Abstract
The riboflavin biosynthesis in bacteria was analyzed using comparative analysis of genes, operons and regulatory elements. A model for regulation based on formation of alternative RNA structures involving the RFN elements is suggested. In Gram-positive bacteria including actinomycetes, Thermotoga, Thermus and Deinococcus, the riboflavin metabolism and transport genes are predicted to be regulated by transcriptional attenuation, whereas in most Gram-negative bacteria, the riboflavin biosynthesis genes seem to be regulated on the level of translation initiation. Several new candidate riboflavin transporters were identified (impX in Desulfitobacterium halfniense and Fusobacterium nucleatum; pnuX in several actinomycetes, including some Corynebacterium species and Strepto myces coelicolor; rfnT in Rhizobiaceae). Traces of a number of likely horizontal transfer events were found: the complete riboflavin operon with the upstream regulatory element was transferred to Haemophilus influenzae and Actinobacillus pleuropneumoniae from some Gram-positive bacterium; non-regulated riboflavin operon in Pyrococcus furiousus was likely transferred from Thermotoga; and the RFN element was inserted into the riboflavin operon of Pseudomonas aeruginosa from some other Pseudomonas species, where it had regulated the ribH2 gene.
INTRODUCTION
Riboflavin (vitamin B2) is an essential component of the basic metabolism, being a precursor of coenzymes flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN). Many microorganisms as well as plants and fungi synthesize riboflavin, but it is not produced by vertebrates.
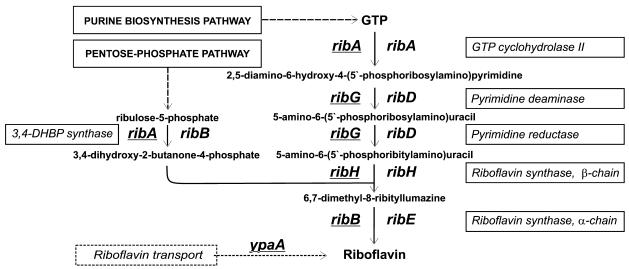
The best studied system of the riboflavin biosynthesis in bacteria is the rib operon of Bacillus subtilis encoding a pyrimidine deaminase/reductase, α-subunit of riboflavin synthase, GTP cyclohydrolase/3,4-dihydroxy 2-butanone 4-phosphate (3,4-DHBP) synthase, and β-subunit of riboflavin synthase (1). These enzymes form a pathway that creates one riboflavin molecule from one molecule of GTP and two molecules of ribulose 5-phosphate (Fig. 1). At the next stage, bifunctional flavokinase/FAD-synthase converts riboflavin to FMN and FAD, which serve as prosthetic groups for many oxidoreductases (1). Riboflavin operons were also studied in Bacillus amyloliquefaciens (2), Actinobacillus pleuropneumoniae (3) and Bartonella species (4). In Photobacterium phosphoreum and Photobacterium leiognathi, the riboflavin genes reside within the lux operon (5,6), whereas in Vibrio fisheri, the pyrimidine deaminase/reductase genes are convergent to the lux operon (7). In contrast to these genomes, the riboflavin biosynthesis genes of Escherichia coli do not form a single operon, but are scattered on the chromosome (8). The operon structures in other genomes were not studied experimentally.
Figure 1.
The riboflavin biosynthesis pathway in bacteria. Bacillus gene names are underlined.
The traditional gene names are different in E.coli and B.subtilis (Fig. 1). The bifunctional enzyme pyrimidine deaminase/reductase RibG and the α-subunit of riboflavin synthase RibB from B.subtilis have their counterparts in E.coli named RibD and RibE, respectively. Moreover, E.coli has two separate genes, ribB and ribA, that encode 3,4-DHBP synthase and GTP cyclohydrolase, respectively, whereas in B.subtilis these functions are encoded by one gene ribA. For consistency, we use the E.coli gene names throughout. Thus, the B.subtilis ribG, ribB and ribA genes are renamed here to ribD, ribE, ribB/A, respectively.
Little is known about the mechanisms of regulation of the bacterial riboflavin genes. Metabolic studies gave no evidence for any regulation of the riboflavin biosynthesis genes in E.coli (8). Based on genetic studies, the regulatory role in B.subtilis had been initially ascribed to the ribC and ribR loci (9,10) and the ribO region located between the promoter and the coding region of the ribGBAH operon (11). Later it has been shown that ribC and ribR encode flavokinase/FAD-synthase and monofunctional flavokinase, respectively (12–14). The riboflavin production is repressed by FMN, but not riboflavin (13,15), which explains why inactivation of ribC and ribR leads to overproduction of riboflavin.
Mutations in the regulatory region ribO release the repression in B.subtilis and B.amyloliquefaciens, and a hypothetical trancription terminator has been observed between this region and the translation start of the first gene in the operon (2,11). A short transcript corresponding to the leader region of the rib operon was identified by northern hybridization analysis (16). It has been suggested that the regulation involves a termination–anti-termination mechanism (2,15). Indeed, this locus is conserved in several bacteria from diverse taxonomic groups (2), and it can fold into a conserved RNA secondary structure with a base stem and four hairpins, named the RFN element (17).
In addition to the riboflavin biosynthesis genes, the RFN element was observed upstream of ypaA genes in several Gram-positive genomes. The product of this gene, YpaA, has five predicted transmebrane segments, which has lead us to the prediction that it is a transporter of riboflavin or related compounds, co-regulated with other riboflavin genes (17). Both these predictions have been verified in experiments. YpaA was shown to transport flavins (18). FMN was shown in a microarray-based experiment to decrease the level of the full-length transcripts of the riboflavin operon and ypaA, and to cause appearance of short attenuator transcripts (15).
The current availability of many complete genomes gives an opportunity to compare genes encoding one metabolic pathway and their regulation in a variety of bacteria. The comparative analysis is a powerful approach to the prediction of the DNA and RNA regulation in bacterial genomes (19). In particular, it has been used to analyze attenuators of transcription of the aromatic amino acid operons in γ-proteobacteria (20), to predict the secondary structure of RNA (21), and to find candidate iron-responsive elements in E.coli (22). In such studies, analysis of complementary substitutions in aligned sequences is used to construct a single conserved structure. Another comparative technique for analysis of gene functions is based on the assumption that functionally coupled genes are often clustered on the chromosome (23). Simultaneous analysis of probable operon structures and regulatory elements is the most effective theoretical method of functional annotation when the standard homology-based methods are insufficient.
