Abstract
We describe a method to monitor rolling-circle replication of circular oligonucleotides in dual-color and in real-time using molecular beacons. The method can be used to study the kinetics of the polymerization reaction and to amplify and quantify circularized oligonucleotide probes in a rolling-circle amplification (RCA) reaction. Modified molecular beacons were made of 2′-O-Me-RNA to prevent 3′ exonucleolytic degradation by the polymerase used. Moreover, the complement of one of the stem sequences of the molecular beacon was included in the RCA products to avoid fluorescence quenching due to inter-molecular hybridization of neighboring molecular beacons hybridizing to the concatemeric polymerization product. The method allows highly accurate quantification of circularized DNA over a broad concentration range by relating the signal from the test DNA circle to an internal reference DNA circle reporting in a distinct fluorescence color.
INTRODUCTION
Real-time measurements of the polymerase chain reaction (PCR) have enabled convenient quantification of target sequences over a wide concentration range by the use of homogenous detection assays for the amplification products (1). Most commonly the assays are based on fluorescence resonance energy transfer (FRET), or fluorescence by compounds that intercalate in nucleic acids.
Hairpin-shaped hybridization probes, so-called molecular beacons, have been used to detect, distinguish or quantify amplification products of DNA or RNA target sequences (2). Molecular beacons are equipped with a fluorophore and a fluorescence quencher at either end, so that the fluorophore is brought close to the quencher when the free probes adopt a stem–loop structure. Upon hybridization to a target nucleic acid molecule, the fluorophore is extended away from the quencher and can emit fluorescence. The stem–loop structure can also be disrupted due to degradation or binding by proteins. This has been utilized to study the activity of single-strand nucleases (3) and single-strand binding proteins (4).
Rolling-circle amplification (RCA) has been applied for sensitive detection of nucleic acid sequences and of proteins (5,6). The method is based on the finding that also small DNA circles can be copied by a rolling-circle replication mechanism (5,7–9), known from the replication of phage genomes. RCA is particularly useful for signal amplification of padlock probes, i.e. linear DNA probes that become circularized upon recognition of a specific nucleic acid sequence (10), since only reacted probes are amplified. Padlock probes have been used to genotype samples of genomic DNA or RNA in solution (5,11–13), or for in situ genotyping of metaphase chromosomes and interphase nuclei (12,14,15). The combination of padlock probe circularization and amplification through RCA has proven useful for genetic analysis (5,16).
The course of RCA reactions has been studied using electrophoresis (5,7–9), or by hybridizing labeled oligonucleotides to RCA products attached to a glass slide (5,6,17,18). Both methods require laborious handling of gels or microscope slides, and the precision of quantification is limited. RCA reactions have also been analyzed using intercalating dyes (19). The major disadvantage of using intercalating dyes is that the nucleic acids present in the sample, as well as any non-specific polymerization products, contribute to the fluorescence. Moreover, only one RCA product can be analyzed per reaction.
Here we describe quantitative, dual-color, real-time monitoring of RCA using a modified molecular beacon design. We show that this isothermal linear signal amplification allows highly accurate quantification of circularized padlock probes.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides were kindly provided by Eurogentec, Belgium. The padlock probes used were: pp90, P-CCT CCCATCATATTAAAGGCTTTCTCTATGTTAAGTGAC CTACGACGATGCTGCTGCTGTACTACTCTTCCTAAG GCATTCTGCAAACAT; pp93, P-CCTCCCATCATATTA AAGGCTTTCTCTATGTTAAGTGACCTACGACCTCAA TGCTGCTGCTGTACTACTCTTCCTAAGGCATTCTGC AAACAT; ppWT, P-CTGCCATCTTAACAAACCCTTTC CTCTATGATTACTGACCTACGACCTCAATGCTG CTGCTGTACTACTCTTCTATGCGATTACCGGGCT and ppMUT, P-CTGCCATCTTAACAAACCCTTTCCTCTATG ATTACTGACCTACGACCTCAATGCACATGTTTG GCTCCTCTTCTATGCGATTACCGGGCC (P = 5′ phosphate). The ligation templates for the padlock probes were t40, (for pp90 and pp93) GCCTTTAATATGATGGG AGGATGTTTGCAGAATGCCTTAG; tWt (for ppWT), GTTTGTTAAGATGGCAGAGCCCGGTAATCG; and tMut (for ppMUT), GTTTGTTAAGATGGCAGGGCCCGGT AATCG. The DNA molecular beacon for detection of pp90 and pp93 RCA products was mbDNAfam (FAM-cgcctcAATGCTGCTGCTGTACTACgaggcg-DABCYL) (the stem part in lower case) and the corresponding 2′-O-Me-RNA molecular beacon was mbRNAhex (HEX-ccucAAUG CUGCUGCUGUACUACgagg-DABCYL). 2′-O-Me-RNA beacons for detection of ppWT and ppMUT RCA products were mbRNAhex and mbRNAfam (FAM-ccucAAUG CACAUGUUUGGCACCgagg-DABCYL), respectively. The stem is 2 bp shorter in the 2′-O-Me-RNA molecular beacons compared with the DNA molecular beacon because of the higher stability of 2′-O-Me-RNA base pairs. The oligonucleotide hybridization targets for the mbRNAhex molecular beacons were NONE (GAAGAGTAGTACAGCAGCA GCATCGTCGTAGGT), FLUO (GAAGAGTAGTACAGC AGCAGCATTGAGGTCGTA) and QUENCH (GAAGACC TCGTAGTACAGCAGCAGCATCGTCGT).
