Abstract
In fragile X syndrome, hypermethylation of the expanded CGG repeat and of the upstream promoter leads to transcriptional silencing of the FMR1 gene. Absence of the FMR1 protein results in mental retardation. We previously proved that treatment with 5-azadeoxycytidine (5-azadC) of fragile X cell lines results in reactivation of the FMR1 gene. We now show that this treatment causes passive demethylation of the FMR1 gene promoter. We employed the bisulfite-sequencing technique to detect the methylation status of individual CpG sites in the entire promoter region, upstream of the CGG repeat. Lymphoblastoid cell lines of fragile X males with full mutations of different sizes were tested before and after treatment with 5-azadC at various time points. We observed that individual cells are either completely unmethylated or not, with few relevant exceptions. We also investigated the extent of methylation in the full mutation (CGG repeat) itself by Southern blot analysis after digestion with methylation-sensitive enzymes Fnu4HI and McrBC and found that the CGG repeat remains at least partially methylated in many cells with a demethylated promoter. This may explain the quantitative discrepancy between the large extent of promoter demethylation and the limited levels of FMR1 transcriptional reactivation estimated by quantitative real-time fluorescent RT–PCR analysis.
INTRODUCTION
The highly polymorphic CGG trinucleotide repeat which is located in the 5′ untranslated region (UTR) of the fragile X mental retardation gene, FMR1, is associated with the disease phenotype when the allele carries more than 200 triplets (full mutation) (1,2). The abnormally expanded CGG repeat of fragile X chromosomes is almost invariably transcriptionally silent (3) and its inactive state has been correlated with the abnormal methylation of the CpG sites of the expanded repeat and—more importantly—of the upstream promoter region of the FMR1 gene (4,5). Rare individuals of normal intelligence were shown to carry a completely or partially unmethylated full mutation and to express the FMR1 protein (FMRP), clearly indicating that the absence of FMRP is the cause of the disease (6,7). We sought to reactivate the fully mutated FMR1 gene in vitro by inducing passive demethylation of the DNA of fragile X lymphoblastoid cell lines (8). Treatment with micromolar concentrations of 5-aza-2-deoxycytidine (5-azadC) for 7 days restored the transcriptional activity, as judged by RT–PCR, and the production of the FMR1 protein in a proportion of cells (8). DNA demethylation of the FMR1 promoter was assayed by Southern blotting after digestion with the methylation-sensitive restriction enzymes EagI, BssHII and SacII, and only with SacII could we confirm that a partial demethylation had occurred. Furthermore, the total amount of FMR1 mRNA (estimated by semi-quantitative RT–PCR) did not exceed 20% of wild-type levels, and the percentage of cells expressing FMRP was even lower (8). It seemed that only a proportion of the cells responded to the treatment, demethylating the FMR1 promoter and resuming transcription. More recently, we observed that histone hyperacetylating drugs sodium butyrate (BA) and 4-phenylbutyrate (4-PBA) synergistically potentiate the FMR1 gene reactivation induced with 5-azadC (9), thus confirming that CpG cytosine methylation and histone deacetylation cooperate in silencing chromatinic domains (10,11). In those experiments, it was also noted that cell lines harboring a shorter CGG expansion (still in the full mutation range) could be reactivated more strongly than those with larger full mutations, suggesting that it may be more difficult to obtain passive demethylation of the FMR1 promoter in the presence of longer CGG repeat tracts.
In order to test if only a small proportion or the majority of treated cells undergo FMR1 promoter demethylation, we set up a bisulfite-sequencing protocol similar to that of Stoeger et al. (12), but with a new reverse primer that allowed testing of an extra 200 bp. We could thus reconstruct the ‘epigenotype’ of individual cells for the whole FMR1 promoter, including all the in vivo footprints first reported by Schwemmle et al. (13) and the transcription start site until the CGG repeat. The four footprints correspond to the binding sites of different transcription factors, as recently reported (14). These results were correlated with the size of the CGG expansion in the tested cell lines and the levels of FMR1 mRNA estimated by real-time fluorescent RT–PCR before and after 5-azadC treatment. We also estimated the methylation status of the CGG repeat by DNA restriction analysis with the methylation-sensitive enzymes Fnu4HI and McrBC.
MATERIALS AND METHODS
Cell cultures and treatment with 5-azadC
Lymphoblastoid cell lines were established by Epstein–Barr virus (EBV) transformation from peripheral blood lymphocytes of male fragile X patients and normal male controls. Cells were grown in RPMI1640 medium with 10% fetal calf serum and penicillin/streptomycin at 37°C with 5% CO2. The medium was changed every 48 h. A 10 mM stock solution of 5-azadC (Sigma) was prepared in sterile water and stored at –80°C in aliquots.
Cells were counted, split and seeded at the initial concentration of 2.5–3 × 105 cells/ml in a total volume of 30 ml per flask. Immediately before use, 3 µl of the 10 mM 5-azadC stock solution was thawed and added daily to the flasks and thoroughly resuspended (final concentration 1 µM), while a control flask was left untreated. Cells were harvested after 3 and 8 days from the start of treatment. In the case of line E3, cells were grown for a further 5 weeks after discontinuing 5-azadC treatment and pellets were prepared every week.
DNA and RNA extraction
Cell pellets for both DNA and RNA extraction were prepared from the same flask of treated or untreated cultures at the different time points. DNA was extracted with a standard salt/chloroform procedure and subsequently employed either for bisulfite sequencing or restriction analysis with methylation-sensitive enzymes. Total RNA was extracted with the single-step acid phenol method, using RNAzol B (Tel-Test, Inc.) and employed for quantitative fluorescent RT–PCR (see below).
