Abstract
Sox-2 is a transcriptional cofactor expressed in embryonic stem (ES) cells as well as in neuronal cells. It has been demonstrated that Sox-2 plays an important role in supporting gene expression in ES cells, especially by forming a complex with embryonic Octamer factor, Oct-3/4. Here, we have analyzed the regulatory regions of the Sox-2 gene and identified two enhancers which stimulate transcription in ES cells as well as in embryonal carcinoma cells. These regulatory regions, which we termed Sox regulatory regions (SRR) 1 and 2, exert their function specifically when cells are in an undifferentiated state. Interestingly, like the regulatory elements of FGF-4 and UTF1 genes, combinatorial action of Octamer and Sox-2 binding sites support the SRR2 activity. However, biochemical analyses reveal that, due to the unique sequence and/or its organization, the SRR2 bears distinct characteristics from those of FGF-4 and UTF1 regulatory elements. That is, unlike the FGF-4 gene enhancer, the SRR2 precludes the binding of the Oct-1–Sox-2 complex. The difference between the SRR2 and UTF1 regulatory element is in the ability of SRR2 to recruit the Oct-6–Sox-2 complex as well as the Oct-3/4–Sox-2 complex. Co-transfection analyses confirm that both complexes are able to stimulate transcription through the SRR2 element.
INTRODUCTION
Early embryogenesis in mammals is marked by numerous critical and unique events including the start of zygotic transcription and formation of extra-embryonic tissues which are distinct from those of other organisms such as Drosophila and Xenopus (1–6). The first overt differentiation step in mammals is the organization of cells to form blastocysts which occurs at ∼3.5 days post coitum in mouse development (7). This blastocyst comprises two distinct kinds of cell lineage. One of them (trophectoderm) generates a major part of the placenta and the extra-embryonic yolk sacs, whereas the other [inner cell mass (ICM)] is the founder tissue of the fetus from which both somatic and germ line tissues are derived. Therefore, ICM is characterized as a pluripotent or omnipotent tissue.
Mouse embryonic stem (ES) cells are established directly from this ICM (7,8). However, unlike other types of primary cultured cell lines, ES cells propagate as permanent cell lines when the cells are cultured under appropriate conditions and, even after extended propagation, ES cells retain the functional properties of ICM cells. Thus, if they are reintroduced into a blastocyst, ES cells resume embryonic development and participate in the formation of a chimeric mouse in which injected ES cells are distributed to essentially any type of organs/cells including germ cells (9). ES cells can also be induced to differentiate in vitro into a range of cell types including cardiomyocytes and neurons, providing an unique in vitro model system for studying the regulation of gene expression during the early developmental stages of mammals (10–13). Embryonal carcinoma (EC) cell lines are also widely used for similar purposes, as these cell lines share important properties with ES cells in terms of the ability to produce a variety of cell types in vitro by induction of differentiation (14) and to participate in regular embryonic development when injected into a normal blastocyst (15). In this context, we have been interested in elucidating the molecular mechanism which governs the above pluripotent properties of these ES and EC cells. To obtain a clue for this purpose, we have previously searched for transcriptional (co)factors expressed rather specifically in early embryonic cells and indeed cloned a cofactor termed UTF1 whose expression is restricted mainly to pluripotent embryonic cells (16). We have also found that UTF1 expression in EC and ES cells is restricted to the pluripotent state of these cells and, therefore, the expression is rapidly extinguished when these cells are induced to differentiate (16,17). In normal mouse embryos, UTF1 mRNA is present in ICM and embryonic ectoderm. During the primitive streak stage, the induction of mesodermal cells is accompanied by the down-regulation of UTF1 expression. Recently, we have found that this unique expression profile of UTF1 gene is supported by a combinatorial action of two embryonic factors, Oct-3/4 and Sox-2 (18). It is known that expression of these two transciptional factors, like that of UTF1, is down-regulated during the primitive streak stage (19–26). However, we have recently noted that, although both UTF1 and Sox-2 genes cease their expression rapidly once the embryonic ectodermal cells are converted to mesodermal cells, expression of Oct-3/4 is rather gradually down-regulated at this stage (M.Nishimoto and A.Okuda, unpublished results). As these results indicate that Sox-2 may be the primary determinant for the UTF1 expression, we have set out to elucidate the mechanism by which the Sox-2 gene is expressed in EC and ES cells specifically when these cells are in the pluripotent state.
Here, we report on the characterization of Sox-2 regulatory regions. Our analyses reveal that the Sox-2 gene bears at least two regulatory regions which work specifically in pluripotent embryonic cells. Interestingly, one of them, like the regulatory regions of fibroblast growth factor (FGF)-4 and UTF1 genes (18,27–30), carries Octamer and Sox-2 binding site-like sequences. Moreover, our analyses reveal that, due to the subtle difference in sequence and/or organization, each regulatory region possesses distinct biochemical characteristics in terms of the specificity of the interaction with Octamer factor-containing complexes.
MATERIALS AND METHODS
Accession number
The nucleotide sequence reported in this paper has been submitted to DDBJ with accession no. AB079241.
Cloning of Sox-2 genomic gene
A mouse (129 line) genomic library was screened with 32P-labeled Sox-2 cDNA (27). Hybridization, washing and Lambda DNA preparation were done as described previously (31).
Plasmid constructions
For constructing tk-Luc reporter plasmids used in Figure 1 bearing a certain portion of Sox-2 flanking region, each genomic DNA fragment was recovered from Lambda phage DNA carrying Sox-2 gene and subcloned into a SalI/BamHI site with the aid of appropriate linkers. Essentially, the same procedure was used to construct tk-Luc reporter plasmid used in Figure 2. However, most of the inserted DNAs were recovered by polymerase chain reaction (PCR) as a SalI/BamHI fragment and then subcloned into the vector. For constructing tk-Luc reporter plasmids bearing wild-type or mutant sequence shown in Figures 3A and 4A, we used our previously established method (32) utilizing overlapping oligonucleotides which carry a portion (from 20 to 30 nt) of Sox regulatory region (SRR) 2. Construction of pCEP4 expression vectors carrying cDNAs for Sox-2 or either one of three Octamer factors (Oct-1, Oct-3/4 and Oct-6) were described previously (18).
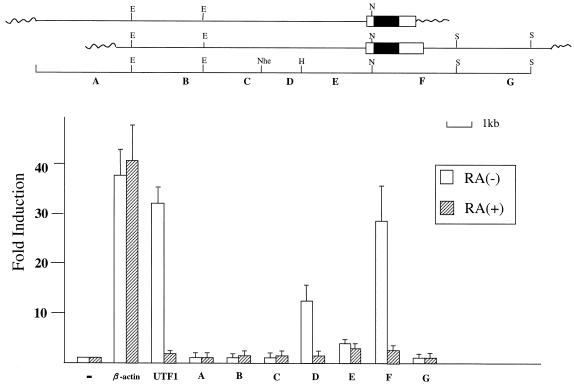
Figure 1.
