Abstract
The A+U-rich element (ARE) in the 3′ non-coding region (3′ NCR) of short-lived cytokine mRNAs binds several regulatory proteins, including hnRNP D/AUF1, which comprises four isoforms of 37, 40, 42 and 45 kDa. ARE-mRNA degradation involves ubiquitin–proteasome activity, and one or more AUF1 proteins are thought to be ubiquitinated. Here we have characterized the mechanism for differential ubiquitination and degradation of the different AUF1 protein isoforms. We demonstrate in an in vitro ubiquitination system that the p37, followed by the p40 protein, are strongly ubiquitinated, whereas the p42 and p45 forms are not. Over expression in cells of enzymes that control the ubiquitin cycle were found to control p37 and p40 AUF1 protein levels through ubiquitination and proteasome activity, but not p42 and p45 forms. The p42 and p45 AUF1 proteins share a C-terminal exon 7 that is not found in the p37/p40 isoforms. Our studies show that exon 7 blocks ubiquitination and rapid degradation of AUF1 proteins, whereas its deletion permits ubiquitination to occur and promotes rapid turnover of AUF1 proteins. Thus, the stabilities of AUF1 isoforms are differentially controlled by insertion of an alternate exon that regulates ubiquitin targeting activity.
INTRODUCTION
The cytoplasmic stability of mammalian cytokine mRNAs is promoted by an A+U-rich element (ARE) that destabilizes the mRNA when present in the 3′ non-coding region (3′ NCR) (reviewed in 1,2–4). The ARE typically consists of two to five copies of the sequence AUUUA (reviewed in 5,6). The canonical ARE found in granulocyte–macrophage colony stimulating factor (GM-CSF) mRNA is sufficient to destabilize a normally stable reporter mRNA if inserted in its 3′ NCR (reviewed in 5,6). A number of proteins bind the ARE and are associated with either stabilization or destabilization of the mRNA (4). The best-characterized ARE-binding protein is HuR (7), which stabilizes ARE-mRNAs in vivo when it is overexpressed (3,8–10), and when added to an in vitro ARE-mRNA decay system (11). HuR is a predominantly nuclear protein, which undergoes rapid nucleo-cytoplasmic shuttling, and can be found bound to polysomes in the cytoplasm, presumably protecting them from rapid degradation (reviewed in 3,12).
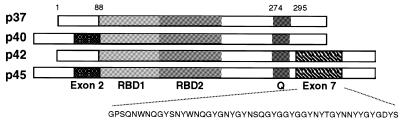
Two ARE-binding proteins facilitate destabilization of the mRNA. Binding to the ARE of the protein known as tristetraprolin promotes rapid decay of tumor necrosis factor-α and GM-CSF mRNAs (13–15). The ARE-binding protein family known as AUF1 or hnRNP D was originally identified as a labile cytoplasmic ARE-mRNA destabilizing activity in an in vitro ARE-mRNA decay system (16). AUF1 consists of four isoforms (p37, p40, p42 and p45), which arise by differential splicing of a single transcript (2,17–19) (Fig. 1). Alternate splicing of the AUF1 transcript produces a p37 core protein, a p40 protein containing an N-terminal 19 amino acid exon 2 insertion, a p42 C-terminal 49 amino acid exon 7 insertion, and a p45 protein containing both exon 2 and exon 7 (2). Differential ARE-RNA-binding characteristics have been ascribed to the different AUF1 isoforms (20), which are found in both the nucleus and cytoplasm (17,21).
Figure 1.
Structure of the four AUF1 protein isoforms. Shown is a diagram of the four isoforms of AUF1 representing the common core sequence containing RNA-binding domains 1 and 2 (RBD1 and RBD2) and a glutamine-rich element (Q), exons 2 and 7. Amino acid positions are provided relative to the core p37 AUF1 sequence. The amino acid sequence of exon 2 and exon 7 is provided below the p45 AUF1 isoform.
Binding of p37 and possibly p40 or p42 AUF1 proteins to the ARE is associated with an increased rate of ARE-mRNA degradation in a variety of studies (reviewed in 2,12,21–25). The mechanism by which AUF1 proteins promote ARE-mRNA decay is not known. One study has found AUF1 proteins in association with the human exosome, a multi-subunit complex involved in ARE-mRNA decay (26), and ARE-mRNA decay has been linked to the activity of the ubiquitin–proteasome network (21,27–29), potentially implicating ubiquitination of one or more AUF1 protein isoforms in an undescribed mechanism that might promote ARE-mRNA decay. However, AUF1 proteins are also reportedly components of a complex that binds to the α-globin 3′ NCR and promotes mRNA stabilization (30). Thus, isoforms of AUF1 in different contexts demonstrate ARE-mRNA destabilizing and stabilizing activities (31). In addition, different AUF1 isoforms may perform functions other than control of mRNA stability. Individual isoforms of AUF1 have been shown to function in a transcription factor complex for c-myc and Epstein–Barr virus promoters (32,33), to bind to nucleolin (34), to bind DNA sequences involved in immunoglobulin heavy chain class switching (35), to interact with the translation initiation complex and heat shock proteins (21), and to bind to telomeric single-stranded DNA (36) and telomerase (37). It is known that one or more AUF1 proteins may be ubiquitinated (21), which could contribute to the different functions of the AUF1 isoforms. We therefore investigated the mechanism for differential ubiquitination and turnover of the AUF1 proteins.