Bisulfite sequencing
A 5 M sodium bisulfite solution (pH 5) was prepared by dissolving 9.5 g of powder in 12 ml of distilled water, 3.5 ml of 2 N NaOH and 2.5 ml of 1 M hydrochinone. After adjusting the pH, water was added to the final volume of 20 ml. Five micrograms of genomic DNA were diluted in 50 µl of water, denatured at 95°C for 10 min, then incubated for 30 min at 37°C with 1.5 µl of 10 N NaOH (final concentration 0.3 N). We then added 310 µl of 5 M sodium bisulfite, 2.5 µl of 0.1 M hydrochinone and 136 µl of water (final volume 500 µl). This mixture was incubated overnight at 55°C under a layer of mineral oil. DNA was then purified with Promega columns, denatured with 0.3 N NaOH for 15 min at 37°C, precipitated with ammonium acetate pH 7 (final concentration 3 M) and four volumes of cold ethanol. After 30 min at –80°C the sample was centrifuged, the pellet washed with 70% ethanol and resuspended in 50 µl of water. The bisulfite-treated DNA of each sample (0.1 µl out of 50 µl) was then amplified in 12 independent PCR reactions of 25 µl in order to minimize the effects of eventual PCR artifacts. PCR reactions were performed as follows: 30 cycles (30 s at 95°C, 30 s at 63°C, 30 s at 72°C) with 10% DMSO, 200 µM dNTPs, 1 U Taq polymerase, 2.5 mM MgCl2, 1 pmol of primers 1F (5′-GGA ATT TTA GAG AGG TC/TG AAT TGG G-3′) and 5-aR (5′-CAC ACC CCC TAA CAA C-3′). A second PCR reaction was then performed with 1 µl of the first reaction as follows: 35 cycles (30 s at 95°C, 30 s at 60°C, 1 min at 72°C) with 200 µM dNTPs, 1 U Taq polymerase, 3 mM MgCl2, 1 pmol of primers 2F (5′-GTT ATT GAG TGT ATT TTT GTA GAA ATG GG-3′) and 5-aR. After the second round of nested PCR, the 12 independent reactions of each sample were pooled, partly evaporated, separated on an agarose gel, and the bands recovered with the Gibco-BRL Concert Rapid Gel extraction system (11456-019). The purified PCR products were then ligated with the TOPO TA cloning kit by Invitrogen (K460001) and used to transform bacterial cells included in the kit. After plating and overnight incubation, colonies were picked and minipreps prepared with Gibco-BRL Concert rapid plasmid miniprep system (11453-016). After a first PCR screen of clones with primers 2F and 5-aR, PCR products were cleaned on spin columns and 3–5 µl out of 30 µl were used for the sequencing reaction. We used the Amersham-Pharmacia Thermosequenase Dye Terminator kit (US79765) with primers M13F and M13R of the TOPO TA vector. Every clone was sequenced in both directions with an ABI 373 machine.
Quantitative RT–PCR analysis
Two micrograms of total RNA were pre-incubated with 0.6 µg of random hexamers (Pharmacia) at 65°C for 10 min. cDNA synthesis was then carried out at 37°C for 120 min in a total volume of 40 µl with 180 U MoMLV-RT and its buffer (Gibco-BRL), 1 mM DTT, 10 U RNase inhibitor (Promega), 0.8 mM each dNTP. Expression of FMR1-specific mRNA was at first determined in a non-quantitative manner as described previously (9), using primers specific for the housekeeping gene hypoxanthine guanine phosphorybosyltransferase (HPRT) as internal control. For a quantitative estimate of the relative FMR1 mRNA levels, we adapted the technique described by Tassone et al. (15), using an ABI 7700 Sequence Detector with dual-labeled TaqMan probes. The FMR1 amplicon is a 89-bp product spanning the junction between exons 13 and 14 of the gene (positions 1432–1520 of GenBank sequence NM_002024). The following primers and TaqMan probe were used: forward, 5′-GGA ACA AAG GAC AGC ATC GC-3′; reverse, 5′-CTC TCC AAA CGC AAC TGG TCT-3′; and TaqMan probe, 5′-(FAM)-AAT GCC ACT GTT CTT TTG GAT TAT CAC CTG AA-(TAMRA)-3′. The relative abundance of FMR1 mRNA was assessed by comparison with the human HPRT mRNA detected with the Pre-Developed TaqMan Assay Reagent ABI 4310890E (huHPRT endogenous control). Each sample was added to the reaction mix and split into four tubes and FMR1 and HPRT reactions were run in parallel. The final reaction volume was 25 µl in the TaqMan Universal PCR Master Mix (ABI 4304437) with 900 nM each primer and 100 nM dual- labeled probe for FMR1. Cycle parameters were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles with 15 s at 95°C denaturation and 1 min at 60°C annealing/ extension. Relative FMR1 levels were calculated as follows: 2–[Δt(fragile X) – ΔCt(control] = 2–ΔΔCt, where ΔCt equals Ct(FMR1) – Ct(HPRT) as discussed in Tassone et al. (15).
Southern blot analysis
Approximately 8 µg of genomic DNA were digested with PstI alone or with PstI and Fnu4HI or McrBC overnight at 37°C. The digested samples were separated on a 0.8% agarose gel with 1× TAE buffer, blotted on the Amersham Hybond N+ nylon membrane and hybridized with the radioactive XhoI–PstI fragment of the StB12.3 XX probe (corresponding to positions 13898–14462 of GenBank sequence L29074). After overnight hybridization and subsequent washing, radioactive filters were exposed to films at –80°C with reinforcing screens before development.
