Abstract
The orthodenticle-related protein (HpOtx) gene derived from the sea urchin Hemicentrotus pulcherrimus encodes two distinct isoforms, HpOtxE and HpOtxL, which are differentially expressed during early embryogenesis and are driven by TATA-less and TATA-containing promoters, respectively. In order to determine if the TATA element is involved in the establishment of the temporally specific expression profile of the HpOtx gene, reporter genes under the control of modified or wild-type HpOtxE/L promoters were introduced into fertilized eggs. When the activities of the different promoter constructs were examined, we found that deletion of the TATA element from the HpOtxL promoter causes early expression, whereas addition of the TATA element to the HpOtxE promoter causes delayed expression. This suppressive action of the TATA element on transcription from the HpOtxE/L promoters requires the presence of upstream CACGTG elements. These results indicate that the presence or absence of the TATA element determines, at least in part, the expression profile of the HpOtxE/L promoters, in concert with the transcription factor(s) that binds to the upstream CACGTG element. Immunoblot and gel retardation analyses suggest that functional interaction between CACGTG binding factor(s) and TATA factor(s) may be regulated by an unidentified third factor(s) during early embryogenesis in the sea urchin.
INTRODUCTION
Most sexually reproducing organisms initiate embryonic development immediately after the formation of a zygote, i.e. fertilization of an egg by a sperm (1). During the very early stages of embryonic development, cell division is under the control of maternal mRNAs stockpiled during oogenesis. As development proceeds, this maternal stock gradually disappears and is replaced by zygotically expressed mRNAs. The specific developmental stage at which this transition from maternal to embryonic control of gene expression occurs appears to be species-specific (reviewed in 2,3). For instance, major zygotic gene activation begins at the two-cell stage in mouse, at stage 7 (approximately 100 cells) in leech, at cycle 7 (90–125 cells) in nematode, and at later stages in Axolotl (cycle 11, corresponding to approximately 2000 cells) and Xenopus (cycle 12, corresponding to approximately 4000 cells) (reviewed in 2). There are some variations in the timing of this transition even within groups of related species such as mammals (reviewed in 3). Although the molecular basis for this species specificity during the maternal-to-zygotic transition is not yet completely understood, it appears that in Xenopus and Drosophila there may be a critical nucleocytoplasmic ratio at which repressive factors are titrated out to allow zygotic gene activation (reviewed in 4). Candidate repressive factors are proposed to be chromatin components in Xenopus (5) and/or specific transcriptional repressors in Drosophila (6). In mouse, on the other hand, the first round of DNA replication, which remodels nucleosome structures, is essential for the expression of several of the earliest embryonic genes, suggesting that zygotic gene activation is prevented by repressive chromatin that constrains the access of transcription factors to binding sites in the one-cell embryo (reviewed in 7–9).
In addition to structural changes in chromatin, the activity of the basal transcriptional apparatus is thought to play a pivotal role in establishing transcriptional competence at a species-specific developmental stage. For instance, the amount of TATA binding protein (TBP) is developmentally regulated in Xenopus. It is barely detectable in oocytes and during early cleavage stages, but accumulates to maximal levels at the onset of the midblastula transition (MBT), the period of major zygotic gene activation (10). In fact, in vitro and in vivo analyses in Xenopus have revealed that TBP is a bona fide rate-limiting factor for basal transcription prior to MBT (10). TFIIB and TFIIF are expressed constitutively during embryogenesis, whereas regulation of phosphorylation of the largest subunit of RNA polymerase II appears to coincide with MBT in Xenopus (10). Similar stage- specific changes in TBP expression and RNA polymerase II modification have been reported in mouse embryos as well (11,12).
A deficiency in co-activator activity is another characteristic feature of the transcriptional apparatus that exists at the very early stages of embryonic development (10,13). In Xenopus, GAL4-VP16 was shown to bind promoter DNA while failing to activate transcription in early embryos (14). Similarly, upstream activating sequences present in the histone H2B promoter do not function normally in cell extracts derived from pre-MBT embryos, although basal transcriptional activity is restored by the addition of TBP (10). Considering that activation can be restored by the addition of HeLa cell extract, the lack of activation in pre-MBT cell extracts is more likely due to a deficiency in certain co-activators than to the presence of one or more dominant repressors (10). In fact, the activation potential, which is normally acquired before the onset of gastrulation in Xenopus, can be precociously conferred by injecting RNA encoding a human co-factor into early embryos (10). Consistent with the idea that the activation potential is achieved at the time of MBT, co-activator activity first appears in two- to four-cell mouse embryos, which is when the major onset of zygotic gene activation occurs in this organism (13,15). Interestingly, activation is TATA element-independent in undifferentiated cells, e.g. two- to eight-cell mouse embryos, but has a strong requirement for the TATA element in differentiated cells, e.g. PMEF primary cells derived from 13-day-old mouse embryos (16). Thus, at least in mouse embryos, the TATA dependence of activated transcription is also developmentally regulated.
Here we examine the effect of the TATA element on the stage-specific expression of the orthodenticle-related protein (HpOtx) gene in the sea urchin Hemicentrotus pulcherrimus, which is particularly well suited for the study of the mechanisms of gene regulation during early stages of development. For instance, the results of reporter assays are highly quantitative since foreign DNA can be readily integrated into chromosomes of a large number of synchronized embryos by the particle gun method (17). HpOtx encodes two distinct isoforms, HpOtxE and HpOtxL, which are expressed at the unhatched blastula and blastula stages, and are driven by TATA-less and TATA-containing promoters, respectively (18). The mRNAs of these two isoforms accumulate in a distinct set of cells during embryogenesis (19), indicating that they regulate a distinct set of genes (20).
An example of the differing functions of the two isoforms of HpOtx can be found in the arylsulfatase (HpArs) gene, which is expressed in a stage- and tissue-specific manner during embryogenesis in H.pulcherrimus (21–23). Cis and trans regulatory elements of this gene have been extensively studied over the past 10 years (reviewed in 24,25). A 229 bp fragment (referred to as the C15 fragment) located in the first intron of HpArs was found to contain strong enhancer activity that appears to be mediated by the coordinated action of HpOtxL and CAAT binding factors (20,26). HpOtxE, which is produced from the same gene as HpOtxL by virtue of differential promoter utilization and alternative splicing (18), cannot activate transcription of the HpArs gene even though the two HpOtx proteins differ only in their extreme N-terminal regions (18). Furthermore, it is also known that orthologous Otx proteins in Strongylocentrotus purputatus play an indispensable role in cell fate decisions (27). Therefore, the HpOtx tandem promoter could provide an ideal system to investigate the molecular mechanisms of how similar but distinct transcription factors with different functions are generated from the same gene during early embryogenesis.
MATERIALS AND METHODS
Embryo culture
Gametes of the sea urchin, H.pulcherrimus, were collected by intracoelomic injection of 0.55 M KCl, washed three times with artificial sea water (ASW) and inseminated (26). Fertilized eggs in which reporter constructs had been introduced (as described below) were cultured at 16°C in a petri dish (6 cm in diameter) with a cell concentration of ∼1.3% (v/v) (∼0.2 ml of eggs suspended in 15 ml of ASW). To prepare nuclear extracts of the different stage embryos, fertilized eggs were cultured at 16°C with gentle aeration at a cell concentration of ∼1% (v/v) (∼1 ml of eggs suspended in 100 ml of ASW) (26).
