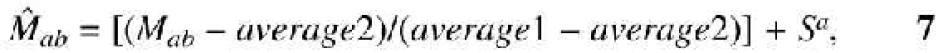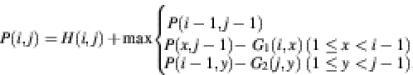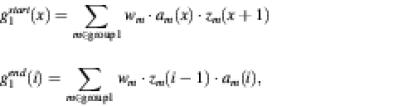Abstract
A multiple sequence alignment program, MAFFT, has been developed. The CPU time is drastically reduced as compared with existing methods. MAFFT includes two novel techniques. (i) Homo logous regions are rapidly identified by the fast Fourier transform (FFT), in which an amino acid sequence is converted to a sequence composed of volume and polarity values of each amino acid residue. (ii) We propose a simplified scoring system that performs well for reducing CPU time and increasing the accuracy of alignments even for sequences having large insertions or extensions as well as distantly related sequences of similar length. Two different heuristics, the progressive method (FFT-NS-2) and the iterative refinement method (FFT-NS-i), are implemented in MAFFT. The performances of FFT-NS-2 and FFT-NS-i were compared with other methods by computer simulations and benchmark tests; the CPU time of FFT-NS-2 is drastically reduced as compared with CLUSTALW with comparable accuracy. FFT-NS-i is over 100 times faster than T-COFFEE, when the number of input sequences exceeds 60, without sacrificing the accuracy.
INTRODUCTION
Multiple sequence alignment is a basic tool in various aspects of molecular biological analyses ranging from detecting key functional residues to inferring the evolutionary history of a protein family. It is, however, difficult to align distantly related sequences correctly without manual inspections by expert knowledge. Many efforts have been made on the problems concerning the optimization of sequence alignment. Needleman and Wunsch (1) presented an algorithm for sequence comparison based on dynamic programming (DP), by which the optimal alignment between two sequences is obtained. The generalization of this algorithm to multiple sequence alignment (2) is not applicable to a practical alignment that consists of dozens or hundreds of sequences, since it requires huge CPU time proportional to NK, where K is the number of sequences each with length N. To overcome this difficulty, various heuristic methods, including progressive methods (3) and iterative refinement methods (4–6), have been proposed to date. They are mostly based on various combinations of successive two-dimensional DP, which takes CPU time proportional to N2.
Even if these heuristic methods successfully provide the optimal alignments, there remains the problem of whether the optimal alignment really corresponds to the biologically correct one. The accuracy of resulting alignments is greatly affected by the scoring system. Thompson et al. (7) developed a complicated scoring system in their program CLUSTALW, in which gap penalties and other parameters are carefully adjusted according to the features of input sequences, such as sequence divergence, length, local hydropathy and so on. Nevertheless, no existing scoring system is able to process correctly global alignments for various types of problems including large terminal extension of internal insertion (8). Considerable improvements in the accuracy have recently been made in CLUSTALW (7) version 1.8, the most popular alignment program with excellent portability and operativity, and T-COFFEE (9), which provides alignments of the highest accuracy among known methods to date.
On the other hand, few improvements have been made successfully to reduce the CPU time, since the proposal of the progressive method by Feng and Doolittle (3). A high-speed computer program applicable to large-scale problems is becoming more important with the rapid increase in the number of protein and DNA sequences. In order to improve the speed of DP, it is effective to use highly homologous segments in the procedure of multiple sequence alignment (10). There are well-known homology search programs, such as FASTA (11) and BLAST (12), based on string matching algorithms.
In this report, we developed a novel method for multiple sequence alignment based on the fast Fourier transform (FFT), which allows rapid detection of homologous segments. In spite of its great efficiency, FFT has rarely been used practically for detecting sequence similarities (13,14). We also propose an improved scoring system, which performs well even for sequences having large insertions or extensions as well as distantly related sequences of similar length. The efficiency (CPU time and accuracy) of the method was tested by computer simulations and the BAliBASE (15) benchmark tests in comparison with several existing methods. These tests showed that the CPU time has been drastically reduced, whereas the accuracy of the resulting alignments is comparable with that of the most accurate methods among existing ones.
METHODS
Group-to-group alignments by FFT
The frequency of amino acid substitutions strongly depends on the difference of physico-chemical properties, particularly volume and polarity, between the amino acid pair involved in the substitution (16). Substitutions between physico-chemically similar amino acids tend to preserve the structure of proteins, and such neutral substitutions have been accumulated in molecules during evolution (17). It is therefore reasonable to consider that an amino acid a is assigned to a vector whose components are the volume value v(a) and the polarity value p(a) introduced by Grantham (18). We use the normalized forms of these values: ![]() and
and ![]() , where an overbar denotes the average over 20 amino acids, and σv and σp denote the standard deviations of volume and polarity, respectively. An amino acid sequence is converted to a sequence of such vectors.
, where an overbar denotes the average over 20 amino acids, and σv and σp denote the standard deviations of volume and polarity, respectively. An amino acid sequence is converted to a sequence of such vectors.
Calculation of the correlation between two amino acid sequences. We define the correlation c(k) between two sequences of such vectors as
c(k) = cv(k) + cp(k),1
where cv(k) and cp(k) are, as defined below, the correlations of volume component and polarity component, respectively, between two amino acid sequences to be aligned. The correlation c(k) represents the degree of similarity of two sequences with the positional lag of k sites. The high value of c(k) indicates that the sequences may have homologous regions.