Padlock probe circularization
Padlock probes (200 nM) were ligated in 10 mM Tris–acetate pH 7.5, 10 mM magnesiumacetate, 50 mM NaCl, 1 mM ATP, 1 µg/µl BSA and 0.2 U/µl T4 DNA ligase (Amersham Pharmacia Biotech, Uppsala, Sweden) at 37°C for 30 min in the presence of 600 nM ligation template.
HPLC characterization of phi29-polymerase-dependent degradation of molecular beacons
Three picomoles of molecular beacons from reactions with or without Φ29 DNA polymerase were analyzed using a LA-Chrom (Merck, Darmstadt, Germany) HPLC system with a Pheromenex Luna 5 µm C-18 column (250 × 2.0 mm). The samples were separated in a 30 min gradient from 13 to 57% Acetonitrile (AcN HPLC grade, Merck) in 0.1 M triethylammoniumacetate buffer. The column was washed for 5 min with 60% AcN and equilibrated for 5 min with 13% AcN before the next sample was injected. Fluorescence was measured at 490 nm excitation and 530 nm emission wavelengths.
Rolling-circle amplification
Polymerization reactions were performed in 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 20 mM (NH4)2SO4, 10 mM dithiothreitol, 0.2 µg/µl BSA, 0.25 mM dNTP and 2 ng/µl Φ29 DNA polymerase (kindly provided by Dr M. Salas) at 37°C, using the ligation template as primer for the polymerization. For real-time monitoring, the RCA was performed in the presence of 100 nM molecular beacon and 300 nM ROX dye. The reactions were followed in a ABI 7700 real-time PCR instrument. Fluorescence values are given as a ratio between the fluorescence emitted by the molecular beacon (FAM or HEX) and the ROX reference dye. The temperature profiles were obtained by sampling fluorescence after temperature increments of 1°C held for 30 s.
Restriction digestion of RCA products
A MseI restriction recognition sequence present once in every monomer of the RCA products was rendered double stranded and digested by adding 360 nM oligonucleotide (TCTATGTTAAGTGACC) and 73 mU/ml MseI (New England Biolabs) to the polymerization reaction. The restriction digestion was performed at 37°C for 30 min.
Quantification of probe circles using an internal reference circle
Polymerization reactions were performed and followed in real-time for 90 min as described except that 1 µg/µl BSA, no ROX reference and 50 nM tWT oligonucleotide was used as primer in a volume of 20 µl. RCA was measured for a serial dilution of circularized ppWT padlock probe, and 30 fmol ppMUT DNA circles as an internal reference. The RCA products from the two DNA circles were detected with the molecular beacons mbRNAfam and mbRNAhex, respectively.