RESULTS
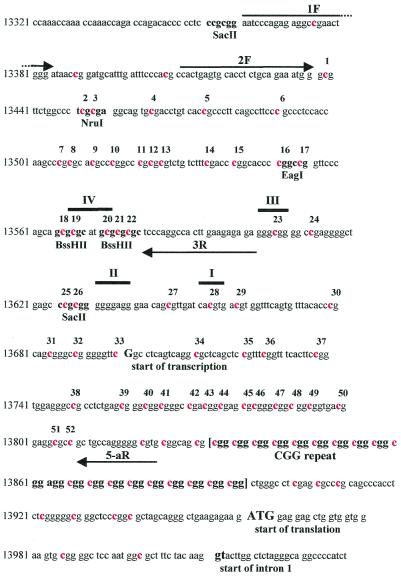
The entire FMR1 promoter region from position –641 to +79, relative to the A of the first codon ATG, is illustrated in Figure 1. The start of transcription [–264 (13698)] and the beginning of intron 1 [+52 (14013)] are indicated below the sequence, while the CGG repeat lies in the 5′ UTR ∼140 bp after the transcription start site and 70 bp before the first ATG. Primers employed by us and by Stoeger et al. (12) are indicated by arrows in the corresponding positions (1F, 2F, 3R and 5-aR) and their sequences, specific for the bisulfite-modified upper strand, are reported in the Materials and Methods. We employed a semi-nested PCR protocol with a first round of amplification using primers 1F and 5-aR, followed by a second round using primers 2F and 5-aR, which gives a final PCR product of 413 bp that was cloned and sequenced in both directions.
Figure 1.
Promoter region of the FMR1 gene (sequence numbering from GenBank L29074). Primers employed by us and others in bisulfite-PCR are indicated by arrows above or below the sequence for forward (1F, 2F) or reverse (3R, 5-aR), respectively. Individual cytosines belonging to CpG sites are indicated in red and those located between primers 2F and 5-aR are numbered from 1 to 52. Restriction sites of methylation-sensitive enzymes SacII, NruI, EagI and BssHII are indicated below the sequence, as well as the start of transcription, of translation and of intron 1. The CGG repeat is within square brackets, downstream of primer 5-aR. The four footprints (IV, III, II and I) reported by Schwemmle et al. (13) and Drouin et al. (17) are indicated with a thick line above the sequence.
As indicated in Figure 1, this PCR product contains 52 potentially methylated CpG sites spanning the in vivo footprints (I–IV) reported by Schwemmle et al. (13), as well as the recognition sites of the NruI, EagI, BssHII and SacII restriction enzymes, commonly employed to assay the methylation status of the FMR1 promoter. It is worth pointing out that Stoeger et al. (12) could analyze only the first 22 CpG sites, comprised within primers 2F and 3R, that span the EagI (CpG 16–17) as well as the two BssHII sites (CpG 18–19 and 20–21). The two BssHII sites overlap with footprint IV, which corresponds to the α-PAL/NRF1 protein binding site. The binding of α-PAL transcription factor has been recently shown to contribute to at least 50% of the total FMR1 promoter activity (14). The rest of the activity appears to be driven by the binding of transcription factors USF1 and USF2 to footprint I, coinciding with an E-box sequence, sometimes referred to as the c-MYC binding site (14). On the contrary, binding of Sp1 to recognition sequences in footprints III and II does not seem to contribute in a relevant manner to FMR1 transcription. Again, primers employed by Stoeger et al. (12) did not allow testing of this portion of promoter sequence including footprints III, II and I. When the DNA of a normal male was tested after bisulfite conversion, all the cytosines were deaminated into uracyl and showed up as thymines in the sequence (data not shown), confirming the efficiency of our bisulfite transformation protocol as well as the observation of Stoeger et al. (12) that the CpG island spanning the FMR1 promoter is not methylated on the active X chromosome.
In our present experiments we treated fragile X cell lines with 1 µM 5-azadC for variable times and extracted genomic DNA and total RNA from the whole culture, as described in the Materials and Methods. DNA was then employed for bisulfite sequencing of the FMR1 promoter region and Southern analysis of the CGG repeat itself, while RNA was used to assay FMR1 gene transcription qualitatively (with a standard RT–PCR) and quantitatively (with the fluorescent real-time PCR system ABI 7700).
Figures 2 and 3 illustrate the results of our bisulfite-sequencing experiments on three cell lines from different fragile X boys with approximately 250 (line E3, Fig. 2), 500 (line S1, Fig. 3) and between 330 and 700 (line S5, Fig. 3) CGG repeats, respectively. Sequencing was performed both in forward and reverse modes with concordant results. On top of each diagram three black bars mark positions 19–21, 23 and 28, which correspond to CpG sites included in footprints IV, III and I (from left to right). As indicated in parentheses below each panel of Figures 2 and 3, a standard RT–PCR was performed in order to quickly verify the transcriptional status of FMR1.
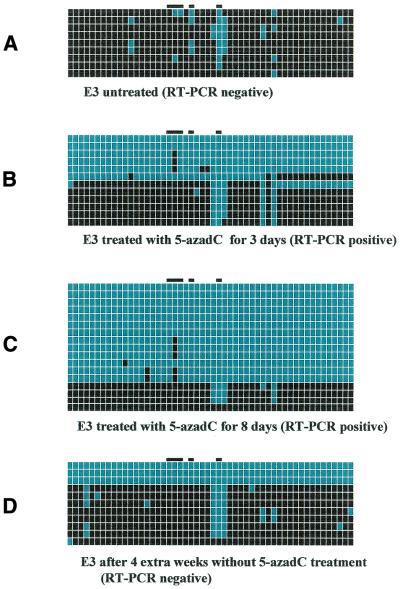
Figure 2.
