggest that this effect takes place mainly at the duplex level, since the particular conformation of the random coils can be considered similar at high temperatures. The presence of the cationic charged chain in the major groove of this duplex causes a reorganization of the groove counterions around the tethered amine group. The chain would displace counterions and water molecules and the amino charge would partially neutralize negative charges. The combined effects would yield a net unscreening of the backbone phosphates that causes a collapse of the sugar– phosphate backbone towards the major groove accompanied by a small and local decrease in base stacking interactions around the amino charge, as proposed by Rouzina and Bloomfield (35). These contributions may be quantified, if we use the counterion release of –2.03 mol Na+/mol duplex obtained for the unfolding of a duplex containing a similar 3-hydroxypropyn-1-yl uridine side chain modification (L.Marky, unpublished results). The effect of the amino charged end is to neutralize through a Coulombic interaction ∼0.17 mol Na+/mol duplex while the propyl chain would displace ∼0.35 mol Na+/mol duplex.
The overall results are consistent with recent results in the literature that discussed the incorporation of an aminopropyl chain in the stem of a DNA hairpin (36). This set of DNA hairpin loops and the set of duplexes in this work have one thing in common in that the aminopropyl chain is located in a similar base pair environment, CGAC/GU*CG. The second row of Table 3 shows a complete differential thermodynamic profile for the hairpin set (36). The incorporation of the cationic chain in the hairpin did not induce a thermal stabilization at a similar salt concentration and did not lower the magnitude of the unfolding enthalpy. In addition, the crossover of their Tm dependence on salt takes place at a lower salt concentration of 31 mM and the release of counterions (36) and water is lowered, as shown in Table 3. The observed differences in these two sets of molecules are explained in terms of their thermal stability; the Tm values of the hairpins are ∼64°C while the duplexes melted in the range 39.7–45.7°C (this work). The 21.3°C increase in the thermal stability of the hairpins may well render their stems more flexible, eliminating any differential contribution from stacking interactions. In addition, it is expected that local distortions would have larger effects in a linear duplex because its shape is better represented as a rod rather than in the ellipsoid shape of a hairpin. Local distortions in a sphere will have little impact, if any, on the global conformation of the molecule, while local distortions in a rod will have larger effects on its global conformation. It is also predicted that shifting the Tm value of the ‘modified’ duplex to higher temperatures, by increasing the salt concentration to 80 mM, would eliminate this differential enthalpy because the increase in salt would strengthen base pair stacking.
Thermodynamic folding profiles at 15°C
Table 2 shows complete thermodynamic profiles for the formation of each duplex at 15°C, a temperature at which both duplexes formed completely. ΔHITC and ΔV are measured directly while the ΔG° and TΔS parameters correspond to those obtained in unfolding experiments but corrected for the endothermic contribution of the disruption of single strand base stacking interactions prior to duplex formation, as previously reported for a variety of duplexes (22,24,25). The formation of each duplex is favorable at this temperature and results from the characteristic compensation of a favorable exothermic heat with an unfavorable entropy contribution. The favorable heats also include endothermic contributions of base–base stacking interactions of the single strands. The unfavorable entropy contributions are consistent with the bimolecular association of two strands and uptake of counterions. However, the release of water (positive ΔV) in the formation of the control duplex contributes with a favorable entropy term while the uptake of water (negative ΔV) by the modified duplex contributes with an unfavorable entropy contribution.
The thermodynamic and hydration contributions for the incorporation of the cationic chain in DNA is best seen by using a thermodynamic cycle, similar to the one used above for the unfolding reactions. This reaction at 15°C is shown below:
control duplex(Na+, H2O) + modified strand(Na+, H2O) + xNa+ + yH2O → modified duplex(Na+, H2O) + control strand (Na+, H2O)
For this reaction we obtained a marginally favorable ΔΔG° of –0.3 kcal/mol resulting from the compensation of an unfavorable ΔΔHITC of +8.8 kcal/mol with a favorable Δ(TΔS) term of +9.1 kcal/mol and the uptake of water molecules, ΔΔV = –51.4 cm3/mol. The differential entropy term is consistent with a net counterion release of 0.6 mol Na+/mol duplex; therefore, the ‘xNa+’ term should be on the right hand side of the above equation.