Identification of two regulatory regions which exert their activities in F9 EC cells in an undifferentiated state-specific manner. Open and filled boxes indicate Sox-2 gene non-coding and coding regions, respectively, while wavy lines represent Lambda phage vector portions. Restriction enzymes are abbreviated as follows: B, BamHI; E, EcoRI; H, HindIII; N, NotI; Nhe, NheI; S, SalI. The Sox-2 gene and its flanking regions (A–G) were individually subcloned into the downstream of the luciferase gene of tk-Luc reporter plasmid. Each plasmid construct was introduced into F9 EC cells together with neomycin-resistant gene which is under the control of the β-actin promoter and an internal control luciferase gene of Renilla reniformis. After selection with G418, approximately 1000 of the drug-resistant colonies were pooled and expanded. Subsequently, the cells were incubated with medium containing charcoal-treated serum supplemented with or without 1 µM retinoic acid. After 48 h, whole-cell extracts were prepared from such treated cells and levels of transcription were determined by the dual-luciferase system according to the manufacturer (Promega). The intrinsic activity of the control tk-Luc plasmid in undifferentiated and differentiated cells is arbitrarily set as one and fold induction due to the presence of each regulatory element was calculated. Data were obtained from five independent experiments with comparable results.
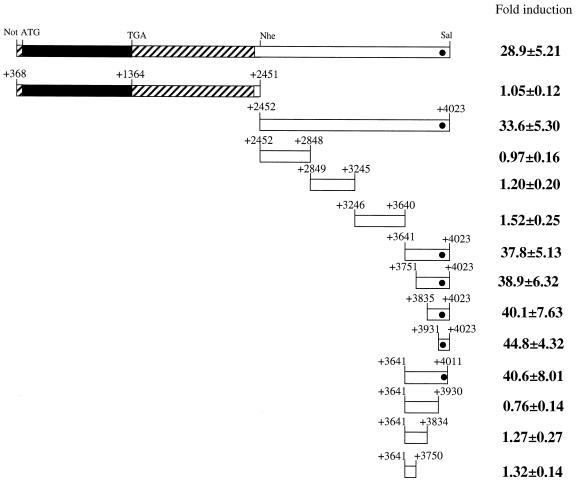
Figure 2.
Localization of the SRR2 core region involved in Sox-2 expression in F9 EC cells. The filled and shaded portions represent Sox-2 coding and non-coding regions, respectively. The filled circle indicates the region which is found to contain the SRR2 core region. The plasmid constructions of the reporter gene bearing the indicated portions of SRR2 were described in Materials and Methods. F9 EC cells were transfected as in Figure 1 with tk-Luc reporter gene carrying portions of SRR2 which are depicted and an internal control luciferase gene of R.reniformis. After 48 h post-transfection, the transcriptional stimulating activity of each deletion mutant of SRR2 was estimated by the dual-luciferase system according to the manufacturer (Promega). The data were obtained as in Figure 1.
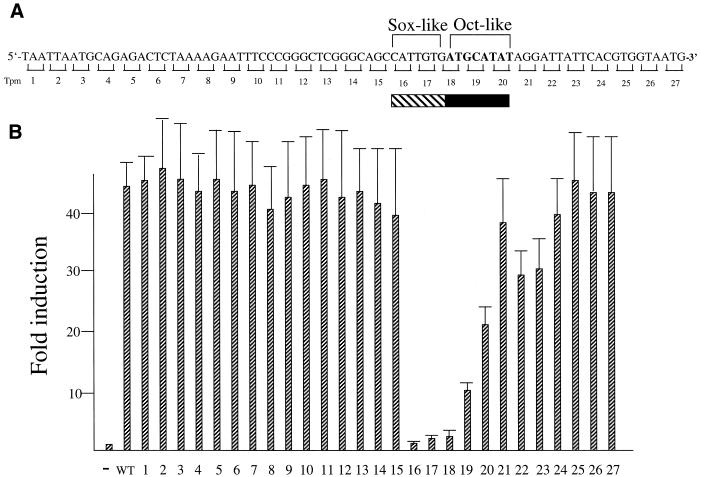
Figure 3.
The Sox-2 site is involved in potentiating SRR2-mediated transcription. (A) The sequence of the SRR2 core region. A synthetic DNA containing wild-type and triple point mutants of the SRR2 sequence was generated by assembly of overlapping oligonucleotides as described in Materials and Methods. The mutants which changed the nucleotides as consecutive triplets (tpm1–27) to non-complementary ones (A ⇔ C, G ⇔ T) are indicated below the sequence. Filled and shaded boxes indicate Octamer and Sox-2 site-like sequences, respectively. (B) Localization of sequence which is crucially involved in SRR2 activity. The tk-Luc reporter gene bearing wild-type or a series of triple point mutants of SRR2 was individually introduced into F9 EC cells together with an internal control luciferase gene of R.reniformis. Forty-eight hours after transfection, the activity of these mutants as well as wild-type SRR2 was evaluated by the dual-luciferase system. The data were obtained as in Figure 1.
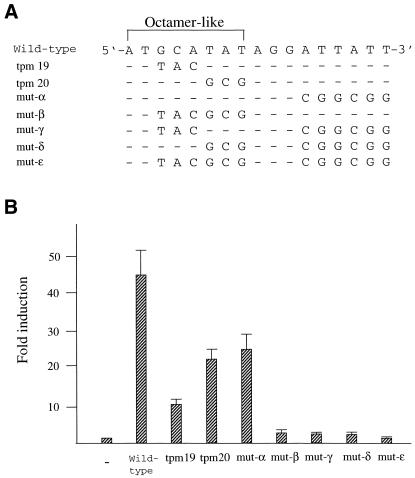
Figure 4.
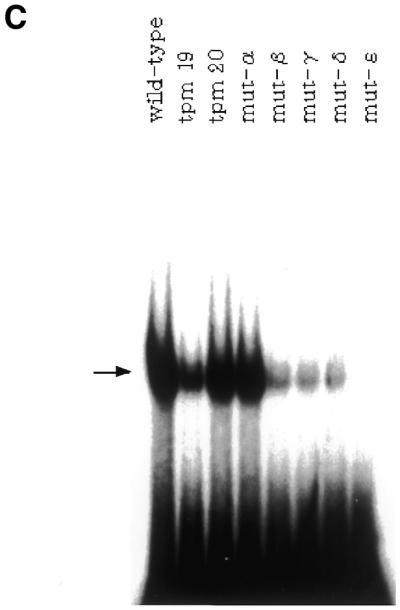
Correlation between the ability of SRR2 to interact with Oct-3/4 protein and its transcriptional stimulating activity in F9 EC cells. (A) The portion of SRR2 sequence containing an Octamer-like element and its immediate downstream sequence. Mutations introduced into the wild-type sequence are indicated. For the tranfection analyses, these mutations were introduced in the context of an 81-bp SRR2 core sequence shown in Figure 3A. However, the probes for the gel-shift analyses shown in (C) lack 21 bp of the 5′ portion of SRR2 sequence (see Materials and Methods for details). (B) The AT-rich sequence located downstream of the Octamer-like sequence is involved in elevating transcriptional stimulating activity of SRR2. The tk-Luc reporter gene bearing wild-type as well as mutated SRR2 sequences are individually introduced into F9 EC cells. Transcriptional stimulating activity of each mutant was obtained as in Figure 1. (C) Effect of mutations on the binding of Oct-3/4 to the SRR2. Radiolabeled DNA probes carrying wild-type and mutated SRR2 sequences shown in (A) were prepared and individually used for gel-shift analyses with whole-cell extracts prepared from COS cells which had been transfected with Oct-3/4 expression vector.