In this study we applied the comparative genomics techniques to identify the riboflavin biosynthetic genes in almost all available bacterial genomes. Analysis of the candidate RFN elements was used to predict the mechanism of regulation on the level of transcription in Gram-positive bacteria, and on the level of translation in most Gram-negative bacteria. Analysis of regulation and positional clustering of genes resulted in identification of a number of new riboflavin-related transporters. Finally, the evolutionary history of the riboflavin operons, involving a number of horizontal transfer events, was elucidated.
MATERIALS AND METHODS
The complete and partial sequences of eubacterial genomes were downloaded from GenBank (24). Preliminary sequence data were obtained also from the WWW sites of The Institute for Genomic Research (http://www.tigr.org), University of Oklahoma’s Advanced Center for Genome Technology (http://www.genome.ou.edu), the Sanger Centre (http://www.sanger.ac.uk), the DOE Joint Genome Institute (http://www.jgi.doe.gov), and the ERGO Database, Integrated Genomics, Inc. (25).
The RNA-PATTERN program (Alexey G. Vitreschak, unpublished data) was used to search for RFN elements. The input RNA pattern described the RNA secondary structure and sequence consensus motifs. The RNA secondary structure was described as a set of the following parameters: the number of helices, lengths of helices, loop lengths, and description of topology of helix pairs. The RNA pattern of the RFN element was constructed using the training set of 20 RFN elements from our previous paper (17). Each genome was scanned with the RFN pattern. The RNA secondary structures of anti-terminators and anti-sequestors were predicted using Zuker’s algorithm of free energy minimization (26) implemented in the Mfold program (http://bioinfo.math.rpi.edu/∼mfold/rna).
The similarity search was done using BLAST (27) and GenomeExplorer (28). Transmembrane segments (TMSs) were predicted using the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html). Multiple sequence alignments were constructed using CLUSTAL X (29). Phylogenetic trees were constructed by the maximum likelihood algorithm implemented in PHYLIP (30) and plotted using the GeneMaster program (A.A.Mironov, unpublished data). Candidate operons were defined as chains of genes transcribed in the same direction such that distance between adjacent genes did not exceed 100 nt.
RESULTS
RFN elements and genes of riboflavin biosynthesis and transport
Scanning of the genomic sequences by RNA-PATTERN trained at known RFN elements identified 61 elements in 49 genomes. Then, a similarity search was used to identify the riboflavin biosynthesis (RB) genes. It showed that riboflavin biosynthesis is a widely distributed metabolic pathway in eubacteria. Only spirochetes, mycoplasmas and rickettsia have neither RB genes nor RFN elements (Table 1). At that, note that the absence of genes can be reliably claimed only for complete genomes. RFN elements were found only upstream of the RB and riboflavin transport genes (Table 1).
Table 1. The operon structures of the riboflavin biosynthesis (RB) and transport genes in eubacteria.
| Genome | AB | RBS operons | Single RBS genes | Riboflavin transporters | ||
|---|---|---|---|---|---|---|
| α-Proteobacteria | ||||||
| Rhodobacter sphaeroides # | RS | ybaD-ribD/ribE2-X-ribBA-ribH-nusB | ||||
| Magnetospirillum magnetotacticum # | MMA | ybaD-ribD-ribE2-ribBA-ribH-nusB | ||||
| Rhodopseudomonas palustris # | RPA | ybaD-ribE2-ribD-ribH-nusB | ribBA | ribH2 | ||
| Mesorhizobium loti | MLO | ybaD-ribD-ribE2-ribH-nusB-rfnT | ribBA | & ^ ribH2 | ||
| Sinorhizobium meliloti | SM | ybaD-ribD-ribE2/ribH-nusB-rfnT | ribBA | & ^ ribH2 | ||
| Agrobacterium tumefaciens | AT | ybaD-ribD-ribE2/ribH-nusB-rfnT | ribBA | & ^ ribB | ||
| Brucella melitensis # | BME | ybaD-ribD-ribE2-ribH-nusB | ribBA | & ^ ribH2 | ||
| Caulobacter crescentus # | CC | ybaD-ribH1-nusB/ribD-ribE-ribBA-ribH2 | ||||
| β-Proteobacteria | ||||||
| (Neisseria) | (NM, NG) | ybaD-ribD/ribA=ribBA/ribH-nusB | ribE | |||
| (Bordetella) | (BP, BPA) | ribBA-ribH-nusB | ribD | ribE | & ^ ribB | |
| (Burkholderia), (Ralstonia) | (BU, BPS); (REU, RSO) | ribD-ribE2=ribBA-ribH-nusB | ribA | & ^ ribB | ||
| γ-Proteobacteria | ||||||
| (Enterobacteriaceae) | (EC, TY, KP, YP) | ybaD-ribD-ribH-nusB | ribA | ribE | & ^ ribB | |
| (Pasteurellaceae) | (HI, VK, AB) | ybaD-ribD/ribH-nusB | ribA | ribE | & ^ ribB | |
| ∼ Haemophilus ducreyi #, Actinobacillus pleuropneumoniae # | DU, AO | &* ribD-ribE-ribBA-ribH | ||||
| Pseudomonas aeruginosa | PA | ybaD-ribD=& ^ ribE2-ribBA-ribH-nusB | ribA | |||
| Pseudomonas fluorescens #, P.syringiae # | PU, Psy | ybaD-ribD-ribE2-ribBA=ribH-nusB | ribA/ribBA | & ^ ribH2 | ||
| Pseudomonas putida # | Ppu | ybaD-ribD-ribE2-ribBA=ribH-nusB | ribA/ribBA | ribE | & ^ ribB | |
| Shewanella putrefaciens # | Spu | ybaD-ribD-ribE2-ribBA-ribH-nusB | ribA | ribE | & ^ ribB | |
| Vibrio cholerae | VC | ybaD-ribD-ribE2-ribBA=ribH-nusB | ribA | & ^ ribB | ||
| Xylella fastidiosa | XFA | ybaD=ribD=ribE2-ribBA-ribH-nusB | ribA | |||
| Acinetobacter calcoaceticus # | AC | ybaD-ribD-X-ribE2/ribBA-ribH] | ribA | & ^ ribB | ||
| Buchnera sp. APS | BUC | mltA-ribH-thiL-ribD-nusB | ribA | ribE | ||