The correlation cv(k) of volume component between sequence 1 and sequence 2 with the positional lag of k sites is defined as
where ![]() and
and ![]() are the volume component of the nth site of sequence 1 with the length of N and that of sequence 2 with the length of M, respectively. Considering N ≃ M in many cases, equation 2 takes O(N2) operations. The FFT reduces the CPU time of this calculation to O(N log N) (19). If V1(m) and V2(m) are the Fourier transform of
are the volume component of the nth site of sequence 1 with the length of N and that of sequence 2 with the length of M, respectively. Considering N ≃ M in many cases, equation 2 takes O(N2) operations. The FFT reduces the CPU time of this calculation to O(N log N) (19). If V1(m) and V2(m) are the Fourier transform of ![]() and
and ![]() , i.e.
, i.e.
it is known that the correlation cv(k) is expressed as
cv(k) ⇔ V1*(m) · V2(m),5
where ⇔ represents transform pairs, and the asterisk denotes complex conjugation.
The correlation cp(k) of polarity component between two sequences
is calculated in the same manner.
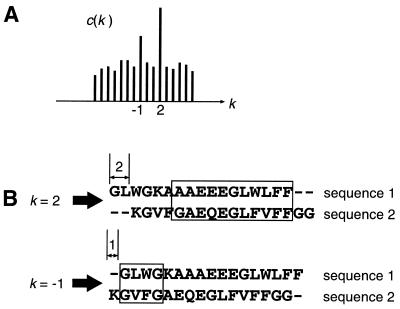
Finding homologous segments. If two sequences compared have homologous regions, the correlation c(k) has some peaks corresponding to these regions (Fig. 1A). By the FFT analysis, however, we can know only the positional lag k of a homologous region in two sequences but not the position of the region. As shown in Figure 1B, to determine the positions of the homologous region in each sequence, a sliding window analysis with the window size of 30 sites is carried out, in which the degree of local homologies is calculated for each of the highest 20 peaks in the correlation c(k). We identify a segment of 30 sites with score value exceeding a given threshold (0.7 per site in our program, see below for details of the scoring system) as a homologous segment. If two or more successive segments are identified as homologous segments, they are combined into one segment of larger length. If the length of the combined segment is longer than 150 sites, the segment is divided into several segments with 150 sites each.
Figure 1.
(A) A result of the FFT analysis. There are two peaks corresponding to two homologous blocks. (B) Sliding window analysis is carried out and the positions of homologous blocks are determined. Note that window size is 30 (see text) but the window size is set to 4 in (B) for simplicity.
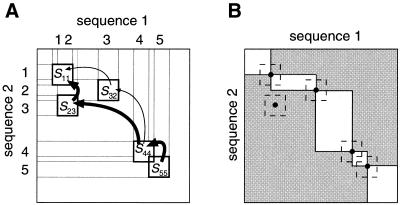
Dividing a homology matrix. To obtain an alignment between two sequences, the homologous segments must be arranged consistently in both sequences. A matrix Sij(1 ≤ i, j ≤ n, n is the number of homologous segments) is constructed in the following manner. If the ith homologous segment on sequence 1 corresponds to the jth homologous segment on sequence 2, Sij has the score value of the homologous segment calculated above; otherwise Sij is set to 0. By applying the standard DP procedure to matrix Sij, we obtain the optimal path, which corresponds to the optimal arrangement of homologous segments. Figure 2A shows an example in which five homologous segments exist. The order of segments in sequence 1 differs from that in sequence 2. The optimal path depends on S23 and S32; if S23 > S32, the path with bold arrows is optimal.
Figure 2.
(A) An example of the segment-level DP; (B) Reducing the area for DP on a homology matrix.
Overall homology matrix is divided into some sub-matrices at the boundary corresponding to the center of homologous segments as illustrated in Figure 2B. As a result, the shaded area in Figure 2B is excluded from the calculation. As many homologous segments are detected by FFT, the CPU time is reduced.
Extension to group-to-group alignments. The procedure described above can be easily extended to group-to-group alignment by considering equations 2 and 6 as a special case with one sequence in each group. These equations are extended to group-to-group alignment by replacing ![]() with
with ![]() , which is the linear combination of the volume components of the members belonging to group 1:
, which is the linear combination of the volume components of the members belonging to group 1:
where wi is the weighting factor for sequence i, which is calculated in the same manner as CLUSTALW (7) for the progressive method, or in the same manner as Gotoh’s (20) weighting system for the iterative refinement method. Similarly, polarity component is calculated as:
This method is applicable to nucleotide sequences by converting a sequence to a sequence of four-dimensional vectors whose components are the frequencies of A, T, G and C at each column, instead of volume and polarity values. In this case, correlation between two nucleotide sequences is:
c(k) = cA(k) + cT(k) + cG(k) + cC(k).
Scoring system
Similarity matrix. In order to increase the efficiency of alignment, the scoring system (similarity matrix and gap penalties) was also modified. Vogt et al. (21) suggested that the Needleman–Wunsch (NW) algorithm performs well with all-positive matrices, in which all elements have positive values. CLUSTALW (7) and other methods use such all-positive matrices by default. Since Vogt et al. (21) examined only the cases in which members of each protein familiy are similar in length, it is not clear whether such all-positive matrices are suitable to various alignment problems, particularly to those of different length. Accordingly, contrary to existing methods, we adopted a normalized similarity matrix 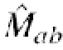 (a and b are amino acids) that has both positive and negative values:
(a and b are amino acids) that has both positive and negative values:
where average1 = ΣafaMaa, average2 = Σa,bfafbMab, Mab is raw similarity matrix, fa is the frequency of occurrence of amino acid a, and Sa is a parameter that functions as a gap extension penalty. Under this similarity matrix 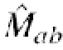 , the score per site between two random sequences is Sa, and the score per site between two identical sequences is 1.0 + Sa. If Sa is much smaller than unity, gaps are scored virtually equivalent to random amino acid sequences.
