Abstract
The zinc finger protein, metal response element-binding transcription factor-1 (MTF-1) regulates the expression of genes in response to metal ions and oxidative stress. The precise mechanisms by which this occurs are not understood. To further examine this problem, mouse MTF-1 was expressed in Saccharomyces cerevisiae and tested for the ability to activate metal response element-driven reporter gene expression. Zinc was an effective inducer of reporter gene expression. In general, the magnitude of zinc induction was dependent on the concentration of zinc in the culture medium, but independent of the amount of MTF-1 expression. Zinc induction also occurred with either integrated or episomal reporter plasmids containing the native mouse metallothionein-I proximal promoter. Deletion of fingers 5 and 6 of MTF-1, which function in a zinc-dependent manner to stabilize the DNA-binding activity of the protein in vitro, did not diminish the zinc induction of either episomal or integrated promoters. However, a Gal4 DNA-binding domain– MTF-1 fusion protein, which binds constitutively to the Gal4-responsive promoter, was not zinc inducible but caused constitutive activation of reporter gene expression. This suggests that zinc activation of the DNA-binding activity of MTF-1 is the rate limiting step in its metalloregulatory function in yeast. In contrast, MTF-1 was not responsive to either cadmium or hydrogen peroxide, suggesting that distinct co-activators or signal transduction cascades not found in yeast are required to mediate MTF-1 activation of gene expression by this toxic metal and by oxidative stress.
INTRODUCTION
DNA motifs termed metal response elements (MRE) (1,2), which are present in multiple copies in the proximal promoters of many metallothionein (MT) genes, represent binding sites for metal response element-binding transcription factor-1 (MTF-1). MTF-1 was first cloned from the mouse (3) and MTF-1 homologs have since been identified in humans (4,5), Drosophila (6), pufferfish (7) and the chicken (H.Jiang and G.K.Andrews, unpublished data). Homozygous disruption of the mouse MTF-1 gene in cultured cells eliminates heavy metal-induced MT gene expression (8), as well as responsiveness to oxidative stresses (9) and hypoxia (10). RNA inhibition experiments suggest that Drosophila MTF-1 mediates copper regulation of these MT genes (6). Thus, several signal transduction pathways impinge on the activities of MTF-1, but the mechanisms by which MTF-1 activates gene expression in response to these diverse signals are not understood.
The importance of this transcription factor is underscored by the fact that the mouse MTF-1 gene is essential for embryonic development (11). Embryos homozygous for MTF-1 null mutations die due to failure of the liver to develop properly. In contrast, development of the nervous system and visceral yolk sac is not impaired in these embryos (11–13). MTF-1 is now known to participate in the regulation of expression of several other genes, including, but not limited to, the zinc transporter-1 gene (14), the γ-glutamylcysteine synthetase heavy chain gene (11) and the placental growth factor gene (15).
MTF-1 is a zinc-finger protein in the Cys2His2 family of transcription factors. Its six zinc fingers have been highly conserved during evolution, although significant divergence has occurred in the remainder of the protein (7). This conservation is consistent with the findings that the zinc finger domain of MTF-1 is critical for both its metalloregulatory and DNA-binding functions in response to zinc (16). Zinc interactions with the zinc-finger domain reversibly modulate the DNA-binding activity of MTF-1 (3,17). There is structural and functional heterogeneity among the MTF-1 zinc fingers (18–22). About half of the fingers exhibit high-affinity zinc-binding sites, while the other half exhibit lower affinity zinc binding (18,19,22). Although functionality has not been convincingly assigned to each of the zinc fingers, current evidence suggests that fingers 2–4 constitute the core DNA-binding fingers, while fingers 1, 5 and 6 may play roles in zinc sensing and further stabilization of DNA binding (18–23).
In contrast to our understanding of the mechanisms by which MTF-1 senses zinc ions, we know very little about how it senses cadmium, copper and oxidative stresses. Zinc, cadmium and a diverse set of signals can regulate the nucleocytoplasmic trafficking of MTF-1 (24,25), although nuclear localization alone does not ensure activation of transcription by MTF-1 (25). Cadmium or copper treatment of mammalian cells causes only a modest increase in DNA-binding activity of MTF-1 in vivo and these metals have no effect in vitro on this activity (26–29). It is conceivable that these metals and oxidants may cause the redistribution of zinc in the cell (30), utilize specific co-activators of MTF-1 and/or activate signal transduction cascades that impinge on MTF-1 to affect MT gene transcription (31). However, mutations in the zinc fingers of MTF-1 which abolish zinc responsiveness also abolish the response to cadmium (23), and a recent study suggests that both zinc and cadmium responsiveness are abrogated by treatment of mammalian cells with inhibitors of protein kinases (31). Thus, the zinc-finger domain of MTF-1 is apparently essential for all of the metalloregulatory functions of MTF-1, and post-translational modifications of the protein may also be important in that regard.
The transactivation domains of MTF-1 are less well characterized than its zinc-finger domain, but it has been suggested that intramolecular interactions are also important for optimal MTF-1 function (32,33). The C-termini of human and mouse MTF-1 contain three transactivation domains which are acidic, proline rich and serine + threonine rich, respectively (32). The VP16 transactivation domain can function with the zinc-finger domain of MTF-1 to produce a metal-responsive factor in transfected cells (30,32), but the transactivation domain of the zinc-finger protein Sp1 cannot (23). Whether metal ions also exert effects on the transcriptional activity of MTF-1 after it binds to DNA is unknown.
The studies reported herein were undertaken to further address the mechanisms of action of MTF-1. Mouse MTF-1 expressed in yeast was shown to function as a zinc sensor. This function was dependent on the zinc-finger domain and reflected the recruitment of MTF-1 to the promoter. In contrast, mouse MTF-1 could not function as a sensor of cadmium or oxidative stresses in this system. Thus, these functions of MTF-1 are clearly separable and distinct from its zinc sensing functions and must involve factors or signal transduction cascades not found in yeast.
MATERIALS AND METHODS
Yeast
Saccharomyces cerevisiae strain ZHy6 was obtained from Dr David Eide (University of Missouri, Columbia, MO). This strain was derived from DY1457 (Matα, ade6, can1, his3, leu2, trp1, ura3) and has a deletion of ZAP1 (ZAP1Δ::TRP1) (34).
Vectors
The yeast expression vectors pVT101u:MTF-1 and pVT-D5,6 were engineered for episomal expression of mouse MTF-1 cDNA and MTF with deletion of zinc fingers 5 and 6, respectively, as described previously (23). Mouse MTF-1 cDNA (3) was cloned into the HindIII and XhoI sites of pVT101u. Subsequently, MTF-1 or MTFΔ5,6 was subcloned from this plasmid into the HindIII and XhoI sites of p406A to create p406A:MTF-1 and p406A:Δ5,6. p406A:GBD-MTF-1 was engineered to express a protein containing the N-terminal 147 amino acids of the DNA-binding domain of Gal4 (GBD) fused to the N-terminus of mouse MTF-1 as follows. MTF-1 cDNA was cloned from pGem7:MTF-1 (9) into the NcoI sites of pAS1, which is downstream of the GBD. The HindIII–SalI fragment of the GBD–MTF fusion protein was subcloned into the HindIII and XhoI sites of pVT101u to create pVT101u;GBD-MTF-1, thereby eliminating the XhoI site. Subsequently, a HindIII–SphI fragment of this plasmid was subcloned into p406A to create p406A:GBD-MTF-1. The yeast reporter gene vector pYEp363:MREd5:βgal was engineered to express β-galactosidase under the control of five tandem copies of mouse MREd, as described previously (35), and for LEU2 auxotrophy. The MREd5:βGeo construct (3023B5) was provided by Dr Richard Palmiter (University of Washington, Seattle, WA). The vector was cleaved with PstI and SacI to liberate the MREd sequences fused to the 5′ half of lacZ. This fragment was cloned into YEp363 (36). The 5′ portion of the plasmid lacZ was replaced by the insert’s lacZ sequence. The yeast reporter gene vectors pYIpl:–150:βgal and pYIpl:–150-Δ:βgal contained the proximal 150 bp of the mouse MT-I promoter, with or without the USF/ARE, respectively, and were engineered for LEU2 auxotrophy. pYIpl and pYEpl were both constructed by cloning a polylinker containing XbaI and HindIII sites into pYIp351 and pYEp363, respectively. The –150 bp or the –150Δ sequence was excised from –150:Luc or –150Δ:Luc (35) with NheI and HindIII and cloned directionally into the XbaI site of pYIpl to create pYIpl:–150:βgal. These same cloning steps were used to clone –150 or –150Δ promoter elements into pYEplink to create pYepl:–150:βgal and pYepl:–150-Δ:βgal. A vector was engineered to express β-galactosidase under the control of five tandem copies of the Gal4 response element (GRE5) as follows: a 227 bp fragment spanning the GRE5 and TATA box of pFR:βgall (Stratagene, Cedar Creek, TX), which contained a PstI site at the 5′ end of the fragment and introduced a HindIII site at the 3′ end, was amplified by PCR. This was cloned into the PstI and HindIII sites of pYEpl to create pYEpl:GRE:βgal.