MATERIALS AND METHODS
Plasmids
Plasmids pcDNA-AUF1 (p37, p40, p42 and p45 isoforms) were a gift of G. Brewer (UMDNJ, NJ). All constructs were confirmed by DNA sequence analysis. Plasmid pPOLYA-(luciferase) T7, used for expression of AUF1 mRNAs and proteins in vitro, was obtained from Promega. Plasmid pUBPY was a gift of G. Draetta (European Institute of Oncology, Italy). Plasmids pUNP and pHA-Ub8 were provided by M. Pagano (NYU School of Medicine, NY). The p45 C-terminal truncation mutants were generated by restriction enzyme digestion of T7 RNA polymerase transcription constructs prior to transcription/translation reactions.
Cell culture and transfection
HeLa cells were propagated at 37°C in 7.5% CO2 using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum (Hyclone), and 50 µg/ml gentamicin sulphate. Transfection of cells was performed using Lipofectamine or Lipofectamine plus reagent (Gibco BRL) according to the manufacturer’s instructions, using 1 µg pGFP and 5 µg of empty plasmid carrier DNA per 5 × 106 cells. Additional constructs included 5 µg of pUBPY, pUNP or pHA-Ub instead of vector DNA. Cells were treated with 20 µM MG132 for up to 8 h (Calbiochem).
[35S]methionine pulse-chase analysis
HeLa cells were incubated for 30 min in Met and Cys-free DMEM followed by addition of 250 µCi per ml Met/Cys mixture (NENLife) for 1 h, with or without addition of 20 µM MG132 during the entire experiment. Following labeling, cells were incubated in complete DMEM without radioactive amino acids for the times shown. Cells were washed in 1× PBS, lysed, and equal protein amounts subjected to immunoprecipitation with anti-AUF1 antibody. Immunoprecipitates were resolved by SDS–PAGE, fluorographed, and quantified by densitometry.
Western blotting and immunoprecipitation
Cells were lysed at 4°C in NP-40 lysis buffer (50 mM Tris pH 7.5, 1 mM DTT, 0.5% NP-40, 250 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 5 mM EDTA, 0.1 mM PMSF, 50 µM E64 Boehringer protease inhibitor complete) by sonication for 5 s at low power and lysates cleared of debris by centrifugation at 10 000 g for 15 min. Immunoprecipitation analysis was performed with equal amounts of soluble protein extract (from 50 to 250 µg). Antibodies consisted of anti-AUF1/hnRNPD (a gift of G. Brewer), anti-HA (Boehringer Mannheim), anti-eIF4E (38), anti-UBPY (a gift of G. Draetta), anti-UNP (a gift of M. Pagano), anti-ubiquitin (ZYMED), anti-p53 (Santa Cruz) and anti-GAPDH (Chemicon). Western blotting was performed according to the enhanced chemiluminescence protocol (Amersham), using horse radish peroxidase- conjugated anti-goat, mouse, rabbit or rat secondary antibody (Santa Cruz). Membranes were exposed to X-ray film from 1 to 20 min to assure linear range detection.
Ubiquitin conjugation assay
The cDNAs for p37, p40, p42 and p45 isoforms of AUF1 were transcribed and translated in vitro using [35S]methionine and the TNT T7 quick coupled Transcription/Translation system (Promega). In vitro translation supernatants corresponding to equal amounts of AUF1 protein isoforms (based on in vitro translation levels) were added to HeLa cell lysate (39) prepared as described next. For studies involving denaturation of AUF1, proteins were first recovered by immunoprecipitation using AUF1 antibodies, and immune pellets were resuspended in ubiquitination buffer (described below) and heated to 100°C for 3 min. HeLa cells were lysed in buffer containing 100 mM Tris pH 8.0, 0.25% NP-40, 50 µM E-64, 1 mM PMSF. Lysates were collected, sonicated, chilled on ice and cleared of debris by centrifugation at 10 000 g for 15 min. Crude HeLa cell lysate was the source for ubiquitination enzymes (El, E2 and E3). Conjugation of ubiquitin to AUF1 was carried out in a total volume of 60 µl as described previously (39,40). Reaction mixtures contained 6 µl of in vitro translated p37, p40, p42 or p45 isoforms of AUF1, 250 µg of crude HeLa cell lysate ± 10 µg of purified ubiquitin (Sigma), 50 mM Tris–HCl pH 8.0, 3 U/ml inorganic pyrophosphate, 2 mM DTT, 50 µM E-64, 0.05 mM CaCl2. Samples were incubated in the presence of 5 mM ATP under ATP regenerating conditions using 5 mM MgCl2, 10 mM phosphocreatine and 0.2 µg/µl of creatine kinase, or upon ATP depletion. To deplete ATP in conjugation assays, crude HeLa cell lysate was incubated with 10 mM 2-deoxyglucose, 0.08 µg/µl hexokinase and 0.02 U/µl apyrase for 30 min at 37°C prior to ubiquitination reactions (40). Ubiquitin enzymes were depleted from the crude HeLa cell lysate by ubiquitin–Sepharose affinity chromatography as described previously (41,42). The cell lysate was incubated with Sepharose-bound ubiquitin for 2 h at 23°C and supernatants depleted of ubiquitin enzymes were collected. EI–E2 ubiquitin conjugating enzymes were inhibited by the addition of 5 mM iodoacetamide to the ubiquitin reactions (40). For denaturation of AUF1, in vitro synthesized AUF1 protein was recovered by immunoprecipitation, denatured at 100°C for 3 min and ice-quenched prior to addition to the ubiquitin reaction. Ubiquitin conjugation reactions were incubated at 37°C for 30 min after which the reaction was stopped with 2× SDS sample buffer. The products of the reaction were analyzed by SDS–PAGE and fluorography.