Construction of reporter plasmids
The HpOtxE promoter fragment (base pairs –461 to +292, numbered with repect to the transcriptional initiation site as +1) (18) was amplified by PCR using the primer pair TK714 and TK715 and the sperm DNA as a template. The PCR product was inserted into the EcoRV site of the pBluescript KSII(+) plasmid (Stratagene) to generate pHpOtxE6. The oligonucleotides used in this study are summarized in Table 1. The SacI–XhoI fragment of pHpOtxE6 containing the HpOtxE promoter was ligated into the corresponding sites of the pGL3-basic vector (Promega), which encodes luciferase as a reporter gene, to generate pM1249. Site-specific mutagenesis (28) was conducted on pM1249 using oligonucleotides TK867 and TK1094 to generate pM1521, a construct with a truncated HpOtxE promoter fragment (base pairs –190 to +180). The TATTCA sequence at base pairs –28 to –23 in the HpOtxE promoter of pM1521 was changed to TATAAA and TATCCA by site-specific mutagenesis using the TK1150 and TK1149 oligonucleotides to generate pM1598 and pM1599, respectively. The CACGTG sequence (E-box) at base pairs –66 to –61 in the HpOtxE promoter of pM1521 and pM1598 was altered to a HindIII site (AAGCTT) by site-specific mutagenesis using the TK1812 oligonucleotide. This generated pM3139 and pM3142, respectively.
Table 1. Oligonucleotides used in this study.
The HpOtxL promoter fragment (base pairs –293 to +282) was amplified by PCR using the primer pair TK712 and TK713 and the sperm DNA as a template. This fragment was inserted into the EcoRV site of pBluescript KSII(+) to generate pHpOtxL14. The KpnI–SmaI fragment of pHpOtxL14 containing the HpOtxL promoter was ligated into the corresponding sites of the pGL3-basic vector to generate pM1248. The TATAAA sequence at base pairs –26 to –21 in the HpOtxL promoter of pM1248 was deleted by site-specific mutagenesis using the oligonucleotide TK804, resulting in pM1293. The CACGTG sequence (E-box) at base pairs –220 to –215 in the HpOtxL promoter of pM1248 and pM1293 was changed to a HindIII site (AAGCTT) by site-specific mutagenesis using the oligonucleotide TK1815 to generate pM3145 and pM3148, respectively.
The XhoI–HindIII fragment containing the H.pulcherrimus arylsulfatase (HpArs) promoter (base pairs –252 to +38) (29) was excised from pAL(ΔXh) and ligated at the same restriction sites (i.e. XhoI and HindIII) in the pGL3-basic vector to generate pM1533. Site-specific mutagenesis was conducted on pM1533 using the oligonucleotide TK2227 to generate pM3571, a construct containing the truncated HpArs promoter fragment (base pairs –168 to +38). The TATAAA sequence at base pairs –32 to –27 in the HpArs promoter of pM3571 was deleted by site-specific mutagenesis using oligonucleotide TK1107 to generate pM3572.
Luciferase assay and RT–PCR analysis
Introduction of reporter plasmids into fertilized eggs was conducted as previously described (17). The pRL-CMV plasmid encoding Renilla reniformis luciferase (Dual-Luciferase Reporter Assay System, Promega) was introduced along with sample DNA (reporter plasmid) as a reference to normalize for transfection efficiency. Bombardment using a particle gun gene delivery system (GIE-III IDERA, Tanaka Co. Ltd) was carried out in triplicate for each DNA on approximately 0.2 million embryos. Bombarded embryos were cultured in a petri dish (6 cm in diameter) and incubated at a constant temperature of 16°C. At the indicated time, aliquots of bombarded embryos were taken from the dishes, collected by centrifugation and stored at –80°C until being used in either luciferase assays (approximately 20 000 embryos × 3) or RT–PCR analyses (approximately 6700 embryos × 3). The luciferase assays were carried out according to the manufacturer’s protocol (Promega).
For RT–PCR analysis, total RNA was prepared from collected embryos using the Isogen RNA extraction system (Wako Pure Chemical Industries, Ltd). Samples were subsequently treated with 0.1 µg/µl DNase I for 18 h at 37°C to degrade contaminating genomic DNA. The 800 bp DNA fragment encoding the region from immediately 3′ of the transcriptional initiation site of the HpOtxE promoter to the middle of the open reading frame (ORF) of firefly luciferase was amplified from 0.3 µg of total RNA using the One Step RNA PCR Kit (AMV) (TaKaRa Biomedicals) and the primers TK1682 and TK1686. As a reference, the 190 bp DNA fragment encoding the 3′-untranslated region (3′-UTR) of the ubiquitin gene was amplified using the primer pair TK1591 and TK1592. The PCR conditions used were 30 min at 50°C for reverse transcription and 2 min at 94°C for denaturation and inactivation of AMV reverse transcriptase, followed by the indicated number of cycles; each cycle comprised 30 s at 94°C, 30 s at 57°C and 90 s at 72°C. PCR amplification was carried out for 28 and 20 cycles for HpOtxE and ubiquitin, respectively. PCR products were resolved on 1.8% agarose gels and stained with ethidium bromide, followed by visualization using a FMBIO II Multi-View image analyzer (TaKaRa Biomedicals).
Isolation of cDNA encoding TBP of H.pulcherrimus
To isolate cDNA clones encoding H.pulcherrimus TBP (HpTBP), we amplified an aliquot of a gastrula cDNA library (30) by PCR using the primers TK753 and TK755. These primers were designed to match perfectly with the nucleotide sequences in the highly conserved region of TBP from other sea urchin species (31). The fragment had an expected size of 345 bp and was subcloned into the EcoRV site of pBluescript KSII(+) (Stratagene). It was confirmed to be a bona fide HpTBP clone by sequence analysis and comparison with other members of the TBP family. The full-length HpTBP cDNA clone, including its long 3′-UTR, was obtained by screening the same gastrula cDNA library using the 345 bp PCR-generated DNA fragment as a probe. The entire clone was sequenced using both universal primers and the specific primers TK848, TK849, TK852, TK853, TK870, TK871, TK872 and TK873. The complete nucleotide sequence of HpTBP cDNA was deposited in GenBank under the accession no. AB074420.