| ε-Proteobacteria | HP, CJ | ribBA-X-ribA/ribH-nusB | ribD | ribE | ||
| The Bacillus/Clostridium group | BS, BA, ZC, BE; SA; LLX; PN; CA, DF | &* ribD-ribE-ribBA-ribH | & ^ ypaA | |||
| ∼ Bacillus halodurans | HD | &* ribD-ribE-ribBA-ribH | None | |||
| ∼ Bacillus amyloliquefaciens | Bam | &* ribD-ribE-ribBA-ribH | ? | |||
| ∼ (Listeria), Streptococcus pyogenes | (LO, LI); ST | None | &* ypaA | |||
| ∼ Enterococcus faecalis #, Streptococcus mutans # | EF, MN | None? | & ^ ypaA | |||
| ∼ Desulfitobacterium halfniense # | DHA | ribD]/[ribE-ribBA-ribH | &* impX | |||
| Actinomycetes | ||||||
| (Mycobacterium) | (MT, ML) | ribE-X-ribBA-ribH | ribD | |||
| Corynebacterium diphtheriae # | DI | ribD-ribE-ribBA-ribH | X-pnuX | |||
| Corynebacterium glutamicum # | GLU | ribD-ribE-ribBA-ribH | & ^ pnuX | |||
| Streptomyces coelicolor # | SX | & ^ ribE-pnuX-ribBA-ribH/ribA-ribD | ||||
| Thermomonospora fusca # | TFU | & ^ ribE-pnuX-ribBA-ribH | ||||
| Atopobium minutum # | AMI | None? | & ^ ypaA | |||
| The Thermus/Deinococcus group | ||||||
| Deinococcus radiodurans | DR | & ^ ribD-ribE-ribBA-ribH | ||||
| Thermus thermophilus # | TQ | & ^ ribD-ribE-ribBA | ribH | |||
| Cyanobacteria | ribBA/ribD | ribE | ribH | |||
| Other groups of eubacteria | ||||||
| Thermotoga maritima | TM | &* ribD-ribE-ribBA-ribH | ypaA | |||
| Fusobacterium nucleatum # | FN | &* ribH-ribD-ribE-ribBA | &* impX | |||
| Chloroflexus aurantiacus # | CAU | & ^ ribD-ribE-ribBA-ribH | ribA | |||
| Aquifex aeolicus | AA | ribF-ribD/ribH-nusB | ribBA | ribE | ||
| (Chlamydia) | (QP, QT) | ybaD/ribE/ribD-ribBA=ribH | ||||
| Archaea | ||||||
| Pyrococcus furiosus | PF | ribBA-ribH-ribD-ribE | ||||
The standard E.coli names of the RB genes are used throughout (see the text for explanation and Fig. 1 for the B.subtilis equivalents). ribBA denotes the fusion gene encoding the protein consisting of two domains, RibB and RibA. Genes forming one candidate operon (with spacers <100 bp) are separated by ‘-’. Larger spacers between genes are marked by ‘=’. Operons from different loci, if shown in one column, are separated by slashes ‘/’. Non-RB genes are shown as X. The predicted RFN elements and possible terminators and sequestors are denoted by ‘&’, ‘*’ and ‘ ^’, respectively. The contig ends are marked by square brackets.
The genome abbreviations are given in column ‘AB’ with unfinished genomes marked by ‘#’. The names of taxonomic groups given in parentheses indicate similar operon structures of the RB genes in all available genomes from the group, the exclusions are listed in the table and marked ‘∼’. Additional genome abbreviations are: Neisseria meningitidis (NG), Neisseria gonorrhoeae (#, NG); Bordetella pertussis (#, BP), Bordetella bronchiseptica (#, BPA); Burkholderia cepacia (#, BU), Burkholderia pseudomallei (#, BPS); Ralstonia eutropha (#, REU), Ralstonia solanacearum (RSO); Escherichia coli (EC), Salmonella typhi (TY), Klebsiella pneumoniae (#, KP), Yersinia pestis (YP); Haemophilus influenzae (HI), Pasteurella multocida (VK), Actinobacillus actinomycetemcomitans (#, AB); Helicobacter pylori (HP), Campylobacter jejuni (CJ); Bacillus subtilis (BS), Bacillus anthracis (#, BA), Bacillus cereus (#, ZC), Bacillus stearothermophilus (#, BE), Staphylococcus aureus (SA), Lactococcus lactis (LLX), Streptococcus pneumoniae (PN), Clostridium acetobutylicum (CA), Clostridium difficile (#, DF); Listeria monocytogenes (LO), Listeria innocua (LI); Mycobacterium tuberculosis (MT), Mycobacterium leprae (ML); Chlamydia pneumoniae (QP), Chlamydia trachomatis (QT).
The RB genes form a single ribDE(B/A)H operon in all complete genomes of the Bacillus/Clostridium group except both Listeria, Enterococcus faecalis and Streptococcus pyogenes. The absence of the riboflavin biosynthetic pathway in the latter bacteria is compensated by the existence of the riboflavin transporters YpaA found in all complete genomes of this group except Bacillus halodurans. The Bacillus/Clostridium group has the most tightly regulated pathway among all considered bacteria, since all RB operons from this group, as well as the transporter genes ypaA, have upstream RFN elements.
A different structure of the RB operon was observed in actinomycetes. In Thermomonospora fusca, this operon consists of ribE, RTFU01116 (named here pnuX, see below), ribB/A and ribH. The upstream region of this operon contains a candidate RFN element. Streptomyces coelicolor has a similar organization of the riboflavin operon and RFN. The pnuX gene is homologous to the nicotinamide mononucleotide transporter pnuC from enterobacteria and encodes a protein with six predicted TMSs. Orthologs of the pnuX gene, RDI02242 and RCGL00070, were detected in two other actinomycetes, Corynebacterium diphtheriae and Coryne bacterium glutamicum. In these genomes pnuX is not clustered with RB genes, but an RFN element was found upstream of pnuX in C.glutamicum. The genome of Atopobium minutum does not contain pnuX; however, it has another transporter gene, ypaA, preceded by an RFN element. Notably, all four RFN elements detected in actinomycetes occur upstream of transporters: pnuX, or a pnuX-containing operon, or ypaA. We propose that pnuX encodes a new type of riboflavin transporter not homologous to ypaA.