, the score per site between two random sequences is Sa, and the score per site between two identical sequences is 1.0 + Sa. If Sa is much smaller than unity, gaps are scored virtually equivalent to random amino acid sequences.
The default parameters of our program are: Mab is the 200 PAM log-odds matrix by Jones et al. (22), fa is the frequency of occurrence for amino acid a calculated by Jones et al. (22), Sop (gap opening penalty, defined below) is 2.4 and Sa is 0.06, for amino acid sequences. For nucleic acid sequences, Mab is the 200 PAM log-odds matrix calculated from Kimura’s two parameter model (23) with transition/transversion ratio of 2.0, fa is 0.25, Sop is 2.4 and Sa is 0.06.
Gap penalty. Homology matrix H(i, j) between two amino acid sequences A(i) and B(j) is constructed from the similarity matrix as 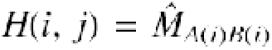 , where i and j are positions in sequences. When two groups of sequences are aligned, homology matrix between group 1 and group 2 is calculated as:
, where i and j are positions in sequences. When two groups of sequences are aligned, homology matrix between group 1 and group 2 is calculated as:
where A(n, i) indicates the ith site of the nth sequence in group 1, B(m, j) is the jth site of the mth sequence in group 2, and wn is the weighting factor, defined previously, for nth sequences.
In the NW algorithm (1), the optimal alignment between two groups of sequences is calculated as:
where P(i,j) is the accumulated score for the optimal path from (1,1) to (i, j), and G1(i, x) and G2(j, y) are gap penalties defined below.
Each group of sequences may contain the gaps already introduced at previous steps. If a gap is newly introduced at the same position as one of such existing gaps, the new gap should not be penalized, because these new and existing gaps are probably resulting from a single insertion or deletion event. Gotoh (6) and Thompson et al. (7) developed position-specific gap penalties depending on the pattern of existing gaps. Our method used in this report is rather simpler than theirs:
G1(i, x) = Sop · {1 – [g1start(x) + g1end(i)]/2},
where Sop corresponds to a gap opening penalty, g1start(x) is the number of the gaps that start at the xth site, and g1end(i) is the number of the gaps that end at the ith site. That is,
where zm(i) = 1 and am(i) = 0, if the ith site of sequence m is a gap; otherwise zm(i) = 0 and am(i) = 1; wm is the weighting factor for sequence m. The other penalty G2(j, y) is calculated in the same manner. Because this formulation is simpler than existing ones (6,7), the CPU time is considerably reduced, but the accuracy of resulting alignments is comparable with that by existing scoring systems (see Results).
Computer programs
We have developed a program package MAFFT, which incorporates new techniques described above. The source code for the FFT algorithm has been taken from Press et al. (19). In MAFFT, the progressive method (3,7) (FFT-NS-1, FFT-NS-2) and the iterative refinement method (4–6) (FFT-NS-i) are implemented with some slight modifications described below.
FFT-NS-1. Using the FFT algorithm and the normalized similarity matrix described above, input sequences are progressively aligned following the branching order of sequences in the guide tree. This method is hereafter referred to as FFT-NS-1. This method requires a guide tree based on the all-pairwise comparison, whose CPU time is O(K2), where K is the number of sequences. Rapid calculation of a distance matrix is important for the case of large K. Thus we adopted the method of Jones et al. (22) with two modifications; 20 amino acids are grouped into six physico-chemical groups (24), and the number Tij of 6-tuples shared by sequence i and sequence j is counted. This value is converted to a distance Dij between sequence i and sequence j as
Dij = 1 – [Tij/min(Tii, Tjj)].
The guide tree is constructed from this distance matrix using the UPGMA method (25).
FFT-NS-2. Input sequences are realigned along the guide tree inferred from the alignment by FFT-NS-1. It is expected that more reliable alignments are obtained on the basis of more reliable guide trees (26). This method is referred to as FFT-NS-2.
FFT-NS-i. An alignment obtained by FFT-NS-2 is subjected to further improvement, in which the alignment is divided into two groups and realigned (4–6). We employ a technique called tree-dependent restricted partitioning (27). This process is repeated until no better scoring alignment is obtained in respect of the score described above. This method is referred to as FFT-NS-i.
To test the effect of the FFT algorithm or the normalized similarity matrix described above, we compared these three methods with several methods in which these newly developed techniques are not used.
NW-NS-1/NW-NS-2. We examined a method that uses the standard NW algorithm, instead of the FFT algorithm, with the normalized similarity matrix described above. This method is referred to as NW-NS-1 or NW-NS-2. Concerning the guide trees, NW-NS-1 and NW-NS-2 are identical to FFT-NS-1 or FFT-NS-2, respectively.
NW-AP-2. To test the effect of the normalized similarity matrix described above, we examined a method with conventional all-positive similarity matrix (21), which is made positive by subtracting the smallest number in the matrix from all elements. This is equivalent to setting Sa in equation 7 to 0.82 for the similarity matrix we use. This method is referred to as NW-AP-2. Except for the similarity matrix, the procedure of NW-AP-2 is identical to that of NW-NS-2.