Cell culture
F9 EC cells, COS cells and E14 ES cells were cultures as described previously (18,19,33). The ZHBTc4 cells, ES cells in which the endogenous Oct-3/4 loci had been double-knocked out, were cultured without feeder cells in LIF-supplemented medium. The ZHBTc4 cells carry an integrated copy of the Oct-3/4 gene whose expression is regulated by tetracycline. Therefore, these cells are induced to differentiate by lowering the level of Oct-3/4 expression in cells with tetracycline as described previously (34).
Gel-shift analysis
Introduction of Octamer expression vectors into COS cells and preparation of whole-cell extracts from such transfected cells were done as described previously (18). Oligonucleotides used as template for generating a probe of the gel-shift assay comprise 60 base nucleotides and the one bearing wild-type SRR2 sequence is 5′-AAGAATTTCCCGGGCTCGGGCAGCCATTGTGATGCATATAGGATTATTCACGTGGTAATG-3′ in which Octamer and Sox-2 site-like sequences are single and double underlined, respectively. All other mutant probes used in Figures 4C and 7 have essentially the same flanking sequences, but carry mutations at indicated nucleotides. Double-stranded 32P-labeled 60 bp probe was prepared to a specific activity of 107 c.p.m./µg by annealing to 3-fold molar excess of primer A (5′-CATTACCACCTGAATAATCCTA-3′) and extending with BcaBest DNA polymerase in the presence of all four unlabeled dNTPs (20 µM each) and [α-32P]dCTP. The gel-shift analyses were performed at room temperature for 25 min with 5 ng of probe. The binding buffer contained 12 mM HEPES–KOH (pH 7.9), 0.5 mM EDTA, 1 mM dithiothreitol, 1 mM MgCl2, 12% glycerol and 60 mM KCl. The reaction mixture (20 µl) contained 1–4 µg of whole-cell extract and 2 µg of sonicated salmon sperm DNA. However, in Figure 6A, lane 9, 12 µg of nuclear extract from E14 ES cells was used. The reaction mixture was loaded onto a 4% gel in 20 mM Tris–acetate (pH 8.0)–0.5 mM EDTA, which had been pre-run at 12 V/cm for 1 h. The gels were electrophoresed at the same voltage until the bromophenol blue dye migrated to the bottom of the gel.
Figure 7.
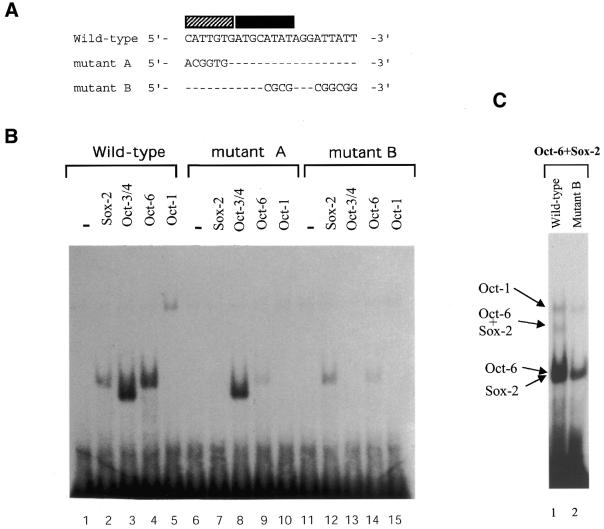
Each Octamer factor shows different sequence requirements of SRR2 for binding. (A) Nucleotide sequences of oligonucleotide probes used for the gel-shift analyses. A portion of the oligonucleotide sequence used for gel-shift analyses is shown. Filled and shaded boxes represent Octamer and Sox-2 site-like sequences, respectively. In the mutant probes, only mutated nucleotides are indicated. In the Oct site mutant, an AT-rich sequence downstream of the Octamer-like sequence is also mutagenized. (B) Effect of mutations of SRR2 sequence on the interaction with Octamer factors. The Oct-1, Oct-3/4 and Oct-6 were individually expressed in COS cells. Whole-cell extracts were prepared from such transfected cells and gel-shift analyses were performed as in Figure 6A using either wild-type or one of two different mutants. (C) The Oct-6 fails to bind to mutant B probe together with Sox-2. Gel-shift analyses were performed with the wild-type (lane 1) or mutant B probe (lane 2). The reaction mixture contains both Oct-6 and Sox-2. Even with long exposure, a band corresponding to the simultaneous binding of Oct-6 and Sox-2 was not detected in lane 2 (data not shown).
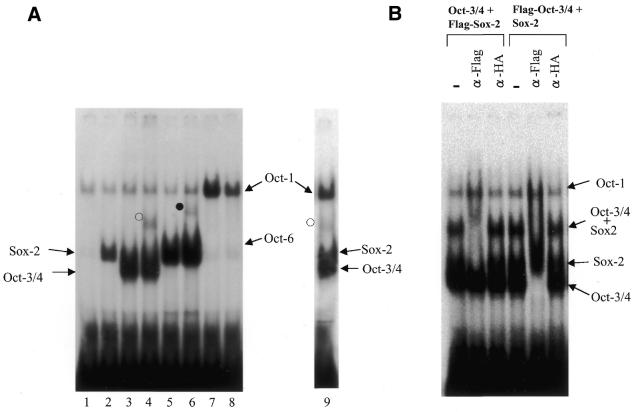
Figure 6.
Gel-shift analyses of SRR2 with Oct and Sox proteins. (A) Both Oct-3/4 and Oct-6, but not Oct-1, are able to form complexes with Sox-2 on SRR2. The Oct-1, Oct-3/4 and Oct-6 were individually expressed in COS cells with or without Sox-2 expression, and gel-shift analyses were performed with whole-cell extracts prepared from such transfected cells by using wild-type SRR2 sequence used in Figure 4C. However, in lane 9, nuclear extract from E14 ES cells was used. The open and filled circles indicate Oct-3/4–Sox-2 and Oct-6–Sox-2 complexes bound to the probe, respectively. The experiments with the specific antibody revealed that bands observed in lanes 1–6 which co-migrate with the bands obtained with exogenously expressed Oct-1 were generated due to the binding of endogenous Oct-1 protein expressed in COS cells (data not shown). (B) Super-shift analyses with antibodies. Gel-shift analyses were done with Oct-3/4 and Sox-2 in which either one of the proteins is Flag-tagged as indicated on top of the panel. The reaction mixture contains no or a certain (anti-Flag or HA) antibody.