RESULTS
Differential ubiquitination of AUF1 proteins in vivo
Previous studies suggested that AUF1 protein promotes ARE-mRNA decay in a proteasome-dependent manner, possibly involving ubiquitination of one or more AUF1 proteins (reviewed in 2,21). We therefore investigated which AUF1 isoforms are targets for ubiquitination. To unambiguously establish whether polyubiquitination of AUF1 proteins occurs in vivo, cells were transfected with a plasmid expressing an octameric polyubiquitin (HA-UB8) that is tagged with hemagglutinin (HA) (43). AUF1 was specifically immunoprecipitated from equal amounts of cell lysates, with or without coexpression of HA-UB8. Proteins were resolved by SDS–PAGE and immunoblotted with antisera against HA. The epitope-tagged octameric ubiquitin has been shown to be conjugated correctly in vivo to target proteins that are degraded by proteasomes (43,44). Addition of the HA octameric ubiquitin results in a large decrease in the electrophoretic mobility of bona fide conjugates, which probably accounts for the significant shift in HA-UB AUF1 migration. Cell lysis was performed in a buffer containing N-ethyl-maleimide (NEM), which inhibits the action of deubiquitinating enzymes (45). The presence of multi-ubiquitin conjugates of AUF1 was readily apparent, consisting of HA-tagged ubiquitin only in the immunoprecipitates of cells expressing HA-UB8 (Fig. 2). Control studies showed that the non-ubiquitinated protein eIF4E was not recognized by anti-HA antibodies and did not demonstrate a slower electrophoretic mobility with expression of HA-UB, excluding non-specific conjugation. Thus, the higher molecular weight conjugates observed with anti-HA antibody indicate that AUF1 isoforms are indeed ubiquitinated in vivo.
Figure 2.
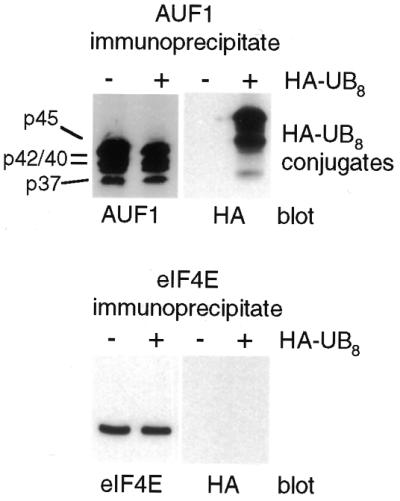
AUF1 is an in vivo substrate for ubiquitination. HeLa cells were transiently transfected with vector HA-UB8, which synthesizes a polypeptide containing eight fused ubiquitins with an N-terminal HA tag, or vector alone. Cells were lysed in NP-40 lysis buffer containing 20 mM NEM, AUF1 protein or translation initiation factor eIF4E were immunoprecipitated from equal amounts of protein lysate using anti-AUF1 antibodies that recognize all four isoforms (p37, p40, p42 and p45), or anti-eIF4E antibodies, and samples were subjected to immunoblot analysis using anti-AUF1, anti-eIF4E or anti-HA antibodies. Polyubiquitin (HA-UB8) conjugates are indicated.