Yeast transfections
Yeast were transformed with episomal reporters and expression plasmids as described previously (23). The plasmids p406A:MTF-1, p406A:Δ5,6 and p406A:GBD-MTF-1 were each linearized with BstBI and 1 µg of gel-purified plasmid was integrated into the URA3 locus of ZHy6 chromatin using the lithium acetate yeast transfection procedure as described previously (23). For integration of pYIpl:–150:βgal and pYIpl:–150-Δ:βgal, the plasmids were linearized with KpnI prior to integration into the LEU2 locus.
Yeast culture/treatment
Co-transformed or singly transformed ZHy6 cells were cultured in yeast peptone dextrose adenine (YPDA) medium and selected for growth in uracil and leucine CM drop-out medium, as discussed below. Transformed colonies were selected, grown overnight in drop-out medium, and glycerol stocks were prepared and frozen at –80°C. Experiments were initiated using colonies of freshly plated cells from the glycerol stocks.
Unless otherwise indicated, cells were cultured in defined zinc-free medium (CM minus zinc; BIO101, Vista, CA) to which zinc was added from a 100 mM ZnCl2 solution in sterile acidified water. For each experiment, the zinc concentration in the medium is indicated in the figure and legend. A single colony of cells was inoculated per 5 ml of medium and incubated overnight at 30°C with agitation (300 r.p.m.). By the next morning the cultures had grown to early to mid log phase (2–8 × 107 cells/ml). Cell concentrations were normalized (2 × 107 cells/ml), cultures were divided into 1 ml aliquots and each sample was manipulated, as described in the figure legend, by the addition of ZnCl2, CdCl2 or freshly prepared H2O2 from 100- to 1000-fold concentrated sterile stock solutions. Incubation was continued at 30°C with constant agitation. For analysis of RNA and protein levels, ∼2 × 108 cells were used.
β-Galactosidase assay
Cells were transferred to a 1.5 ml microfuge tube and collected by centrifugation at 15 000 r.p.m. for 6 s. The cell pellet was resuspended in 1 ml of cold wash buffer (100 mM Na2HPO4, pH 7.0, 10 mM KCl, 1 mM MgCl2), collected by centrifugation, as above, and frozen at –80°C. A cell lysate was prepared by glass bead disruption of the cells. The frozen cell pellet was thawed in 800 µl of wash buffer plus 200 µl of acid-washed glass beads (0.4 mm diameter) and vortexed vigorously five times for 30 s each. The beads were allowed to settle and the extract was recovered. β-Galactosidase activity was measured using the microtiter plate assay as described previously (35). Protein content was determined using the Protein Assay Reagent (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
Western blotting
The washed and frozen yeast pellet (∼1.5 × 108 cells) was resuspended in 300 µl of cold 10% trichloroacetic acid and acid-washed glass beads (200 µl) were added. The mixture was vortexed vigorously for 5 min and the extract was recovered from the beads. Precipitated proteins were recovered by centrifugation at 10 000 g for 20 min. The pellet was suspended in 1 ml of cold acetone and the precipitated proteins were recovered by centrifugation, as above. The protein pellet was suspended in 100 µl of SDS sample buffer without reducing agent or dye and heated at 100°C to dissolve the pellet. The pH of the extract was adjusted to neutral by the addition of 1 M Tris base and the extract was clarified of insoluble material by centrifugation at 17 000 g for 5 min. Protein concentration of the extract was determined using the BCA Protein Assay Reagent (Pierce, Rockford, IL). Proteins were reduced by the addition of 5% β-mercaptoethanol and 50 µg were separated by 10% SDS–PAGE (37). Proteins were transferred to nitrocellulose membranes. The membranes were blocked overnight at 4°C and probed with MTF-1 antisera (1:5000 dilution) for 1 h at room temperature as described previously (29). Membranes were then incubated with secondary antibody conjugated to horseradish peroxidase for 30 min at room temperature and developed by chemiluminescence and exposed to hyperfilm ECL. Equal protein loading and transfer was verified visually by staining membranes with Ponceau’s solution.
The polyclonal antiserum against purified bacterial recombinant mouse MTF-1 (rabbit anti-mMTF-1) fused to glutathione S-transferase was raised in rabbits (Covance Research Products, Denver, CO) and purified by protein A chromatography followed by removal of glutathione S-transferase antibodies (10).
Recombinant mouse MTF-1 was synthesized in vitro using the TnT coupled reticulocyte lysate transcription/translation system (TnT lysate), as described in detail previously (27).
Immunoprecipitation/western blotting
MTF-1 or GBD–MTF-1 was immunoprecipitated from trans fected ZHy6 cells. Approximately 5 × 108 cells in late log phase, previously transfected with reporter plasmids and either p406A:MTF-1 or p406A:GBD-MTF-1 or reporter plasmids alone, were lysed with 200 µl of glass beads plus 500 µl of lysis buffer [20 mM HEPES pH 7.9, 10 mM MgCl2, 400 mM KCl, 1 mM dithiothreitol, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN)] by vortexing as described in ‘Western blotting’ above. Lysates were separated from beads, clarified by centrifugation at 12 000 g for 10 min at 4°C, concentrated as described previously (17) and protein concentrations determined using the Bradford protein assay (Bio-Rad). Rabbit anti-mMTF-1 or normal rabbit IgG were conjugated to Actigel (Sterogene, Carlsbad, CA) as per the manufacturer’s protocol and coupling efficiency estimated by elution from beads in 5% β-mercaptoethanol/1× SDS sample buffer at 95°C for 5 min and Coomassie Blue staining of SDS–PAGE gels containing resolved eluates. Lysates (700 µg) in IP buffer (lysis buffer containing 150 mM KCl and 0.5% NP-40) were precleared for 1 h with 40 µl of normal IgG–Actigel (1 µg antibody), centrifuged briefly and immunoprecipitated with 40 µl of normal IgG–Actigel or rabbit anti-mMTF-1–Actigel. Immunoprecipitates were washed three times in IP buffer and once in IP buffer without detergent. Proteins were eluted from the Actigel in reducing SDS buffer as described above, separated by 6% SDS–PAGE, and western blotting with rabbit anti mMTF-1 was carried out as described above.
RNA extraction and northern blot analysis
RNA was extracted in guanidine isothiocyanate-containing buffers using the MIDI RNA isolation kit essentially according to the manufacturer’s instructions (Qiagen, Valencia, CA). Briefly, the washed and frozen yeast pellet (∼2.5 × 108 cells) was thawed in 600 µl of guanidine isothiocyanate containing 500 µl of acid-washed glass beads. The mixture was vortexed vigorously five times for 45 s each and the extract was collected from the beads. The extract was processed as described in the kit. Residual DNA was removed from the RNA (∼50 µg in 220 µl of water) by precipitation of the RNA once with 3 M ammonium acetate at –2°C, followed by a final ethanol precipitation, as described previously (38).
Total RNA (1 µg in 5 µl) was denatured and size separated by electrophoresis in a 0.75% agarose–formaldehyde gel. RNA was transferred and crosslinked to nylon membranes, and northern blots were prehybridized, hybridized and washed as described previously (35). Hybrids were detected by autoradiography at –70°C with intensifying screens and quantitated by radioimage analysis (Molecular Dynamics, Sunnyvale, CA). In each experiment, a duplicate gel was stained with acridine orange to verify the integrity and equal loading of RNA.
β-Galactosidase cDNA was cloned into a pGEM vector (Promega Corp., Madison, WI) and was used as a template for the synthesis of 32P-labeled cRNA probe as described previously (35).