RESULTS
Identification of two regulatory regions which boost the level of transcription in F9 EC cells in an undifferentiated state-specific manner
Sox genes encode a group of proteins that carry a DNA-binding HMG domain and additional domains involved in transcriptional regulation (35,36). They are expressed in various phases of embryonic development. One such factor termed Sox-2 is expressed in pluripotent embryonic cells and neuronal cells and plays specific biological roles in these cells (25,27). Recently, it was demonstrated that the 5′-flanking region (–528 to +238) of the Sox-2 gene is able to support the expression of the reporter gene in EC cells preferentially when the cells are in an undifferentiated state (37). Later, we confirmed these data. However, we also note that the transcriptional stimulating activity of this region is significantly weaker than the enhancer of the gene encoding an embryonic transcriptional coactivator UTF1, although the amount of Sox-2 mRNA appears to be equivalent to that of UTF1 mRNA in ES and EC cells. Moreover, we found that the 5′-flanking region (–528 to +238) of the Sox-2 gene retains its significant activity even when the EC and ES cells are induced to differentiate. This property is quite distinct from that of UTF1 enhancer which loses its entire activity upon differentiation of these cells. Therefore, we assumed that the Sox-2 gene might possess additional regulatory region(s) which are implicated in its expression in pluripotent embryonic cells. To identify such region(s), we have first isolated the chromosomal DNA carrying the Sox-2 gene including 5′- and 3′-flanking regions and examined whether any unidentified regulatory region(s) are located in that DNA fragment. Namely, the genomic DNA bearing the Sox-2 gene was cut down into small pieces, whose length is ∼1–2 kb, with the aid of restriction enzymes, and each DNA fragment was subcloned into ptk-Luc reporter plasmid which bears the promoter region, but lacks any putative enhancer region. Subsequently, a series of constructed plasmids was individually introduced into F9 EC cells together with the pSV2/Neo-bearing neomycin-resistant gene and stable transformants were obtained. These stable transformants were treated with or without retinoic acid and examined whether any introduced genomic DNA activates the promoter in the EC cells in an undifferentiated state-specific manner. As shown in Figure 1, at least two of the DNA fragments (fragments D and F) were able to confer the expected transcriptional enhancing activity on the reporter plasmid and we termed these regulatory regions as SRR1 and 2, respectively. These regulatory regions gave a much more profound effect on the transcription than the previously reported region (37) which is located within fragment E. We also note that SRR2 shows almost equivalent activity to that of the UTF1 regulatory region which we have recently identified (18) in terms of the magnitude and its specificity. Therefore, we have decided to characterize this SRR2 in more detail.
Localization of the core region of the SRR2
As shown in Figure 2, a series of deletion mutants was constructed using fragment F (Fig. 1) as a starting material and their transcriptional enhancing activities were examined as above by transient transfection assay in F9 EC cells. These analyses delineated the SRR2 region within the 81 nt (+3831 to +4011).
SRR2 contains Octamer and Sox-2 binding elements
Figure 3A shows the sequence which is implicated to contain the core sequence element(s) of SRR2. We note that the SRR2 carries Octamer and Sox-2 binding sequences. However, both sequences are not identical to their corresponding consensus sequences. Therefore, we examined the cis-active SRR2 sequence in a systematic manner instead of focusing only on these elements. That is, in addition to the wild-type SRR2 sequence, we made a series of mutants in which consecutive triple nucleotides are changed to non-complementary ones and examined their transcriptional enhancing activities. As shown in Figure 3B, these analyses revealed that the mutants bearing mutations within the Sox-2 site-like sequence do not show any transcriptional stimulating activity. However, these analyses also showed that mutations in the Octamer-like sequence do not result in complete loss of the SRR2 activity. In particular, tpm20 is able to raise the level of transcription more than 20-fold. We also found that mutations located outside of two possible elements do not give any significant effect on transcription. These results indicate that Sox-2 is involved in the SRR2-mediated transcriptional stimulation. However, these results cast a doubt on the assumption that Octamer factors including Oct-3/4 play a pivotal role in supporting SRR2 activity.
Correlation between the ability of the SRR2 to activate transcription and to recruit Oct-3/4
Analyses with triple point mutants indicate that the Octamer factors may not contribute to the SRR2 activity significantly. However, this interpretation of the data is based on the assumption that mutants (tpm19, 20) bearing mutations within the Octamer-like elements would be profoundly impaired in terms of the ability to recruit certain Octamer factors. Therefore, it is important to determine whether this is indeed the case. It is known that the Octamer sequence comprises two parts in which the first 4 nt (5′-ATTA/T) are recognized by the POU-homeo domain and the remaining 4 nt (5′-GCAT-3′) are recognized by the POU-specific domain. It has also been shown that the relative position and orientation of these two sequence units can be changed without losing the ability to bind to Octamer factors (38). We note that the AT-rich sequence is present immediately downstream of the Octamer-like sequence and mutations of this portion moderately attenuate the transcriptional stimulating activity of the SRR2 (see the activities obtained with tpm22 and 23). Therefore, it is possible to assume that this AT-rich portion may serve as the binding site for the POU-homeo domain of the factors, especially when the first 4 nt of the Octamer-like sequence are mutated. To address this question, we made an additional series of mutants shown in Figure 4A and examined their ability to activate transcription in F9 cells as above. As shown in Figure 4B, mutation of the downstream AT-rich portion by itself gave a moderate, but not profound effect on the level of transcription. Indeed, mutant-α bearing this mutation showed equivalent activity to those of tpm19 and 20 in terms of activating transcription. However, like the case of mutant β, γ and δ, a combination of two mutations resulted in almost complete loss of the activity. We examined the ability of these mutants to bind Oct-3/4 with gel-shift analyses. As shown in Figure 4C, first lane, the DNA fragment bearing the wild-type SRR2 sequence gave a strong signal on the gel and we confirmed that this band reflects the binding of Oct-3/4 to the probe with the experiments using anti-Oct-3/4 antibody (data not shown). We also performed the gel-shift analyses using mutant DNAs and found that there is an excellent correlation between the ability of DNA to stimulate transcription and to interact with Oct-3/4. Thus, these results indicate that recruitment of Octamer factors is essential for SRR2 to exert its function. In the case of tpm19 and 20, the AT-rich sequence located downstream of the Octamer-like sequence plays an important role in recruiting Octamer factors such as Oct-3/4 to exert the activity.
Extinction of Oct-3/4 is accompanied by the loss of the transcriptional stimulating activity of SRR2
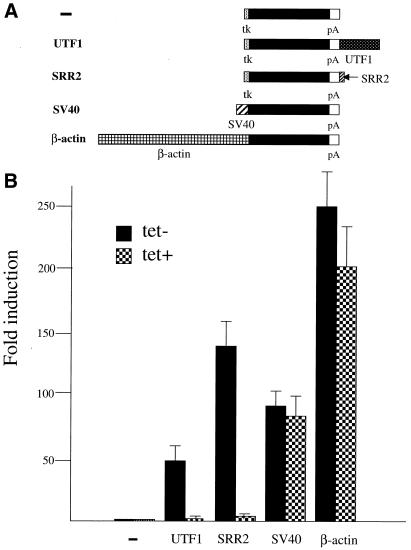
As to the organization of the regulatory elements of genes expressed in EC and ES cells, there are two other examples, i.e. FGF-4 and UTF1 genes, whose enhancers comprise Octamer and Sox-2 binding sequences (18,27–30). Thus, SRR2 is the third example. In the case of FGF-4 and UTF1 enhancers, it was demonstrated that Oct-3/4, but not Oct-1, plays a central role in supporting their function. Therefore, we have determined whether the SRR2 activity is also absolutely dependent on the Oct-3/4. To test this, we used genetically manipulated ES cell line ZHBTc4 in which the Oct-3/4 gene shows tetracycline-regulated expression (34). We individually introduced five different luciferase reporter genes shown in Figure 5A in these cells and determined the strength of these promoter/enhancer activities in the presence or absence of tetracycline by dual-luciferase assay. These analyses revealed that SRR2 and UTF1 enhancers exert their activity when the ES cells are maintained in pluripotent states due to the expressed Oct-3/4. However, these enhancers have lost their activity almost completely when the ES cells are cultured in the presence of tetracycline (Fig. 5B). These data are reminiscent of those obtained with other Oct-3/4-dependent regulatory regions such as those of FGF-4, Rex-1 and 052 retrotransposon (39). However, marked inhibitions were not observed when control enhancer/promoters derived from SV40 and β-actin genes were used (Fig. 5B). Thus, these results indicate that the SRR2 is able to exert its activity in ES cells as well as in F9 EC cells. More importantly, the activity is, like the case of UTF1 enhancer, dependent on the presence of Oct-3/4 in ES cells, although we cannot rule out the possibility that loss of the enhancer activity is some secondary effect due to the loss of Oct-3/4 expression in ES cells. In addition to Oct-3/4, ES cells express Oct-1 abundantly, but the expression level of Oct-6 in the cell is very low. Therefore, the above results do not necessarily rule out the possibility that Oct-6 supports the Sox-2 expression through SRR2 in Oct-6-expressing embryonic ectoderm.