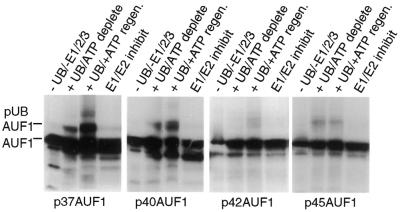
Using an in vitro ubiquitin conjugation system (39–42) it was next examined whether all four isoforms of AUF1 undergo ubiquitination, or whether it occurs differentially. In the first step of the ubiquitination pathway, ubiquitin is activated in an ATP-dependent reaction by the ubiquitin-activating enzyme E1. The activated ubiquitin molecule is then transferred from E1 by one of many E2 ubiquitin-conjugating enzymes to a member of the ubiquitin-protein ligase family, known as E3, to which the substrate protein is specifically bound. A polyubiquitin chain is synthesized by transfer of additional ubiquitin moieties to the previously conjugated molecule. The in vitro ubiquitination system reliably reproduces the hierarchical cascade and target selectivity found in vivo. The cDNAs of p37, p40, p42 and p45 isoforms of AUF1 were transcribed and translated in vitro, and supernatants corresponding to similar amounts of AUF1 proteins (based on in vitro translation levels confirmed by autoradiography; Fig. 3), were added to HeLa cell extracts containing ubiquitin conjugating enzymes E1, E2 and E3 in the presence of exogenous ubiquitin, and an ATP regenerating system (39,40). Ubiquitination of AUF1 protein was determined by generation of electrophoretically higher molecular weight forms in an ATP-dependent manner (39,40). The p37 AUF1 isoform was the most strongly ubiquitinated (Fig. 3), which was specific as it was inhibited upon depletion of all ubiquitinating enzymes, inactivation of the ATP regenerating system or inactivation of E1/E2 ubiquitinating enzymes. The p40 AUF1 isoform was also targeted for ubiquitination, but at levels reduced 3–4-fold from that of p37 AUF1 (Fig. 3). In vivo, the p42 and p45 isoforms of AUF1 are the most abundant and, accordingly, they were only weak targets for ubiquitination in vitro. Thus, the results of in vitro ubiquitination are consistent with the in vivo steady-state levels of the different AUF1 isoforms (16,21). These results indicate that the p37 and p40 isoforms of AUF1 are significant targets for ubiquitination.
Figure 3.
In vitro ubiquitination of AUF1 isoforms. Individual AUF1 protein isoform mRNAs corresponding to p37, p40, p42 or p45 were transcribed and translated in vitro in a rabbit reticulocyte lysate with [35S]methionine, equal labeled amounts of each AUF1 isoform were mixed with HeLa cell extract containing ubiquitin conjugating enzymes E1, E2 and E3 in the presence or absence of exogenous ubiquitin and an ATP regenerating system (40,41). Ubiquitin conjugation reactions were analyzed by SDS–PAGE and fluorography. Ubiquitination of AUF1 was detected by slower electrophoretic mobility, and results were quantified by digital densitometry. Typical results are shown.
Regulation of ubiquitin addition reaction controls p37 and p40 AUF1 protein levels in vivo
Deubiquitinating enzymes function as negative or positive regulators of the ubiquitin system, inhibiting or activating proteasome-mediated proteolytic degradation of proteins (46). One class of deubiquitinating enzymes facilitates the recycling of the intracellular pool of ubiquitin conjugated to degraded peptide fragments, in turn accelerating ubiquitin addition to target proteins and their degradation by proteasomes (47–50). The deubiquitinating enzyme UBPY acts in this manner, enhancing ubiquitin-mediated proteolytic degradation (45,51). Another class of deubiquitinating enzymes removes polyubiquitinated chains from proteins before degradation, inhibiting protein degradation by proteasomes when overexpressed (52,53). The UNP protein acts in this manner (54,55).
Studies presented above demonstrated that p37 and p40 AUF1 proteins were preferentially ubiquitinated. We therefore investigated whether the forward ubiquitin addition reaction or the reverse ubiquitin removal reaction on AUF1 proteins controls their abundance and stability in vivo, and therefore their ability to participate in mRNA decay. A UBPY or UNP expression vector was cotransfected into cells, and we examined the level of AUF1 proteins, translation factor eIF4E (a stable non-ubiquitinated protein; 56), and cellular p53 protein, which is ubiquitinated and degraded in proteasomes. Equal amounts of protein extracts from control and UBPY transfected cells were resolved by SDS–PAGE, transferred to nitrocellulose and subjected to immunoblot analysis (Fig. 4). Cells expressing UBPY showed significantly reduced levels of the p37 isoform (8–10-fold), and slightly reduced (by <2-fold) levels of either p40 or p42 (which cannot be electrophoretically resolved). There was no change in steady-state levels of eIF4E. Cellular p53 protein was reduced in abundance by almost 10-fold with UBPY overexpression. It should be noted that these data likely represent an underestimate of the effect of UBPY expression, as 60% of the cells were transfected (data not shown). Human UBPY protein was weakly detectable in non-transfected cells compared with transfected cells. Overexpression of the deubiquitinase UNP, which impairs protein ubiquitination and turnover (54,55), increased the level of p37 AUF1 by 5-fold, with no change in eIF4E levels. UNP moderately increased cellular p53 protein levels by ∼3–4-fold.
Figure 4.