Two RFN elements were found in Fusobacterium nucleatum. The first one is located upstream of the ribHDE(B/A) operon, whereas the second one precedes a new gene encoding a hypothetical protein with nine candidate TMSs. This gene, named impX, is not similar to any known protein and has only one ortholog in a Gram-positive bacterium from the Bacillus/Clostridium group, Desulfitobacterium halfniense. This ortholog is also RFN-regulated. Thus, we predict that ImpX is one more new riboflavin transporter.
Genomes of all cyanobacteria and chlamydia as well as the genome of Aquifex aeolicus have a complete set of RB genes but no RFN elements. Thermotoga maritima, Chloroflexus aurantiacus, Deinococcus radiodurans and Thermus thermophilus have a single RFN element upstream of the ribDE(B/A)H operon, the structure of the operon is similar to that in B.subtilis. The only exception is T.thermophilus where ribH is a separate gene without an RFN element. Thermotoga maritima has ypaA which is not preceded by an RFN element.
Most proteobacteria have some redundancy of the RB genes due to paralogs of the ribH, ribB/A and ribE genes. Moreover, some genomes contain not only the fused ribB/A gene, but also additional single ribB or ribA genes. The genomes of all proteobacteria, except rickettsia, have several single RB genes as well as at most one probable RB operon which usually is preceded by ybaD and followed by nusB genes.
The most tightly RFN-regulated RB genes in proteobacteria are ribB and ribH2. ribB is always a single gene and in all cases it has an upstream RFN element. The ribH2 gene, which is paralogous to ribH, was found in some α-proteobacteria and Pseudomonas species. ribH2 as a single gene is always regulated by an RFN element with only one exception in Rhodopseudomonas palustris. Phylogenetic analysis of the RB protein sequences reveals two examples of possible horizontal transfer of the ribDE(B/A)H operon from the Bacillus/Clostridium group to two genomes of Pasteur ellaceae, Haemophilus ducreyi and A.pleuropneumoniae (see below). In both cases the RFN element preceding the RB operon is also well conserved. In general, the RFN elements were found in the genomes of almost all proteobacteria. The exceptions are Xylella fastidiosa, both Neisseria, Caulobacter crescentus, ε-proteobacteria (Helicobacter pylori and Campylobacter jejuni) and some unfinished genomes from the α-proteobacteria group.
The last gene of the hypothetical RB operon ybaD-ribDEH-nusB-mlr8412 in Mesorhizobium loti encodes a hypothetical transmembrane protein with 11 predicted TMSs. This gene is similar to transporters from the MFS family and has orthologs with the same operon structure in two other rhizobium genomes, Sinorhizobium meliloti and Agro bacterium tumefaciens. Possibly, mlr8412 encodes a new type of riboflavin transporter in Rhizobiaceae, and we tentatively name it rfnT.
Possible attenuation mechanism for the RFN-mediated regulation
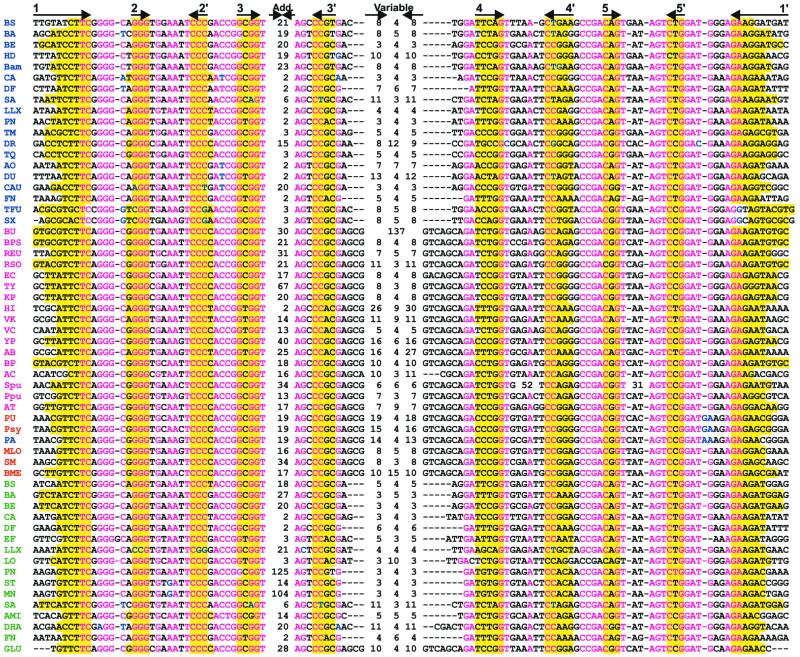
The alignment of 61 RFN elements confirms a high degree of conservation of the RFN primary and secondary structure (Fig. 2). The improved secondary structure of the RFN element is shown in Figure 3. The RFN element consists of five conserved helices, one variable stem–loop, and one facultative additional stem–loop. The lengths of the latter two hairpins are very variable and depend on the taxonomy. The maximal observed length of additional stem–loops exceeds 100 nt. The length of the variable stem–loop varies from 10 to 137 nt. All other stem–loops and internal loops in the RFN secondary structure are highly conserved, the only exception being the long loops in the fourth and fifth helices in Shewanella putrefaciens. These loops of 55 and 35 nt, respectively, can form additional stem–loops.
Figure 2.
Multiple alignment of 58 RFN elements from eubacteria. The first column contains the genome abbreviations (see Table 1). The riboflavin operons, single ribH2 genes, single ribB genes and possible riboflavin transporters are marked in blue, red, magenta and green, respectively. The complementary stems of the RNA secondary structure are shown by arrows in the upper line. Base-paired positions are highlighted by the yellow background. Conserved positions and non-consensus nucleotides are shown in red and blue, respectively. Black indicates non-conserved positions. The lengths of additional (Add.) and variable stem–loops are given.
Figure 3.
The conserved structure of the RFN element. Upper case letters, invariant (absolutely conserved) positions; lower case letters, strongly conserved positions. Dashes and asterisks indicate obligatory and facultative base pairs, respectively. Degenerate positions: R = A or G; Y = C or U; K = G or U; B = not A; V = not U. N, any nucleotide; X, any nucleotide or deletion.
The RFN elements can be classified into two major types based on the existence of two conserved fragments, AGCG and GTCAGCA, located in the branching loops adjacent to the variable helix. The RFN elements occurring in all proteobacteria (excluding H.ducreyi and A.pleuropneumoniae, see below) have these conserved sequences, whereas RFN elements without sequences are observed in the Gram-positive bacteria and other taxonomic groups, the only exception being the RFN element upstream of the pnuX gene in C.glutamicum, that belongs to the proteobacterial type.