Figure 5.
The extinction of Oct-3/4 is accompanied by loss of SRR2 activity in ES cells. (A) Schematic representation of five different lucifease reporter plasmids bearing distinct regulatory regions. The filled box represents the luciferase reporter gene. tk and pA represent the thymidine kinase gene promoter (–109 to +51) and poly(A) addition signal from SV40 virus, respectively. UTF1, SV40 and β-actin indicate the UTF1 regulatory element-containing 1.2 kb BamHI/SalI genomic DNA fragment (18), the SV40 early promoter, and the human β-actin promoter, respectively. (B) SRR2 and the UTF1 regulatory element require Oct-3/4 expression to exert their activity in ES cells. The five different plasmids bearing the luciferase reporter gene were individually introduced into ZHBTc4 ES cells together with the Puromycin-resistant gene and an internal control luciferase gene of R.reniformis by a lipofection method. After selection with Puromycin, approximately 1000 of the drug-resistant colonies were pooled and expanded. Subsequently, the cells were incubated with medium with or without tetracycline (1 µg/ml). After 48 h, whole-cell extracts were prepared from such treated cells and levels of transcription were determined by the dual-luciferase system according to the manufacturer (Promega). The data were obtained as in Figure 1.
The Oct-3/4 and Oct-6, but not Oct-1, can form a complex with Sox-2 on SRR2
We have previously reported that the UTF1 gene enhancer selectively recruits the complex comprising Oct-3/4 and Sox-2 and precludes the binding of a complex containing Oct-1 or Oct-6 (18). Furthermore, we have shown that these properties are dictated by the unique ability of Oct-3/4 to bind to a variant of the Octamer motif present in the UTF1 regulatory element. As the Octamer sequence of the SRR2 is also different from the consensus sequence, we assumed that the SRR2 may also have a similar biochemical property. To address this question, three different Octamer factor expression vectors were individually introduced into COS cells with or without Sox-2 expression vector, and whole-cell extracts were prepared for the gel-shift analyses. As shown in Figure 6A, lanes 2 and 3, Sox-2 or Oct-3/4 alone each generated a specific band in the gel, indicating that both Oct-3/4 and Sox-2 can bind to the SRR2 independently. When we used the extract containing both Oct-3/4 and Sox-2, a novel slow migrating band was generated (lane 4) in addition to that obtained with Oct-3/4 alone. We assumed that, from its slow migrating character, the band reflected the simultaneous binding of Oct-3/4 and Sox-2 and we confirmed that this is indeed the case (see below). However, the efficiency of the Oct-3/4–Sox-2 complex formation on SRR2 appears to be less significant compared with the case of FGF-4 and UTF1 regulatory elements, and we will discuss these data later (see Discussion). We also performed the gel-shift analyses with different sets of proteins and found that Oct-6 can also bind to the probe (lane 5). Furthermore, the protein gave a novel band in the gel in the presence of Sox-2 which migrates more slowly than that observed in lane 5 (lane 6), indicating that Oct-6 is also able to bind to the probe together with Sox-2. As to the Oct-1, we confirmed that the probe interacts with the protein (lane 7). However, unlike other Octamer factors, no novel band was obtained even in the presence of Sox-2. These results indicate that Oct-1 displays different DNA-binding specificity from that of Oct-3/4 and Oct-6, and the protein appears not to bind to the SRR2 element together with Sox-2. As all the binding studies were done with COS cell extract with overexpressed Oct and Sox proteins, we also examined whether expected protein–DNA complexes are generated with ES cell nuclear extract. Due to the low expression level of Oct-6 in ES cells, we can expect four bands which reflect the binding of Oct-3/4, Sox-2 or Oct-1 alone, or simultaneous binding of Oct-3/4 and Sox-2 to the probe. As shown in Figure 6A, lane 9, ES cell extract indeed gave bands in the expected positions of the gel, although the probe bound only to Sox-2 and that associated with Oct-3/4 alone were, like the case in lane 4, not well resolved in the gel. To confirm that the slow migrating band observed in lane 4 represents the Oct-3/4–Sox-2 complex bound to the probe, we have done the experiments using antibodies (Fig. 6B). For the first three lanes, gel-shift analyses were done with wild-type Oct-3/4 and Flag-tagged Sox-2 proteins and reactions were done with or without the specific antibody. As shown in lane 2, anti-Flag, but not unrelated (anti-HA) antibody, affect the mobility of only the slow migrating band. We have also done the experiments with Flag-tagged Oct-3/4 and wild-type Sox-2 proteins. As shown in the last three lanes, the addition of anti-Flag, but not anti-HA antibody, results in super-shifting the slow migrating band in addition to the band obtained with Oct-3/4 alone. Taken together, these results confirm that both Oct-3/4 and Sox-2 proteins are involved in the formation of the slow migrating band. We have also done the same experiments with the set of Oct-6 and Sox-2 proteins and reached essentially the same conclusion (data not shown).
As to the inability of SRR2 to interact with Oct-1 and Sox-2 simultaneously, we have previously had a similar or related observation with the experiments using the UTF1 regulatory element bearing a complete Sox-2 binding site and variant Octamer sequence (5′-ACTAGCAT-3′) (18). That is, certain Octamer factors such as Oct-1 do bind to the UTF1 regulatory element, but utilize the AT-rich Sox-2 binding sequence instead of using the deviated portion of the Octamer sequence. Therefore, such Octamer factors fail to bind to the UTF1 regulatory element together with Sox-2, but compete with this protein for their binding to the element. We assumed that the apparent failure of SRR2 for simultaneous association with Oct-1 and Sox-2 may also be due to the requirement of Oct-1 with the Sox-2 site for its binding. To address this question, we prepared two mutant probes termed mutant A and mutant B as shown in Figure 7A. The mutant A carries the mutation in the Sox-2 site, while mutant B bears the mutation in part of the Octamer-like sequence in which the portion recognized by the POU-specific domain remains intact. In the mutant B, we also put a mutation in the AT-rich sequence located downstream of the Octamer-like sequence, as this portion appears to function as part of the Octamer-like sequence (see Fig. 4). Using these probes, gel-shift analyses were performed with the extracts containing either one of the Octamer factors or Sox-2 protein. As shown in Figure 7B, exogenously expressed Octamer factors as well as Sox-2 protein produced specific bands in the gel with the probe bearing the wild-type SRR2 sequence as expected. However, when the experiments were done with mutant A probe, binding of Oct-1 as well as Sox-2 to the probe was completely eliminated and the level of the binding of Oct-6 was also significantly reduced, although no obvious effect of this mutation was evident about the binding of Oct-3/4. The gel-shift analyses with mutant B showed that binding of Sox-2 was not affected as expected. However, the binding of Oct-1 and Oct-3/4 proteins failed to bind to this mutant probe, while Oct-6 could bind to this probe, albeit less efficiently compared with the wild-type sequence. In summary, Oct-3/4 absolutely requires the portion mutagenized in probe B, while Oct-1 protein further requires the Sox-2 site for its binding. The Oct-6 protein can bind to SRR2 when either one of the AT-rich portions remains intact. Indeed, we found that the Oct-6 fails to bind to the SRR2 when the mutations of two probes are combined (data not shown). These results are in line with the notion that the Octamer factors which can bind without utilizing the Sox-2 site are able to form a complex with Sox-2 on SRR2. To further ascertain the above assumption that binding of Oct-6 to the mutant B probe is to the remaining AT-rich element within the Sox binding site, we examined whether Oct-6 cannot bind to this mutant B probe together with Sox-2. As shown in Figure 7C, lane 2, we found that this is indeed the case.