Isolation of a cDNA encoding the N-terminal region of upstream stimulatory factor (USF) of H.pulcherrimus
To isolate cDNA clones encoding H.pulcherrimus USF (HpUSF), the 420 bp DNA fragment corresponding to the region from 7 to 147 amino acids of the S.purputatus USF (SpUSF) was amplified from 0.3 µg of total RNA using the One Step RNA PCR Kit (AMV) (TaKaRa Biomedicals) and the primers TK2396 and TK2398, which were designed to perfectly match the nucleotide sequences of SpUSF. The fragment with the expected size of 420 bp was subcloned into the EcoRV site of pBluescript KSII(+) (Stratagene) and was confirmed to be a bona fide HpUSF clone by sequence analysis and sequence comparison with other members of the USF family. To isolate cDNA clones including the entire 5′-UTR, the 5′-rapid amplification of cDNA end (5′-RACE) experiment was performed using the 5′-Full RACE Core Set Kit (TaKaRa Biomedicals) and the primers TK2457, TK2458/2461 and TK2459/2460 as RT-primer, first primer pair and second primer pair, respectively. The second-round PCR product was subcloned into pBluescript KSII(+) and sequenced with universal primers for verification. As a result, the DNA fragment containing the 45 bp of the 5′-UTR as well as the coding region from 1 to 21 amino acids of HpUSF were obtained and then combined with the sequences of the RT–PCR products described above. TK2396 was found to perfectly match not only the nucleotide sequence of SpUSF but also that of HpUSF. The nucleotide sequence of this partial HpUSF cDNA clone was deposited in GenBank under the accession no. AB082982.
Preparation of recombinant proteins, and GST-pulldown and gel retardation assays
An ∼750 bp NdeI–BamHI fragment obtained from PCR using the primers TK850 and TK851 was ligated into the corresponding restriction sites of a pET28a expression vector (Novagen). This generated pM1541, which was used to express His-tagged HpTBP in bacterial cells. Recombinant HpTBP protein was prepared according to the protocol used previously to obtain yeast TBP protein (32). Recombinant yeast TFIIA and TAND were prepared as previously described (32). GST-pulldown and gel retardation assays were also performed as described previously (32).
To prepare recombinant HpUSF protein, an ∼420 bp NdeI–BamHI fragment, obtained by PCR using the primers TK3442 and TK3441, was ligated into the corresponding restriction sites of the pET28a expression vector (Novagen). This generated pM3988, which was then used to express the His-tagged N-terminal region (9–147 amino acids) of HpUSF in bacterial cells. Note that the most C-terminal six amino acid residues (142–147 amino acids; QPSGAA) were derived from a primer corresponding to the same region of SpUSF. It is currently unknown whether HpUSF has the same sequence or not. Recombinant HpUSF protein was prepared according to the protocol used to obtain HpTBP protein.
Antibody production, preparation of SDS lysates from embryo cultures and immunoblot analysis
Anti-HpTBP and -HpUSF polyclonal antibodies were prepared by injecting gel-purified recombinant HpTBP and HpUSF (9–147 amino acids) proteins into New Zealand White rabbits as previously described (33). Preparation of SDS-lysed embryo cultures was carried out using a slight modification of the method described in Edelmann et al. (31). Approximately 10 000 embryos were removed from a 1% (1 packed ml per 100 ml of ASW) culture at 0, 8, 11, 14, 25, 35, 45, 63 and 70 h post-fertilization and collected by microcentrifugation at 4°C for 10 s. Embryo pellets were washed once each with Ca2+- and Mg2+-free ASW (500 mM NaCl, 9 mM KCl, 30 mM Na2SO4, 2 mM NaHCO3, pH 8.0) (31) and STE buffer (150 mM NaCl, 10 mM Tris–HCl pH 7.4, 1 mM EDTA, 1 mM EGTA) (31). Washed embryos were boiled in 500 µl of SDS lysis buffer (0.5% SDS, 50 mM Tris–HCl pH 7.4, 2 mM PMSF, 0.7 µg/ml bestatin, 4 mM benzamidine, 80 µg/ml pepstatin A, 1 µg/ml leupeptin, 0.05 µg/ml antipain) for 10 min and stored at –80°C until use. For immunoblot analysis, 20 µl of SDS-lysed sample (corresponding to approximately 400 embryos) was separated on a 12.5% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane and probed with anti-HpTBP and anti-HpUSF antibodies as described previously (33).
Preparation of stage-specific nuclear extracts and detection of E-box binding activities
Nuclei and nuclear extracts were prepared from embryos at the hatched blastula, mesenchyme blastula and gastrula stages according to the method described by Sakamoto et al. (26). Gel retardation assays were conducted as described previously (34) with a few modifications. Briefly, four pairs of single-stranded oligonucleotides, i.e. TK3449/3450, TK3451/3452, TK3453/3454 and TK3455/3456, were annealed to generate four double-stranded oligonucleotides containing the wild-type E-box from HpOtxL, the mutated E-box from HpOtxL, the wild-type E-box from HpOtxE and the mutated E-box from HpOtxE, respectively. These double-stranded oligonucleotides were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase and purified by the spun column technique. Gel retardation assays were performed in a solution containing 20 mM HEPES pH 7.6, 5 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol (DTT), 0.1% Triton X-100, 10% glycerol and 100 µg/ml of poly(dI-dC) as non-specific competitor. Two micrograms of nuclear extract was added to each assay and incubated for 10 min at 4°C prior to the addition of 1–2 ng of radiolabeled probe. The reaction mixture was then incubated for 30 min at 15°C. The DNA–protein complex was resolved from free probe on a 4% polyacrylamide gel in 0.5× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0) containing 5% glycerol and 0.5 mM DTT. The gel was dried and visualized by autoradiography.
RESULTS
A role for the TATA element in the temporally specific expression of HpOtx mRNAs
Two proteins, HpOtxE and HpOtxL, are produced from HpOtx by virtue of differential promoter utilization and alternative splicing (18) (Fig. 1). Based on several reports that TATA and/or initiator elements determine the timing of expression of tandem promoters during development (35,36), we examined the contribution of core promoter structures to the temporal specificity of the expression from the HpOtxE/L promoters. A canonical TATA element (TATAAA) was found in the HpOtxL promoter but not in the HpOtxE promoter (Fig. 1) (18). Thus, we first tested whether the presence of the TATA element contributes to the later expression of the HpOtxL promoter and, conversely, if its absence allows earlier expression of the HpOtxE promoter. The HpOtxL promoter fragment (base pairs –293 to +282) (Fig. 1) with or without the TATAAA sequence was fused to the firefly luciferase reporter gene and subsequently introduced into fertilized eggs along with a reference plasmid encoding the Renilla luciferase gene (Fig. 2A). Firefly luciferase activity normalized to that of Renilla luciferase showed that the deletion of the TATAAA sequence from the HpOtxL promoter induced earlier expression (Fig. 2A) without shifting the transcriptional initiation site (data not shown). The results were reproduced in several independent experiments (Fig. 3A and data not shown). When we conducted similar experiments using an HpArs promoter fragment (base pairs –160 to +38), the opposite effect was obtained when the TATAAA sequence was deleted, in that expression from the HpArs promoter was decreased and/or delayed (Fig. 2B). Therefore, the role of the TATA element in suppressing early expression appears not to be a universal function but rather one that is specific to the HpOtxL promoter.
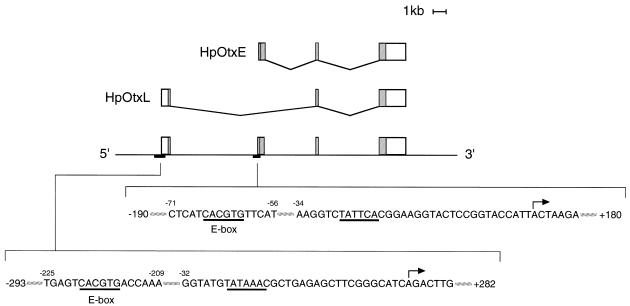
Figure 1.