Alcohol dehydrogenase (ADH) assay
Yeast cells (∼2–8 × 107 cells) at mid log phase growth were collected by centrifugation at 15 000 r.p.m. for 6 s in a microcentrifuge and washed twice with 1 ml each of cold 16 mM phosphate buffer, pH 8.8. Cell pellets were frozen and stored at –80°C until assayed. Cells were thawed and disrupted by repeated vortexing with glass beads in 300 µl of 16 mM phosphate buffer, pH 8.8, 10 µM EDTA and 20 µM PMSF. The extract was clarified by centrifugation at 13 000 r.p.m. for 10 s and the supernatant was assayed for ADH activity and protein content. ADH activity was quantitated by monitoring the rate of reduction of NAD (change in A340), as described previously (39). Reactions were carried out in 3 ml containing 16 mM ethanol, 1.6 mM NAD, 16 mM phosphate buffer, pH 8.8, 10 µM EDTA. Purified yeast ADH (Sigma Chemical Co., St Louis, MO) was used as the kinetic standard and activities in extracts (25–100 µl were assayed) are expressed in units of ADH activity per µg protein in the extract.
RESULTS
Mouse MTF-1 expressed in yeast activates MRE-driven reporter gene expression in response to zinc
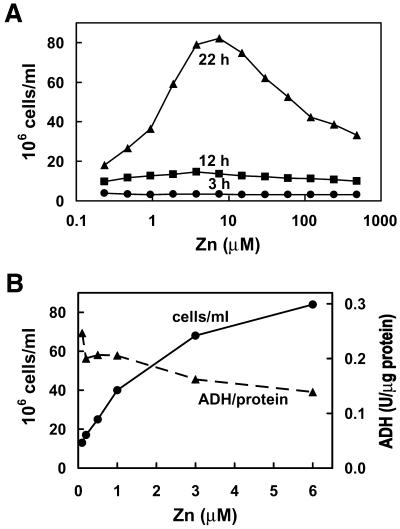
The Saccharomyces cerevisiae strain ZHy6 was used in these studies because these cells cannot readily adapt to changes in zinc concentration in the environment, which allowed more experimental control of the zinc status of the culture. ZHy6 cells have a mutation in ZAP1. In yeast, ZRT1 encodes the high affinity zinc transporter (40) and its transcription is regulated by the transcription factor Zap1p (34), the activity of which is inhibited by zinc. Loss of function of ZAP1 results in severely attenuated zinc transporter gene expression and increased dependence of these cells on zinc levels in the culture medium. As shown in Figure 1, optimal growth of ZHy6 occurred when the culture medium contained 3–8 µM ZnCl2. Under these conditions the cells underwent four or five cell divisions within 18 h. This growth rate was reduced to three cell divisions in medium containing only 0.25 µM zinc. In addition, this growth rate was reduced in medium containing >10 µM zinc (Fig. 1A), which reflects a zinc shock effect due to the lack of expression of the vaculolar zinc transporter that buffers cytosolic zinc and is regulated by Zap1p (41).
Figure 1.
Effects of zinc on the rate of growth and the specific activity of the zinc-dependent enzyme ADH in ZHy6 cells. (A) ZHy6 cells were inoculated into medium (2 000 000 cells/ml) containing the indicated concentrations of zinc and incubated at 30°C for up to 22 h. Cell density was monitored every 3 h (only three time points are shown) by measuring the A600 of the culture and assuming that 0.1 A600 = 3 000 000 cells/ml. (B) ZHy6 cells were cultured overnight in medium containing the indicated concentrations of zinc and the cell density in the culture and the specific activity of ADH were measured. ADH activity in extracts was quantitated by monitoring the rate of reduction of NAD (A340) compared with that of purified yeast ADH. ADH activity (units) was normalized to protein content in the extract.
As another measure of the zinc status of the ZHy6 cells, the specific activity of the zinc-dependent ADH was measured. Under these culture conditions, the specific activity of ADH was actually reduced slightly when cells were cultured in zinc-replete (3–6 µM) medium compared with zinc-limiting (0.1–0.5 µM) medium (Fig. 1B). Thus, the specific activity of ADH is not reduced under these zinc-limiting conditions, but the growth rate and total protein per cell (data not shown) are significantly reduced. ZHy6 cells can grow in medium containing 0.25–480 µM ZnCl2 and, based on growth rates and ADH activity, it is possible to define culture conditions which are moderately zinc limiting, zinc normal or moderately zinc excess.
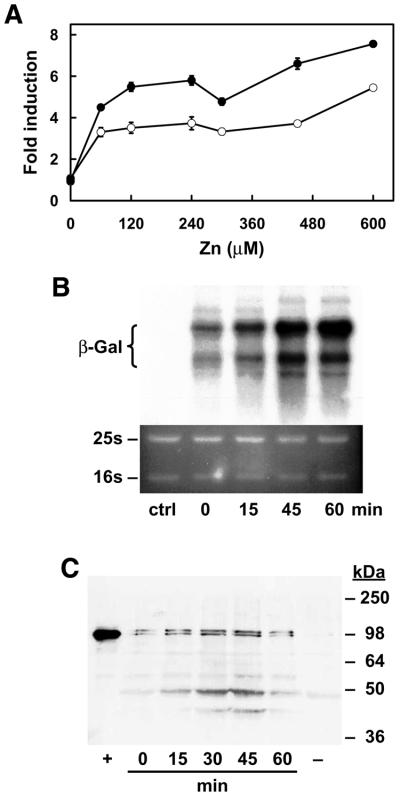
To determine if mouse MTF-1 can function in ZHy6 cells, this strain was co-transfected with an episomal mouse MTF-1 expression vector and an episomal MREd5:β-gal reporter vector and the cells were assayed for zinc induction of β-galactosidase activity (Fig. 2A), for zinc induction of β-galactosidase mRNA (Fig. 2B) and for levels of immunoreactive mouse MTF-1 (Fig. 2C). Although not shown, transfection of the expression vector alone or the reporter gene alone resulted in only background levels of β-galactosidase activity. Several (8) co-transfected colonies were selected for analysis and all yielded results similar to those obtained using the two representative colonies shown in Figure 2A. Co-transfected cells were cultured in CM drop-out medium containing 2.5 µM zinc and at mid log phase they were exposed to increased zinc in the medium. Peak β-galactosidase activity (4- to 8-fold induction) was detected 8–12 h after addition of zinc (60 µM) and β-galactosidase mRNA increased 5-fold within 1 h. In contrast, in cells transfected with the reporter gene alone, zinc had no effect on β-galactosidase activity (data not shown). Thus, this induction was dependent on MTF-1. Western blotting (Fig. 2C) confirmed MTF-1 immunoreactive protein which co-migrated with recombinant mouse MTF-1 (∼100 kDa) in the co-transfected cells and the relative amount of MTF-1 remained unchanged in cells treated for 1 h with zinc.
Figure 2.
Mouse MTF-1 expressed in yeast activates MRE-driven β-galactosidase gene expression in response to exogenous zinc. (A) ZHy6 cells were co-transfected with a mouse MTF-1 episomal expression vector and an MREd5:βgal episomal reporter vector. Colonies were selected for growth on leucine and uracil CM drop-out plates. The initial zinc concentration in the medium was 2.5 µM. Co-transfected cells (results from two different colonies are shown) were grown to mid log phase (18 h) in medium containing 2.5 µM zinc and then incubated for 8 h in the same medium containing the indicated concentrations of additional zinc. Data are expressed as fold induction of β-galactosidase specific activity (units/mg protein ± SEM) relative to that in control cultures. (B) Northern blotting was used to detect β-galactosidase mRNA in co-transfected or control (ctrl) ZHy6 cells. RNA was isolated from co-transfected cells [colony represented by the closed circles in (A)] that were incubated in medium containing 60 µM zinc for up to 1 h. RNA (1 µg) was fractionated by formaldehyde–0.75% agarose gel electrophoresis, blotted to a nylon membrane and hybridized with a radiolabeled β-galactosidase cRNA probe. Hybrids were detected by autoradiography (top) and quantitated by radioimage analysis. Acridine orange staining of the gel (bottom) was used to confirm equal loading of each RNA sample. The mobilities of 25S and 16S rRNA are indicated to the left. (C) Western blotting was used to detect mouse MTF-1 in extracts from the same co-transfected yeast used in (B). Trichloroacetic acid-precipitable proteins were washed in acetone, dissolved in SDS loading buffer and applied (50 µg) to a SDS–10% polyacrylamide gel. After electrophoresis, proteins were electroblotted to a nitrocellulose membrane and MTF-1 was detected using rabbit anti-mouse MTF-1 antisera and ECL detection. Lane 1 (+) contains recombinant mouse MTF-1 synthesized in a coupled in vitro transcription–translation system. The relative mobilities (kDa) of protein standards are shown to the left.