The reactions were monitored in an ABI 7700 real-time PCR instrument. Instead of analyzing the data with the SDS v1.7 software package provided with the instrument, we used a simplified multi-component analysis to resolve the overlapping fluorescence spectra of the two fluorophores used. Individual fluorescence spectra were recorded from the two molecular beacons hybridizing to RCA products at 37°C to determine the relative contribution of the two fluorophores (fFAM and fHEX) at the optimal emission wavelength of respective fluorophore (FAM = λ1 and HEX = λ2). For each sample position in the ABI 7700 instrument a background spectrum was obtained in order to determine the background fraction at the two wavelengths (Bkgλ1 and Bkgλ2). To determine the signal from the two molecular beacons (SFAM and SHEX), we assumed the following relationships between the signals from the two molecular beacons and the measured fluorescence at respective wavelengths with 90 s interval (Fλ1 and Fλ2): Fλ1 = fFAMλ1 × SFAM + fHEXλ1 × SHEX + Bkgλ1, and Fλ2 = fFAMλ2 × SFAM + fHEXλ2 × SHEX + Bkgλ2. The ratio of the maximal slope of HEX and FAM signals as a function of time (ΔmaxHEX/ΔmaxFAM) was used as a measure of the amount of ppWT probe circles relative to the amount of reference ppMUT circles present in the RCA reaction. With this procedure we obtained more reproducible quantification, with minimal influence from well-to-well differences in background. To further minimize the contribution from instrument fluctuations the fluorescence values were averaged over scanning windows of five consecutive measurements. The maximum increase of fluorescence was similarly determined from average of scanning windows of five consecutive changes in fluorescence.
RESULTS
Protection of molecular beacons from exonucleolytic degradation
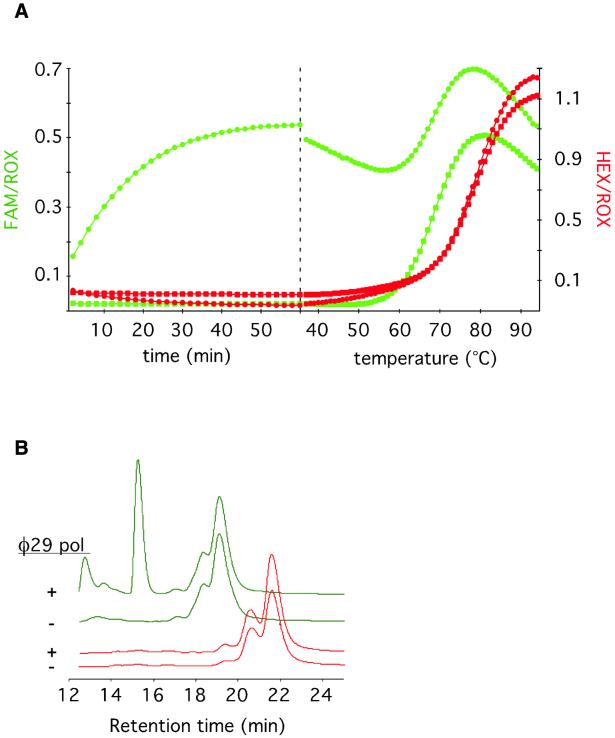
In early attempts to detect RCA products using DNA molecular beacons, we observed a non-specific accumulation of fluorescence. We suspected that this could be due to 3′ exonucleolytic degradation of the molecular beacon, separating the DABCYL quencher from the fluorophore at either end of the molecules. We therefore designed a molecular beacon composed of 2′-O-Me-RNA residues, to render the probes resistant to nuclease degradation (20). The nuclease resistance was tested in an experiment where 2′-O-Me-RNA (mbRNAhex) or DNA (mbDNAfam) molecular beacons were labeled with distinct fluorophores, and added to the same tube in the presence or absence of Φ29 DNA polymerase (Fig. 1A). The non-specific accumulation of fluorescence from 2′-O-Me-RNA molecular beacons was negligible compared with that from DNA molecular beacon. Analysis of the fluorescence temperature profiles shows that in the absence of Φ29 DNA polymerase both molecular beacons are quenched at temperatures below the melting temperatures (Tm) of their respective stems. After a 1 h incubation in the presence of Φ29 DNA polymerase, the DNA molecular beacon emits fluorescence at temperatures below its Tm, indicating that the stem has been disrupted, while the 2′-O-Me-RNA molecular beacon was unaffected by the Φ29 DNA polymerase treatment.
Figure 1.
Non-specific accumulation of fluorescence due to exonucleolytic degradation of molecular beacons can be avoided by replacing all DNA residues of the molecular beacon with 2′-O-Me-RNA residues. (A) The DNA molecular beacon mbDNAfam labeled with a FAM fluorophore (green) and the 2′-O-Me-RNA molecular beacon mbRNAhex labeled with a HEX fluorophore (red) were added to the same test tube in the presence (circles) or absence (squares) of Φ29 DNA polymerase. The left portion of the graph shows the variation of fluorescence over time at 37°C. The right portion shows the variation of fluorescence in response to increasing temperature. This temperature profile gives an indication whether the stem parts of the molecular beacons are intact at the end of the 60 min incubation. The different molecular beacons have slightly different melting characteristics because of different hybrid stability of their respective stem sequences. (B) HPLC chromatograms showing the fluorescence from mbDNAfam (green line) and mbRNAfam (red line) molecular beacons before and after a 1 h treatment with Φ29 DNA polymerase at 37°C.