RESULTS
Computer simulations
In order to evaluate the performance of the present methods, we have conducted computer simulations focusing on the CPU time and the accuracy. Using the sequences generated by a simulation program ROSE (28), the CPU times of the present methods and two existing methods, CLUSTALW version 1.82 and T-COFFEE, were compared for the various length and the various numbers of sequences. Two types of sequence sets were used; one is composed of highly conserved sequences with ∼35–85% identities (average distance is 100 PAM), and the other is a group of distantly related sequences with ∼15–65% identities (average distance is 250 PAM). We also estimated the order of CPU time [Y of O(XY), where X is the length or the number of input sequences] by the power regression analysis.
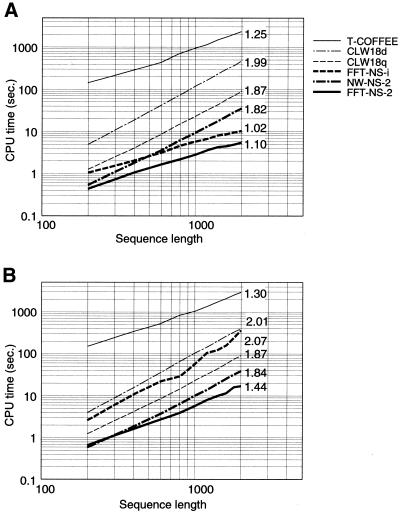
Figure 3 shows the dependence of CPU time on sequence length. The regression coefficient of each method is also shown. The standard NW-based methods, CLUSTALW and NW-NS-2, require the CPU time proportional to the square of sequence length (the regression coefficients are close to 2 for both methods) independently of the degrees of sequence similarities, as expected. In contrast, the CPU times of FFT-based methods, FFT-NS-2 and FFT-NS-i, depend on the degree of similarities of input sequences; the CPU times of FFT-NS-2 and FFT-NS-i are virtually proportional to the sequence length for highly conserved sequences (regression coefficients are close to 1 in Fig. 3A), whereas the CPU time of FFT-NS-2 is close to that of NW-NS-2 for distantly related sequences (Fig. 3B).
Figure 3.
The plot of CPU time versus the average lengths of input sequences for three methods described in the text, FFT-NS-2, FFT-NS-i and NW-NS-2, and two existing methods, CLUSTALW and T-COFFEE. The average percent identities among input sequences are ∼35–85% (A) and ∼15–65% (B). The number of sequences is 40. The regression coefficient calculated from the power regression analysis is shown for each method. For all cases, default parameters were used, except for CLUSTALW, in which both cases default setting (CLW18d) and ‘quicktree’ option (CLW18q) were examined. All of the calculations were performed on a Linux operating system (Intel Xeon 1.7 GHz with 1 GB of memory). The gcc version 2.96 compiler was used with the optimization option ‘-O3’.
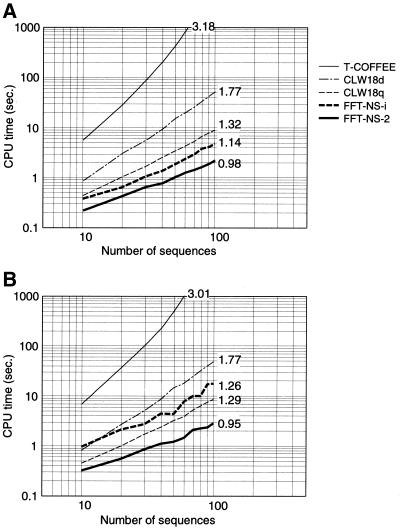
Figure 4A and B show the dependence of CPU times on the number (K) of input sequences. The time consumption of T-COFFEE is O(K3) for alignments of relatively large number of sequences, as Notredame et al. (9) estimated. CLUSTALW (default), which requires the all-pairwise comparison by the standard NW algorithm, consumes O(K2) CPU time. Other methods require CPU times of approximately O(K).
Figure 4.
The plot of CPU time versus the number of input sequences for three methods described in the text, FFT-NS-2 and FFT-NS-i, and two existing methods, CLUSTALW and T-COFFEE. The average percent identities among input sequences are ∼35–85% (A) and ∼15–65% (B). The average length of input sequences is 300. The regression coefficient calculated from the power regression analysis is shown for each method. For all cases, default parameters were used, except for CLUSTALW, in which both cases default setting (CLW18d) and ‘quicktree’ option (CLW18q) were examined. All of the calculations were performed on a Linux operating system (Intel Xeon 1.7 GHz with 1 GB of memory). The gcc version 2.96 compiler was used with the optimization option ‘-O3’.
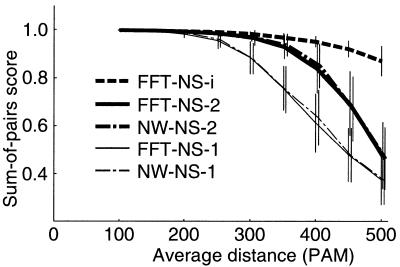
To test the accuracy, five newly developed methods, FFT-NS-1, FFT-NS-2, NW-NS-1, NW-NS-2 and FFT-NS-i, were applied to the sequences of various homology levels generated by ROSE (28). The accuracy of each method was measured by sum-of-pairs score, where a reconstructed alignment is compared with the simulated (‘correct’) alignment and the ratio of correctly aligned pairs is calculated from all possible pairs (8). The simulations were repeated 100 times and averaged for each method (Fig. 5).