These results demonstrate that pre-existing mouse MTF-1 can rapidly induce MRE-driven β-galactosidase gene expression in yeast cells exposed to excess zinc in the culture medium.
Zinc-responsive MTF-1 activity in yeast is modulated by the availability of zinc in the culture medium
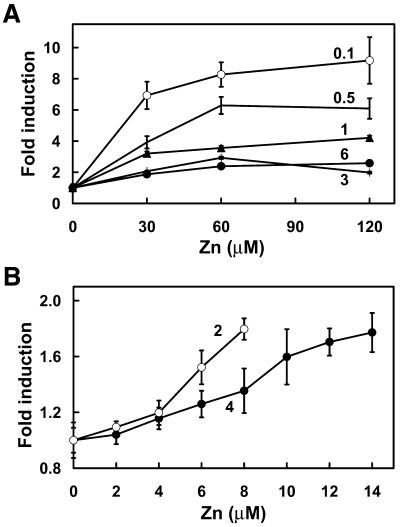
To better define the role of zinc in this system, the effects of initial zinc concentration in the culture medium on subsequent induction of β-galactosidase by exposure to increased ZnCl2 concentrations was examined. Zinc induction of β-galactosidase was significantly enhanced (up to 9-fold induction) in cells cultured under zinc-limiting conditions and less responsive (2-fold induction) in those cultured under zinc-replete conditions (Fig. 3A). The amount of β-galactosidase activity was normalized to the total protein in each culture because the addition of zinc to zinc-limited cells caused a significant increase in the total amount of protein in the culture during the 8 h exposure period. Zinc induction of β-galactosidase activity was not detected in cultures grown under zinc-excess conditions (>15 µM ZnCl2) (data not shown). The concentration– response curve for exogenous zinc induction of β-galactosidase activity was maximal at ∼60 µM zinc (Fig. 3A). However, induction of β-galactosidase activity was measurable in response to as little as a 3-fold increase in zinc concentration in the medium (Fig. 3B) and the concentration– response curve for zinc induction was shifted to the right in cells exposed to subtly higher initial zinc concentrations in the growth medium.
Figure 3.
Zinc-responsive MTF-1 activity in yeast is modulated by the availability of zinc in the culture medium. Co-transfected ZHy6 cells (Fig. 3, closed circles) were incubated to mid log phase in medium containing the indicated concentrations of zinc (µM concentrations above each line on the graphs). The indicated concentration of additional zinc was added to each culture and the incubation was continued for 8 h. (A) Cells were grown in medium containing 0.1–6 µM zinc and then exposed to 30–120 µM zinc. (B) Cells were grown in medium containing 2 or 4 µM zinc and then exposed to 2–14 µM zinc. The specific activity of β-galactosidase was determined and compared with that of the control culture, as described in the legend to Figure 2.
Zinc induction of reporter gene expression is not dependent on the levels of MTF-1 accumulated in yeast
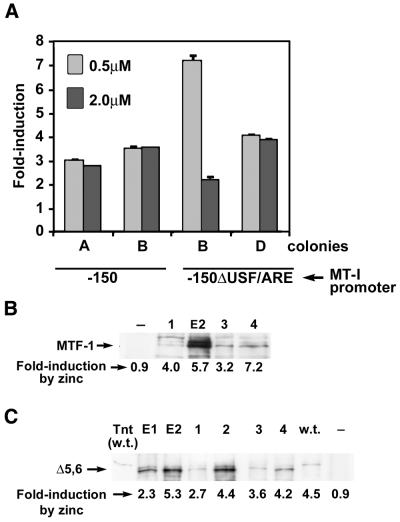
In order to examine the possibility that the zinc responsiveness of MTF-1 in these yeast simply reflects the existence of a large pool of inactive MTF-1 due to high level overexpression of this protein, several ZHy6 clones transfected with integrated or episomal MTF-1 expression vectors were analyzed by western blotting for MTF-1 levels and for zinc induction of β-galactosidase activity, as described above. In these experiments, cells were cultured in medium containing 0.5 µM zinc and then exposed to 60 µM zinc. In general, those cells containing the episomal expression vector also contained much higher levels of MTF-1 than did those containing a single integrated expression vector. However, β-galactosidase induction by zinc (relative to basal level) did not correlate with the levels of MTF-1 in these yeast (Fig. 4B).
Figure 4.
Zinc-responsive MTF-1 activity in yeast is not dependent on high amounts of MTF-1 accumulated in these cells and can occur through the native mouse MT-I promoter integrated into chromatin and in the absence of zinc fingers 5 and 6. Mouse MTF-1 or MTFΔ5,6 expression vectors were integrated into the URA3 locus of ZHY6 cells, followed by integration of a β-galactosidase reporter gene driven by the native (–150 bp) or mutated (–150ΔUSF/ARE) mouse MT-I promoter (35) into the LEU2 locus. (A) Yeast were transfected with the integrating MTF-1 expression vector and the indicated integrating MT-I promoter-driven reporter genes. Two different colonies for each reporter were grown to mid log phase in medium containing 0.5 or 2.0 µM zinc and then incubated for 8 h in the same medium containing 60 µM zinc. Data are expressed as the fold induction of β-galactosidase specific activity (units/mg protein ± SEM) relative to that in control cultures which had been maintained in the initial zinc-containing medium. There was no zinc induction of reporter gene expression in the absence of MTF-1. (B) Western blotting, as described in the text and legend to Figure 2, was used to monitor the relative levels of MTF-1 accumulated in transfected yeast and this was correlated with the levels of zinc induction of reporter gene expression for each colony. MTF-1 was expressed from an integrated vector (lanes 1, 3 and 4) or from an episomal vector (lane E2). The reporter gene vectors were –150:βgal (lanes 1 and 3) or –150ΔUSF/ARE:βGal (lanes 2 and 4). Cells were grown to mid log phase in medium containing 0.5 µM zinc and then induced with 60 µM zinc as in (A). The fold induction by zinc of β-galactosidase reporter gene expression for each colony is shown below each lane. (C) Western blotting was used to monitor the relative levels of MTF-1 lacking zinc fingers 5 and 6 (MTFΔ5,6) that accumulated in the yeast and this was correlated with the levels of zinc induction of the integrated –150:βgal reporter gene in each colony. Lanes E1 and E2, episomal expression vector; lanes 1–4, integrated expression vector; lane w.t., native MTF-1 expressed from an integrated vector [see (B), lane 3]; lane Tnt (w.t.), recombinant mouse MTF made in a TnT lysate (1 µl), as described in the legend to Figure 2; lane –, –150:βgal colony lacking MTF-1. All experiments were repeated twice with the same results.
Mouse MTF-1 expressed in yeast can interact with an integrated, intact metallothionein-I promoter to activate gene expression in response to zinc
In order to determine whether the zinc responsiveness of mouse MTF-1 in yeast is influenced by packaging of the reporter gene promoter into chromatin and to examine whether MTF-1 can function using the authentic mouse MT-I promoter in yeast, a linearized vector (–150:βgal) containing a 216 bp fragment of the mouse MT-I promoter (–150 to +66 bp) upstream of the β-galactosidase reporter gene was integrated into the LEU2 locus. This region of the MT-I promoter contains all (5) of the functional MREs, as well as other promoter elements. In these experiments, MTF-1 was expressed from an integrated expression vector. Induction of β-galactosidase in these two colonies (–150:MTF) by zinc (60 µM) was ∼3-fold, regardless of whether the cells had been grown in 0.5 or 2.0 µM zinc (Fig. 4A). Integration of the MREd5:βgal reporter gene also attenuated the zinc response of the reporter (data not shown). Overall, these results demonstrate that mouse MTF-1 can function in a zinc-dependent manner to activate the intact, chromatin-packaged mouse MT-I promoter in yeast.
Previous studies from our laboratory suggested that another element in the MT-I promoter, a composite element consisting of a binding site for upstream stimulatory factor overlapping an antioxidant response element (USF/ARE) located at –89 to –101 bp, may participate in the basal expression, as well as cadmium and oxidative stress induction, of the mouse MT-I promoter (9,35). To determine if the USF/ARE element affects MTF-1 induction of β-galactosidase in yeast, a reporter vector in which the USF/ARE was deleted from the proximal MT-I promoter (–150Δ:βgal) was integrated into the yeast genome and examined for zinc inducibility. Deletion of the USF/ARE had no reproducible effect on its zinc induction. Furthermore, strains without MTF-1 (–150 or –150Δ) were refractile to induction by zinc and the USF/ARE did not significantly contribute to cadmium or oxidative stress induction (data not shown; Fig. 6).