FMR1 promoter region of fragile X cell line E3 (full mutation of 250 CGGs). Every line corresponds to bisulfite sequencing of an individual cell. From left to right, black or blue positions correspond to the 52 methylated or unmethylated CpG sites, respectively. Sites 19–21, 23 and 28 are marked by a black bar on the top of the diagram and are included in footprints IV, III and I, respectively. (A) The untreated E3 cell line. (B and C) The same cell line after 3 and 8 days of 5-azadC treatment (1 µM), respectively, and (D) 4 weeks after suspending the 5-azadC treatment. The presence or absence of FMR1 mRNA is indicated below each panel.
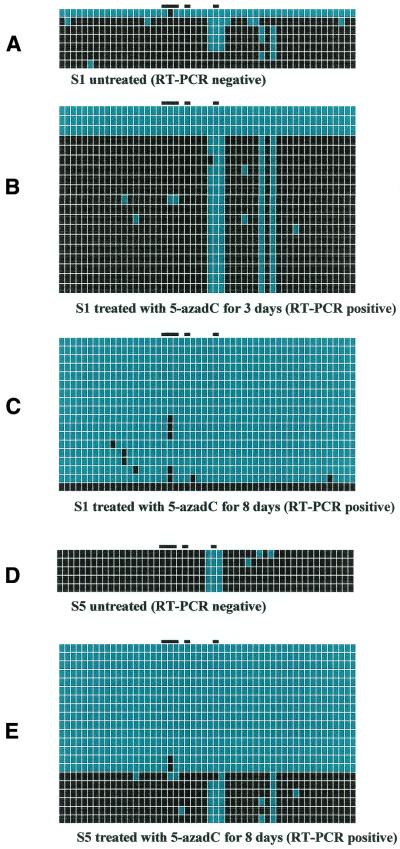
Figure 3.
FMR1 promoter region of fragile X cell lines S1 (full mutation of approximately 500 CGGs) and S5 (full mutation with multiple bands of approximately 330–700 CGGs). Every line corresponds to bisulfite sequencing of an individual cell. From left to right, black or blue positions correspond to the 52 methylated or unmethylated CpG sites, respectively. Sites 19–21, 23 and 28 are marked by a black bar on the top of the diagram and are included in footprints IV, III and I, respectively. (A) The untreated S1 cell line. (B and C) The same cell line after 3 and 8 days of 5-azadC treatment (1 µM), respectively. (D) The untreated S5 cell line and (E) cell line S5 after 8 days of 5-azadC treatment (1 µM). The presence or absence of FMR1 mRNA is indicated below each panel.
Figure 2A depicts the sequences of nine independent cells from the untreated E3 lymphoblastoid line (approximately 250 CGG repeats), which demonstrate the almost complete methylation of the FMR1 promoter with the notable exception of position 28, which is unmethylated in all but one cell. After 3 days of 1 µM 5-azadC treatment (Fig. 2B), RT–PCR became positive and almost all 52 CpGs tested were now unmethylated in five out of 12 cells, with the partial exception of position 20. Surprisingly, in the rest of the otherwise methylated cells, positions 27–29 as well as positions 36 and 38 were also unmethylated. After 8 days of 1 µM 5-azadC treatment (Fig. 2C), an even larger proportion of cells (13 out of 17) were unmethylated, again with the above mentioned exceptions of position 20 (relatively methylated) and positions 27–29, 36 and 38 (relatively unmethylated). 5-azadC treatment was stopped after day 8, but we kept culturing the E3 cell line and extracting RNA and DNA every week. The RT–PCR remained positive for three more weeks and FMR1 mRNA disappeared at the fourth week. Figure 2D illustrates the FMR1 promoter methylation status at that time point when no more mRNA was detected: three out of 11 cells were unmethylated, the other eight being almost completely methylated with the exception of positions 27–29.
Figure 3A shows that, with the exception of one cell, the FMR1 promoter of line S1 (approximately 500 CGGs) was methylated before 5-azadC treatment (again positions 27–29 being relatively unmethylated). After 3 days of 1 µM 5-azadC treatment (Fig. 3B), RT–PCR was positive but, in contrast to what we observed with line E3, only three out of 19 cells were unmethylated. Positions 27–29, as well as 36 and 38, were again the exception. After 8 days of 5-azadC treatment (Fig. 3C), all but one cell were unmethylated (only position 20 was still methylated in five out of 17 cells).
Finally, Figure 3D illustrates five methylated cells of lymphoblastoid line S5 (between 330 and 700 CGG repeats) before treatment. RT–PCR was negative, although CpG sites 27–29 are unmethylated. After 8 days of 5-azadC (Fig. 3E), 15 out of 21 cells were almost completely unmethylated (with the exception of position 20 in two cells). The other cells remained methylated with the ‘usual’ exception of CpG sites 27–29 and 38.
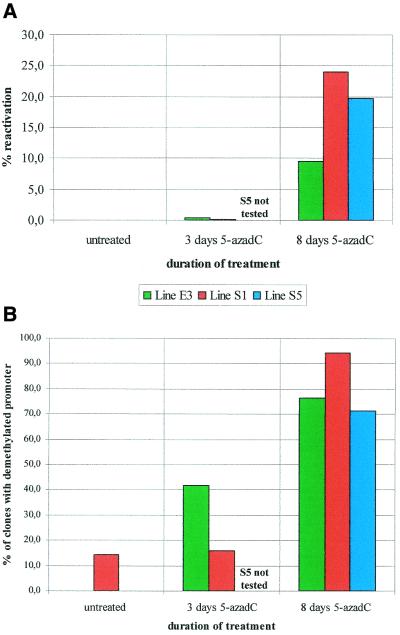
Figure 4A summarizes the results of the quantitative RT–PCR experiments performed with the ABI 7700 real-time fluorescent system. This technique, adapted by Tassone et al. (15) for the FMR1 locus, is currently considered to be as accurate as northern analysis to quantify the amount of a specific mRNA and it requires far less RNA to perform. Furthermore, each measurement reported in Figure 4A is obtained by averaging the measurements of the four independent PCR reactions performed for every sample. Figure 4B illustrates the proportion of cells with a demethylated promoter, deduced from Figures 2 and 3. We observe that after 3 days of 5-azadC treatment, only 0.4 and 0.1% of the wild-type mRNA levels are detected in the E3 and S1 cell lines, respectively, although 42 and 16% of these treated fragile X cells have an unmethylated promoter.