Figure 5.
The plot of sum-of-pairs score (8) versus the average distance of input sequences for five methods, FFT-NS-1, FFT-NS-2, FFT-NS-i, NW-NS-1 and NW-NS-2. The number of input sequences is 40, and sequence lengths are 200 sites on average. Vertical lines indicate the standard deviations of the scores. For all cases, default parameters were used.
The accuracy of FFT-based methods (FFT-NS-1 and FFT-NS-2) is almost equivalent to that of standard NS-based methods (NW-NS-1 and NW-NS-2). This result indicates that the FFT algorithm does not sacrifice the accuracy. FFT-NS-2 performs better than FFT-NS-1 as expected. FFT-NS-i has an advantage in accuracy over FFT-NS-1 and FFT-NS-2 for distantly related sequences.
Benchmarks using BAliBASE
Thompson et al. (8) have published a systematic comparison of widely distributed alignment programs using the BAliBASE benchmark alignment database (15), a database of ‘correct’ alignments based on three-dimensional structural superimpositions. The BAliBASE database is categorized into five different types of references. The first category is made up of phylogenetically equidistant members of similar length. In the second category, each alignment contains up to three orphan sequences with a group of close relatives. The third category contains up to four distantly related groups, while the fourth and fifth categories involve long terminal and internal insertions, respectively. These references will be referred to as categories 1–5 hereafter.
We have applied four methods described in Methods, NW-AP-2, NW-NS-2, FFT-NS-2 and FFT-NS-i, to this database to compare their efficiencies with those of five existing methods, DIALIGN (29,30), PIMA (31), CLUSTALW (7) version 1.82, PRRP (32) and T-COFFEE (9). The sum-of-pairs scores (see above) and the column scores [the ratio of correctly aligned columns (8)] were calculated and averaged in each category. Wilcoxon matched-pair signed-rank test and t-test were carried out to test the significance of the difference in the accuracy of each method. These tests give P-values, which is the probability that the observed differences may be due to chance.
Table 1 shows the results of this benchmark test together with the CPU time of each method for performing this test. Unlike the simulation above, FFT-NS-2 (FFT-based method) takes CPU time almost equivalent to NW-NS-2. This is because the FFT algorithm is not efficient for distantly related sequences like these tests. NW-NS-2 takes less CPU time than CLUSTALW does, possibly because of the simple calculation procedure of the former. FFT-NS-i takes less CPU time than T-COFFEE does.
Table 1. Sum-of-pairs scores and column scores of various alignment methods for the BAliBASE benchmark tests.
| Method | CPU time (s) | Cat. 1 | Cat. 2 | Cat. 3 | Cat. 4 | Cat. 5 | Average1 | Average2 |
|---|---|---|---|---|---|---|---|---|
| Progressive methods | ||||||||
| PIMA | 1116 | 0.825/0.737 | 0.751/0.127 | 0.525/0.262 | 0.700/0.480 | 0.788/0.555 | 0.772/0.558 | 0.718/0.432 |
| CLW18d | 2202 | 0.871/0.792 | 0.856/0.329 | 0.754/0.490 | 0.745/0.417 | 0.852/0.617 | 0.844/0.639 | 0.816/0.529 |
| CLW18q | 1657 | 0.871/0.790 | 0.859/0.334 | 0.763/0.473 | 0.728/0.402 | 0.887/0.709 | 0.847/0.644 | 0.824/0.542 |
| NW-AP-2 | 250 | 0.842/0.746 | 0.833/0.268 | 0.770/0.443 | 0.703/0.311 | 0.851/0.667 | 0.821/0.593 | 0.800/0.487 |
| NW-NS-2 | 243 | 0.849/0.761 | 0.844/0.334 | 0.779/0.486 | 0.797/0.532 | 0.951/0.826 | 0.845/0.652 | 0.844/0.588 |
| FFT-NS-2 | 227 | 0.849/0.761 | 0.844/0.334 | 0.779/0.486 | 0.797/0.532 | 0.951/0.826 | 0.845/0.652 | 0.844/0.588 |
| Iterative refinement methods and T-COFFEE | ||||||||
| DIALIGN2-1 | 18132 | 0.792/0.681 | 0.814/0.219 | 0.673/0.327 | 0.818/0.615 | 0.938/0.840 | 0.801/0.584 | 0.807/0.536 |
| PRRP | 9782 | 0.871/0.793 | 0.860/0.354 | 0.823/0.569 | 0.663/0.275 | 0.885/0.742 | 0.845/0.646 | 0.820/0.547 |
| T-COFFEE | 12065 | 0.876/0.797 | 0.856/0.343 | 0.777/0.497 | 0.811/0.555 | 0.961/0.901 | 0.865/0.683 | 0.856/0.619 |
| FFT-NS-i | 1466 | 0.864/0.787 | 0.853/0.363 | 0.789/0.518 | 0.799/0.534 | 0.956/0.835 | 0.857/0.675 | 0.852/0.607 |
| Number of alignments | 82 | 23 | 12 | 15 | 12 | 144 | – | |
Categories (Cat.) 1–5 correspond to five different categories of alignments described in the text. Two types of scores averaged across alignments of each category are shown in each column, separated by a slash (the average of sum-of-pairs scores/the average of column scores). Average1 gives the average score across all the 144 alignments. Since the number of alignments differs for different categories ranging from 12 to 82, another type of average score (Average2), i.e. the score averaged across the five categories, was calculated for each method after Notredame et al. (9). Total CPU time for computing all 144 alignments is also shown for each method. For all cases, default parameters were used, except for CLUSTALW, in which both cases default setting (indicated by CLW18d) and ‘quicktree’ option (CLW18q) were examined. All calculations were performed on a Sun Ultra/2 workstation (UltraSPARC 168 MHz with 128 MB of memory, Solaris 2.6). The gcc version 2.8.1 compiler was used with the optimization option ‘-O3’, except for DIALIGN, for which the pre-compiled version by the authors was used.