Figure 6.
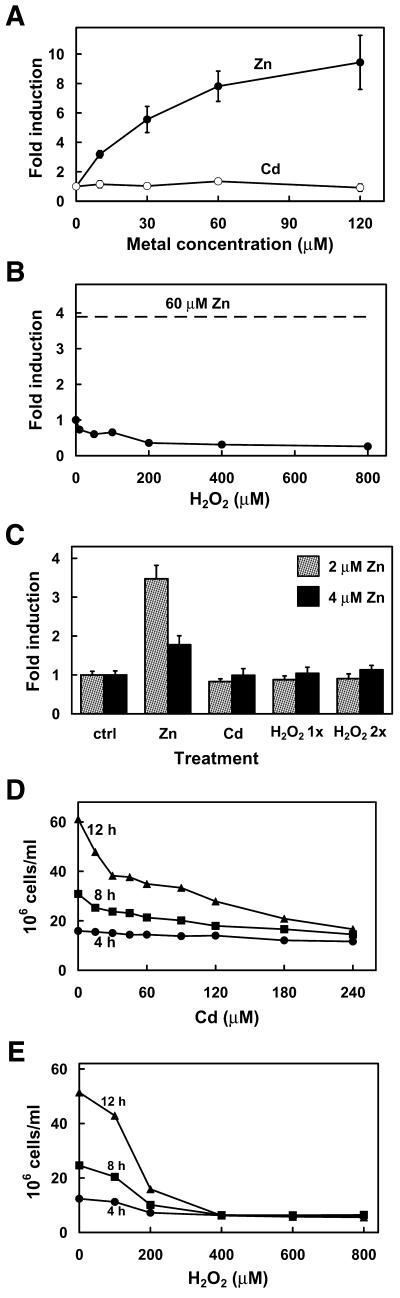
Mouse MTF-1 expressed in yeast does not activate gene expression in response to toxic concentrations of cadmium or hydrogen peroxide. Co-transfected cells (Fig. 1, closed circles) were grown to mid log phase in medium containing 1 µM (A and B) or the indicated concentrations of zinc (C). In (A) and (B) the culture was then adjusted to the indicated final concentrations of zinc, cadmium or hydrogen peroxide and the incubation was continued for 8 h. In (C) cells were treated with zinc (60 µM), cadmium (60 µM) or hydrogen peroxide (100 µM) (H2O2 1×). A second addition of the same amount of hydrogen peroxide was performed at 4 h (H2O2 2×) and cultures were assayed at 8 h. Data are expressed as the fold induction of β-galactosidase specific activity (units/µg protein ± SEM) relative to that in control cultures. In (D) and (E) a mid log culture was diluted 1:10 in CM (2.5 µM zinc) drop-out medium containing the indicated concentrations of cadmium (D) or H2O2 (E) and cell density was monitored for up to 12 h. These same experiments were carried out on ZHY6 cells containing integrated MTF-1 expression vector and the integrated –150:βgal reporter vector (see Fig. 4), with essentially the same results.
It was noted during these experiments that some colonies exhibited a much more robust zinc induction when grown in medium containing 0.5 µM zinc (Fig. 4, colony C), as was demonstrated earlier (Fig. 3). However, this was not always the case (Fig. 4, colony D, and Fig. 5, colonies 4 and 5) in colonies containing the integrated reporter vectors. The reason for this inconsistency is not clear, but may reflect some genetic variation among the selected colonies. Although not shown here, we have found that expression of MTF-1 in the yeast strain DY1457, which has a functional zinc-regulated transporter system, results in zinc-responsive reporter gene expression which is also not effected by the amount of zinc in the culture medium.
Figure 5.
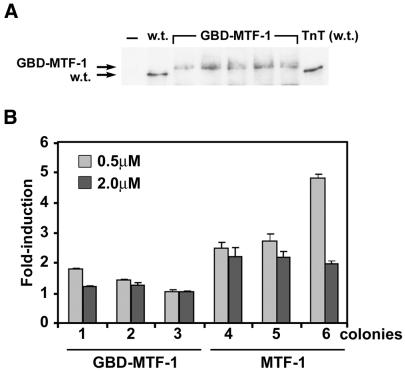
A GBD–MTF fusion protein exerts constitutive activity, but not zinc inducibility, on a promoter containing multiple GREs in yeast. An expression vector encoding the yeast GBD fused to the N-terminus of mouse MTF-1 (GBD–MTF-1) was integrated into the ZHy6 genome, followed by integration of a β-galactosidase reporter gene under control of five tandem copies of the GRE, as described in Materials and Methods. (A) Western blotting of immunoprecipitates was used to monitor the relative levels of the GBD–MTF-1 fusion protein accumulated in the yeast. Cell extracts (700 µg) were immunoprecipitated using anti-MTF-1 antibody and the immunoprecipitates were analyzed by western blotting. Lane w.t., native MTF-1, also immunoprecipitated from 700 µg of extract, expressed from an integrated vector (see Fig. 4B, lane 3, and 4C, w.t.); lanes GBD–MTF-1, five randomly selected colonies of yeast containing the integrated GBD–MTF-1 expression vector; lane TnT (w.t.), recombinant mouse MTF made in a TnT lysate (1 µl), as described in the legend to Figure 2. (B) Representative colonies expressing the GBD–MTF-1 fusion protein (lanes 1–3) or native MTF (lanes 4–6) were grown in medium containing the indicated concentrations of zinc and then exposed to excess zinc for 8 h before measuring β-galactosidase activity, as described in the legend to Figure 4. Data are expressed as the fold induction of β-galactosidase specific activity (units/mg protein ± SEM) relative to that in control cultures.
To further examine the mechanisms of action of MTF-1 in yeast, we tested the ability of mouse MTF-1 with zinc fingers 5 and 6 deleted (MTFΔ5,6) to confer zinc-mediated induction of gene expression in ZHY6 cells. Our previous studies demonstrated that MTF-1 with or without fingers 5 and 6 could activate an episomal MREd5:βgal reporter vector in a zinc-dependent manner in these yeast (23). Herein, we examined the ability of this finger deletion mutant of MTF-1 to activate β-galactosidase expression from the integrated intact MT-I promoter (–150) (Fig. 4C). Deletion of fingers 5 and 6 of MTF-1 had no effect on zinc induction of the integrated MT-I promoter. Therefore, the biological function of these zinc fingers of MTF-1 is not revealed in this experimental system.
A GBD–MTF-1 fusion protein does not confer zinc-inducibility on a GRE-driven reporter gene
Studies on the mechanisms of action of mouse MTF-1 suggest that the DNA-binding domain plays a central role in metal and oxidative stress induction of gene expression (16). Whether zinc also affects MTF-1 functions after it binds to DNA or in addition to DNA-binding activation remains unclear. To begin to address this issue here, an expression vector which encodes the GBD of Gal4 (147 residues) fused in-frame to mouse MTF-1 (GBD–MTF) was examined for its ability to respond to zinc by activation of a promoter consisting of five copies of a GRE immediately upstream of a TATA box and the β-galactosidase reporter gene. Expression of GBD–MTF in these cells was verified by immunoprecipitation and western blotting (Fig. 5A).
Expression of GBD–MTF led to a 9-fold increase in β-galactosidase activity [control = 38 ± 14 mAU/min/µg protein (n = 8); GBD–MTF = 340 ± 168 mAU/min/µg protein (n = 22)] in these yeast, which was not further increased by treatment with zinc (Fig. 5B). The large standard error of the mean in these measurements reflects the variation in absolute levels of β-galactosidase among the individual colonies. β-Galactosidase levels were also variable among colonies containing the MRE/MT reporter vectors, but were generally higher in colonies expressing higher levels of wild-type MTF-1 (Figs 2 and 4) without diminishing zinc induction. However, in the experiments shown in Figure 5, the levels of GBD–MTF were relatively low (Fig. 5A) and were similar to those of MTF-1 found in the colony labeled w.t. and also shown in Figure 4B (lane 3) and Figure 4C (lane labeled w.t.). Basal β-galactosidase levels [79 ± 50 mAU/min/µg protein (n = 20)] in these cells were about 4.3 times lower than those found in cells expressing the GBD–MTF fusion protein. Furthermore, zinc treatment increased β-galactosidase levels in the MTF-1-expressing cells 3.2-fold (Fig. 4B) and 4.3-fold (Fig. 4C) in separate experiments. These results suggest that the GBD–MTF fusion protein is able to efficiently transactivate gene expression in a zinc-independent manner. Thus, the rate limiting step in zinc responsiveness of MTF-1 in yeast is the zinc-dependent recruitment of the transactivation domains to the promoter rather than an effect exerted after DNA binding. Furthermore, these studies suggest that the Gal4 zinc-finger DNA-binding domain is constitutively active in these yeast, in contrast to the MTF-1 zinc finger domain.