Figure 4.
Quantitative estimate of FMR1 mRNA levels by real-time fluorescent RT–PCR (A), compared with the proportion of cells with a demethylated FMR1 promoter (B). Note that line S5 was not tested after 3 days of 5-azadC.
The FMR1 mRNA levels eventually reached 9.5 and 24% of wild-type after 8 days of 5-azadC in lymphoblastoid lines E3 and S1, respectively, in the face of a much larger proportion of cells with an unmethylated promoter (76 and 94%, respectively). Also, lymphoblastoid line S5 reached 20% of wild-type mRNA levels after 8 days of treatment, although >70% of the cells appeared to have a demethylated promoter.
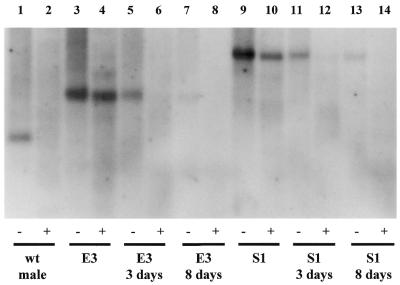
Bisulfite sequencing is not technically possible across the CG-rich full mutation; therefore, we decided to employ restriction analysis with PstI and the methylation-sensitive enzymes Fnu4HI (Fig. 5) or McrBC (data not shown) in order to test the methylation status of the CGG repeat. We employed as probe the 562 bp XhoI–PstI fragment, starting 6 bp downstream of the CGG repeat. Figure 5 illustrates the results with the Fnu4HI enzyme, which cleaves only the unmethylated GCNGC sequence (4). Fnu4HI can thus cut at each CGG repeat unit, provided that cytosines are not methylated. In fact, the normal band present in the normal male cut with PstI alone (lane 1) contains the CGG repeat and disappears when Fnu4HI is also added (lane 2). On the contrary, the expanded bands of fragile X lines E3 and S1 (lanes 3 and 9) are resistant to Fnu4HI because their CGG repeat is methylated (lanes 4 and 10). However, even if cytosines become unmethylated in few CGG units with 5-azadC treatment, we would expect to see the expanded band disappear exactly as it does in lanes 6, 8, 12 and 14 (compared with lanes 5, 7, 11 and 13). McrBC endonuclease cleaves DNA containing two half-sites (G/A)mC with a methylated cytosine, separated by 40–80 bp, and will not act upon unmethylated DNA. Therefore, McrBC will cut the PstI fragments containing a methylated full mutation and will not cut normal unmethylated samples (16). After FMR1 gene reactivation, we observed that McrBC continues to cleave the DNA, indicating that the CGG repeat is still at least partially methylated in many cells after 5-azadC treatment (data not shown). Unlike bisulfite sequencing, which provides information on single alleles, Southern blotting does not, given that digestion with methylation-sensitive enzymes is performed on genomic DNA. Still, we can conclude from the Southern experiments that the CGG expansion remains partially methylated in many cells with an unmethylated promoter.
Figure 5.
Indirect assessment of the extent of methylation in the expanded CGG repeat by Southern blot analysis, using as probe the XhoI–PstI fragment corresponding to positions 13898–14462 of GenBank sequence L29074. Odd and even lanes correspond to DNA samples digested with PstI alone (–) or PstI and Fnu4HI (+), respectively. Lanes 1–2 correspond to a normal male. Lanes 3–4 and 9–10 correspond to the untreated E3 and S1 fragile X cell lines, respectively. Lanes 5–6 and 11–12 correspond to the E3 and S1 cell lines treated for 3 days with 5-azadC (1 µM), while lanes 7–8 and 13–14 correspond to the same fragile X lines treated with 5-azadC (1 µM) for 8 days.
DISCUSSION
The structural features of the FMR1 promoter—high GC content, presence of numerous Sp1 sites, lack of a TATA box—are typical of a housekeeping gene (17), as confirmed by the ubiquitous distribution of FMR1 protein in most adult and fetal tissues (18,19). Genomic footprint analysis of the FMR1 promoter revealed four sites of protein–DNA interaction (13,17) where different transcription factors bind (see Fig. 1). The footprints are usually absent in fragile X males, as the FMR1 gene is inactive.
Kumari and Usdin (14) were able to show in murine brain that the α-PAL/NRF1 factor binds footprint IV (including CpG sites 18–22), while the USF1 and USF2 factors (but not c-MYC, nor CREB) bind footprint I (including CpG site 28). Footprints III and II, corresponding to GC-boxes of Sp1 (including CpG site 23) and Sp1-like type (no CpG sites), are probably bound by Sp3 (but not Sp1) in lymphoid cells and contribute less to the activity of the FMR1 promoter (14). In transient transfection experiments conducted in rat PC12 cells, constructs with a mutation in either the α-PAL or the E-box site had just 20% of promoter activity left, while constructs with both mutated sites had no residual activity at all. Constructs with a mutation in the GC-box of footprints III and II had 55 and 80% residual promoter activity, respectively (14).