The samples were also analyzed with HPLC to investigate whether the Φ29 DNA polymerase-dependent accumulation of fluorescence was due to exonucleolytic degradation of the DNA molecular beacon (Fig. 1B). As expected from the temperature profiles, the HPLC traces of the Φ29 DNA polymerase treated and untreated 2′-O-Me-RNA beacons were identical, whereas additional peaks appeared from the DNA molecular beacon treated with Φ29 DNA polymerase compared with the untreated DNA molecular beacon. The new peaks are less hydrophobic than the original material, which is consistent with loss of the hydrophobic quencher and perhaps one or more nucleotides at the 3′ end. The degradation products emit more fluorescence than the corresponding decrease of fluorescence from the original material, which provides further evidence that the quencher has been physically separated from the fluorophore.
Inter-molecular quenching of molecular beacons
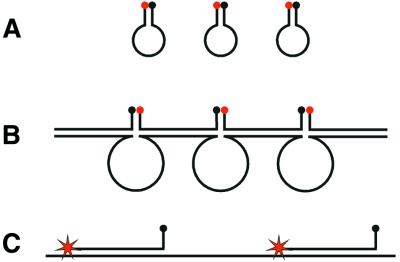
Despite the chemical modification of the molecular beacons we were still unable to obtain reliable fluorescence signals from RCA products using a conventional molecular beacon design. We hypothesized that this could be due to hybridization between stem segments of neighboring molecular beacons when hybridizing to an RCA product. Thereby the fluorescence quencher and the fluorophore of the molecular beacons would be brought close together by inter- rather than intra-molecular hybridization (Fig. 2). We therefore redesigned the original pp90 padlock probe to a new pp93 version that generated an RCA product which included the complement of one of the stem sequences of the molecular beacon. With this modification the molecular beacons hybridize to the RCA product with one of the stem arms in addition to the loop part. We expected that this would prevent inter-molecular molecular beacon quenching.
Figure 2.
A schematic representation of the expected intra- and inter- molecular hairpin structures that quench fluorescence from molecular beacons when free in solution, or when hybridized to RCA products, and how the inter-molecular quenching structures can be avoided by modifying the molecular beacon design. (A) Molecular beacons form intra-molecular stem–loop structures when they are free in solution. (B) When the molecular beacons hybridize with a concatemeric RCA product, they can also hybridize to neighboring molecular beacons, thereby forming the stems in inter-molecular stem–loop structures, where the loops are formed by the RCA product. (C) The inter-molecular stem–loop structure can be avoided by introducing a stem-complementary sequence in the RCA product.
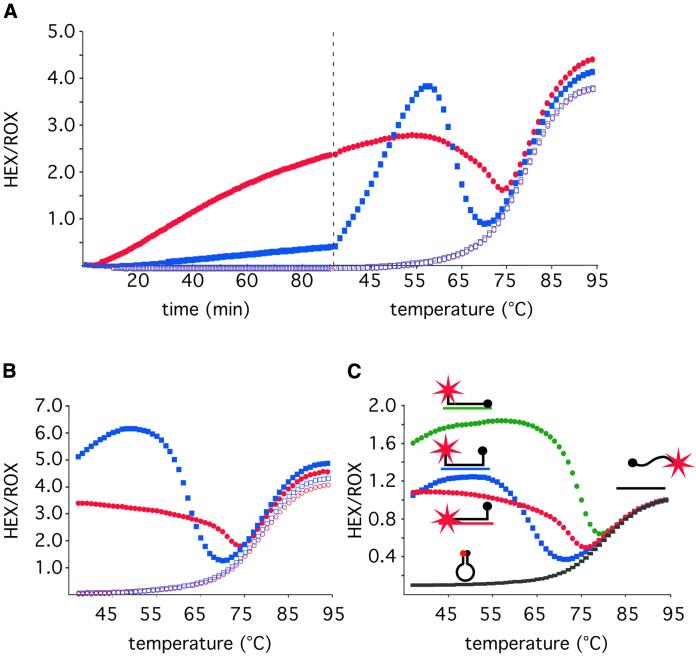
Figure 3A (left) demonstrates the results of an experiment where the mbRNAhex molecular beacon was used to follow in real-time RCA reactions of either of the two padlock probe versions. The molecular beacon signal was ∼6-fold stronger in the RCA reaction with the modified pp93 padlock probe compared with the original pp90 version. The temperature profiles of the two RCA products support the notion that inter-molecular stems were formed between neighboring molecular beacons hybridizing to the RCA product. The quenched inter-molecular structures formed in the RCA product from the original version of the padlock probe melt at a temperature ∼35°C lower than that required to melt the intra-molecular hairpins of molecular beacons that do not hybridize to a target sequence (Fig. 3A, right). The lower Tm of the hairpin structure formed by the RCA products compared with the one formed by free molecular beacons is probably a consequence of increased entropy due to the larger loops formed by the RCA products.