Mouse MTF-1 expressed in yeast does not activate gene expression in response to toxic concentrations of cadmium or hydrogen peroxide
Cadmium and oxidative stress activation of MRE-driven gene expression is dependent on MTF-1 (8) in mouse cells, and previous studies suggest that mutations in MTF-1 which result in the loss of responsiveness to zinc also result in the loss of responsiveness to cadmium and oxidative stresses. However, cadmium has little effect on the DNA-binding activity of MTF-1 in vitro or in vivo. Therefore, it is unclear whether these inducers utilize common or distinct mechanisms for activation of MTF-1. To examine this issue, MTF-1 was expressed in yeast and its ability to induce reporter gene expression in response to toxic levels of cadmium and hydrogen peroxide was examined. Neither cadmium nor hydrogen peroxide was able to induce reporter gene expression in the co-transfected ZHy6 cells (Fig. 6). This was observed using a wide range of concentrations of these agents and using cells which had been cultured under zinc-limiting as well as zinc-replete conditions (compare Fig. 6A and B with C). Furthermore, higher initial concentrations of zinc in the culture medium (8 and 16 µM) did not enable cadmium activation of the reporter gene (data not shown). Growth of the culture was monitored to confirm that these concentrations of cadmium and hydrogen peroxide were biologically active under these experimental conditions. The lowest concentrations of cadmium (15–30 µM) and hydrogen peroxide (100 µM) examined were mildly toxic and slowed cell growth, whereas higher concentrations of these agents were more toxic and prevented growth of the cells (Fig. 6D and E). Although the USF/ARE promoter element participates in cadmium and oxidative stress induction of the mouse MT-I gene in mouse cells (9,35), the intact MT-I promoter (–150) was not responsive to cadmium or hydrogen peroxide in these yeast (data not shown). Therefore, these results reveal that distinct mechanisms are involved in MTF-1-mediated gene expression in response to zinc versus cadmium and hydrogen peroxide.
DISCUSSION
These studies were undertaken to address the mechanisms of activation of the transcription factor MTF-1. This zinc-finger protein is highly conserved during evolution and serves to activate a group of protective genes in response to a diverse set of inducers, including essential and non-essential metals and oxidants. Furthermore, this protein serves an essential function during embryonic development of the mouse. In these studies it was shown that mouse MTF-1 when expressed in yeast cells is capable of acting as a zinc-sensing metalloregulatory protein. This finding is consistent with previous studies which demonstrated that this factor directly and reversibly interacts with zinc and that these interactions reversibly modulate its DNA-binding activity.
Mutational analysis of the zinc-finger domain of MTF-1 suggests that a subset of the zinc fingers function as zinc sensors. Our previous studies demonstrated that deletions of fingers 5 and 6 did not diminish the metalloregulatory functions of MTF-1 measured using episomal reporter genes. Deletion of those fingers also does not diminish the ability of MTF-1 to activate the native MT-I promoter integrated and packaged into chromatin in the yeast genome. Thus, these experiments were uninformative with regard to the in vivo functions of these highly conserved zinc fingers, but suggest that they are not alone responsible for the metal-sensing functions of MTF-1. The magnitude of zinc induction of reporter gene expression in this yeast system (ZHy6 cells) was generally modest (2- to 4-fold) unless the cells were first starved of zinc. In mammalian cells, MTF-1 mediates a 10- to 20-fold induction of expression of the endogenous MT-I gene under zinc-replete culture conditions. Thus, these zinc fingers of MTF-1 may be essential for optimal function on the native chromatin template, but not on the yeast chromatin template. The structure of yeast chromatin differs significantly from that of mammalian chromatin (42). Experiments are underway to examine that possibility.
The results presented herein suggest that the activitiy of MTF-1 is not constitutive but rather is zinc-dependent in yeast. This argues against a zinc-dependent inhibitor of MTF-1 function (30). These results further indicate that in yeast, zinc may not significantly affect the functions of MTF-1 other than its DNA-binding activity. Previous studies indicated that the DNA-binding domain and the transactivation domains of MTF-1 both contribute to optimal metal responsiveness of the protein in mammalian cells (4,33). Thus, metal ions could be envisioned to cause not only changes in the conformation of the DNA-binding domain, but also in the transactivation domain(s) which is essential for metal responsiveness.
Examination of the full-length MTF-1 peptide fused with the GBD provided an approach to test this possibility in this heterologous system. The finding that the GBD–MTF fusion protein constitutively activates transcription under zinc-limiting conditions suggests that a metal-induced alteration in the transactivation domains is not essential for the fundamental metalloregulatory function of this protein. However, this conclusion must be made with some caution. In the absence of extensive mutagenesis experiments, it is not possible to conclude unequivocally which region(s) of the GBD–MTF fusion protein exerted the transactivation function, although the simplest explanation of these results is that the MTF-1 transactivation domains served this function in the fusion protein. Furthermore, it is possible that fusion of the GBD to the N-terminus of MTF-1 could result in a conformational change in the protein which, in turn, diminishes a potential zinc dependence of the transactivation domain(s). However, the N-terminal region of MTF-1 is not conserved during evolution and is variable in structure, which suggests that the addition of the GBD would not have a profound effect on MTF-1 conformation. Finally, the levels of reporter gene expression in zinc-limited cells expressing the GBD–MTF fusion protein compared with those in zinc-treated cells expressing MTF-1 were very similar. This suggests that the transactivation of transcription by both of these proteins was similar in efficacy. However, these studies do not exclude the possibility that the optimal function of MTF-1 in the context of the higher eukaryotic cell involves modifications of the transactivation domains. Overall, these results are consistent with a model in which zinc activation of the DNA-binding activity of MTF-1 is a pivotal step which serves to tether its transactivation domains to the promoter, resulting in increased transcription.
The zinc-sensing and DNA-binding functions of MTF-1 are both located, at least in part, within the N-terminal zinc fingers 1–4, but recent studies suggest that phosphorylation/dephosphorylation of MTF-1 also plays an essential role in its metalloregulatory functions (31,43,44). Although sites of metal-dependent phosphorylation of MTF-1 have not been identified, multiple protein kinase pathways appear to play a role in its activation of MT-I gene expression without effecting its DNA-binding activity (43). MTF-1 contains a serine + threonine-rich transactivation domain (32) and several potential sites of phosphorylation. Such post- translational modifications may serve to potentiate the metalloregulatory functions of MTF-1 in mammalian cells. As mentioned above, the magnitude of induction of reporter gene expression by MTF-1 in this yeast system was attenuated relative to its activity on the native mouse MT-I gene. Recent studies suggest the existence of a zinc-sensing receptor on mammalian cells which may trigger the release of calcium in the cell (45,46), which, in turn, may activate protein kinase cascades (47). Given the fundamental roles of zinc in cell proliferation and differentiation, it is conceivable that yeast cells could also possess a zinc-responsive kinase or phosphatase that acts on mouse MTF-1. Our results clearly suggest, however, that zinc and cadmium must utilize distinct signaling cascades to modulate the transactivation functions of MTF-1.
A novel finding of these studies was that, in contrast to results obtained in mammalian cells, hydrogen peroxide and cadmium did not cause mouse MTF-1 to activate reporter gene expression in this yeast system. Factors in addition to MTF-1 are required to activate MT gene expression in response to these agents. Previous studies suggested that activation of the mouse MT gene by hydrogen peroxide is mediated, in part, by a transient increase in ‘free’ zinc which serves as a second messenger to activate MTF-1 DNA-binding activity (16). In mouse cells, hydrogen peroxide treatment causes a rapid and transient increase in DNA-binding activity of MTF-1 (9). The source of the zinc released during oxidative stress is unknown. Mammalian cells can store zinc in vesicles (48) and alter their uptake and efflux of zinc (49). Oxidative stress may cause the release of bound intracellular zinc by oxidation of thiols. Nitric oxide induces zinc release from mammalian MT (50), and oxidized glutathione, which is increased during oxidative stress, has also been shown to mobilize zinc from mammalian MT (51). However, the mouse MT-I gene remains responsive to oxidative stress and cadmium in mouse cells which lack functional MT-I and MT-II proteins (G.K.Andrews, unpublished data), and yeast cells can also sequester zinc in vesicles and regulate zinc uptake and efflux (41,52). A possible explanation for this observation is that relative to the yeast cell, mammalian cells contain large amounts of zinc (200 µM), the overwhelming majority of which is probably bound to a myriad of low and high affinity binding sites (53). Oxidative stress may release or redistribute zinc from many sites within this large pool in mammalian cells. It is also possible that oxidative stress may activate signal transduction cascades that impinge on MTF-1 function in mammalian cells but that are not present in yeast (54).