Schematic representation of the HpOtx gene and the two mRNAs generated from it by differential promoter utilization and alternative splicing (18). The rectangles above the straight line with 5′ and 3′ demarcations indicate exons, and shaded areas within them correspond to ORFs. The double inclined lines connecting the rectangles depict the splicing pattern of the intervening introns. The nucleotide sequences around the E-box (CACGTG) and the transcriptional initiation site (marked with arrows) are shown for both HpOtxL and HpOtxE.
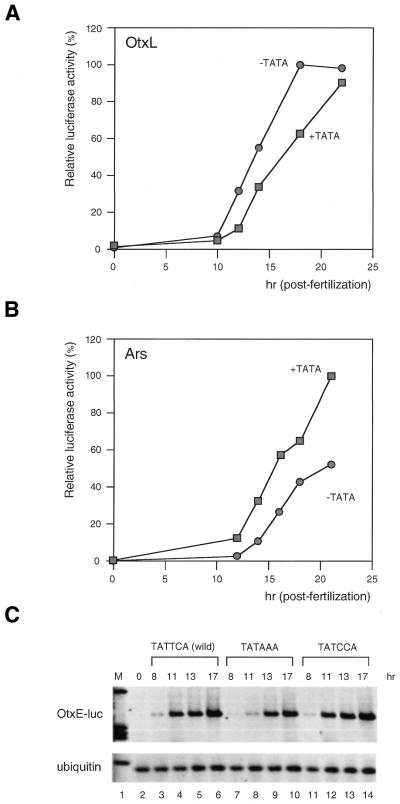
Figure 2.
The effect of the TATA element on the temporal expression profile of three promoters in H.pulcherrimus. (A) Firefly luciferase activities at different times after fertilization with the wild-type HpOtxL promoter (closed rectangle) or with the mutant HpOtxL promoter (closed circle) lacking the TATAAA sequence (transfections were conducted at time 0). Raw values were normalized to the activity of co-transfected Renilla luciferase and expressed as a percentage of the maximal value which was taken to be 100%. (B) Relative luciferase activities under the control of the HpArs promoter with (wild-type, closed rectangle) and without (closed circle) the TATAAA sequence. All procedures are the same as represented in (A). (C) RT–PCR analysis was used to measure the amount of firefly luciferase mRNA transcribed under the control of the HpOtxE promoter containing the TATTCA sequence (wild-type, lanes 3–6), TATAAA sequence (lanes 7–10), or TATCCA sequence (lanes 11–14) at the indicated post-fertilization time. Lane 2 represents the amount of the wild-type HpOtxE-luciferase construct mRNA immediately after transfection. Results obtained from the mutated HpOtxE-luciferase constructs were similar to those of the wild-type (lane 2). Ubiquitin mRNA, which is ubiquitously expressed, was used as an internal standard for all the RNA samples. M (lane 1) shows the migration pattern of molecular weight markers.
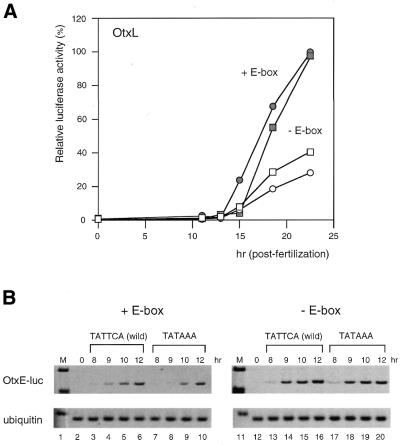
Figure 3.
Functional interaction between the TATA and upstream CACGTG sequences elicits the temporally specific expression from the HpOtxL and HpOtxE promoters. (A) Relative luciferase activities from intact HpOtxL promoters with (wild-type, closed rectangle) and without (closed circle) the TATAAA sequence as well as under the control of an HpOtxL promoter lacking the E-box and containing (open rectangle) or lacking (open circle) the TATAAA sequence. Note that the effect of the TATA element on the temporal expression profile is reversed when the CACGTG element (E-box) is deleted. (B) RT–PCR analysis was undertaken to measure the amount of luciferase mRNA transcribed under the control of an HpOtxE promoter with the TATTCA sequence (wild-type, lanes 3–6) or the TATAAA sequence (lanes 7–10), as represented in Figure 2C. Results of similar analyses in the absence of the E-box are shown in lanes 11–20.
We reasoned that if the TATA element were necessary for the temporally specific expression profile of the HpOtxE/L promoters, the addition of a TATAAA sequence to the HpOtxE promoter would delay its expression. However, in our experiments, luciferase gene expression from the HpOtxE promoter (base pairs –190 to +180) (Fig. 1) was not significantly affected by the presence of a TATA element (data not shown). The sensitivity of the dual-luciferase assay method may not be sufficient to detect small differences in initiation of gene expression because it may take several hours to generate luciferase proteins from a reporter gene. Therefore, we decided to measure the amounts of luciferase mRNA directly using quantitative RT–PCR (Fig. 2C). The increased sensitivity of this method allowed us to observe in several independent experiments that substitution of the wild-type TATTCA sequence at base pairs –28 to –23 by TATAAA caused delayed expression of the HpOtxE promoter (Figs 2C and 3B, and data not shown). This effect is intrinsic to the TATA element since the TATCCA sequence cannot act as an effective substitute (Fig. 2C, lanes 11–14). The level of ubiquitously expressed ubiquitin mRNA (37), used as a control, was constant in all of the RT–PCR experiments (Fig. 2C).
Functional interaction between TATA and upstream CACGTG elements
We have demonstrated that the presence of the TATA element delays expression from the HpOtxE/L promoters but not from the HpArs promoter (Fig. 2). These results indicate that the TATA element cannot be the only factor determining the expression profile of the HpOtx promoters. Thus, we assumed that promoter-specific transcription factors and their functional interaction with TATA-binding factors (TFIID or TBP) direct the temporally specific expression of these promoters. Considering that in most cases the TATA element affects transcription positively (38), it is likely that certain factors specifically bound to the HpOtxE/L promoters suppress the function of TFIID (or TBP) at an earlier stage of embryogenesis. To identify such factors, we first performed a computer-aided database search (Y.Akiyama, National Institute of Advanced Industrial Science and Technology, Japan, http://www.cbrc.jp/research/db/TFSEARCHJ.html) (39) for transcription factor binding sites that are shared by the HpOtxE and HpOtxL promoters but not by the HpArs promoter. This screen identified only the CACGTG element, located at base pairs –220 to –215 of the HpOtxL promoter and at base pairs –66 to –61 of the HpOtxE promoter, as a candidate (Fig. 1). The CACGTG element, also referred to as the E-box (40), has been known to act as the binding site for a family of transcription factors containing a basic/helix–loop– helix/leucine-zipper (b/HLH/Z) motif. The b/HLH/Z family includes various transcription factors such as USF, Myc, Mad, Max, Mxi1, TFEB, TFE3 and AP4 (reviewed in 41).