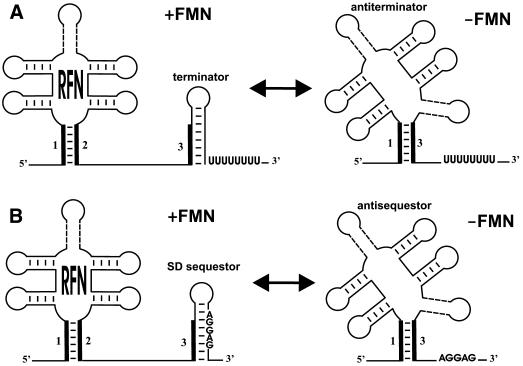
Recently, it was shown that FMNs regulate expression of the RB operon and ypaA in B.subtilis (15). We propose here a possible mechanism of the FMN-mediated regulation via the RFN element (Fig. 4).
Figure 4.
The predicted mechanism of the RFN-mediated regulation of riboflavin genes: (A) transcription attenuation; (B) translation attenuation.
Downstream of all RFN elements, there are potential hairpins that are either followed by runs of thymidines (and thus are candidate terminators) or overlap the translation start region of the first gene in the operon (and thus are candidate sequestors). Moreover, all RFN elements are capable of forming alternative structures, in which the base stem of RFN interacts with the regulatory hairpin (terminator or sequestor). This leads to formation of a structure alternative to the regulatory hairpin, similar to transcriptional and translational attenuation by competing RNA structures. Thus, two different types of regulation are suggested, attenuation of transcription via an anti-termination mechanism and attenuation of translation by sequestering of the Shine– Dalgarno (SD) box.
In Gram-positive bacteria, T.maritima and C.aurantiacus, terminator-like RNA structures are located between the predicted RFN element and the start of translation of RB genes. We found complementary fragments of RNA sequences that partially overlap both the first helix of RFN and the left stem of the terminator (Figs 4A and 5A). Furthermore, these complementary fragments always form the main helix of a new, more stable alternative secondary structure with ΔG smaller than ΔG of the RFN element. We predict that this structure functions as an anti-terminator, alternative to both the RFN element and the terminator. Thus, the RFN element is the predicted anti-antiterminator that in the repressing conditions of excess FMN prevents formation of the anti-terminator hairpin. Then, the terminator forms and transcription is preliminarily terminated. Without FMN, non-stabilized RFN is replaced by the more energy-favorable anti-termination conformation that allows the transcription read-through. In this model RFN acts as an anti-antiterminator.
Figure 5.
The conserved RNA elements in the upstream regions of the RFN-regulated genes: (A) the RFN elements and potential terminators; (B) the RFN elements and potential SD-sequestors. The yellow background indicates the first stem of the RFN element. The blue background indicates the proposed teminator/SD-sequestor. Mangenta text indicates the main stem of the anti-terminator or anti-sequestor. Arrows in the upper line show the complementary stems of these RNA secondary structures. The SD-box and the start codon are shown in green and blue, respectively. The pink background indicates additional helices in the loop of the SD-sequestor. The color code of the genome abbreviations in the first column is as in Figure 3. The presence of two terminators in the upstream region of the S.pneumoniae riboflavin operon is marked by an asterisk. The distinct groups of conserved sequestors from γ- and β-proteobacteria are marked.
In other cases, mostly in Gram-negative bacteria, the RNA hairpins downstream of the RFN element sequester the ribosome-binding site (the SD-box). In most cases we have found a highly conserved sequence, GCCCTGA, which overlaps the proposed sequestor hairpin and is complementary to the base stem of the RFN element (Figs 4B and 5B). These two complementary sequences always form the stem of the RNA secondary structure, anti-sequestor, which is more stable than the RFN element. The proposed mechanism of translational regulation of the RB operons is similar to the termination–anti-termination mechanism described above, but includes the SD-sequestor instead of the terminator. In the repressing conditions, RFN prevents formation of the anti-sequestor, and the SD-sequestor structure represses the initiation of translation. In the de-repressing conditions, RFN is replaced by the anti-sequestor that releases the SD-box and allows for initiation of translation.
We have observed two main arrangements of sequestors with respect to the SD-box and the start codon. The highly conserved sequestors in ribB in γ-proteobacteria overlap both the start codons and the SD-boxes, whereas in ribB of β-proteobacteria, sequestors overlap only the SD-boxes.
Analysis of the 5′-non-coding RNA regions of the ypaA genes reveals two possibilities for the regulation. In most cases the predicted terminator hairpin overlaps the SD-box of the ypaA gene. Therefore, this hairpin can function both as a terminator and a sequestor. The RFN elements of ribD of T.thermophilus and ypaA of A.minutum overlaps the SD-boxes directly. In these cases we predict that RFN regulates translation without additional RNA elements. In the presence of FMN, the stabilized RFN element represses initiation of translation. Conversely, RFN is not stable in the absence of FMN which results in opening of the SD-box and releasing from repression.
The left stem of the main helix of the predicted anti-terminator overlaps either the left or the right part of the base stem of the RFN element (Fig. 5A). In the first case, the anti-terminator is formed by the spacer between RFN and the terminator with intact RFN hairpins. At that, the spacer potentially folds into an additional secondary structure. In the second case, which is predominant for the ypaA genes, the alternative secondary structure of the anti-terminator only partially overlaps the RFN element and the terminator.
Paralogs and horizontal transfer
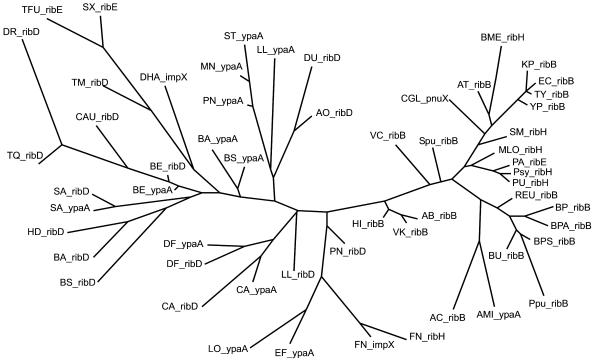
Comparison of RB protein phylogenetic trees with the standard trees for ribosomal proteins reveals some unusual branches. The most interesting observation is the likely horizontal transfer of the RB operon from the Bacillus/Clostridium group to two proteobacterial genomes. For instance, the RibH proteins from H.ducreyi and A.pleuropneumoniae cluster with RibH from the Bacillus/Clostridium group (Fig. 6). The same holds for other phylogenetic trees (data not shown). Moreover, the RB operon structure in these two proteobacteria differs from that of other proteobacteria, and is similar to the operon structure observed in the Bacillus/Clostridium group. Finally, the RFN elements upstream of these operons are of the Gram-positive type.