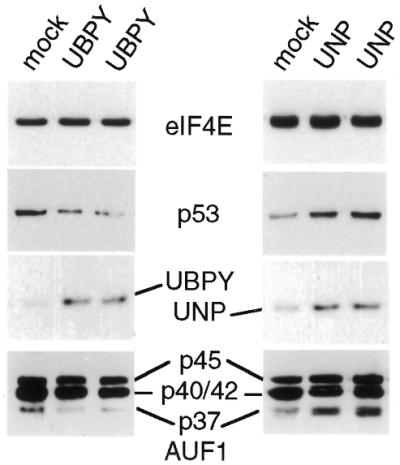
Control of AUF1 protein levels by the ubiquitination reaction. Overexpression of deubiquitinating enzyme UBPY facilitates removal of polyubiquitin from degraded peptides and accelerates protein degradation by enhancing new rounds of ubiquitination. Overexpression of deubiquitinating enzyme UNP inhibits ubiquitin-dependent protein degradation by prematurely removing polyubiquitin from undegraded proteins. HeLa cells were transiently transfected with a UBPY or UNP expression vector or vector alone. Equal amounts of protein extracts were resolved by gel electrophoresis and immunoblotted with antisera to AUF1, eIF4E, p53 and UBPY or UNP. Lanes marked UBPY or UNP represent duplicate samples.
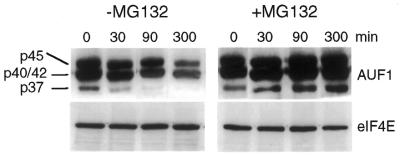
Studies next determined whether the differential ubiquitination of the four AUF1 isoforms corresponds to different stabilities and turnover rates in proteasomes. Cells were labeled with [35S]Met-Cys for 1 h and pulse-chase half-life analysis was carried out on endogenous AUF1 proteins. AUF1 protein levels were determined by immunoprecipitation of AUF1, SDS–PAGE, fluorography and densitometry (Fig. 5). The p37 and p45 isoforms of AUF1 were easily resolved, but the p40 and p42 forms typically overlap and cannot be fully separated. The p37 isoform of AUF1 displayed a short half-life, averaging a t1/2 for decay of 20 min (Fig. 5). Addition of the proteasome inhibitor MG132 to the labeling and chase reactions stabilized the p37 isoform to a half-life of almost 3 h, slightly less than the p45 isoform. These data indicate an absolute requirement for proteasome activity in rapid degradation of p37, and probably p40, AUF1 isoforms. The p45 AUF1 isoform displayed a longer half-life (t1/2 ∼8 h), which was only slightly changed by addition of MG132. The p40/42 AUF1 band demonstrated a 2.5-fold reduction by 90 min of chase, which corresponded to some loss of the bottom half of the band. We presume that this reflects loss of the p40 isoform, with a t1/2 for decay of ∼60 min, although it could also include some loss of the p42 AUF1 isoform. Addition of MG132 prevented turnover of the p40/p42 isoform of AUF1, also consistent with the essential requirement for ubiquitination and proteasome-targeted degradation. There was no change in the stability of eIF4E, which is a stable protein.
Figure 5.
Half-life analysis of AUF1 protein isoforms. HeLa cells were metabolically labeled with [35S]Met-Cys for 1 h followed by a chase with unlabeled medium for the times shown. When added, proteasome inhibitor MG132 was present throughout labeling and chase. AUF1 or eIF4E were immunoprecipitated from equal amounts of cell lysate, resolved by SDS–PAGE, fluorographed, and quantified by densitometry. Decay profiles were calculated by densitometry of autoradiograms and data plotted as log10 relative protein changes from time 0.
Ubiquitination of AUF1 is suppressed by exon 7
Studies were conducted to determine whether the poor ubiquitination of the p42 and p45 AUF1 isoforms is related to the insertion of exon 7, which is common to both isoforms and excluded from p37 and p40 forms that undergo rapid degradation in a ubiquitin–proteasome dependent manner and are strong targets for ubiquitination. To determine whether exon 7 suppresses in cis the ability of AUF1 to be ubiquitinated, we in vitro translated a mutant of the p45 protein deleted in amino acids 290 to the C-terminus (p45ΔCTD) which includes exon 7 (Fig. 6). The wild-type p45, p45ΔCTD and wild-type p37 AUF1 proteins were synthesized by in vitro translation in the presence of [35S]methionine, and equal amounts were added to an in vitro ubiquitination system (Fig. 6). As observed earlier, native p45 AUF1 is only very poorly ubiquitinated relative to the p37 AUF1 isoform. However, in vitro ubiquitination of p45ΔCTD was quite strong compared with wild-type p45 AUF1, and approximately the same as that of wild-type p37 AUF1. To determine whether loss of exon 7 or the entire CTD unmasks the ability to ubiquitinate AUF1, a deletion mutant was translated in vitro that lacked the C-terminal half of the CTD but retained exon 7 (p45Δ1/2CTD), and used to program the in vitro ubiquitination system. In vitro ubiquitination analysis of p45Δ1/2CTD showed that it was suppressed to the same low level as that of the wild-type p45 AUF1 protein. Thus, these results indicate that exon 7 suppresses in cis the ubiquitination of AUF1 protein isoforms.