Cooperativity between the Octamer factors and Sox-2 protein on the SRR2
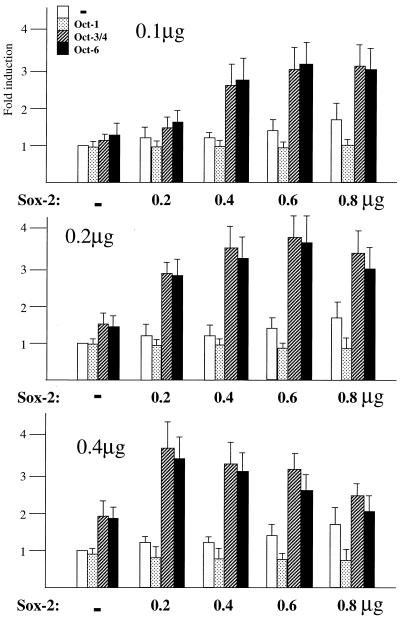
We have previously reported that the UTF1 regulatory element specifically recruits the Oct-3/4–Sox-2 complex and precludes the binding of the Oct-1–Sox-2 or Oct-6–Sox-2 complex. Therefore, only Oct-3/4 is able to support the transcriptional activation of the UTF1 regulatory element (18). However, the SRR2 can serve as the binding site for the Oct-6–Sox-2 complex as well as the Oct-3/4–Sox-2 complex. Therefore, we examined whether both complexes are able to boost the level of transcription through SRR2, or the Oct-3/4-containing complex specifically supports the function of the region. For this purpose, we introduced a reporter gene carrying wild-type SRR2 sequence together with Octamer factor and Sox-2 expression vectors by transient transfection into COS cells in which Sox-2 mRNA is not present and SRR2 does not show any transcriptional stimulating activity. As shown in Figure 8, increasing amounts of Sox-2 expression vector elevated the level of transcription especially when certain amounts of Oct-3/4 or Oct-6 expression vector were cotransfected. However, large amounts of Octamer factors and/or Sox-2 expression vectors resulted in an attenuation of the induction, probably due to the squelching effects of these proteins. The observed transcriptional activation appears to occur through the SRR2 element, as no significant elevation of transcriptional level was observed when the experiments were done with the reporter gene which lacks the SRR2 (data not shown). The stimulatory effects of these ectopically expressed Sox-2 and Octamer factors on SRR2 are clearly much lower than those observed in F9 EC and ES cells. We also encountered similar observations with the experiments using the UTF1 regulatory element (18). Thus, these results indicate that certain cofactor(s) which augment the level of transcriptional stimulating activities of these Oct and Sox proteins are present in EC and ES cells, but not in non-EC cells such as COS cells. We also performed the experiments with Oct-1 expression vector and found that no obvious activation was observed with this vector even when the Sox-2 expression vector was cotransfected. Thus, these results indicate that Oct-3/4–Sox-2 and Oct-6–Sox-2 complexes, but not Oct-1–Sox-2 complex, are able to augment the level of SRR2-mediated transcriptional activation.
Figure 8.
Both Oct-3/4 and Oct-6, but not Oct-1, are able to potentiate transcription through SRR2. COS cells were transfected with 0.1, 0.2 or 0.4 µg of Oct-1, Oct-3/4 or Oct-6 expression vectors, and the increasing amounts of Sox-2 vector as indicated. In addition, an internal control luciferase gene (2 µg) of R.reniformis and the tk-luc reporter plasmid (2 µg) bearing SRR2 were also introduced. After 48 h post-transfection, transcriptional level was estimated by the dual-luciferase system according to the manufacturer (Promega). The data were obtained as in Figure 1.
DISCUSSION
Understanding the molecular basis of the pluripotent properties of ES cells as well as EC cells will be a key step for elucidating the molecular events governing the early developmental stages of mammals (10–13,34). Furthermore, these studies may allow to provide a molecular tool to examine the degree of pluripotency of each ES cell line. Such a tool appears to be important especially when ES cells are used to generate certain specific cells for a therapeutic purpose. Recent reports underscore the importance of the Oct-3/4– Sox-2 complex in terms of maintaining the pluripotent state of ES cells (18,27–30,40). Therefore, it appears to be important to isolate target genes whose activities are controlled by this complex. However, we also consider that it is equally important to know how the expression of the Sox-2 gene is controlled in EC and ES cells, as these analyses may uncover the regulatory hierarchy among embryonic factors and help unravel broader aspects of the regulatory network operating in pluripotent embryonic cells. In this context, we have searched the regulatory elements in the vicinity of the Sox-2 gene which exert their activity in F9 EC cells specifically when the cells are in an undifferentiated state. As described in this paper, these analyses have led to the identification of two regulatory elements, termed SRR1 and SRR2, which meet these criteria. Recently, Wiebe et al. (37) have shown that the region around the major transcriptional start site of mouse Sox-2 gene has an ability to potentiate the level of transcription. They also demonstrate that the region shows relatively higher activity when the cells are in an undifferentiated state. We confirmed their data. At the same time, we found that this region displays much weaker activity than the SRR1 and SRR2. Moreover, we found that the region retains the significant activity even when F9 cells are in differentiated states. These characteristics are in marked contrast to those of SRR1 and SRR2 which lose their function almost completely when F9 cells are induced to differentiate by using retinoic acids. Therefore, we assume that expression of the Sox-2 gene in EC cells is supported in major part by SRR1 and SRR2, and the contribution of the region which was previously identified (37) is probably very little. Expression of Sox-2 is not restricted to early embryonic tissues such as ICM and embryonic ectoderm, but the gene is known to be expressed panneurally (41). Recently, Zappone et al. (25) have identified the regulatory region in the 5′-flanking region which supports expression of the Sox-2 gene in telencephalon. Of note, this regulatory region and SRR1 appear to be closely located and indeed are not distinguishable at present. Therefore, it will be interesting to determine whether the telencephalic regulatory element and SRR1 are distinct regulatory elements or a single regulatory region, i.e. SRR1 exerts the activity both in telencephalon and ES cells. Although we do not know which is the case at present, we assume that the transcriptional stimulating activity of the 5′-flanking region of the Sox-2 gene in ES cells, which Zappone et al. (25) have observed, reflects the activity of SRR1.