Figure 6.
The maximum likelihood phylogenetic tree of bacterial riboflavin synthases encoded by the ribH gene. The genome abbreviations are listed in Table 1. The separate branch including the ribH2 genes from some α-proteobacteria and Pseudomonas species is shown by bold lines. The ribH genes likely to be horizontally transferred are boxed.
Another example of possible horizontal transfer is the RB operon of F.nucleatum. Again, the RB proteins of this bacterium cluster with the Bacillus/Clostridium group (e.g. in the RibH tree, Fig. 6). Furthermore, the new RFN-regulated transporter ImpX from F.nucleatum has only one ortholog in D.halfniense, the latter belonging to the Bacillus/Clostridium group.
Finally, the RB operon observed in Pyrococcus furiosus also seems to be transferred, probably from T.maritima, as the P.furiosus RB proteins consistently cluster with those of T.maritima (e.g. in the RibH tree, Fig. 6), whereas closely related Pyrococcus horicoshii and Pyrococcus abyssi have no RB genes.
Some α-proteobacteria and Pseudomonas species have two ribH paralogs, ribH in the RB operon and a single gene ribH2 (Table 1). Interestingly, most single ribH2 genes, in contrast to ribH, are regulated by the RFN element. The RibH2 proteins form a separate group in the RibH phylogenetic tree. The same was observed for the ribE gene in proteobacteria. Some genomes, in particular E.coli, have a single ribE gene. Other genomes either have a related, non-orthologous gene in the RB operon, named ribE2, or both ribE2 and ribE (Table 1). Unlike single ribH2 genes, single ribE genes are not regulated by RFN. Finally, most proteobacteria contain single ribB and ribA genes as well as a fused ribB/A gene which is usually located in the RB operon. The single ribA gene is never regulated by RFN, whereas the single ribB gene in all cases is RFN-regulated.
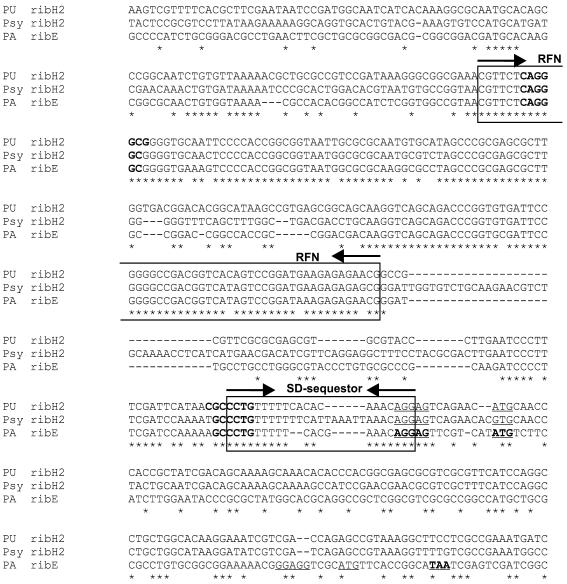
As mentioned above, the riboflavin operons in proteobacteria are not regulated by RFN, with the only exception being P.aeruginosa. In this genome we have observed a possible insertion of the RFN element inside the ybaD-ribD-ribE2-rib(B/A)-ribH-nusB operon upstream of the ribE2 gene. Strikingly, the alignment of the upstream sequences of the ribE2 gene from P.aeruginosa and the ribH2 genes from other Pseudomonas species shows that both RFN and the predicted sequestor are highly conserved (Fig. 7). This highly conserved region overlaps with the ribH2 start codon. However, the sequestor does not overlap the SD-box of the ribE2 gene in P.aeruginosa, as the ribE2 start codon lies ∼100 nt downstream. A possible explanation of this mysterious case of the RFN-mediated regulation is that regulation is carried out via translation of the leader peptide. Such a mechanism was described for regulation of the erythromycin resistance genes in B.subtilis (31). Another possibility is that we observe the result of a recent horizontal transfer, which will eventually lead to formation of a functional regulation cassette or will be eliminated. The latter possibility seems to be corroborated by the phylogenetic tree of RFN elements (Fig. 8). Indeed, the RFN element of ribE from P.aeruginosa tightly clusters with the RFN elements of ribH genes from other Pseudomonas species.
Figure 7.
The multiple alignment of the ribH2 upstream regions from P.fluorescens (PU) and P.syringiae (Psy) and the ribE2 upstream region from P.aeruginosa (PA). The highly conserved RFN elements and the predicted SD-sequestors are boxed. The main stems of the predicted anti-sequestors are shown in bold-face. The predicted SD-boxes and start codons of ribH2 and ribE are underlined. The predicted SD-box, start and stop codons of a possible short leader ORF upstream of ribE are set in bold-face and underlined.
Figure 8.
The maximum likelihood phylogenetic tree of the RFN elements. The names of the first genes of the RFN-regulated operons are given. The genome abbreviations are listed in Table 1.
Consistent with the structural analysis (above), the tree of RFN elements can be roughly divided into two main branches corresponding to Gram-positive and Gram-negative types. Another noteworthy feature of this tree is that in five of nine genomes containing two RFN elements, these elements form a single branch. On the other hand, no consistent clustering of functionally similar RFN elements (those upstream of the rib operons in the Bacillus/Clostridium genomes and those upstream of the ypaA genes) was observed. Thus, it is likely that the evolution of the RFN elements involved several independent genome-specific duplications.
DISCUSSION
Using the global analysis of RFN elements in available bacterial genomes, we have found that this conserved RNA regulatory element is widely distributed in eubacteria. Analysis of the operon structure shows that RFN predominantly regulates single RB genes (ribB or ribH2) in proteobacteria and the RB operon in most Gram-positive bacteria. Thus, in contrast to only one regulated step of riboflavin biosynthesis in the former taxonomic group, the complete riboflavin biosynthetic pathway is under FMN-mediated regulation in the latter group of bacteria. Another phylogenetic observation is that all observed RFN elements are divided into two major groups based on conservation of the fragment close to the variable stem–loop. Moreover, single RB genes seem to be regulated on the level of translation, whereas the RB operons are predicted to be regulated on the level of transcription. As a result, Gram-negative and Gram-positive bacteria significantly differ in the RFN element structure, the target of RFN-mediated regulation, and the predicted mechanism of the regulation.