Next we tested whether the upstream CACGTG element (E-box) was involved in the establishment of the HpOtxE/L expression profile. When the E-box was mutated, the deletion of the TATA element decreased and/or delayed expression of the HpOtxL promoter (Fig. 3A). In contrast, the same TATA deletion induced early expression of the intact HpOtxL promoter (Fig. 3A). This indicates that the difference was not due to the condition of the embryos used in this particular experiment. Thus, removal of the upstream CACGTG element appeared to reverse the function of the TATA sequence. It is noteworthy that the expression profile of the HpOtxL promoter lacking an E-box is very similar to that of the HpArs promoter with regard to the effect of the deletion of the TATA element (Figs 2B and 3A). A similar functional interaction between the TATA element and E-box was also observed for the HpOtxE promoter (Fig. 3B), in that the delayed expression of the HpOtxE promoter caused by insertion of the TATA element (Figs 2C and 3B, lanes 1–10) was negated by mutation of the E-box (Fig. 3B, lanes 11–20). These results suggest that as yet unidentified b/HLH/Z transcription factor(s) that are specifically bound to the CACGTG element may communicate with TFIID (or TBP) to direct the temporally specific expression of the HpOtxE/L promoters.
Isolation and characterization of TBP from H.pulcherrimus
cDNA clones encoding sea urchin TBP have been isolated from two related species, S.purputatus and Lytechinus variegatus, by Childs and co-workers (31). It has been shown that the amount of TBP protein is relatively constant throughout embryogenesis in S.purputatus (31). Our observations that TATA utilization by the HpOtxE/L promoters is developmentally regulated by an upstream CACGTG element raises the possibility that the amount of TBP might be different in H.pulcherrimus embryos than in S.purputatus, even though they are taxonomically very closely related.
To address this issue, we isolated the cDNA encoding H.pulcherrimus TBP (HpTBP) by PCR amplification and cDNA library screening. The longest cDNA we obtained was 2137 bp. The ORF encoded a protein of 250 amino acids with an estimated molecular weight of 27 kDa (Fig. 4A). Although the cDNA encoding S.purputatus TBP (SpTBP) has been shown to have a long 5′-UTR (466 bp) (31), we found that HpTBP cDNA has a much shorter 5′-UTR (105 bp). However, HpTBP does have a longer 3′-UTR (1302 bp) (data not shown). The amino acid sequence of HpTBP is 99, 98 and 82% identical to those of S.purputatus (Sp), L.variegatus (Lv) and Saccharomyces cerevisiae (Sc), respectively, within the C-terminal highly conserved region, and 82, 73 and 6.7% identical within the N-terminal non-conserved region (Fig. 4B). Phylogenic analysis (42) confirmed that HpTBP is more closely related to SpTBP than to LvTBP and ScTBP (data not shown).
Figure 4.
(A) Amino acid sequence alignment of HpTBP with those of TBPs from other sea urchin species (Sp, S.purputatus; Lv, L.variegatus) and yeast (Sc, S.cerevisiae). An asterisk denotes the boundary between the N-terminal non-conserved region and the C-terminal highly conserved region. Dashes indicate gaps introduced to maintain optimal alignment. Shading indicates identical residues. (B) Schematic representation of the comparison of the four TBPs. The C-terminal highly conserved regions are shaded. The percentage of identical residues when compared with HpTBP are calculated individually for the N- and C-terminal regions and are shown in each rectangle.
To further verify that the isolated cDNA encodes a bona fide HpTBP, we examined its TATA binding activity (Fig. 5A, left) and its ability to interact with yeast TFIIA (Fig. 5A, right) as well as with the N-terminal domain of yeast TAF145 (TAND) (33) (Fig. 5B). We verified that the recombinant HpTBP protein was able to bind to the TATA element of the adenovirus major late (AdML) promoter (Fig. 5A, left) and to TAND (Fig. 5B) as strongly as yeast TBP. In addition, HpTBP can form a complex with yeast TFIIA on the AdML promoter, although the affinity of HpTBP for yeast TFIIA appears to be much weaker than that of yeast TBP (Fig. 5A, right). These observations indicate that the isolated cDNA encodes a functional H.pulcherrimus TBP.
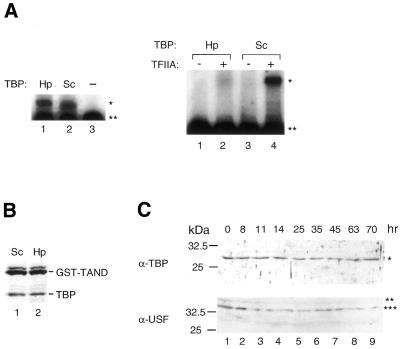
Figure 5.
In vitro and in vivo characterization of HpTBP. (A) Gel mobility shift analyses of TBP–TATA element interactions (left) and TFIIA– TBP–promoter DNA interactions (right). Hp and Sc indicate the source of TBP as H.pulcherrimus and S.cerevisiae, respectively. Adenovirus major-late promoter (–119 to +61) was used as a probe. The amount of protein used for each assay was 20 ng of TBP (left), 10 ng of TBP and 12 ng of yeast TFIIA (right). The positions of the TBP–DNA complex and TFIIA–TBP–DNA complex are indicated by an asterisk on the right. A double asterisk denotes the position of the free probe. (B) Interaction of TBP with GST–TAND. GST–TAND fusion proteins were incubated with an equimolar amount of TBP. Complexes were obtained by incubating the proteins with glutathione Sepharose beads, and washing them extensively with buffer containing 0.2 M KCl. The bound proteins were then analyzed by SDS–PAGE and Coomassie blue staining. (C) Immunoblot analysis of extracts prepared from whole embryos collected at indicated times post- fertilization. Anti-HpTBP and -HpUSF polyclonal antibodies were used as primary antibodies. A single asterisk denotes the position of HpTBP; double and triple asterisks denote the positions of alternative forms of HpUSF-like factors. The size markers are shown on the left. (D) Amino acid sequence alignment of the N-terminal region of HpUSF with those of corresponding regions of USFs from other sea urchin species (Sp, S.purputatus; Lv, L.variegatus). Shading indicates identical residues.
We generated polyclonal antibodies against HpTBP to compare the expression level at each stage of embryogenesis. Immunoblot analysis showed that HpTBP levels were almost constant throughout embryogenesis, at least when calculated on a single-embryo basis (Fig. 5C, upper panel). This is similar to what is observed in S.purputatus (31). A polyclonal antibody against yeast TBP recognized SpTBP as a closely spaced doublet at, and after, the blastula stage, indicating that SpTBP is post-translationally modified during this period of embryogenesis. However, our antibody did not reveal any such modification of HpTBP (Fig. 5C, upper panel).