The accuracy of NW-AP-2, which contains neither the improved scoring system described above nor the FFT algorithm, is comparable with that of the previous version (1.7) of CLUSTALW (data not shown). By using the improved scoring system shown in equation 7, NW-NS-2 and FFT-NS-2 perform considerably better than NW-AP-2. T-COFFEE marked the highest average accuracy, but the accuracy of FFT-NS-i is comparable with that of T-COFFEE. P-values by Wilcoxon matched-pair signed-rank test are 0.13 for sum-of-pairs score and 0.43 for column score, and P-values by t-test are 0.10 for sum-of-pairs score and 0.23 for column score. Thus the difference is not significant.
Applications to the LSU rRNA and RNA polymerase sequences
BAliBASE is biased toward alignments composed of a small number of short sequences; the number of sequences in each alignment is 9.2 and sequence length is 251.1 on average. To illustrate the power of our approach to practical sequence analyses, we selected two examples of relatively large data sets: the nucleotide sequences of LSU rRNA and the amino acid sequences of the RNA polymerase largest subunit.
LSU rRNA. The Ribosomal Database Project (RDP-II) (33) contains 72 LSU rRNA sequences from Bacteria, Archaea and Eucarya. This alignment was used as a reference alignment. We also use another reference alignment of 59 sequences in which fragment sequences were excluded from the full 72 sequences set (the reference alignments are available at http://www.biophys.kyoto-u.ac.jp/∼katoh/align/example/lsu). The CPU times and the sum-of-pairs and column scores (8) of NW-AP-2, NW-NS-2, FFT-NS-2 and FFT-NS-i were compared with those of two existing methods, CLUSTALW (version 1.82) and T-COFFEE using these two data sets (Table 2). The FFT-based methods (FFT-NS-2 and FFT-NS-i) are efficient for such relatively large data sets.
Table 2. Comparison of several methods using the LSU rRNA sequences.
| Method | CPU time (s) | Sum-of-pairs score | Column score |
|---|---|---|---|
| 72 sequences × 1305–5183 sites | |||
| CLW18d | 1998 | 0.692 | – |
| CLW18q | 600.2 | 0.597 | – |
| NW-AP-2 | 197.0 | 0.796 | – |
| NW-NS-2 | 205.2 | 0.770 | – |
| FFT-NS-2 | 73.39 | 0.769 | – |
| FFT-NS-i | 251.8 | 0.781 | – |
| 59 sequences × 2810–5183 sites | |||
| T-COFFEE | 35 860 | 0.806 | 0.559 |
| CLW18d | 1523 | 0.754 | 0.411 |
| CLW18q | 395.6 | 0.643 | 0.315 |
| NW-AP-2 | 153.7 | 0.823 | 0.482 |
| NW-NS-2 | 159.8 | 0.793 | 0.463 |
| FFT-NS-2 | 51.09 | 0.794 | 0.468 |
| FFT-NS-i | 181.7 | 0.817 | 0.552 |
The CPU time and the sum-of-pairs and column scores (8) of each method are shown for the full LSU data set (72 sequences) retrieved from RDP-II (33), which includes some fragment sequences, and a subset composed of 59 sequences without fragment sequences. In the case of the full LSU data set, the column score cannot be calculated because of fragment sequences, and the test of T-COFFEE is aborted due to a memory shortage. The default parameters were used for all methods, except for CLUSTALW, in which default setting (CLW18d) and ‘quicktree’ option (CLW18q) were examined. All calculations were performed on a Linux operating system (Intel Xeon 2 GHz with 4 GB of memory). The gcc compiler (version 2.96) was used with the optimization option ‘-O3’.
The largest subunit of RNA polymerase. We used a reference alignment of the largest subunit sequences of RNA polymerase by Iwabe et al. (34), which includes 11 highly conserved blocks. Two data sets, one (large) composed of 76 sequences and the other (small) composed of 24 sequences, were compiled. Both of them contain amino acid sequences from Bacteria, Archaea and three major classes (I, II and III) from Eucarya (the reference alignments are available at http://www.biophys.kyoto-u.ac.jp/∼katoh/align/example/rpol). Table 3 shows the CPU time and the number of correctly detected conserved blocks of sequences by six methods: NW-AP-2, FFT-NS-2, NW-NS-2, FFT-NS-i, CLUSTALW version 1.82 and T-COFFEE. T-COFFEE, FFT-NS-2, FFT-NS-i and NW-NS-2 successfully detected all of the 11 blocks, although the CPU times differ for different methods. The CPU time of FFT-NS-2 (FFT-based method) is about one-third of that of NW-NS-2 (standard NW-based method).