Figure 3.
Comparison of real-time RCA of circularized padlock probes that generate quenching or non-quenching products, and demonstration of the intra-molecular quenching structure by varying the temperature or by restriction digestion of the RCA products. (A) A real-time fluorescence measurement (left) was followed by a temperature gradient to study the temperature dependence of the fluorescence (right). Analysis of the fluorescence temperature profile demonstrates the inter-molecular quenching structure. RCA was performed of padlock probes subjected to ligation (filled symbols) or no ligation (open symbols). The RCA reactions were templated by 10 fmol of either the pp90 (blue squares) or the pp93 (red circles) padlock probes, producing quenching and non-quenching RCA products, respectively. In the left portion of the graph the fluorescence from the mbRNAhex molecular beacon was followed in real-time at 37°C. The right portion shows the fluorescence temperature profile of the products at the end of the RCA. (B) Temperature profiles of the products generated in (A) after restriction digestion. (C) Temperature profiles of the mbRNAhex molecular beacon in presence of no hybridization target (black rectangles), or hybridization targets that in addition to the loop sequence also hybridizes to the quencher-carrying stem sequence (QUENCH, green circles), the fluorophore-carrying stem sequence (FLUO, red circles), or none of the stem sequences (NONE, blue squares).
To further verify that the reduced signal from molecular beacons hybridizing to the original RCA product design was due to inter-molecular quenching of the molecular beacons rather than differences in the amount of RCA product, the RCA products were monomerized by adding an oligonucleotide complementary to a MseI restriction enzyme recognition sequence in the RCA product, along with the corresponding restriction enzyme. Fluorescence temperature profiles of the restriction-digested products were recorded after a 30 min incubation at 37°C (Fig. 3B). After restriction digest, the molecular beacons hybridizing to the quenched version of the RCA product emitted 10-fold more fluorescence at 37°C compared with when hybridizing to the non-digested product, while the fluorescence from molecular beacons hybridizing to the non-quenched version of the RCA product was only marginally affected by the restriction digestion.
Notably, the restriction-digested originally quenched product, fluoresce 2-fold stronger than the corresponding non-quenched product at 50°C. This phenomenon could not be explained by differences in restriction digestion efficiencies or in molar amounts of the two products, as judged by gel electrophoresis analysis of the restriction-digested products (data not shown). The molecular beacon hybridizes to the two RCA products in a slightly different manner. In the original design both the quencher and the fluorophore are connected to stem sequences that do not hybridize to the target sequence, while in the non-quenched version, the stem sequence that carries the fluorophore hybridizes to the target (Fig. 2). To investigate whether this physical difference could explain the difference in fluorescence, we synthesized three oligonucleotide hybridization targets to which the molecular beacon would hybridize in three different ways. To the first target the molecular beacon could hybridize only with the loop segment, as a traditional molecular beacon. To the other two targets, either the quencher stem or the fluorophore stem could hybridize in addition to the loop sequence. Figure 3C shows the fluorescence temperature profiles of the mbRNAhex molecular beacon hybridizing to the three different targets. Evidently the target that hybridizes to the fluorophore-carrying stem yields less fluorescence than the one hybridizing to the quencher-carrying stem. The decreased fluorescence from molecular beacons with hybridized fluorophore stem may be due to quenching caused by the bases in the target sequence (21).