The abnormal methylation of the CGG repeat and of the upstream promoter has been associated with silencing of the FMR1 gene in fragile X patients (3,5,20), as well as on the inactive X chromosome in normal females (4). Transfection of reporter constructs confirmed that methylation abolishes the activity of the FMR1 promoter (21,22). Furthermore, we demonstrated in vitro the possibility of reactivating methylated fragile X mutations after a demethylating treatment with 5-azadC (8). 5-azadC is incorporated in DNA and becomes covalently bound with the maintenance DNA methyltransferase (DNMT1) as this enzyme tries to methylate the trivalent nitrogen in position 5 of the pyrimidine ring (23). DNMT1 molecules are irreversibly blocked as adducts on the DNA (24), thus allowing passive demethylation to take place as cells divide. We also showed that addition of histone hyperacetylating drugs potentiates the effect of 5-azadC in reactivating the FMR1 gene (9), as it happens for several cancer genes (10). Our results support the notion that DNA methylation leads to histone deacetylation (25) and chromatin silencing, via binding of methylcytosine binding proteins such as MeCP2 and recruiting of a multiprotein complex (11).
However, histone hyperacetylating drugs alone had almost no effect, indicating that methylation must be silencing the FMR1 gene also in alternative ways. Kumari and Usdin (14) proved that methylation of CpG sites 18–20 (Fig. 1) reduces binding of α-PAL to 45% of wild-type levels, while methylation of CpG site 28 in the E-box reduces binding of USF1 and USF2 to 80%. Therefore, we now have evidence that the fully mutated FMR1 gene is silenced in at least two ways: (i) by indirectly causing a local chromatin modification that reduces the access of the transcriptional machinery to the promoter region; (ii) by direct interfering with the binding of transcription factors.
De novo methylation of fragile X full mutations is probably established during early embryogenesis (26), possibly in an effort to stabilize the expanded CGG repeat that acts like a parasitic sequence element from a genomic perspective (27). Rare individuals with an unmethylated full mutation have been described and presumably escaped such de novo methylation (7,28). These individuals have no global impairment of DNA methylation (16) and are intellectually normal or ‘high functioning’ males. Schwemmle (29) confirmed the presence of footprints in one such individual, suggesting normal transcription. Probably the unmethylated CGG expansion does not impede transcription per se, neither in the premutation (15) nor in the full mutation range (30,31). Actually, it is likely that FMR1 mRNA levels are directly correlated with the size of the CGG expansion in the effort of compensating a relative translational deficiency (32,33). Anyhow, sufficient amounts of FMR1 protein must be produced to account for the normal intellect of these rare males with unmethylated full mutations.
With the present series of experiments we intended to refine our understanding of the reactivation process by analyzing the methylation status of individual CpG sites in the FMR1 promoter before and after 5-azadC treatment, compared with the FMR1 mRNA levels quantified with the ABI 7700 system. We optimized a bisulfite-sequencing technique, similar to that of Stoeger et al. (12) and of Genc et al. (34). However, we used different primers and were able to analyze >400 bp of sequence immediately upstream of the CGG repeat (Fig. 1), including all the four binding sites for transcription factors. Panels in Figures 2 and 3 corresponding to untreated fragile X lines (Figs 2A, and 3A and D) demonstrate that the FMR1 promoter is completely methylated in fragile X cells with the relevant exception of CpG sites 27, 28 and 29, located in footprint I (see untreated S5 cell line in Fig. 3D). This observation possibly reflects a continued binding of transcription factors USF1 and USF2, which are relatively unaffected by methylation (14) and seem to still have access to their recognition sequence, though they are clearly unable to drive transcription by themselves, with the rest of the promoter fully methylated.
After 3 days of 5-azadC treatment (Figs 2B and 3B), we observed that a proportion of cells have a completely demethylated promoter, while others are still methylated, though positions 27–29, 36 and 38 are always unmethylated. Demethylation of CpG sites 36 and 38 may reflect the assembly of the transcriptional machinery on yet inactive alleles, downstream of the promoter (mRNA transcription starts at CpG site 33). It seems that 5-azadC does not induce a partial or random demethylation of just some CpG sites, but a sequential and thorough demethylation of the entire promoter, explaining why we observed with immunocytochemistry a clear reappearance of FMRP in a proportion of cells (8). After 8 days of 5-azadC treatment (Figs 2C, and 3C and E), a large proportion (70–90%) of cells are unmethylated. However, the CpG in position 20 is sometimes still methylated, thus impeding the binding of α-PAL to footprint IV and reducing transcriptional efficiency by >50% (14). It is worth pointing out that passive demethylation occurs equally on both DNA strands, as demonstrated by the concordant results of forward and reverse bisulfite sequencing.
Figure 4 shows that the amount of FMR1 mRNA does correlate with the proportion of cells with an unmethylated promoter, although we would have expected higher levels of mRNA, considering the extensive demethylation demonstrated with the bisulfite sequencing experiments. In previous experiments with a semi-quantitative radioactive RT–PCR technique with a low number of cycles (9), we had reported levels of FMR1 mRNA not exceeding 20–25% of that of a wild-type cell line after 8 days of 1 µM 5-azadC treatment. Taking into account the observations of Tassone et al. (30), confirmed by Kenneson et al. (33), that unmethylated full mutations are actually being transcribed at 5–8-fold higher levels relative to normal cells, we have to suspect that actually only 5% of our treated cells were actually reactivated. This fits well with the limited number of cells positive for FMRP that we observed by immunocytochemistry (8), but leaves us with the problem of explaining the discrepancy between the large number of cells with a demethylated promoter after 8 days of 5-azadC treatment and the lower number of reactivated cells, as judged by mRNA and protein levels.