Isolation of the USF from H.pulcherrimus involved in vitro E-box binding activity
Since we did not observe any significant changes in the amount or modification state of HpTBP, we also examined whether CACGTG binding factors were developmentally regulated. The major protein in sea urchin extracts capable of binding to the CACGTG element has been shown to be USF (34). Sea urchin USF is known to be required for transcription of genes such as Spec (43) and U6 (34). Importantly, human USF can increase the rate or stability of TFIID binding to the promoter and thereby stimulate transcription (44–46). To obtain an anti-HpUSF antibody, we first isolated a partial cDNA clone encoding the N-terminal half (1–141 amino acids) of HpUSF using the RT–PCR method. The amino acid sequence of HpUSF within this region is 97 and 83% identical to those of SpUSF (43) and LvUSF1 (47), respectively (Fig. 5D). Thus, HpUSF is more similar to SpUSF than to LvUSF1, as was observed for TBP (Fig. 4). Immunoblot analysis using a polyclonal antibody against the N-terminal half of HpUSF proteins demonstrated that two forms of the HpUSF-like factor were expressed almost constantly at all of the stages examined (Fig. 5C). The smaller form was always more abundantly expressed than the larger form. It is noteworthy that two distinct forms of USF with similar molecular sizes to those of HpUSF-like factors were also expressed constantly throughout development in another sea urchin, Lytechinus pictus (47). Intriguingly, as was observed for HpUSF-like factors, the smaller form, i.e. LvUSF1, was more abundantly expressed than the larger form, i.e. LvUSF2 (47).
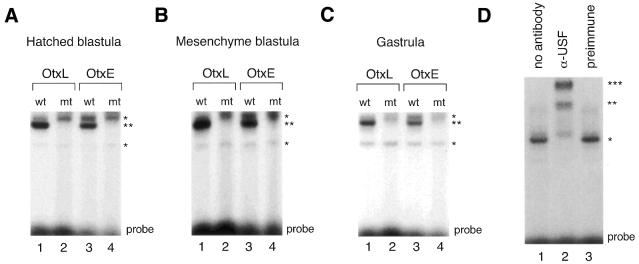
Next, we tested for E-box binding activity and for any changes in E-box binding patterns during the course of embryogenesis using probes from HpOtxE and HpOtxL promoter regions and USF1 consensus oligonucleotides (48) (Fig. 6). Gel retardation analysis was performed using nuclear extracts prepared from embryos at the hatched blastula stage, mesenchyme blastula stage and gastrula stage, corresponding to ∼12–14, 24 and 32 h after fertilization, respectively. E-box specific DNA binding activity was detected in all three nuclear extracts at similar levels with oligonucleotides (22 bp) derived from OtxE/L promoters containing the wild-type E-box sequence (Fig. 6A–C), or with USF1 consensus oligonucleotides (23 bp; purchased from Santa Cruz Biotechnology) (data not shown). The positions of the specifically retarded bands on the EMSA gel were apparently the same irrespective of the development stage and amounts of protein used in the assays (data not shown), suggesting that only one or a very restricted number of related proteins of similar size act as E-box binding factors during embryogenesis of H.pulcherrimus. To determine if HpUSF was the factor that bound to these three E-box sequences, antiserum specific for HpUSF was used. Addition of antisera against HpUSF to DNA-binding reaction mixtures containing nuclear extracts prepared at the mesenchyme blastula and wild-type OtxL probe produced supershifts of the retarded bands (Fig. 6D, lane 2), indicating that HpUSF was mainly responsible for the E-box binding activity expressed at this stage. Similar results were obtained with nuclear extracts prepared at other stages or with other E-box containing probes (data not shown). Therefore, we conclude that neither the amount nor the DNA binding activity of HpUSF(s) changed significantly during H.pulcherrimus embryogenesis, suggesting that HpUSF expression or DNA binding activity is not tightly regulated. Further analyses will be required to determine if any other regulatory function of HpUSF(s), e.g. interaction with basal transcriptional machineries, is involved in the temporally specific expression of the HpOtx gene.
Figure 6.
E-box binding activities detected in stage-specific nuclear extracts. (A) Gel mobility shift analysis was performed using nuclear extracts (2 µg) prepared from embryos at the hatched blastula stage and double-stranded oligonucleotide extending either from base pairs –229 to –208 of the HpOtxL promoter (OtxL) or from base pairs –75 to –54 of the HpOtxE promoter (OtxE) as probe. ‘wt’ and ‘mt’ indicate wild-type and mutant probes, respectively; the latter probe contains the mutant AAGCTT sequence while the former probe contains the wild-type CACGTG sequence. The positions of the most prominently shifted bands of the non-specific and specific E-box probes are marked by single and double asterisks, respectively, on the right. ‘Probe’ at the bottom denotes the position of the free probe. (B) Gel mobility shift analysis was conducted as represented in (A) except that nuclear extracts prepared at the mesenchyme blastula stage were employed. (C) Gel mobility shift analysis was conducted as represented in (A) except that nuclear extracts prepared at the gastrula stage were employed. (D) Immunoshift analysis was performed. The wild-type OtxL probe was incubated with either nuclear extract (2 µg) alone prepared from embryos at the mesenchyme blastula stage (lane 1), or together with antiserum directed against recombinant HpUSF (lane 2) or with preimmune serum (lane 3).
DISCUSSION
In this study, we demonstrated that the functional interaction between TATA and upstream CACGTG elements contributes to establishing the temporally specific expression profile of the tandem promoters of the HpOtx gene. Immunoblot analysis showed that the amount of TBP is relatively constant throughout embryogenesis, at least when calculated on a per embryo basis. However, since the number of cells increases dramatically during embryogenesis (>1 × 103-fold), the amount of TBP per cell should be greatly reduced at later stages (31). This may explain why a promoter that should be activated at a relatively late stage (e.g. HpArs) requires the TATA element for efficient transcription (Fig. 2B). Although the expression of both HpOtxE and HpOtxL is delayed by the presence of the TATA element, there is a clear difference in the onset of gene expression between these two promoters with regard to the effect of E-box deletion. Deletion of the E-box induced earlier expression of the HpOtxE promoter irrespective of the presence of the TATA element (Fig. 3B), while the same deletion delayed expression of the HpOtxL promoter (Fig. 3A). This may be due to a difference in core promoter structure and/or promoter-specific transcription factors that still remain active on HpOtxE/L promoters lacking the E-box. For instance, the HpOtxE core promoter may be intrinsically pre-programmed to be expressed much earlier than the HpOtxL core promoter and E-box binding factors can delay the expression of the former but not of the latter.
In order to compare the expression of HpUSF at different developmental stages, we also isolated a cDNA clone encoding the N-terminal region of HpUSF and generated a specific polyclonal antibody against the expressed polypeptide. Similar to previously reported LvUSFs (47), two different sizes of HpUSF were expressed continuously throughout development with the smaller form always being expressed in higher amounts compared with the larger form (Fig. 5C). These observations are consistent with the gel retardation analyses which showed that the levels of the retarded E-box bands supershifted by anti-HpUSF antisera (Fig. 6D and data not shown) remained relatively constant during the three developmental stages (Fig. 6A–C). Hence, we assume that some other regulatory function of HpUSF required after its binding to the E-box and/or some other unidentified but minor E-box binding factor(s) must direct the faithful and temporally specific expression of the HpOtx gene in cooperation with the TATA factor. Further analysis will be required to clarify this point.