Treatment of cells with cadmium, in contrast to oxidative stress, does not result in a large increase in the DNA-binding activity of MTF-1 (26–29). Therefore, it is thought that cadmium may potentiate the transactivation activity of MTF-1 without increasing the amount of MTF-1 bound to the promoter. The finding that cadmium does not activate MTF-1 in yeast is consistent with that concept. As mentioned above, perhaps these yeast lack a cadmium-‘activated’ kinase found in mammalian cells. Cadmium can stimulate myosin light chain kinase (55), protein kinase C (56), mitogen-activated protein kinase (57,58) and c-Jun N-terminal kinase (59). This yeast system should provide an approach to identifying other factors which impinge on the activation of gene expression by MTF-1.
Whether MTF-1 senses changes in free zinc ions and/or zinc–protein complexes in these experiments is not known. Copper trafficking in yeast is tightly controlled (60,61) and the metallochaperone yCCS binds intracellular copper and delivers this metal specifically to the antioxidant enzyme copper-zinc superoxide dismutase (61). Free copper is essentially undetectable in yeast and yCCS is essential to activate copper-zinc superoxide dismutase (60). Similarly, recent studies of bacteria suggest that no free zinc ions exist in the cell and that zinc-sensing metalloregulatory proteins have femtomolar binding constants (53,62,63). Direct metalloregulation of MTF-1 in yeast suggests that zinc presentation to MTF-1 may not be metallochaperone-specific. This is consistent with the concept that ‘labile’ zinc directly and reversibly interacts with the zinc finger domain of MTF-1.
In summary, the studies reported herein demonstrate that mouse MTF-1 functions as a direct sensor of changes in zinc levels in yeast. MTF-1 activation of gene expression by zinc does not require unique complexes not also found in yeast. In contrast, additional factors are required to mediate cadmium and hydrogen peroxide activation of MTF-1.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Jim Geiser and Steve Eklund for excellent technical assistance and to Dr L. J. Martins for the construction of the MTF-1 expression vector and the MRE-driven β-galactosidase reporter gene vector. This work was supported, in part, by NIH grant ES05704 to G.K.A. and by NIH grant ES03817 to D.R.W. P.J.D. was supported, in part, by NIH training grant T32 ES07079 and by individual grant NRSA F32 HD40705.
REFERENCES
- 1.Stuart G.W., Searle,P.F., Chen,H.Y., Brinster,R.L. and Palmiter,R.D. (1984) A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc. Natl Acad. Sci. USA, 81, 7318–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuart G.W., Searle,P.F. and Palmiter,R.D. (1985) Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature, 317, 828–831. [DOI] [PubMed] [Google Scholar]
- 3.Radtke F., Heuchel,R., Georgiev,O., Hergersberg,M., Gariglio,M., Dembic,Z. and Schaffner,W. (1993) Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J., 12, 1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugnera E., Georgiev,O., Radtke,F., Heuchel,R., Baker,E., Sutherland,G.R. and Schaffner,W. (1994) Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res., 22, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otsuka F., Iwamatsu,A., Suzuki,K., Ohsawa,M., Hamer,D.H. and Koizumi,S. (1994) Purification and characterization of a protein that binds to metal responsive elements of the human metallothionein IIA gene. J. Biol. Chem., 269, 23700–23707. [PubMed] [Google Scholar]
- 6.Zhang B., Egli,D., Georgiev,O. and Schaffner,W. (2001) The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol., 21, 4505–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auf der Maur A., Belser,T., Elgar,G., Georgiev,O. and Schaffner,W. (1999) Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol. Chem., 380, 175–185. [DOI] [PubMed] [Google Scholar]
- 8.Heuchel R., Radtke,F., Georgiev,O., Stark,G., Aguet,M. and Schaffner,W. (1994) The transcription factor MTF-I is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J., 13, 2870–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton T.P., Li,Q.W., Bittel,D., Liang,L.C. and Andrews,G.K. (1996) Oxidative stress activates metal-responsive transcription factor-1 binding activity—occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J. Biol. Chem., 271, 26233–26241. [DOI] [PubMed] [Google Scholar]
- 10.Murphy B.J., Andrews,G.K., Bittel,D., Discher,D.J., McCue,J., Green,C.J., Yanovsky,M., Giaccia,A., Sutherland,R.M., Laderoute,K.R. et al. (1999) Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res., 59, 1315–1322. [PubMed] [Google Scholar]
- 11.Günes Ç., Heuchel,R., Georgiev,O., Müller,K.H., Lichtlen,P., Blüthmann,H., Marino,S., Aguzzi,A. and Schaffner,W. (1998) Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J., 17, 2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichtlen P., Georgiev,O., Schaffner,W., Aguzzi,A. and Brandner,S. (1999) The heavy metal-responsive transcription factor-1 (MTF-1) is not required for neural differentiation. Biol. Chem., 380, 711–715. [DOI] [PubMed] [Google Scholar]
- 13.Andrews G.K., Lee,D.K., Ravindra,R., Lichtlen,P., Sirito,M., Sawadogo,M. and Schaffner,W. (2001) The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J., 20, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmade S.J., Ravindra,R., Daniels,P.J. and Andrews,G.K. (2000) The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem., 275, 34803–34809. [DOI] [PubMed] [Google Scholar]
- 15.Green C.J., Lichtlen,P., Huynh,N.T., Yanovsky,M., Laderoute,K.R., Schaffner,W. and Murphy,B.J. (2001) Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res., 61, 2696–2703. [PubMed] [Google Scholar]
- 16.Andrews G.K., (2000) Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol., 59, 95–104. [DOI] [PubMed] [Google Scholar]
- 17.Dalton T.D., Bittel,D. and Andrews,G.K. (1997) Reversible activation of the mouse metal response element-binding transcription factor-1 DNA binding involves zinc interactions with the zinc-finger domain. Mol. Cell. Biol., 17, 2781–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X.H., Agarwal,A. and Giedroc,D.P. (1998) Structural and functional heterogeneity among the zinc fingers of human MRE-binding transcription factor-1. Biochemistry, 37, 11152–11161. [DOI] [PubMed] [Google Scholar]
- 19.Chen X.H., Chu,M.H. and Giedroc,D.P. (1999) MRE-binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity and specificity of the metal-response element complex. Biochemistry, 38, 12915–12925. [DOI] [PubMed] [Google Scholar]
- 20.Koizumi S., Suzuki,K., Ogra,Y., Gong,P. and Otuska,F. (2000) Roles of zinc fingers and other regions of the transcription factor human MTF-1 in zinc-regulated DNA binding. J. Cell Physiol., 185, 464–472. [DOI] [PubMed] [Google Scholar]
- 21.Giedroc D.P., Chen,X., Pennella,M.A. and LiWang,A.C. (2001) Conformational heterogeneity in the C-terminal zinc fingers of human MTF-1: an NMR and zinc-binding study. J. Biol. Chem., 276, 42322–42332. [DOI] [PubMed] [Google Scholar]
- 22.Apuy J.L., Chen,X., Russell,D.H., Baldwin,T.O. and Giedroc,D.P. (2001) Ratiometric pulsed alkylation/mass spectrometry of the cysteine pairs in individual zinc fingers of MRE-binding transcription factor-1 (MTF-1) as a probe of zinc chelate stability. Biochemistry, 40, 15164–15175. [DOI] [PubMed] [Google Scholar]
- 23.Bittel D.C., Smirnova,I. and Andrews,G.K. (2000) Functional heterogeneity in the zinc fingers of the metalloregulatory transcription factor, MTF-1. J. Biol. Chem., 275, 37194–37201. [DOI] [PubMed] [Google Scholar]
- 24.Apostolova M.D., Chen,S.L., Chakrabarti,S. and Cherian,M.G. (2001) High-glucose-induced metallothionein expression in endothelial cells: an endothelin-mediated mechanism. Am. J. Physiol. Cell Physiol., 281, C899–C907. [DOI] [PubMed] [Google Scholar]