The physiological significance of the presence of two regulatory regions (SRR1 and SRR2) which direct the expression of a single gene in identical cells is not known at present. However, there are ample examples which resemble this case. For example, lens specific δ-crystallin gene carries two distinct regulatory elements, one of which exerts its action by utilizing L-Maf (42) and the other is controlled by a Pax-6–Sox-2 complex (43). In each case, it is assumed that two regulatory regions synergistically function to support the high level of gene expression in particular cells. We have performed detailed biochemical analyses of the SRR2, as this regulatory region exerts a stronger activity than the SRR1. These analyses revealed that, like the case of the regulatory elements of FGF-4 and UTF1 genes (18,27–30), the SRR2 possesses Octamer and Sox-2 binding sequences, although both of them are not identical to their corresponding consensus sequences. However, transient transfection analyses coupled to mutagenesis analyses unequivocally demonstrate that the SRR2 is under the control of the complex. Thus, these results provide additional evidence that the Oct-3/4–Sox-2 complex plays a pivotal role in supporting the gene expression in early embryonic cells and we assume that a substantial number of other target genes of this complex remain to be identified. The fact that the Sox-2 protein is involved in its expression is particularly interesting, as ES cells have many unique characteristics such as their rapid growth rate and the ability to propagate permanently without converting to cancerous cells (7). We assume that this positive auto-regulatory loop of the Sox-2 protein may be at least in part involved in these biological properties of ES cells.
As above, our analyses demonstrate the similarity of the SRR2 to the regulatory elements of UTF1 and FGF-4 genes. However, we also found that each regulatory element has distinct biochemical properties in terms of the specificity of the association with Octamer-containing complexes. As to the FGF-4 regulatory element, it has been shown that the element is able to serve as the binding site for both Oct-3/4–Sox-2 and Oct-1–Sox-2 complexes (27–30) and we found that the Oct-6–Sox-2 complex is also able to bind to the element (data not shown). However, we have previously shown that the UTF1 regulatory element specifically recruits the Oct-3/4– Sox-2 complex and precludes the binding of other Octamer factor-containing complexes (18). Our biochemical analyses also demonstrate that this unique property is dictated in the variant Octamer sequence present in the UTF1 regulatory element. The SRR2 also bears the non-consensus Octamer sequence, but our analyses reveal that this regulatory region possesses different biochemical properties, being able to associate with the Oct-6–Sox-2 complex as well as the Oct-3/4–Sox-2 complex, although the Oct-1–Sox-2 complex fails to interact with the SRR2. Moreover, our analyses reveal that both Oct-3/4–Sox-2 and Oct-6–Sox-2 complexes are able to augment the transcriptional stimulating activity of the SRR2. Thus, it is assumed that, in Oct-6-expressing embryonic ectoderm, both Oct-6–Sox-2 and Oct-3/4–Sox-2 complexes support Sox-2 expression through SRR2. During the course of these analyses, we noted that, unlike the case of FGF-4 and UTF1 regulatory elements, binding of Sox-2 and Oct-3/4 on SRR2 does not seem to be cooperative as judged from the in vitro binding assay. However, SRR2 displays equivalent activity to other Oct and Sox binding sites containing regulatory elements. Therefore, it is possible to speculate that an additional cofactor is present in vivo which facilitates the binding of Oct and Sox proteins on SRR2 and is involved in potentiating the SRR2 activity. The above analyses also indicated that each Octamer factor possesses a unique DNA-binding specificity. Recently, we have systematically determined potential binding sites for each Octamer factor and found that all these factors are able to interact with a broader range of sequences deviated from the consensus Octamer sequence (S.Miyagi and A.Okuda, unpublished results).
So far, several Oct-3/4 target genes have been identified including those of UTF1, Sox-2, FGF-4, Rex-1 and Osteopontin (18,27,40,44,45). To evaluate the importance of these genes including the Sox-2 gene for maintaining ES cells in the pluripotent state, we have expressed these genes in Oct-3/4 double-knock-out ES cells (ZHBTc4 cells) and examined whether such cells are able to retain ES cell-specific properties even in the absence of Oct-3/4, although newly discovered putative Oct-3/4 target genes identified by Du et al. (46) and those cloned by Saijoh et al. (47) have not been examined yet. These analyses showed that such ES cells are rapidly differentiated by adding tetracycline to the medium (H.Niwa, unpublished results), indicating that these Oct-3/4 target genes are not sufficient for preventing induction of differentiation in ES cells and certain critical Oct-3/4 target gene(s) remain to be identified. Obviously, it is crucial to clone and analyze such genes towards the complete understanding of the pluripotent properties of ES cells at a molecular level, and it is probably important to be aware that such target genes do not necessarily carry the consensus Octamer element but, like the case of UTF1 and Sox-2 genes, may possess variant sequences in the regulatory regions.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are indebted to Daisuke Horiuchi for his technical assistance. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology, Japan. This work was especially supported by Grant for the Promotion of the Advancement of Education and Research in Graduate Schools of the Ministry. A.O. and M.N. were also supported by The Mochida Memorial Foundation for Medical and Pharmaceutical Research and Kato Memorial Bioscience Foundation, respectively.
DDBJ/EMBL/GenBank accession no. AB079241
REFERENCES
- 1.Dooley T.P., Miranda,M., Jones,N.C. and DePamphilis,M.L. (1989) Transactivation of the adenovirus EIIa promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development, 107, 945–956. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert S.F., (1991) Developmental Biology, 3rd Edn. Sinauer Associates, Sunderland, MA.