In other organisms, there are several genes like HpOtx that are known to be controlled by tandem promoters, one containing a TATA element and the other lacking this element. The Saccharomyces cerevisiae HIS3 gene is under the control of TR (consensus TATA) and TC (non-consensus TATA) proximal elements which direct transcriptional initiation at +13 and +1, respectively (reviewed in 49). Under conditions of constitutive expression, transcription is initiated with equal efficiency from both sites (50,51). However, a transcriptional activator, Gcn4, can stimulate transcription of this gene almost exclusively from the +13 site (50,51), indicating that Gcn4 activates a TATA binding factor (TFIID or TBP) in a core promoter-specific manner. The Drosophila melanogaster alcohol dehydrogenase (Adh) gene is also regulated by tandem promoters. The distal promoter contains a non-consensus TATA element and is used primarily in early- to mid-stage embryos as well as in adult flies. The proximal promoter has a consensus TATA element that is active during late embryonic and early- to mid-stage larval development (reviewed in 35). In this case, however, the initiation element appears to be more important for differential promoter utilization than the TATA element itself (35). Considering that the initiation element is also recognized by TFIID through its subunits TAF250 and TAF150 (35,52), the functional interaction between promoter-specific factors and TFIID may precisely direct the stage-specific expression of the tandem promoters of the Adh gene. The mouse gene encoding translation initiation factor eIF-1A is expressed preferentially by a proximal TATA-containing promoter in fully-grown oocytes, whereas in blastocysts it is expressed almost exclusively by a distal TATA-less promoter (36). These observations suggest that tandem promoters carrying different core promoter elements (e.g. TATA, initiator) are commonly used to direct environmentally, temporally and/or spatially specific expression of the same gene in a wide variety of organisms from yeast to mammals. Recent studies demonstrate that there are an increasing number of core promoter recognition factors besides TFIID, including TFTC (53), TRF1 (54) and TAC (55). In addition, there is some evidence for the existence of tissue-specific isoforms of TFIID (56–58). Therefore, it is likely that distinct TFIID-like complexes are involved in the expression of the HpOtxE/L promoters under physiological conditions. Our future goal is to determine the identities of the TATA and CACGTG binding factors that coordinately regulate the expression of the HpOtxE/L promoters.
ACKNOWLEGEMENTS
We would like to thank Takae Kiyama and Kazuko Takata for technical assistance and helpful discussions. We also thank Professor Hiraku Shimada for his general support and encouragement. This study was supported by grants from the Ministry of Education, Science, and Culture of Japan, and from the Asahi Glass Foundation, the NAITO Foundation, the Sumitomo Foundation and the NOVARTIS Foundation (Japan) for the Promotion of Science.
DDBJ/EMBL/GenBank accession nos AB074420 and AB082982
REFERENCES
- 1.Gilbert S.F., (1997) Developmental Biology, 5th Edn. Sinauer Associates Inc., Sunderland, MA.
- 2.Andeol Y., (1994) Early transcription in different animal species: implication for transition from maternal to zygotic control in development. Roux’s Arch. Dev. Biol., 204, 3–10. [DOI] [PubMed] [Google Scholar]
- 3.Schultz R.M., (1993) Regulation of zygotic gene activation in the mouse. Bioessays, 15, 531–538. [DOI] [PubMed] [Google Scholar]
- 4.Thompson E.M., Legouy,E. and Renard,J.P. (1998) Mouse embryos do not wait for the MBT: chromatin and RNA polymerase remodeling in genome activation at the onset of development. Dev. Genet., 22, 31–42. [DOI] [PubMed] [Google Scholar]
- 5.Prioleau M.N., Huet,J., Sentenac,A. and Mechali,M. (1994) Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell, 77, 439–449. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard D.K., and Schubiger,G. (1996) Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev., 10, 1131–1142. [DOI] [PubMed] [Google Scholar]
- 7.Davis W. Jr, and Schultz,R.M. (1997) Role of the first round of DNA replication in reprogramming gene expression in the preimplantation mouse embryo. Mol. Reprod. Dev., 47, 430–434. [DOI] [PubMed] [Google Scholar]
- 8.Forlani S., Bonnerot,C., Capgras,S. and Nicolas,J.F. (1998) Relief of a repressed gene expression state in the mouse 1-cell embryo requires DNA replication. Development, 125, 3153–3166. [DOI] [PubMed] [Google Scholar]
- 9.Schultz R.M., Davis,W.,Jr, Stein,P. and Svoboda,P. (1999) Reprogramming of gene expression during preimplantation development. J. Exp. Zool., 285, 276–282. [DOI] [PubMed] [Google Scholar]
- 10.Veenstra G.J., Destree,O.H. and Wolffe,A.P. (1999) Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol. Cell. Biol., 19, 7972–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellier S., Chastant,S., Adenot,P., Vincent,M., Renard,J.P. and Bensaude,O. (1997) Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J., 16, 6250–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worrad D.M., Ram,P.T. and Schultz,R.M. (1994) Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development, 120, 2347–2357. [DOI] [PubMed] [Google Scholar]
- 13.Lawinger P., Rastelli,L., Zhao,Z. and Majumder,S. (1999) Lack of enhancer function in mammals is unique to oocytes and fertilized eggs. J. Biol. Chem., 274, 8002–8011. [DOI] [PubMed] [Google Scholar]
- 14.Prioleau M.N., Buckle,R.S. and Mechali,M. (1995) Programming of a repressed but committed chromatin structure during early development. EMBO J., 14, 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majumder S., Zhao,Z., Kaneko,K. and DePamphilis,M.L. (1997) Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO J., 16, 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majumder S., and DePamphilis,M.L. (1994) TATA-dependent enhancer stimulation of promoter activity in mice is developmentally acquired. Mol. Cell. Biol., 14, 4258–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akasaka K., Nishimura,A., Hijikata,K., Iuchi,Y., Morokuma,J., Takahashi,M., Morikawa,H. and Shimada,H. (1995) Introduction of DNA into sea urchin eggs by particle gun. Mol. Mar. Biol. Biotechnol., 4, 255–261. [PubMed] [Google Scholar]
- 18.Kiyama T., Akasaka,K., Takata,K., Mitsunaga-Nakatsubo,K., Sakamoto,N. and Shimada,H. (1998) Structure and function of a sea urchin orthodenticle-related gene (HpOtx). Dev. Biol., 193, 139–145. [DOI] [PubMed] [Google Scholar]
- 19.Mitsunaga-Nakatsubo K., Akasaka,K., Sakamoto,N., Takata,K., Matsumura,Y., Kitajima,T., Kusunoki,S. and Shimada,H. (1998) Differential expression of sea urchin Otx isoform (HpOtxE and HpOtxL) mRNAs during early development. Int. J. Dev. Biol., 42, 645–651. [PubMed] [Google Scholar]
- 20.Kiyama T., Sasai,K., Takata,K., Mitsunaga-Nakatsubo,K., Shimada,H. and Akasaka,K. (2000) CAAT sites are required for the activation of the H. pulcherrimus Ars gene by Otx. Dev. Genes. Evol., 210, 583–590. [DOI] [PubMed] [Google Scholar]