The ΔΔHITC value of +8.8 kcal/mol indicates base stacking and hydration differences not only between the two duplexes but also between the ‘control’ and ‘modified’ strands, because its value is 4.3 kcal higher than the ΔΔHcal term obtained from the unfolding profiles. Therefore, this contribution of 4.3 kcal corresponds to the incorporation of the aminopropyl chain in the single strand at 15°C, which yields increases in both base stacking interactions and electrostricted hydration in the ‘modified’ single strand. These effects are consistent with the presence of a tethered amino charge that directly or indirectly neutralizes charge on the phosphate backbone, allowing closer proximity of the aromatic bases. The remaining 4.5 kcal corresponds to differences in base pair stacking and hydration contributions among the two duplexes.
The localized action of the aminopropyl chain also results in an increased differential release of counterions at 15°C. As low temperatures may decrease duplex flexibility, the interactions of the cationic side chain and the DNA grooves are expected to be more localized at 15°C. Therefore, the effect of the cationic chain is more pronounced at this temperature, yielding perhaps larger differences in stacking interactions among the duplexes.
At 15°C the ΔΔV of –51.4 cm3/mol is in good agreement with a ΔΔV of –55.0 cm3/mol estimated from absolute measurements of the apparent molar volume of all species involved in this reaction and assuming that all strands have similar van der Waals volumes. The negative sign of this term indicates that there is a net uptake of water molecules for this reaction. Furthermore, the sign of the ΔΔHITC–Δ(TΔS) compensation is opposite to the sign of the ΔΔV term, which empirically indicates an uptake of structural water (19,26,37). This ΔΔV term overrides the small release of electrostricted water obtained from the unfolding profiles, but at 15°C includes two hydration contributions: the differential hydration between the modified and control strands and the differential hydration of the two duplexes. The values of the apparent molar volume for each species indicated that the modified strand and duplex are more hydrated than the control strand and duplex, respectively. Therefore, the net uptake of structural water is due to the uptake of water by the modified duplex that overrides the release of water by the modified strand. Unfortunately, a calculation of the number of water molecules involved cannot be done because we do not know the molar volume of structural water.
The overall uptake of structural water by the modified duplex is consistent with the decrease in stacking interaction indicated by the lower unfolding enthalpy of this duplex. This suggests that the incorporation of an aminopropyl chain in a DNA duplex induces a higher exposure of non-polar groups to the solvent. One possibility is that the observed decrease in stacking interactions is due to a DNA deformation induced by the presence of the cationic side chain that partially neutralizes negative charges generating a small and local helical bend, as has been shown by gel mobility studies (6).
The greater exposure of non-polar groups or aromatic bases to the solvent, with the concomitant immobilization of structural water, is consistent with previous investigations of bent duplexes from in laboratory (19,26,37). These systems include duplexes with AT tract sequences (19) and duplexes containing benzo[a]pyrene (26) or cisplatin (37) adducts. The presence of a bend in these systems has been confirmed indirectly by gel mobility studies (38–40) or directly by NMR and X-ray crystallographic investigations (41,42).
Furthermore, these results are in good agreement with a recent NMR solution investigation of the [d(CGCGAA TU*CGCG)]2 duplex containing an aminopropyl deoxyuridine at each CGAA/TU*CG base pair stack environment (31). NMR analysis of this modified Dickerson dodecamer indicates a classical B-DNA structure with normal Watson–Crick base pairing interactions: no significant deviations for 1H chemical shifts were observed with the 3-aminopropyl side chain (31). The sequential NOE connectivities were observed for all base steps and no NOE peaks exhibited unusual intensities. The observation of the imino resonance of the 3-aminopropyl deoxyuridine nucleotide is evidence that the residue is Watson–Crick hydrogen bonded and stacked in the helix. However, rMD calculations based on the NMR data and electrostatic footprinting (28,29) predict that the ammonium group is proximate to the electronegative center at the O6 position of G10, which requires the modified dodecamer to be bent. The back calculation of NOE data using the bent duplex indicated that the bent structure is consistent with all the available NOE data.