Table 3. Comparison of several methods using the largest subunit sequences of RNA polymerase.
| Method | CPU time (s) | Number of correctly aligned blocks |
|---|---|---|
| 76 sequences × 1182–2890 sites | ||
| CLW18d | 675.5 | 10 |
| CLW18q | 159.4 | 10 |
| NW-AP-2 | 54.95 | 8 |
| NW-NS-2 | 59.30 | 11 |
| FFT-NS-2 | 18.15 | 11 |
| FFT-NS-i | 173.1 | 11 |
| 24 sequences × 1206–2890 sites | ||
| T-COFFEE | 745.3 | 11 |
| CLW18d | 100.1 | 9 |
| CLW18q | 50.78 | 9 |
| NW-AP-2 | 20.79 | 10 |
| NW-NS-2 | 22.77 | 11 |
| FFT-NS-2 | 7.150 | 11 |
| FFT-NS-i | 46.00 | 11 |
The CPU time and the number of correctly aligned blocks by each method are shown for a large data set composed of 76 sequences and a small data set composed of 24 sequences. In the case of the large data set, the test of T-COFFEE was aborted due to a memory shortage. For all cases, default parameters were used, except for CLUSTALW, in which both cases default setting (CLW18d) and ‘quicktree’ option (CLW18q) were examined. All calculations were performed on a Linux operating system (Intel Xeon 1.7 GHz with 1 GB of memory). The gcc compiler (version 2.96) was used with the optimization option ‘-O3’.
DISCUSSION
It has been supposed that appropriate alignment algorithm depends on the nature of the sequences to be aligned (8,35); the NW algorithm produces accurate and reliable alignments for references 1, 2 and 3 in BAliBASE, whereas the Smith–Waterman (SW) algorithm (36), a method for detecting local homology, is successful for categories 4 and 5. It may be quite impractical to select properly these different algorithms, depending on the nature of input sequences; actual sequence data contain various types of sequences, i.e. fragment sequences, fusion proteins, orphan sequences, over-representation of some members and so on.
On the basis of such considerations, Notredame et al. (9) formulates a combination of NW and SW alignment procedures in T-COFFEE. This attempt is successful in improving the accuracy at the sacrifice of the computational simplicity. Thus, this method may be applicable to short and small data sets like those in BAliBASE as Karplus and Hu (37) pointed out. In contrast, the present methods employ a simple NW algorithm (NW-NS-2) or a more rapid algorithm based on FFT (FFT-NS-2 and FFT-NS-i). Nevertheless, the BAliBASE benchmark tests show that the present methods with the normalized similarity matrix perform well also for categories 4 and 5. As a result, the accuracy of FFT-NS-i is comparable with that of T-COFFEE. This result indicates that the accuracy of alignments can be considerably improved without complicating any computational process, contrary to the conventional thought that a combination of the NW and SW algorithms was necessary for computing high-quality alignments (8,9,35). The improvement in accuracy was achieved simply by normalizing the similarity matrix.
This suggests the importance of parameter choice as Thompson et al. (7,8) pointed out. However, there is a large difference between their strategy and ours. The scoring system used in CLUSTALW is complicated and time consuming; many parameters in the scoring system dynamically vary depending on input sequences. In contrast, the present scoring system is simple; the similarity matrix is fixed for any input sequences, and even extension gap penalty is not explicitly contained in the DP algorithm. Nevertheless, the accuracy of NW-NS-2/FFT-NS-2 is comparable with that of CLUSTALW.
In all cases tested above, the present methods consume generally less CPU time than existing methods of comparable accuracy do. It is remarkable that the order of CPU time is reduced from O(N2) to O(N) by the FFT algorithm for highly conserved sequences (Fig. 3A), where N is sequence length. Such a rapid multiple alignment method is suitable for automated high-throughput analysis of genomic sequences. At the same time, biologists’ expertise is still of particular importance and, consequently, a user-friendly alignment workbench is required, which provides easy access to the various information collected by database searches, alignment analyses and the predictions obtained by non-homology methods (38). The method presented here is also useful as a core component of such an integrated alignment workbench.
The MAFFT program package is freely available at http://www.biophys.kyoto-u.ac.jp/∼katoh/programs/align/mafft. It has been tested on the Linux operating system. A graphical user interface, written by H. Suga, K. Katoh, Y. Yamawaki, K. Kuma, D. Hoshiyama, N. Iwabe and T. Miyata, on the X Window System is also available at http://www.biophys.kyoto-u.ac.jp/∼katoh/programs/align/xced.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs N. Iwabe, H. Suga and D. Hoshiyama for helpful comments. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Needleman S.B., and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 2.Sankoff D., and Cedergren,R.J. (1983) Simultaneous comparison of three or more sequences related by a tree. In Sankoff,D. and Kruskal,J.B. (eds), Time Warps, String Edits and Macromolecules: The Theory and Practice of Sequence Comparison. Addison-Wesley, London, UK, pp. 253–264.