Figure 6.
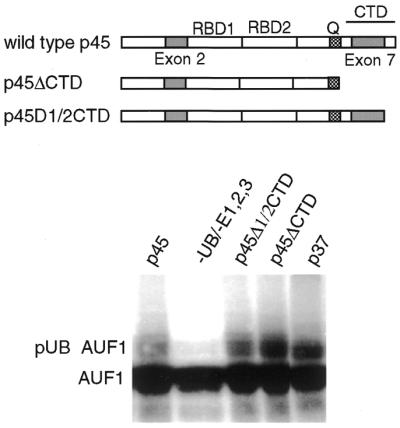
In vitro ubiquitination of AUF1 controlled by exon 7. p45 and p37 AUF1 protein isoform mRNAs were transcribed and translated in vitro in a rabbit reticulocyte lysate with [35S]methionine, equal labeled amounts of each AUF1 isoform were immunoprecipitated with antibodies to AUF1 and mixed with the HeLa cell ubiquitin conjugating system. The p45ΔCTD mutant was deleted of the entire C-terminus, including exon 7. The p45Δ1/2CTD mutant was deleted of the C-terminal half of the CTD but retains exon 7. Ubiquitin conjugation reactions were analyzed by SDS–PAGE and fluorography. Ubiquitination of AUF1 was detected by slower electrophoretic mobility, and results were quantified by digital densitometry. Typical results are shown.
DISCUSSION
We previously obtained evidence that one or more isoforms of AUF1 are likely ubiquitinated in vivo. In this report we therefore investigated which AUF1 protein isoforms are ubiquitinated, the mechanism for differential ubiquitination of highly related AUF1 proteins and whether ubiquitination is associated with differential instability. We found that the p37, and to some extent the p40, isoform of AUF1 is a predominant substrate for ubiquitin conjugation. The overexpression of UBPY, which is a post-proteasomal deubiquitinating enzyme (45), strongly and selectively reduced the abundance of the AUF1 p37 isoform, followed by the p40 protein, which is also consistent with their ubiquitin-dependent turnover rates in proteasomes as shown here as well. Conversely, overexpression of deubiquitinating enzyme UNP, which removes polyubiquitin chains from ubiquitinated proteins prior to proteasome degradation (54,55,57), resulted in a partial increased accumulation of p37 and p40 AUF1 protein and increased stability.
Previous studies implicated the p37 isoform of AUF1 as an important link that integrates the ubiquitin–proteasome network with rapid decay of ARE-mRNAs (2,21,29). Collectively with the correlation found here for AUF1 ubiquitination and degradation, it is possible that the p37 and p40 AUF1 isoforms couple ubiquitination and proteasome activity to degradation of ARE-mRNAs by participating as substrates in the decay reaction. It is notable that the rate of p37, and possibly p40, AUF1 protein turnover by the ubiquitin–proteasome system correlates with the rate of ARE-mRNA turnover, which also requires functional ubiquitin and proteasome systems for rapid decay (21,29). Future studies need to elucidate, at a molecular level, how the rapid turnover of ubiquitinated p37 AUF1 is linked to the rapid and selective degradation of ARE-mRNAs, and particularly whether it involves preferential degradation of poly(A)-binding protein on ARE-mRNAs, which has been shown to complex with AUF1 (21).
As for the selective ubiquitination of the p37 and p40 AUF1 isoforms, we noted that both isoforms differ from the p42 and p45 isoforms in lacking C-terminal exon 7. Mutagenesis of the C-terminal region of p45, which deleted exon 7 sequences, strongly increased the ability of the protein to become a ubiquitination substrate. Collectively, these data indicate that the 49 amino acid sequence that comprises exon 7 suppresses ubiquitination at an upstream site through conformational masking and possibly by sequence-specific effects. Although the ubiquitination target site has not been mapped in AUF1, our results indicate that it is likely part of the core sequence found in all four isoforms. Computer sequence analysis using the PEST-find algorithm implicates several possible sequences common to all four isoforms that could be recognition sites for E3 ligases, between position 180 and 250 relative to the p37 sequence, although none has fully definitive consensus scores (G.Laroia and R.J.Schneider, unpublished results). Exon 7 is inserted at position 294 relative to the p37 sequence. Reported differences in the strength of ARE binding and RNA sequences bound by the different AUF1 isoforms is consistent with altered AUF1 conformation mediated by exon 7 (2). Thus, it is certainly with precedent that exon 7 alters the conformation of core p37 AUF1 protein which in turn could suppress ubiquitination. Ubiquitin-related SUMOylation of topoisomerase II isoenzymes by SUMO-1 is thought to be controlled by protein conformational changes (58), which establishes a precedent for ubiquitin-like target recognition by differential protein conformation. The composition of exon 7 is unusual, averaging 53% glycine and tyrosine residues. While a glycine–tyrosine-rich sequence has not been previously associated with suppression of ubiquitination in cis in other proteins, there has also not been a specific analysis performed to date for elements that suppress ubiquitin target recognition, apart from phosphorylation-related events (46). However, polyamine analogs have been shown to induce a conformational alteration in the spermidine N1-acetyltransferase protein, masking its ubiquitination and blocking degradation by proteasomes (59). It is therefore novel, but potentially not unique, that altered protein conformation of AUF1 isoforms p42 and p45, mediated by the sequence of exon 7, mediates suppression of ubiquitination.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institutes of Health (GM60428) to R.J.S.