To understand how the aminopropyl-induced bending of the DNA would affect the hydrophobic surface exposed to solvent, the accessible surfaces of the NMR structures (linear and bent) were determined. The surface areas for the bent (constrained) and linear (unconstrained) NMR structures were 3631 and 3587 Å2, respectively. This gives an increase of 25 Å2 per modified residue since there are two cationic side chains in the self-complementary duplex. The same type of surface analysis was done using the same DNA structures except that the aminopropyl side chain was replaced by a hydrogen atom to eliminate any effect due to the location of the side chain in the major groove. In this case, the bent structure has ∼33 Å2 more surface area per modified residue than the linear structure. Therefore, the bent structure from NMR is consistent with the conclusion from the thermodynamic studies that the modified DNA is associated with more structured (hydrophobic) water.
CONCLUSIONS
We have used a combination of spectroscopy, calorimetry and density techniques to investigate the overall energetics and hydration properties for the incorporation of a cationic aminopropyl chain in a DNA duplex. This modification induces DNA deformation as measured by gel mobility and NMR solution studies (6,31). In low salt buffer and relative to the control duplex, our results demonstrate that the single incorporation of a cationic chain yielded a more stable duplex, which unfolds with a lower endothermic heat, lower release of counterions and a lower release of electrostricted water. Thus, this stabilization is entropy driven as a result of an electrostatic effect because it gradually disappears as the salt concentration is increased. Complete thermodynamic profiles for the formation of each duplex were obtained at 15°C by mixing their respective complementary strands. The favorable formation of each duplex yielded the typical compensation of a favorable enthalpy with an unfavorable entropy contribution, which is consistent with an uptake of counterions. However, the isothermal profiles yielded a differential enthalpy of 8.8 kcal/mol, which is 4.3 kcal/mol higher than the differential enthalpy observed in the unfolding profiles. This shows that the presence of the aminopropyl chain in the DNA major groove induces an increase in base stacking interactions in the modified single strand and a decrease in base stacking interactions in the modified duplex. Furthermore, the folding of the ‘control’ duplex releases water while the ‘modified’ duplex immobilizes water molecules. The main effects for the incorporation of the cationic chain into a DNA duplex are best understood from inspection of a thermodynamic cycle, in which the formation of the control duplex is used as the reference reaction. We obtained a marginally favorable ΔΔG° term, a positive ΔΔHITC–Δ(TΔS) compensation, negative ΔΔV and a net release of counterions. The opposite signs of the differential enthalpy–entropy compensation and differential volume change terms indicate a net uptake of structural water, which is consistent with a higher exposure of non-polar groups to the solvent. Therefore, the incorporation of the cationic chain in the major groove of a DNA duplex displaces counterions and screens the local phosphates, inducing a greater exposure of the aromatic bases to the solvent and suggesting the presence of a small local bend in the modified duplex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Don Zebolsky (Creighton University) for providing facilities for osmometry. This work was supported by grants GM42223 (L.A.M.) and CA76049 (B.G.) from the National Institutes of Health, Cancer Center Support Grant CA36727 from the National Cancer Institute and a Blanche Widaman Fellowship (A.M.S.) from UNMC.
REFERENCES
- 1.Daune M., (1999) Molecular Biophysics, translated by Duffin,W.J. Oxford University Press, New York, NY.