The exact mechanism of regulation is not clear. It is known that FMNs can specifically bind to RNA aptamers (32). The FMN-mediated regulation of the RB genes apparently requires high conservation of the sequence and the structure of RFN due to possible FMN binding to this site. Preliminary experimental data seem to confirm involvement of the transcriptional and translational attenuation in the regulation of RB gene expression in B.subtilis and E.coli, respectively, although this model seems to be insufficient to explain all observations (A. S. Mironov, personal communication). Indeed, a number of other factors are known to be involved in the riboflavin regulation in a variety of bacteria. In particular, the ribA gene of E.coli is regulated by the soxRS locus, which is responsible for the superoxide stress response (33), and ribBA of H.pylori is regulated by the iron utilization repressor FUR (34,35). Together with observations of Fur-regulation of the superoxide stress-related genes sodA in E.coli (36), fumC and sodA in P.aeruginosa (37), sodA and sodB in Pseudomonas putida (38) and, vice versa, the regulation of the fur gene by SoxRS in E.coli (39) and regulation of siderophore biosynthesis by oxidative stress in Azotobacter vinelandii (40), this establishes an interconnection between response to the superoxide stress, iron metabolism and flavinogenesis, supported also by the observed co-regulation of the latter two systems in the yeast Pichia guilliermondii (41).
One of the remaining open problems is the meaning of positional clustering of ybaD and nusB genes with riboflavin operons in proteobacteria. The hypothetical protein YbaD contains a Zn-ribbon domain and is highly conserved in bacteria. Zn-ribbons participate in various functions, in particular DNA or RNA binding and redox reactions (42). The RNA-binding protein NusB is involved in anti-termination of E.coli ribosomal RNA operons and lambdoid phage genes (43,44). Their functional relationship with the riboflavin biosynthesis is not clear, although is has been suggested that YbaD is the riboflavin repressor (45). At that, it might be relevant that the ybaD- and nusB-containing RB operons are not regulated by RFN with only one exception in P.aeruginosa resulting from horizontal transfer. However, other genes in the same genomes are often regulated by RFN.
There are four riboflavin-related transporters. One of them is ypaA in Gram-positive bacteria from the Bacillus/Clostridium group, A.minutum (an actinomycete) and T.maritima. It is always a single gene preceded by RFN elements in all genomes excluding T.maritima. Its specificity for riboflavin and co-regulation with the RB genes was predicted in our previous paper (17) and confirmed in experiments (15,18). Two newly identified transporters, pnuX from actinomycetes and impX from F.nucleatum and Gram-positive bacterium D.halfniense are always regulated by RFN, the former as a single gene or within an operon, and the latter in both cases as a single gene. The PnuX protein is homologous to the mononucleotide transporter PnuC from enterobacteria. One more candidate transporter is rfnT from Rhizobiaceae. However, this prediction is less certain than that for other transporters, as it is based solely on positional clustering.
Finally, this study has demonstrated that the evolutionary history of the RB genes involves a number of horizontal transfer events both of the structural genes and the regulatory RFN element. These events manifest in protein phylogenetic trees, operon structures and the RFN element architecture.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Andrei Osterman for attention, advice and encouragement, Ekaterina Panina, Iain Anderson and Svetlana Gerdes for useful discussions, and D. A. Perumov and A. S. Mironov for sharing preliminary experimental data. This study was partially supported by grants from INTAS (99-1476) and the Howard Hughes Medical Institute (55000309).
REFERENCES
- 1.Perkins J.B., and Pero,J.G. (2001) Vitamin biosynthesis. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and Its Relatives: From Genes to Cells. American Society for Microbiology, Washington, DC, pp. 279–293.
- 2.Gusarov I.I., Kreneva,R.A., Podcharniaev,D.A., Iomantas,I.V., Abalakina,E.G., Stoinova,N.V., Perumov,D.A. and Kozlov,I.I. (1997) Riboflavin biosynthetic genes in Bacillus amyloliquefaciens: primary structure, organization and regulation of activity. Mol. Biol., 31, 446–453. [PubMed] [Google Scholar]
- 3.Fuller T.E., and Mulks,M.H. (1995) Characterization of Actinobacillus pleuropneumoniae riboflavin biosynthesis genes. J. Bacteriol., 177, 7265–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bereswill S., Hinkelmann,S., Kist,M. and Sander,A. (1999) Molecular analysis of riboflavin synthesis genes in Bartonella henselae and use of the ribC gene for differentiation of Bartonella species by PCR. J. Clin. Microbiol., 37, 3159–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C.Y., O’Kane,D.J. and Meighen,E.A. (1994) Riboflavin synthesis genes are linked with the lux operon of Photobacterium phosphoreum. J. Bacteriol., 176, 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J.W., Chao,Y.F. and Weng,S.F. (2001) Riboflavin synthesis genes ribE, ribB, ribH, ribA reside in the lux operon of Photobacterium leiognathi. Biochem. Biophys. Res. Commun., 284, 587–595. [DOI] [PubMed] [Google Scholar]
- 7.Lee C.Y., Szittner,R.B., Miyamoto,C.M. and Meighen,E.A. (1993) The gene convergent to luxG in Vibrio fischeri codes for a protein related in sequence to RibG and deoxycytidylate deaminase. Biochim. Biophys. Acta, 1143, 337–339. [DOI] [PubMed] [Google Scholar]
- 8.Bacher A., Eberhardt,S. and Richter,G. (1994) Biosynthesis of riboflavin. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 657–664.