REFERENCES
- 1.Mitchell P., and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- 2.Guhaniyogi J., and Brewer,G. (2001) Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C.M., and Steitz,J.A. (2001) HuR and mRNA stability. Cell Mol. Life Sci., 58, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz C.J., Wormington,M. and Peltz,S.W. (2001) The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell. Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- 5.Chen C.-Y.A., and Shyu,A.-B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- 6.Wilson G.M., and Brewer,G. (1999) The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol., 62, 257–291. [DOI] [PubMed] [Google Scholar]
- 7.Myer V.E., Fan,X.C. and Steitz,J.A. (1997) Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J., 16, 2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng S.S., Chen,C.Y., Xu,N. and Shyu,A.B. (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X.C., and Steitz,J.A. (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean J.L., Wait,R., Mahtani,K.R., Sully,G., Clark,A.R. and Saklatvala,J. (2001) The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol., 21, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallouzi I.E., Brennan,C.M., Stenberg,M.G., Swanson,M.S., Eversole,A., Maizels,N. and Steitz,J.A. (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl Acad. Sci. USA, 97, 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carballo E., Lai,W.S. and Blackshear,P.J. (2000) Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood, 95, 1891–1899. [PubMed] [Google Scholar]
- 14.Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoecklin G., Ming,X.F., Looser,R. and Moroni,C. (2000) Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol., 20, 3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brewer G., (1991) An A+U rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol., 11, 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W., Wagner,B.J., Ehrenman,K., Schaefer,A.W., DeMaria,C.T., Crater,D., DeHaven,K., Long,L. and Brewer,G. (1993) Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol., 13, 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dempsey L.A., Li,M.J., DePace,A., Bray-Ward,P. and Maizels,N. (1998) The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics, 49, 378–384. [DOI] [PubMed] [Google Scholar]
- 19.Wagner B., DeMaria,C.T., Sun,Y., Wilson,G.M. and Brewer,G. (1998) Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four isoforms. Genomics, 48, 195–202. [DOI] [PubMed] [Google Scholar]
- 20.Kajita Y., Nakayama,J.-I., Aizawa,M. and Ishikawa,F. (1995) The UUAG-specific RNA binding protein, heterologous nuclear ribonucleoprotein D0. J. Biol. Chem., 270, 22167–22175. [DOI] [PubMed] [Google Scholar]
- 21.Laroia G., Cuesta,R. and Schneider,R.J. (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- 22.Loflin P., Chen,C.Y. and Shyu,A.B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pende A., Tremmel,K.D., DeMaria,C.T., Blaxall,B.C., Minobe,W.A., Sherman,J.A., Bisognano,J.D., Bristow,M.R., Brewer,G. and Port,J.D. (1996) Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J. Biol. Chem., 271, 8493–8501. [DOI] [PubMed] [Google Scholar]
- 24.DeMaria C.T., and Brewer,G. (1996) AUF1 binding affinity to A+U rich elements correlates with rapid mRNA degradation. J. Biol. Chem., 271, 12179–12184. [DOI] [PubMed] [Google Scholar]
- 25.Sirenko O.I., Lofquist,A.K., DeMaria,C.T., Morris,J.S., Brewer,G. and Haskill,S. (1997) Adhesion-dependent regulation of an A+U rich element-binding activity associated with AUF1. Mol. Cell. Biol., 17, 3898–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C.Y., Gherzi,R., Ong,S.E., Chan,E.L., Raijmakers,R., Pruijn,G.J., Stoecklin,G., Moroni,C., Mann,M. and Karin,M. (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107, 451–464. [DOI] [PubMed] [Google Scholar]
- 27.Schmid H.-P., Pouch,M.-N., Petit,F., Dadet,M.-H., Badaoui,S., Boissonnet,G., Buri,J., Norris,V. and Briand,Y. (1995) Relationships between proteasomes and RNA. Mol. Biol. Rep., 21, 43–47. [DOI] [PubMed] [Google Scholar]
- 28.Pouch M.-N., Petit,F., Buri,J., Briand,Y. and Schmid,H.-P. (1995) Identification and initial chracterization of a specific proteasome (prosome) associated RNase activity. J. Biol. Chem., 270, 22023–22028. [DOI] [PubMed] [Google Scholar]