- 2.Lunger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 3.Strauss J.K., and Maher,J.L.,III (1994) DNA bending by asymmetric phosphate neutralization. Science, 266, 1829–1834. [DOI] [PubMed] [Google Scholar]
- 4.Maher J.L. III, (1998) Mechanisms of DNA bending. Curr. Opin. Chem. Biol., 2, 688–694. [DOI] [PubMed] [Google Scholar]
- 5.Manning G., Ebralidse,K.K., Mirzabekov,A.D. and Rich,A. (1989) An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J. Biomol. Struct. Dyn., 6, 877–889. [DOI] [PubMed] [Google Scholar]
- 6.Strauss J.K., Roberts,C., Nelson,M.G., Switzer,C. and Maher,J.L.,III (1996) DNA bending by hexamethylene-tethered ammonium ions. Proc. Natl Acad. Sci. USA, 93, 9515–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss J.K., Prakash,T.P., Roberts,C., Switzer,C. and Maher,J.L.,III (1996) DNA bending by a phantom protein. Chem. Biol., 3, 671–678. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto H., Nelson,M.G. and Switzer,C. (1993) Formation of chimeric duplexes between zwitterionic and natural DNA. J. Org. Chem., 58, 4194–4195. [Google Scholar]
- 9.Hashimoto H., Nelson,M.G. and Switzer,C. (1993) Zwitterionic DNA. J. Am. Chem. Soc., 115, 7128–7134. [Google Scholar]
- 10.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 11.Marky L.A., Blumenfeld,K.S., Kozlowski,S. and Breslauer,K.J. (1983) Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers, 22, 1247–1257. [DOI] [PubMed] [Google Scholar]
- 12.Marky L.A., and Breslauer,K.J. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- 13.Rentzeperis D., Kharakoz,D.P. and Marky,L.A. (1991) Coupling of sequential transitions in a DNA double hairpin: energetics, ion binding and hydration. Biochemistry, 30, 6276–6283. [DOI] [PubMed] [Google Scholar]
- 14.Spink C.H., and Chaires,J.B. (1999) Effects of hydration, ion release and excluded volume on the melting of triplex and duplex DNA. Biochemistry, 38, 496–508. [DOI] [PubMed] [Google Scholar]
- 15.Millero F.J., (1972) Partial molal volumes of electrolytes in aqueous solutions. In Horn,R.A. (ed.), Water and Aqueous Solutions. Wiley-Interscience, New York, NY, pp. 519–595.
- 16.Kankia B.I., and Marky,L.A. (1999) DNA, RNA and DNA/RNA oligomer duplexes: a comparative study of their stability, heat, hydration and Mg2+ binding properties. J. Phys. Chem. B, 103, 8759–8767. [Google Scholar]
- 17.Shiio H., Ogawa,T. and Yoshihashi,H. (1955) Measurement of the amount of bound water by ultrasonic interferometer. J. Am. Chem. Soc., 77, 4980–4982. [Google Scholar]
- 18.Buckin V.A., Kankiya,B.I., Sarvazyan,A.P. and Uedaira,H. (1989) Acoustical investigation of poly(dA)·poly(dT), poly[d(A-T)]·poly[d(A-T)], poly(A)·poly(U) and DNA hydration in dilute aqueous solutions. Nucleic Acids Res., 17, 4189–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marky L.A., and Kupke,D.W. (2000) Enthalpy-entropy compensations in nucleic acids: contribution of electrostriction and structural hydration. Methods Enzymol., 323, 419–441. [DOI] [PubMed] [Google Scholar]
- 20.King E.J., (1969) Volume changes for ionization of formic, acetic and butyric acids and the glycinium ion in aqueous solution at 25.deg. J. Phys. Chem., 73, 1220–1232. [Google Scholar]
- 21.Conway B.E., (1981) Ionic Hydration in Chemistry and Biophysics. Elsevier, New York, NY.