- 3.La Thangue N.B. and Rigby,P.W.J. (1987) An adenovirus E1A-like transcription factor is regulated during the differentiation of murine embryonal carcinoma stem cells. Cell, 49, 507–513. [DOI] [PubMed] [Google Scholar]
- 4.Majunder S., Zhao,Z., Kaneko,K. and DePamphilis,M.L. (1997) Developmental acquisition of enhancer function requires coactivator activity. EMBO J., 16, 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko K.J., and DePamphilis,M.L. (1998) Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev. Genet., 22, 43–55. [DOI] [PubMed] [Google Scholar]
- 6.Vassiler A., Kaneko,K.J., Shu,H., Zhao,Y. and DePamplilis,M.L. (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev., 15, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan B., Beddington,R., Constantini,F. and Lacy,E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Martin G.R., (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA, 78, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner R.L., and Rossant,J. (1979) Investigation of the fate of 4–5 day postcoitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol., 52, 141–152. [PubMed] [Google Scholar]
- 10.Keller G.M., (1995) In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol., 7, 862–869. [DOI] [PubMed] [Google Scholar]
- 11.Okabe S., Forsberg-Nilsson,K., Spiro,A.C., Segal,M. and McKay,R.D. (1996) Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev., 59, 89–102. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita J., Itoh,H., Hirashima,M., Ogawa,M., Nishikawa,S., Yurugi,T., Naito,M., Nakao,K. and Nishikawa,S. (2000) Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature, 408, 92–96. [DOI] [PubMed] [Google Scholar]
- 13.Lumelsky N., Blondel,O., Laeng,P., Velasco,I., Ravin,R. and McKay,R. (2001) Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science, 292, 1389–1394. [DOI] [PubMed] [Google Scholar]
- 14.Ziman M.R., Thomas,M., Jacobsen,P. and Beazley,L. (2001) A key role for Pax7 transcripts in determination of muscle and nerve cells. Exp. Cell Res., 268, 220–229. [DOI] [PubMed] [Google Scholar]
- 15.Mintz B., and Illmensee,K. (1975) Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc. Natl Acad. Sci. USA, 72, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda A., Fukushima,A., Nishimoto,M., Yamagishi,T., Orimo,A., Nabeshima,Y., Kuro-o,M., Nabeshima,Y., Boon,K., Keaveney,M., Stunnenberg,H.G. and Muramatsu,M. (1998) UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J., 17, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukushima A., Okuda,A., Nishimoto,M., Seki,N., Hori,T. and Muramatsu,M. (1998) Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. J. Biol. Chem., 273, 25840–25849. [DOI] [PubMed] [Google Scholar]
- 18.Nishimoto M., Fukushima,A., Okuda,A. and Muramatsu,M. (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol., 19, 5453–5465. [Erratum (2001) Mol. Cell. Biol., 21, 978.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto K., Okazawa,H., Okuda,A., Sakai,M., Muramatsu,M. and Hamada,H. (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell, 60, 461–472. [DOI] [PubMed] [Google Scholar]
- 20.Rosner M.H., Vigano,M.A., Ozato,K., Thimmons,P.M., Poirier,F., Rigby,P.W.J. and Staudt,L.M. (1990) A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature, 345, 686–692. [DOI] [PubMed] [Google Scholar]
- 21.Scholer H.R., Ruppert,S., Suzuki,N., Chowdhury,K. and Gruss,P. (1990) New type of POU domain in germ line-specific protein Oct-4. Nature, 344, 435–439. [DOI] [PubMed] [Google Scholar]
- 22.Nichols J., Zevnik,B., Anastassiadis,K., Niwa,H., Klewe-Nebenius,D., Chambers,I., Scholer,H.R. and Smith,A.G. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct-4. Cell, 95, 379–391. [DOI] [PubMed] [Google Scholar]
- 23.Wood H.B., and Episkopou,V. (1999) Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev., 86, 197–201. [DOI] [PubMed] [Google Scholar]
- 24.Barnea E., and Bergman,Y. (2000) Synergy of SF1 and RAR in activation of Oct-3/4 promoter. J. Biol. Chem., 275, 6608–6619. [DOI] [PubMed] [Google Scholar]
- 25.Zappone M., Galli,R., Catena,R., Meani,N., Biasi,S.D., Mattei,E., Tiveron,C., Vescovi,A.L., Lovell-Badge,R., Ottolenghi,S. and Nocolis,S.K. (2000) Sox-2 regulatory sequences direct expression of a β-geo transgene to telecephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development, 127, 2367–2382. [DOI] [PubMed] [Google Scholar]
- 26.Fuhrmann G., Chung,A.C., Jackson,K.J., Hummelke,G., Baniahmad,A., Sutter,J., Sylvester,S., Scholer,H.R. and Cooney,A.J. (2001) Mouse germline restriction of Oct-4 expression by germ cell nuclear factor. Dev. Cell, 1, 377–387. [DOI] [PubMed] [Google Scholar]
- 27.Yuan H., Corbi,N., Basilico,C. and Dailey,L. (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev., 9, 2635–2645. [DOI] [PubMed] [Google Scholar]
- 28.Ambrosetti D.C., Basilico,C. and Dailey,L. (1997) Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein–protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol., 17, 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrosetti D.C., Scholer,H.R., Dailey,L. and Basilico,C. (2000) Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem., 275, 23387–23397. [DOI] [PubMed] [Google Scholar]
- 30.Dailey L., and Basilico,C. (2001) Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell Physiol., 186, 315–328. [DOI] [PubMed] [Google Scholar]
- 31.Okuda A., Sakai,M. and Muramatsu,M. (1987) The structure of the rat glutathione S-transferase P gene and related pseudogenes. J. Biol. Chem., 262, 3858–3863. [PubMed] [Google Scholar]
- 32.Okuda A., Imagawa,M., Maeda,Y., Sakai,M. and Muramatsu,M. (1989) Structural and functional analysis of an enhancer GPEI having a phorbol 12-O-tetradecanoate 13-acetate responsive element-like sequence found in the rat glutathione transferase P gene. J. Biol. Chem., 264, 16919–16926. [PubMed] [Google Scholar]
- 33.Okuda A., Imagawa,M., Sakai,M. and Muramatsu,M. (1990) Functional cooperativity between two TPA responsive elements in undifferentiated F9 embryonic stem cells. EMBO J., 9, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa H., Miyazaki,J. and Smith,A.G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genet., 24, 372–376. [DOI] [PubMed] [Google Scholar]
- 35.Pevny L.H., and Lovell-Badge,R. (1997) Sox genes find their feet. Curr. Opin. Genet. Dev., 7, 338–344. [DOI] [PubMed] [Google Scholar]
- 36.Kamachi Y., Uchikawa,M. and Kondoh,H. (2000) Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet., 16, 182–187. [DOI] [PubMed] [Google Scholar]
- 37.Wiebe M.S., Wilder,P.J., Kelly,D. and Rizzino,A. (2000) Isolation, characterization, and differential expression of the murine Sox-2 promoter. Gene, 246, 383–393. [DOI] [PubMed] [Google Scholar]
- 38.Herr W., and Cleary,M.A. (1995) The POU domain: versatility in transcriptional regulation by a flexible two in one DNA-binding domain. Genes Dev., 9, 1679–1693. [DOI] [PubMed] [Google Scholar]
- 39.Niwa H., Masui,S., Chambers,I., Smith,A.G. and Miyazaki,J. (2002) Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol., 22, 1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Botquin V., Hess,H., Fuhrmann,G., Anastassiandis,C., Gross,M.K., Vriend,G. and Scholer,H.R. (1998) New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev., 12, 2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collignon J., Sockanathan,S., Hocker,A., Cohen-Tannoudji,M., Norris,D., Rastan,S., Stevanovic,M., Goodfellow,P.N. and Lovell-Badge,R. (1996) A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development, 122, 509–520. [DOI] [PubMed] [Google Scholar]
- 42.Ogino H., and Yasuda,K. (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science, 280, 115–118. [DOI] [PubMed] [Google Scholar]
- 43.Kamachi Y., Uchikawa,M., Tanouchi,A., Sekido,R. and Kondoh,H. (2001) Pax-6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev., 15, 1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosler B.A., LaRosa,G.J., Grippo,J.F. and Gudas,L.J. (1989) Expression of REX-1, a gene containing zinc finger motifs is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol. Cell. Biol., 9, 5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Shushan E., Thompson,J.R., Gudas,L.J. and Bergman,Y. (1998) Rex-1, a gene encoding a transcription factor expressed in the early embryo, regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol., 18, 1866–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Z., Cong,H. and Yao,Z. (2001) Identification of putative downstream genes of Oct-4 by suppression-subtractive hybridization. Biochem. Biophys. Res. Commun., 282, 701–706. [DOI] [PubMed] [Google Scholar]
- 47.Saijoh Y., Fujii,H., Meno,C., Sato,M., Hirota,Y., Nagamatsu,S., Ikeda,M. and Hamada,H. (1996) Identification of putative downstream genes of Oct-3, a pluripotent cell-specific transcription factor. Genes Cells, 1, 239–252. [DOI] [PubMed] [Google Scholar]