- 29.Laroia G., and Schneider,R.J. (2002) Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl Acad. Sci. USA, 99, 1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiledjian M., DeMaria,C.T., Brewer,G. and Novick,K. (1997) Identification of AUF1 (heterogenous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol., 17, 4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu N., Chen,C.Y. and Shyu,A.B. (2001) Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol., 21, 6960–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuentes-Panana E.M., Peng,R., Brewer,G., Tan,J. and Ling,P.D. (2000) Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol., 74, 8166–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brys A., and Maizels,N. (1994) LR1 regulates c-myc transcription in B-cell lymphomas. Proc. Natl Acad. Sci. USA, 91, 4915–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanakahi L.A., Dempsey,L.A., Li,M.J. and Maizels,N. (1997) Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc. Natl Acad. Sci. USA, 94, 3605–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dempsey L.A., Hanakahi,L.A. and Maizels,N. (1998) A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem., 273, 29224–29229. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eversole A., and Maizels,N. (2000) In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol., 20, 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feigenblum D., and Schneider,R.J. (1996) Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol. Cell. Biol., 16, 5450–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stancovski I., Gonen,H., Orian,A., Schwartz,A.L. and Ciechanover,A. (1995) Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol. Cell. Biol., 15, 7106–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hershko A., Heller,H., Elias,S. and Ciechanover,A. (1985) Components of ubiquitin-protein ligase system. J. Biol. Chem., 258, 8206–8214. [PubMed] [Google Scholar]
- 41.Haas A.L., Warms,J.V., Hershko,A. and Rose,I.A. (1982) Ubiquitin-activating enzyme. J. Biol. Chem., 257, 2543–2548. [PubMed] [Google Scholar]
- 42.Pickart C.M., and Rose,I.A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem., 260, 1573–1581. [PubMed] [Google Scholar]
- 43.Treier M., Staszewski,L.M. and Bohmann,D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell, 78, 787–798. [DOI] [PubMed] [Google Scholar]
- 44.Diehl J.A., Zindy,F. and Sherr,C.J. (1997) Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev., 11, 957–972. [DOI] [PubMed] [Google Scholar]
- 45.Naviglio S., Mattecucci,C., Matoskova,B., Nagase,T., Nomura,N., Di Fiore,P.P. and Draetta,G.F. (1998) UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J., 17, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson K.D., (2000) Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol., 11, 141–148. [DOI] [PubMed] [Google Scholar]
- 47.Amerik A., Swaminathan,S., Krantz,B.A., Wilkinson,K.D. and Hochstrasser,M. (1997) In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J., 16, 4826–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hadari T., Warms,J.V., Rose,I.A. and Hershko,A. (1992) A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem., 267, 719–727. [PubMed] [Google Scholar]
- 49.Papa F.R., and Hochstrasser,M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature, 366, 313–319. [DOI] [PubMed] [Google Scholar]
- 50.Stein R.L., Chen,Z. and Melandri,F. (1995) Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry, 34, 12616–12623. [DOI] [PubMed] [Google Scholar]
- 51.Piotrowski J., Beal,R., Hoffman,L., Wilkinson,K.D., Cohen,R.E. and Pickart,C.M. (1997) Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem., 272, 23712–23721. [DOI] [PubMed] [Google Scholar]
- 52.Kalderon D., (1996) Protein degradation: de-ubiquitinate to decide your fate. Curr. Biol., 6, 662–665. [DOI] [PubMed] [Google Scholar]
- 53.Rolfe M., Chiu,M.I. and Pagano,M. (1997) The ubiquitin-mediated proteolytic pathway as a therapeutic area. J. Mol. Med., 75, 5–17. [DOI] [PubMed] [Google Scholar]
- 54.Gupta K., Copeland,N.G., Gilbert,D.J., Jenkins,N.A. and Gray,D.A. (1993) Unp, a mouse gene related to the tre oncogene. Oncogene, 8, 2307–2310. [PubMed] [Google Scholar]
- 55.Gupta K., Chevrette,M. and Gray,D.A. (1994) The Unp proto-oncogene encodes a nuclear protein. Oncogene, 9, 1729–1731. [PubMed] [Google Scholar]
- 56.Huang J., and Schneider,R.J. (1991) Adenovirus inhibition of cellular protein synthesis involves inactivation of cap binding protein. Cell, 65, 271–280. [DOI] [PubMed] [Google Scholar]
- 57.Gray D.A., Inazawa,J., Gupta,K., Wong,A., Ueda,R. and Takahashi,T. (1995) Elevated expression of Unph, a proto-oncogene at 3p21.3, in human lung tumors. Oncogene, 10, 2179–2183. [PubMed] [Google Scholar]
- 58.Mao Y., Desai,S.D. and Liu,L.F. (2000) SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem., 275, 26066–26073. [DOI] [PubMed] [Google Scholar]
- 59.Coleman C.S., and Pegg,A.E. (2001) Polyamine analogues inhibit the ubiquitination of spermidine/spermine N1-acetyltransferase and prevent its targeting to the proteasome for degradation. Biochem. J., 358, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]