- 9.Kreneva R.A., and Perumov,D.A. (1990) Genetic mapping of regulatory mutations of Bacillus subtilis riboflavin operon. Mol. Gen. Genet., 222, 467–469. [DOI] [PubMed] [Google Scholar]
- 10.Solov’eva I.M., Iomantas,Iu.A., Kreneva,R.A., Kozlov,Iu.I. and Perumov,D.A. (1997) Cloning of ribR, an additional regulatory gene of the Bacillus subtilis riboflavin operon. Genetika, 33, 739–743. [PubMed] [Google Scholar]
- 11.Kil Y.V., Mironov,V.N., Gorishin,I.Yu., Kreneva,R.A. and Perumov,D.A. (1992) Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. Mol. Gen. Genet., 233, 483–486. [DOI] [PubMed] [Google Scholar]
- 12.Gusarov I.I., Kreneva,R.A., Rybak,K.V., Podcherniaev,D.A., Iomantas,Iu.V., Kolibaba,L.G., Polanuer,B.M., Kozlov,Iu.I. and Perumov,D.A. (1997) Primary structure and functional activity of the Bacillus subtilis ribC gene. Mol. Biol., 31, 820–825. [PubMed] [Google Scholar]
- 13.Mack M., van Loon,A.P., Hohmann,H.P. (1998) Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol., 180, 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solovieva I.M., Kreneva,R.A., Leak,D.J. and Perumov,D.A. (1999) The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology, 145, 67–73. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.M., Zhang,S., Saha,S., Santa Anna,S., Jiang,C. and Perkins,J. (2001) RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol., 183, 7371–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azevedo V., Sorokin,A., Ehrlich,S.D. and Serror,P. (1993) The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci. Mol. Microbiol., 10, 397–405. [DOI] [PubMed] [Google Scholar]
- 17.Gelfand M.S., Mironov,A.A., Jomantas,J., Kozlov,Y.I. and Perumov,D.A. (1999) A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet., 15, 439–442. [DOI] [PubMed] [Google Scholar]
- 18.Kreneva R.A., Gelfand,M.S., Mironov,A.A., Iomantas,I.A., Kozlov,I.I., Mironov,A.S. and Perumov,D.A. (2000) Study of the phenotypic occurrence of ypaA gene inactivation in Bacillus subtilis. Genetika, 36, 1166–1168. [PubMed] [Google Scholar]
- 19.Gelfand M.S., Novichkov,P.S., Novichkova,E.S. and Mironov,A.A. (2000) Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform., 1, 357–371. [DOI] [PubMed] [Google Scholar]
- 20.Panina E.M., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2001) Regulation of aromatic amino acid biosynthesis in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol., 3, 529–543. [PubMed] [Google Scholar]
- 21.Eddy S.R., and Durbin,R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dandekar T., Beyer,K., Bork,P., Kenealy,M.R., Pantopoulos,K., Hentze,M., Sonntag-Buck,V., Flouriot,G., Gannon,F. and Schreiber,S. (1998) Systematic genomic screening and analysis of mRNA in untranslated regions and mRNA precursors: combining experimental and computational approaches. Bioinformatics, 14, 271–278. [DOI] [PubMed] [Google Scholar]
- 23.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overbeek R., Larsen,N., Pusch,G.D., D’Souza,M., Selkov,E.Jr, Kyrpides,N., Fonstein,M., Maltsev,N. and Selkov,E. (2000) WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res., 28, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyngso R.B., Zuker,M. and Pedersen,C.N. (1999) Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics, 15, 440–445. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S., Madden,T., Schaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mironov A.A., Vinokurova,N.P. and Gelfand,M.S. (2000) GenomeExplorer: software for analysis of complete bacterial genomes. Mol. Biol., 34, 222–231. [Google Scholar]
- 29.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein J., (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol., 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 31.Hue K.K., and Bechhofer,D.H. (1991) Effect of ermC leader region mutations on induced mRNA stability. J. Bacteriol., 173, 3732–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann T., and Patel,D.J. (2000) Adaptive recognition by nucleic acid aptamers. Science, 287, 820–825. [DOI] [PubMed] [Google Scholar]
- 33.Koh Y.S., Choih,J., Lee,J.H. and Roe,J.H. (1996) Regulation of the ribA gene encoding GTP cyclohydrolase II by the soxRS locus in Escherichia coli. Mol. Gen. Genet., 251, 591–598. [DOI] [PubMed] [Google Scholar]
- 34.Fassbinder F., van Vliet,A.H., Gimmel,V., Kusters,J.G., Kist,M. and Bereswill,S. (2000) Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp). FEMS Microbiol. Lett., 184, 225–229. [DOI] [PubMed] [Google Scholar]
- 35.Worst D.J., Gerrits,M.M., Vandenbroucke-Grauls,C.M. and Kusters,J.G. (1998) Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol., 180, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan H.M., and Schrum,L.W. (1994) Roles of manganese and iron in the regulation of the biosynthesis of manganese-superoxide dismutase in Escherichia coli. FEMS Microbiol. Rev., 14, 3153–3123. [DOI] [PubMed] [Google Scholar]
- 37.Hassett D.J., Howell,M.L., Ochsner,U.A., Vasil,M.L., Johnson,Z. and Dean,G.E. (1997) An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol., 179, 1452–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y.C., Miller,C.D. and Anderson,A.J. (1999) Transcriptional regulation by iron of genes encoding iron- and manganese-superoxide dismutases from Pseudomonas putida. Gene, 239, 129–135. [DOI] [PubMed] [Google Scholar]
- 39.Zheng M., Doan,B., Schneider,T.D. and Storz,G. (1999) OxyR and SoxRS regulation of fur. J. Bacteriol., 181, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tindale A.E., Mehrotra,M., Ottem,D. and Page,W.J. (2000) Dual regulation of catecholate siderophore biosynthesis in Azotobacter vinelandii by iron and oxidative stress. Microbiology, 146, 1617–1626. [DOI] [PubMed] [Google Scholar]
- 41.Fedorovich D., Protchenko,O. and Lesuisse,E. (1999) Iron uptake by the yeast Pichia guilliermondii. Flavinogenesis and reductive iron assimilation are co-regulated processes. Biometals, 12, 295–300. [DOI] [PubMed] [Google Scholar]
- 42.Aravind L., and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nodwell J.R., and Greenblatt,J. (1993) Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell, 72, 261–268. [DOI] [PubMed] [Google Scholar]
- 44.Luttgen H., Robelek,R., Muhlberger,R., Diercks,T., Schuster,S.C., Kohler,P., Kessler,H., Bacher,A. and Richter,G. (2002) Transcriptional regulation by antitermination. Interaction of RNA with NusB protein and NusB/NusE protein complex of Escherichia coli.J. Mol. Biol., 316, 875–885. [DOI] [PubMed] [Google Scholar]
- 45.Wolf Y.I., Rogozin,I.B., Kondrashov,A.S. and Koonin,E.V. (2001) Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res., 11, 356–372. [DOI] [PubMed] [Google Scholar]