- 22.Zieba K., Chu,T.M., Kupke,D.W. and Marky,L.A. (1991) Differential hydration of dA.dT base pairing and dA and dT bulges in deoxyoligonucleotides. Biochemistry, 30, 8018–8026. [DOI] [PubMed] [Google Scholar]
- 23.Holbrook J.A., Capp,M.W., Saecker,R.M. and Record,M.T.,Jr (1999) Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry, 38, 8409–8422. [DOI] [PubMed] [Google Scholar]
- 24.Vesnaver G., and Breslauer,K.J. (1991) The contribution of DNA single-stranded order to the thermodynamics of duplex formation. Proc. Natl Acad. Sci. USA, 88, 3569–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentzeperis D., Kupke,D.W. and Marky,L.A. (1994) Differential hydration of dA.dT base pairs in parallel-stranded DNA relative to antiparallel DNA. Biochemistry, 33, 9588–9591. [DOI] [PubMed] [Google Scholar]
- 26.Marky L.A., Rentzeperis,D., Luneva,N.P., Cosman,M., Geacintov,N.E. and Kupke,D.W. (1996) Differential hydration thermodynamics of stereoisomeric DNA-benzo[a]pyrene adducts derived from diol epoxide enantiomers with different tumorigenic potentials. J. Am. Chem. Soc., 118, 3804–3810. [Google Scholar]
- 27.Ahmadian M., Zhang,P. and Bergstrom,D.E. (1998) A comparative study of the thermal stability of oligodeoxyribonucleotides containing 5-substituted 2′-deoxyuridines. Nucleic Acids Res., 26, 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang G., Encell,L., Switzer,C. and Gold,B. (1995) The role of electrostatics in the sequence selective reaction of charged alkylating agents with DNA. J. Am. Chem. Soc., 117, 10135–10136. [Google Scholar]
- 29.Dande P., Liang,G., Chen,F.-X., Roberts,C., Nelson,M.G., Hashimoto,H., Switzer,C. and Gold,B. (1997) Regioselective effect of zwitterionic DNA substitutions on DNA alkylation: evidence for a strong side chain orientation preference. Biochemistry, 36, 6024–6032. [DOI] [PubMed] [Google Scholar]
- 30.Heystek L.E., Zhou,H.-q., Dande,P. and Gold,B. (1998) Control over the localization of positive charge in DNA: the effect on duplex DNA and RNA stability. J. Am. Chem. Soc., 120, 12165–12166. [Google Scholar]
- 31.Li Z., Huang,L., Dande,P., Gold,B. and Stone,M.P. (2002) Structure of tethered cationic 3-aminopropyl chain incorporated into an oligodeoxynucleotide: evidence for 3′-orientation in the major groove accompanied by DNA bending. J. Am. Chem. Soc., in press. [DOI] [PubMed] [Google Scholar]
- 32.Gasan A.I., Maleev,V.Y. and Semenov,M.A. (1990) Role of water in stabilizing the helical biomacromolecules DNA and collagen. Stud. Biophys., 136, 171–178. [Google Scholar]
- 33.Kankia B.I., and Marky,L.A. (2001) Folding of the thrombin aptamer into a G-quadruplex with Sr2+: stability, heat and hydration. J. Am. Chem. Soc., 123, 10799–10804. [DOI] [PubMed] [Google Scholar]
- 34.Krakauer H., (1972) A calorimetric investigation of the heats of binding of Mg++ to poly A, to poly U and to their complexes. Biopolymers, 11, 811–828. [DOI] [PubMed] [Google Scholar]
- 35.Rouzina I., and Bloomfield,V.A. (1998) DNA bending by small, mobile multivalent cations. Biophys. J., 74, 3152–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto A.M., Kankia,B.I., Dande,P., Gold,B. and Marky,L.A. (2001) Incorporation of a cationic aminopropyl chain in DNA hairpins: thermodynamics and hydration. Nucleic Acids Res., 29, 3638–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kankia B.I., Kupke,D.W. and Marky,L.A. (2001) The incorporation of a platinated cross-link into duplex DNA yields an uptake of structural water. J. Phys. Chem. B, 105, 11402–11405. [Google Scholar]
- 38.Alessi K., (1995) Thermodynamics of deoxyoligonucleotides containing A·T base pairs. Doctoral dissertation, New York University, New York, NY.
- 39.Mao B., (1994) Studies of site-specific and stereospecific BPDE-N2-dG oligodeoxynucleotide adducts by gel electrophoresis. Doctoral dissertation, New York University, New York, NY.
- 40.Hagerman P.J., (1985) Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry, 24, 7033–7041. [DOI] [PubMed] [Google Scholar]
- 41.Gelasco A., and Lippard,S.J. (1998) NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry, 37, 9230–9239. [DOI] [PubMed] [Google Scholar]
- 42.Takahara P.M., Rosenzweig,A.C., Frederick,C.A. and Lippard,S.J. (1995) Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature, 377, 649–652. [DOI] [PubMed] [Google Scholar]