- 21.Akasaka K., Akimoto,Y., Sato,M., Hirano,H. and Shimada,H. (1990) Histochemical detection of arylsulfatase activity in sea urchin embryos. Dev. Growth Differ., 32, 293–298. [DOI] [PubMed] [Google Scholar]
- 22.Akasaka K., Ueda,T., Higashinakagawa,T., Yamada,K. and Shimada,H. (1990) Spatial patterns of arylsulfatase mRNA expression in sea urchin embryo. Dev. Growth Differ., 32, 9–13. [DOI] [PubMed] [Google Scholar]
- 23.Akasaka K., and Shimada,H. (2001) Body plan of sea urchin embryo an ancestral type animal. Zoolog. Sci., 18, 757–770. [Google Scholar]
- 24.Akasaka K., Nishimura,A., Takata,K., Mitsunaga,K., Mibuka,F., Ueda,H., Hirose,S., Tsutsui,K. and Shimada,H. (1999) Upstream element of the sea urchin arylsulfatase gene serves as an insulator. Cell. Mol. Biol. (Noisy-le-grand), 45, 555–565. [PubMed] [Google Scholar]
- 25.Ogawa M., Akasaka,K., Mitsunaga-Nakatsubo,K. and Shimada,H. (2000) Sox regulates transcription of the sea urchin arylsulfatase gene. Dev. Growth Differ., 42, 429–435. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto N., Akasaka,K., Mitsunaga-Nakatsubo,K., Takata,K., Nishitani,T. and Shimada,H. (1997) Two isoforms of orthodenticle-related proteins (HpOtx) bind to the enhancer element of sea urchin arylsulfatase gene. Dev. Biol., 181, 284–295. [DOI] [PubMed] [Google Scholar]
- 27.Li X., Wikramanayake,A.H. and Klein,W.H. (1999) Requirement of SpOtx in cell fate decisions in the sea urchin embryo and possible role as a mediator of beta-catenin signaling. Dev. Biol., 212, 425–439. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 29.Morokuma J., Akasaka,K., Mitsunaga-Nakatsubo,K. and Shimada,H. (1997) A cis-regulatory element within the 5′ flanking region of arylsulfatase gene of sea urchin, Hemicentrotus pulcherrimus. Dev. Growth Differ., 39, 469–476. [DOI] [PubMed] [Google Scholar]
- 30.Ishii M., Mitsunaga-Nakatsubo,K., Kitajima,T., Kusunoki,S., Shimada,H. and Akasaka,K. (1999) Hbox1 and Hbox7 are involved in pattern formation in sea urchin embryos. Dev. Growth Differ., 41, 241–252. [DOI] [PubMed] [Google Scholar]
- 31.Edelmann L., Zheng,L., Wang,Z.F., Marzluff,W., Wessel,G.M. and Childs,G. (1998) The TATA binding protein in the sea urchin embryo is maternally derived. Dev. Biol., 204, 293–304. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi A., Miyake,T., Ohyama,Y., Kawaichi,M. and Kokubo,T. (2001) Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J. Biol. Chem., 276, 395–405. [DOI] [PubMed] [Google Scholar]
- 33.Kotani T., Miyake,T., Tsukihashi,Y., Hinnebusch,A.G., Nakatani,Y., Kawaichi,M. and Kokubo,T. (1998) Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem., 273, 32254–32264. [DOI] [PubMed] [Google Scholar]
- 34.Li J.M., Parsons,R.A. and Marzluff,W.F. (1994) Transcription of the sea urchin U6 gene in vitro requires a TATA-like box, a proximal sequence element and sea urchin USF, which binds an essential E box. Mol. Cell. Biol., 14, 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen S.K., and Tjian,R. (1995) TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell, 82, 565–575. [DOI] [PubMed] [Google Scholar]
- 36.Davis W. Jr, and Schultz,R.M. (2000) Developmental change in TATA-box utilization during preimplantation mouse development. Dev. Biol., 218, 275–283. [DOI] [PubMed] [Google Scholar]
- 37.Nemer M., Rondinelli,E., Infante,D. and Infante,A.A. (1991) Polyubiquitin RNA characteristics and conditional induction in sea urchin embryos. Dev. Biol., 145, 255–265. [DOI] [PubMed] [Google Scholar]
- 38.Burley S.K., and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- 39.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxevanis A.D., and Vinson,C.R. (1993) Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev., 3, 278–285. [DOI] [PubMed] [Google Scholar]
- 41.Ferre-D’Amare A.R., Pognonec,P., Roeder,R.G. and Burley,S.K. (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J., 13, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozlowski M.T., Gan,L., Venuti,J.M., Sawadogo,M. and Klein,W.H. (1991) Sea urchin USF: a helix-loop-helix protein active in embryonic ectoderm cells. Dev. Biol., 148, 625–630. [DOI] [PubMed] [Google Scholar]
- 44.Workman J.L., Roeder,R.G. and Kingston,R.E. (1990) An upstream transcription factor, USF (MLTF), facilitates the formation of preinitiation complexes during in vitro chromatin assembly. EMBO J., 9, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokubo T., Takada,R., Yamashita,S., Gong,D.-W., Roeder,R.G., Horikoshi,M. and Nakatani,Y. (1993) Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J. Biol. Chem., 268, 17554–17558. [PubMed] [Google Scholar]
- 46.Chiang C.M., and Roeder,R.G. (1995) Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science, 267, 531–536. [DOI] [PubMed] [Google Scholar]
- 47.George J.M., Seid,C.A., Lee,H. and Tomlinson,C.R. (1996) Two distinct forms of USF in the Lytechinus sea urchin embryo do not play a role in LpS1 gene inactivation upon disruption of the extracellular matrix. Mol. Reprod. Dev., 45, 1–9. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman P.W., and Chernak,J.M. (1995) DNA binding and regulatory effects of transcription factors SP1 and USF at the rat amyloid precursor protein gene promoter. Nucleic Acids Res., 23, 2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyer V., and Struhl,K. (1995) Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol., 15, 7059–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struhl K., (1986) Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell. Biol., 6, 3847–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponticelli A.S., and Struhl,K. (1990) Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol. Cell. Biol., 10, 2832–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chalkley G.E., and Verrijzer,C.P. (1999) DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250–TAF(II)150 complex recognizes the initiator. EMBO J., 18, 4835–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wieczorek E., Brand,M., Jacq,X. and Tora,L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- 54.Holmes M.C., and Tjian,R. (2000) Promoter-selective properties of the TBP-related factor TRF1. Science, 288, 867–870. [DOI] [PubMed] [Google Scholar]
- 55.Mitsiou D.J., and Stunnenberg,H.G. (2000) TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAalphabeta precursor and the TFIIAgamma subunit. Mol. Cell, 6, 527–537. [DOI] [PubMed] [Google Scholar]
- 56.Georgieva S., Kirschner,D.B., Jagla,T., Nabirochkina,E., Hanke,S., Schenkel,H., de Lorenzo,C., Sinha,P., Jagla,K., Mechler,B. et al. (2000) Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol., 20, 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freiman R.N., Albright,S.R., Zheng,S., Sha,W.C., Hammer,R.E. and Tjian,R. (2001) Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science, 293, 2084–2087. [DOI] [PubMed] [Google Scholar]
- 58.Hiller M.A., Lin,T.Y., Wood,C. and Fuller,M.T. (2001) Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev., 15, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]