- 25.Saydam N., Georgiev,O., Nakano,M.Y., Greber,U.F. and Schaffner,W. (2001) Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem., 276, 25487–25495. [DOI] [PubMed] [Google Scholar]
- 26.Koizumi S., Yamada,H., Suzuki,K. and Otsuka,F. (1992) Zinc-specific activation of a HeLa cell nuclear protein which interacts with a metal responsive element of the human metallothionein-IIA gene. Eur. J. Biochem., 210, 555–560. [DOI] [PubMed] [Google Scholar]
- 27.Bittel D., Dalton,T., Samson,S., Gedamu,L. and Andrews,G.K. (1998) The DNA-binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J. Biol. Chem., 273, 7127–7133. [DOI] [PubMed] [Google Scholar]
- 28.Koizumi S., Suzuki,K., Ogra,Y., Yamada,H. and Otsuka,F. (1999) Transcriptional activity and regulatory protein binding of metal-responsive elements of the human metallothionein-IIA gene. Eur. J. Biochem., 259, 635–642. [DOI] [PubMed] [Google Scholar]
- 29.Smirnova I.V., Bittel,D.C., Ravindra,R., Jiang,H. and Andrews,G.K. (2000) Zinc and cadmium can promote the rapid nuclear translocation of MTF-1. J. Biol. Chem., 275, 9377–9384. [DOI] [PubMed] [Google Scholar]
- 30.Palmiter R.D., (1994) Regulation of metallothionein genes by heavy metals appears to be mediated by a zinc-sensitive inhibitor that interacts with a constitutively active transcription factor, MTF- 1. Proc. Natl Acad. Sci. USA, 91, 1219–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaRochelle O., Gagne,V., Charron,J., Soh,J.W. and Seguin,C. (2001) Phosphorylation is involved in the activation of metal-regulatory transcription factor 1 in response to metal ions. J. Biol. Chem., 276, 41879–41888. [DOI] [PubMed] [Google Scholar]
- 32.Radtke F., Georgiev,O., Müller,H.-P., Brugnera,E. and Schaffner,W. (1995) Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res., 23, 2277–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller H.P., Brugnera,E., Georgiev,O., Badzong,M., Müller,K.H. and Schaffner,W. (1995) Analysis of the heavy metal-responsive transcription factor MTF-1 from human and mouse. Somat. Cell Mol. Genet., 21, 289–297. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H., and Eide,D.J. (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalton T.P., Palmiter,R.D. and Andrews,G.K. (1994) Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic Acids Res., 22, 5016–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers A.M., Tzagoloff,A., Kinney,D.M. and Lusty,C.J. (1986) Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene, 45, 299–310. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U.K., (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 38.Andrews G.K., Huet,Y.M., Lehman,L.D. and Dey,S.K. (1987) Metallothionein gene regulation in the preimplantation rabbit blastocyst. Development, 100, 463–469. [DOI] [PubMed] [Google Scholar]
- 39.Racker E., (1950) Crystalline alcohol dehydrogenase from Bakers’ yeast. J. Biol. Chem., 184, 313–319. [PubMed] [Google Scholar]
- 40.Davis W. Jr, De Sousa,P.A. and Schultz,R.M. (1996) Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev. Biol., 174, 190–201. [DOI] [PubMed] [Google Scholar]
- 41.MacDiarmid C.W., Gaither,L.A. and Eide,D. (2000) Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J., 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory P.D., (2001) Transcription and chromatin converge: lessons from yeast genetics. Curr. Opin. Genet. Dev., 11, 142–147. [DOI] [PubMed] [Google Scholar]
- 43.Saydam N., Adams,T.K., Steiner,F., Schaffner,W. and Freedman,J. (2002) Regulation of metallothionein transcription by the metal responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J. Biol. Chem., 277, 20438–20445. [DOI] [PubMed] [Google Scholar]
- 44.Yu C.W., Chen,J.H. and Lin,L.Y. (1997) Metal-induced metallothionein gene expression can be inactivated by protein kinase C inhibitor. FEBS Lett., 420, 69–73. [DOI] [PubMed] [Google Scholar]
- 45.Hershfinkel M., Moran,A., Grossman,N. and Sekler,I. (2001) A zinc-sensing receptor triggers the release of intracellular Ca2+ and regulates ion transport. Proc. Natl Acad. Sci. USA, 98, 11749–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maret W., (2001) Crosstalk of the group IIa and IIb metals calcium and zinc in cellular signaling. Proc. Natl Acad. Sci. USA, 98, 12325–12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beyersmann D., Block,C. and Malviya,A.N. (1994) Effects of cadmium on nuclear protein kinase C. Environ. Health Perspect., 102 (suppl. 3), 177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmiter R.D., Cole,T.B. and Findley,S.D. (1996) ZnT-2, a mammalian protein that confers resistance to zinc by facilitating vesicular sequestration. EMBO J., 15, 1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 49.McMahon R.J., and Cousins,R.J. (1998) Mammalian zinc transporters. J. Nutr., 128, 667–670. [DOI] [PubMed] [Google Scholar]
- 50.Aravindakumar C.T., Ceulemans,J. and De Ley,M. (1999) Nitric oxide induces Zn2+ release from metallothionein by destroying zinc-sulphur clusters without concomitant formation of S-nitrosothiol. Biochem. J., 344, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maret W., (1995) Metallothionein/disulfide interactions, oxidative stress and the mobilization of cellular zinc. Neurochem. Int., 27, 111–117. [DOI] [PubMed] [Google Scholar]
- 52.Eide D.J., (1998) The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr., 18, 441–469. [DOI] [PubMed] [Google Scholar]
- 53.Outten C.E., and O’Halloran,T.V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science, 292, 2488–2492. [DOI] [PubMed] [Google Scholar]
- 54.Whisler R.L., Goyette,M.A., Grants,I.S. and Newhouse,Y.G., (1995) Sublethal levels of oxidant stress stimulate multiple serine/threonine kinases and suppress protein phosphatases in Jurkat T cells. Arch. Biochem. Biophys., 329, 23–35. [DOI] [PubMed] [Google Scholar]
- 55.Chao S.H., Bu,C.H. and Cheung,W.Y. (1995) Stimulation of myosin light-chain kinase by Cd2+ and Pb2+. Arch. Toxicol., 69, 197–203. [DOI] [PubMed] [Google Scholar]
- 56.Tang N., and Enger,M.D. (1993) Cd2+-induced c-myc mRNA accumulation in NRK-49F cells is blocked by the protein kinase inhibitor H7 but not by HA1004, indicating that protein kinase C is a mediator of the response. Toxicology, 81, 155–164. [DOI] [PubMed] [Google Scholar]
- 57.Alam J., Wicks,C., Stewart,D., Gong,P.F., Touchard,C., Otterbein,S., Choi,A.M.K., Burow,M.E. and Tou,J.S. (2000) Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells—role of p38 kinase and Nrf2 transcription factor. J. Biol. Chem., 275, 27694–27702. [DOI] [PubMed] [Google Scholar]
- 58.Ding W., and Templeton,D.M. (2000) Activation of parallel mitogen-activated protein kinase cascades and induction of c-fos by cadmium. Toxicol. Appl. Pharmacol., 162, 93–99. [DOI] [PubMed] [Google Scholar]
- 59.Matsuoka M., and Igisu,H. (1998) Activation of c-Jun NH2-terminal kinase (JNK/SAPK) in LLC-PK1 cells by cadmium. Biochem. Biophys. Res. Commun., 251, 527–532. [DOI] [PubMed] [Google Scholar]
- 60.Rae T.D., Schmidt,P.J., Pufahl,R.A., Culotta,V.C. and O’Halloran,T.V. (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science, 284, 805–808. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt P.J., Rae,T.D., Pufahl,R.A., Hamma,T., Strain,J., O’Halloran,T.V. and Culotta,V.C. (1999) Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem., 274, 23719–23725. [DOI] [PubMed] [Google Scholar]
- 62.Hitomi Y., Outten,C.E. and O’Halloran,T.V. (2001) Extreme zinc-binding thermodynamics of the metal sensor/regulator protein, ZntR. J. Am. Chem. Soc., 123, 8614–8615. [DOI] [PubMed] [Google Scholar]
- 63.Outten C.E., Tobin,D.A., Penner-Hahn,J.E. and O’Halloran,T.V. (2001) Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry, 40, 10417–10423. [DOI] [PubMed] [Google Scholar]