Though our experiments with Fnu4HI (Fig. 5) and McrBC provide just a partial answer, we speculate that a large proportion of the cells with a demethylated promoter still harbor a (partially) methylated CGG repeat that either blocks or slows down transcription, even if transcription factors may bind the unmethylated promoter. Treatment with 5-azadC may succeed in demethylating the upstream promoter in most cells, but the expanded CGG repeat may ‘resist’ demethylation by attracting the few DNMT1 molecules still active in the nucleus. Therefore, methylation of the expanded CGG repeat and the binding of proteins such as the p20 CGGBP described by Muller-Hartmann et al. (35), MeCP2 itself or other methyl-DNA binding proteins, may be very important in determining the transcriptional status of the FMR1 gene. However, under other circumstances, e.g. in the case of the lung tumor with a premutation described by de Graaff et al. (36), abundant FMRP was produced and although the promoter seemed partly methylated, the premutation itself may well have been unmethylated. A combination of footprinting analysis, quantitative RT–PCR and RNA in situ hybridization studies (the latter allowing the study of individual cells) may help to settle this issue.
We had previously suggested that the size of the full mutation may be inversely correlated to the level of 5-azadC reactivation (9). However, there seems to be no such direct relationship if one considers that the E3 line (250 CGG repeats) is more unmethylated than the S1 line (500 CGG repeats) after 3 days, but these proportions invert after 8 days of 5-azadC treatment, with E3 eventually producing less than half FMR1 mRNA compared with S1. Finally, we must consider the methylation status of line E3 after 4 weeks without 5-azadC (Fig. 2D), when the RT–PCR becomes negative. Although most of the cells are again methylated (with the exception of positions 27–29), three out of 11 are still completely unmethylated. We suggest that in these cells the CGG repeat may be methylated and—as stated above—effectively suppresses transcription. The reactivated cells may have been remethylated by a de novo methyltransferase, starting from the CGG repeat and eventually spreading to the promoter. However, there is no evidence that de novo methyltransferases are expressed in adult cells after embryogenesis and we strongly suspect that many of the reactivated cells eventually died by apoptosis in the first 2–3 weeks after suspending 5-azadC because of the many DNMT1-DNA adducts.
In conclusion, a complex picture emerges from our present study, suggesting—among other things—that cytosine methylation of the FMR1 promoter and of the fully expanded CGG repeat is lost at a different rate in the presence of demethylating agents.
Acknowledgments
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Steve Warren for kindly providing the fragile X lymphoblastoid cell lines S1 and S5. This work was financially supported by the FRAXA Foundation, SIGMA-TAU and Associazione Anni Verdi to G.N.
REFERENCES
- 1.Verkerk A.J.M.H., Pieretti,M., Sutcliffe,J.S., Fu,Y.H., Kuhl,D.P.A., Pizzuti,A., Reiner,O., Richards,S., Victoria,M.F., Zhang,F. et al. (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell, 65, 905–914. [DOI] [PubMed] [Google Scholar]
- 2.Fu Y.H., Kuhl,D.P., Pizzuti,A., Pieretti,M., Sutcliffe,J.S., Richards,S., Verkerk,A.J., Holden,J.J., Fenwick,R.G.,Jr, Warren,S.T. et al. (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell, 67, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 3.Pieretti M., Zhang,F., Fu,Y.H., Warren,S.T., Oostra,B.A., Caskey,C.T. and Nelson,D.L. (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell, 66, 817–822. [DOI] [PubMed] [Google Scholar]
- 4.Hansen R.S., Gartler,S.M., Scott,C.R., Chen,S.H. and Laird,C.D. (1992) Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum. Mol. Genet., 1, 571–578. [DOI] [PubMed] [Google Scholar]
- 5.Sutcliffe J.S., Nelson,D.L., Zhang,F., Pieretti,M., Caskey,C.T., Saxe,D. and Warren,S.T. (1992) DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet., 1, 397–400. [DOI] [PubMed] [Google Scholar]
- 6.Hagerman R.J., Hull,C.E., Safanda,J.F., Carpenter,I., Staley,L.W., O’Connor,R.A., Seydel,C., Mazzocco,M., Snow,K., Thibodeau,S.N. et al. (1994) High functioning fragile X males: demonstration of an unmethylated fully expanded FMR1 mutation associated with protein expression. Am. J. Med. Genet., 51, 298–308. [DOI] [PubMed] [Google Scholar]
- 7.Smeets H.J., Smits,A.P., Verheij,C.E., Theelen,J.P., Willemsen,R., van de Burgt,I., Hoogeveen,A.T., Oosterwijk,J.C. and Oostra,B.A. (1995) Normal phenotype in two brothers with a full FMR1 mutation. Hum. Mol. Genet., 4, 2103–2108. [DOI] [PubMed] [Google Scholar]
- 8.Chiurazzi P., Pomponi,M.G., Willemsen,R., Oostra,B.A. and Neri,G. (1998) In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum. Mol. Genet., 7, 109–113. [DOI] [PubMed] [Google Scholar]
- 9.Chiurazzi P., Pomponi,M.G., Pietrobono,R., Bakker,C.E., Neri,G. and Oostra,B.A. (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum. Mol. Genet., 8, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 10.Cameron E.E., Bachman,K.E., Myohanen,S., Herman,J.G. and Baylin,S.B. (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet., 21, 103–107. [DOI] [PubMed] [Google Scholar]
- 11.Razin A., (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoeger R., Kajimura,T.M., Brown,W.T. and Laird,C.D. (1997) Epigenetic variation illustrated by DNA methylation patterns of the fragile-X gene FMR1. Hum. Mol. Genet., 6, 1791–1801. [DOI] [PubMed] [Google Scholar]