- 3.Feng D.F., and Doolittle,R.F. (1987) Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol., 25, 351–360. [DOI] [PubMed] [Google Scholar]
- 4.Barton G.J., and Sternberg,M.J. (1987) A strategy for the rapid multiple alignment of protein sequences. Confidence levels from tertiary structure comparisons. J. Mol. Biol., 198, 327–337. [DOI] [PubMed] [Google Scholar]
- 5.Berger M.P., and Munson,P.J. (1991) A novel randomized iterative strategy for aligning multiple protein sequences. Comput. Appl. Biosci., 7, 479–484. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh O., (1993) Optimal alignment between groups of sequences and its application to multiple sequence alignment. Comput. Appl. Biosci., 9, 361–370. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson J.D., Plewniak,F. and Poch,O. (1999) A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res., 27, 2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 10.Delcher A.L., Kasif,S., Fleischmann,R.D., Peterson,J., White,O. and Salzberg,S.L. (1999) Alignment of whole genomes. Nucleic Acids Res., 27, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson W.R., and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J., Sawyer,S. and Kochin,R. (1982) An efficient method for matching nucleic acid sequences. Nucleic Acids Res., 10, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajasekaran S., Jin,X. and Spouge,J.L. (2002) The efficient computation of position-specific match scores with the fast Fourier transform. J. Comput. Biol., 9, 23–33. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J.D., Plewniak,F. and Poch,O. (1999) BAliBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics, 15, 87–88. [DOI] [PubMed] [Google Scholar]
- 16.Miyata T., Miyazawa,S. and Yasunaga,T. (1979) Two types of amino acid substitutions in protein evolution. J. Mol. Evol., 12, 219–236. [DOI] [PubMed] [Google Scholar]
- 17.Kimura M., (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- 18.Grantham R., (1974) Amino acid difference formula to help explain protein evolution. Science, 185, 862–864. [DOI] [PubMed] [Google Scholar]
- 19.Press W.H., Teukolsky,S.A., Vetterling,W.T. and Flannery,B.P. (1995) Numerical Recipes in C: The Art of Scientific Computing, 2nd Edn. Cambridge University Press, Cambridge, UK.
- 20.Gotoh O., (1995) A weighting system and algorithm for aligning many phylogenetically related sequences. Comput. Appl. Biosci., 11, 543–551. [DOI] [PubMed] [Google Scholar]
- 21.Vogt G., Etzold,T. and Argos,P. (1995) An assessment of amino acid exchange matrices in aligning protein sequences: the twilight zone revisited. J. Mol. Biol., 249, 816–831. [DOI] [PubMed] [Google Scholar]
- 22.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci., 8, 275–282. [DOI] [PubMed] [Google Scholar]
- 23.Kimura M., (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16, 111–120. [DOI] [PubMed] [Google Scholar]
- 24.Dayhoff M.O., Schwartz,R.M. and Orcutt,B.C. (1978) A model of evolutionary change in proteins. In Dayhoff,M.O. and Ech,R.V. (eds), Atlas of Protein Sequence and Structure. National Biomedical Research Foundation, MD, pp. 345–352.
- 25.Sokal R.R., and Michener,C.D. (1958) A statistical mehod for evaluating systematic relationships. University of Kansas Scientific Bulletin, 28, 1409–1438. [Google Scholar]
- 26.Tateno Y., Ikeo,K., Imanishi,T., Watanabe,H., Endo,T., Yamaguchi,Y., Suzuki,Y., Takahashi,K., Tsunoyama,K., Kawai,M., Kawanishi,Y., Naitou,K. and Gojobori,T. (1997) Evolutionary motif and its biological and structural significance. J. Mol. Evol., 44 (Suppl. 1), S38–S43. [DOI] [PubMed] [Google Scholar]
- 27.Hirosawa M., Totoki,Y., Hoshida,M. and Ishikawa,M. (1995) Comprehensive study on iterative algorithms of multiple sequence alignment. Comput. Appl. Biosci., 11, 13–18. [DOI] [PubMed] [Google Scholar]
- 28.Stoye J., Evers,D. and Meyer,F. (1997) Generating benchmarks for multiple sequence alignments and phylogenetic reconstructions. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 303–306. [PubMed] [Google Scholar]
- 29.Morgenstern B., Dress,A. and Werner,T. (1996) Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl Acad. Sci. USA, 93, 12098–12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgenstern B., (1999) DIALIGN2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 31.Smith R.F., and Smith,T.F. (1992) Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modelling. Protein Eng., 5, 35–41. [DOI] [PubMed] [Google Scholar]
- 32.Gotoh O., (1996) Significant improvement in accuracy of multiple protein sequence alignments by iterative refinement as assessed by reference to structural alignments. J. Mol. Biol., 264, 823–838. [DOI] [PubMed] [Google Scholar]
- 33.Maidak B.L., Cole,J.R., Lilburn,T.G., Parker,C.T.,Jr, Saxman,P.R., Farris,R.J., Garrity,G.M., Olsen,G.J., Schmidt,T.M. and Tiedje,J.M. (2001) The RDP-II (ribosomal database project). Nucleic Acids Res., 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwabe N., Kuma,K., Kishino,H., Hasegawa,M. and Miyata,T. (1991) Evolution of RNA polymerases and branching patterns of the three major groups of archaebacteria. J. Mol. Evol., 32, 70–78. [DOI] [PubMed] [Google Scholar]
- 35.McClure M.A., Vasi,T.K. and Fitch,W.M. (1994) Comparative analysis of multiple protein-sequence alignment methods. Mol. Biol. Evol., 11, 571–592. [DOI] [PubMed] [Google Scholar]
- 36.Smith T.F., and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 37.Karplus K., and Hu,B. (2001) Evaluation of protein multiple alignments by SAM-T99 using the BAliBASE multiple alignment test set. Bioinformatics, 17, 713–720. [DOI] [PubMed] [Google Scholar]
- 38.Lecompte O., Thompson,J.D., Plewniak,F., Thierry,J. and Poch,O. (2001) Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene, 270, 17–30. [DOI] [PubMed] [Google Scholar]