- 13.Schwemmle S., de Graaff,E., Deissler,H., Glaser,D., Wohrle,D., Kennerknecht,I., Just,W., Oostra,B.A., Dorfler,W., Vogel,W. et al. (1997) Characterization of FMR1 promoter elements by in vivo-footprinting analysis. Am. J. Hum. Genet., 60, 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari D., and Usdin,K. (2001) Interaction of the transcription factors USF1, USF2, and α-Pal/Nrf-1 with the FMR1 promoter. Implications for Fragile X mental retardation syndrome. J. Biol. Chem., 276, 4357–4364. [DOI] [PubMed] [Google Scholar]
- 15.Tassone F., Hagerman,R.J., Taylor,A.K., Gane,L.W., Godfrey,T.E. and Hagerman,P.J. (2000) Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet., 66, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burman R.W., Yates,P.A., Green,L.D., Jacky,P.B., Turker,M.S. and Popovich,B.W. (1999) Hypomethylation of an expanded FMR1 allele is not associated with a global DNA methylation defect. Am. J. Hum. Genet., 65, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drouin R., Angers,M., Dallaire,N., Rose,T.M., Khandjian,W. and Rousseau,F. (1997) Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum. Mol. Genet., 6, 2051–2560. [DOI] [PubMed] [Google Scholar]
- 18.Devys D., Lutz,Y., Rouyer,N., Belloq,J.P. and Mandel,J.L. (1993) The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nature Genet., 4, 335–340. [DOI] [PubMed] [Google Scholar]
- 19.Khandjian E.W., Fortin,A., Thibodeau,A., Tremblay,S., Cote,F., Devys,D., Mandel,J.L. and Rousseau,F. (1995) A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum. Mol. Genet., 4, 783–789. [DOI] [PubMed] [Google Scholar]
- 20.Oberlé I., Rousseau,F., Heitz,D., Kretz,C., Devys,D., Hanauer,A., Boué,J., Bertheas,M.F. and Mandel,J.L. (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science, 252, 1097–1102. [DOI] [PubMed] [Google Scholar]
- 21.Hwu W.L., Lee,Y.M., Lee,S.C. and Wang,T.R. (1993) In vitro DNA methylation inhibits FMR-1 promoter. Biochem. Biophys. Res. Commun., 193, 324–329. [DOI] [PubMed] [Google Scholar]
- 22.Sandberg G., and Schalling,M. (1997) Effect of in vitro promoter methylation and CGG repeat expansion on FMR-1 expression. Nucleic Acids Res., 25, 2883–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juttermann R., Li,E. and Jaenisch,R. (1994) Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl Acad. Sci. USA, 91, 11797–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson-Grusby L., Laird,P.W., Magge,S.N., Moeller,B.J. and Jaenisch,R. (1997) Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl Acad. Sci. USA, 94, 4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffee B., Zhang,F., Warren,S.T. and Reines,D. (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature Genet., 22, 98–101. [DOI] [PubMed] [Google Scholar]
- 26.Malter H.E., Iber,J.C., Willemsen,R., de Graaff,E., Tarleton,J.C., Leisti,J., Warren,S.T. and Oostra,B.A. (1997) Characterization of the full fragile X syndrome mutation in fetal gametes. Nature Genet., 15, 165–169. [DOI] [PubMed] [Google Scholar]
- 27.Bestor T.H., and Tycko,B. (1996) Creation of genomic methylation patterns. Nature Genet., 12, 363–367. [DOI] [PubMed] [Google Scholar]
- 28.de Vries B.B., Jansen,C.C., Duits,A.A., Verheij,C., Willemsen,R., van Hemel,J.O., van den Ouweland,A.M., Niermeijer,M.F., Oostra,B.A. and Halley,D.J. (1996) Variable FMR1 gene methylation of large expansions leads to variable phenotype in three males from one fragile X family. J. Med. Genet., 33, 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwemmle S., (1999) In vivo footprinting analysis of the FMR1 gene: proposals concerning gene regulation in high-functioning males. Am. J. Med. Genet., 84, 266–267. [PubMed] [Google Scholar]
- 30.Tassone F., Hagerman,R.J., Loesch,D.Z., Lachiewicz,A., Taylor,A.K. and Hagerman,P.J. (2000) Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet., 94, 232–236. [DOI] [PubMed] [Google Scholar]
- 31.Tassone F., Hagerman,R.J., Taylor,A.K. and Hagerman,P.J. (2001) A majority of fragile X males with methylated full mutation alleles have significant levels of FMR1 messenger RNA. J. Med. Genet., 38, 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Y., Zhang,F., Lokey,L.K., Chastain,J.L., Lakkis,L., Eberhart,D. and Warren,S.T. (1995) Translational suppression by trinucleotide repeat expansion at FMR1. Science, 268, 731–734. [DOI] [PubMed] [Google Scholar]
- 33.Kenneson A., Zhang,F., Hagedorn,K. and Warren,S.T. (2001) Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet., 10, 1449–1454. [DOI] [PubMed] [Google Scholar]
- 34.Genc B., Muller-Hartmann,H., Zeschnigk,M., Deissler,H., Schmitz,B., Majewski,F., von Gontard,A. and Doerfler,W. (2000) Methylation mosaicism of 5′-(CGG)(n)-3′ repeats in fragile X, premutation and normal individuals. Nucleic Acids Res., 28, 2141–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller-Hartmann H., Deissler,H., Naumann,F., Schmitz,B., Schroer,J. and Doerfler,W. (2000) The human 20-kDa 5′-(CGG)(n)-3′-binding protein is targeted to the nucleus and affects the activity of the FMR1 promoter. J. Biol. Chem., 275, 6447–6452. [DOI] [PubMed] [Google Scholar]
- 36.de Graaff E., Willemsen,R., Zhong,N., de Die-Smulders,C.E., Brown,W.T., Freling,G. and Oostra,B. (1995) Instability of the CGG repeat and expression of the FMR1 protein in a male fragile X patient with a lung tumor. Am. J. Hum. Genet., 57, 609–618. [PMC free article] [PubMed] [Google Scholar]