Quantitative analysis of circularized padlock probes
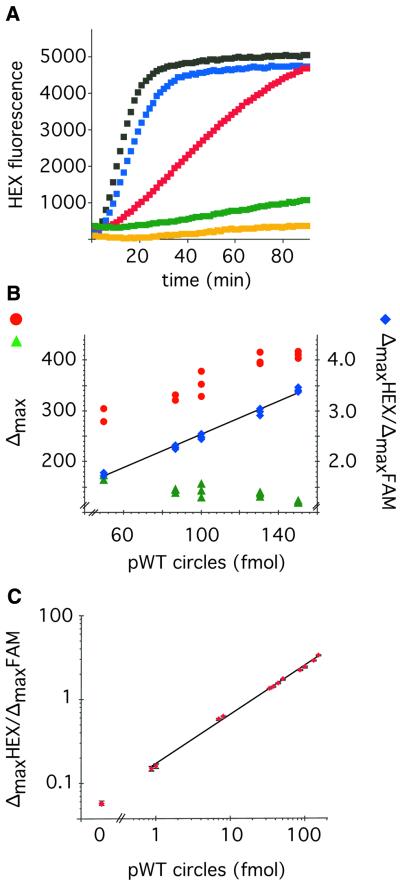
As the RCA is a linear signal amplification reaction, it should be possible to measure accurately the amount of circles present in a given sample. To investigate this we made a dilution series of preformed ppWT probe circles, and used these as templates in an RCA reaction. Each reaction also contained a fixed amount of circularized ppMUT probe serving as an internal quantification reference to standardize quantifications in separate experiments. The RCA of the ppWT was monitored for HEX fluorescence using the mbRNAhex molecular beacon, while the reference probe circle templates an RCA that was monitored using the mbRNAfam molecular beacon labeled with a separate fluorophore (FAM). The increase in HEX and FAM fluorescence was followed in real-time and the maximal slope from the two molecular beacons (ΔmaxHEX and ΔmaxFAM) was determined in each reaction. Figure 4A shows the increase in HEX fluorescence from a subset of these reactions. The slope of the fluorescence increase was steeper for RCA reactions templated by higher concentrations of probe circles. As demonstrated in Figure 4B the ΔmaxHEX signal increases in proportion to the number of probe circles in the reaction. By normalizing the ΔmaxHEX signal to the ΔmaxFAM signal in the same reaction, the proportionality in signal response to the number of probe circles in the reaction improved, and the experimental variability was reduced. Figure 4C shows that the ΔmaxHEX/ΔmaxFAM signal ratio is proportional to the amount of probe circles in the reaction over a >100-fold concentration range with an average CV of 1.7% (based on 12 triplicate measurements with a range of 0.27–3%).
Figure 4.
Real-time quantification of probe circles using RCA and molecular beacons. (A) HEX generated fluorescence from mbRNAhex molecular beacons plotted against time from RCA reactions containing 0 (orange), 0.87 (green), 8 (red), 50 (blue) and 100 (black) fmol ppWT probe circles, respectively. (B) ΔmaxHEX (red circles) and ΔmaxFAM (green triangles) signal values from the mbRNAhex and mbRNAfam molecular beacons, respectively, plotted against the amount of ppWT probe circles present in triplicate RCA reactions containing 30 fmol ppMUT reference circles. The corresponding ΔmaxHEX/ΔmaxFAM signal ratios were calculated for the individual reactions and plotted as blue diamonds. (C) The ΔmaxHEX/ΔmaxFAM signal ratio is plotted against the amount of ppWT probe circles (HEX signal) present in RCA reactions containing 30 fmol ppMUT reference circles (FAM signal). The data points are from reactions containing 0.87, 1, 7, 8, 33, 38, 44, 50, 87, 100, 130 and 150 fmol of ppWT probe circles. The error bars represent the standard deviation of triplicate reactions at each concentration of ppWT probe circles.
DISCUSSION
We show herein that molecular beacons can be used to monitor RCA reactions in real-time, allowing highly quantitative measurements of the number of circles templating the RCA. However, the molecular beacon design had to be modified in two ways to obtain reliable results.
First, the molecular beacon had to be resistant to the 3′ exonucleolytic activity of the polymerase. In this study we used the Φ29 DNA polymerase as it is a very efficient RCA polymerase (9), but it also has a very strong 3′ exonucleolytic activity (22). We found that by replacing all DNA residues in the molecular beacon with 2′-O-Me-RNA residues, the degradation of molecular beacons was avoided. More recently, we have found that it is sufficient to replace the three 3′-most residues in a 5 bp stem with 2′-O-Me-RNA to obtain full protection of molecular beacons. We have further found that also DNA molecular beacons with a 5 bp stem is degraded by the exonucleolytic activity of the polymerase, so the protection of the molecular beacons seems to be due to the 2′-O-Me-RNA residues as such, and not due to the shorter stem used in the modified molecular beacons compared with the DNA molecular beacons (F.Dahl, unpublished results).
Secondly, one of the stem sequences should be designed to hybridize to the RCA product in order to avoid inter-molecular quenching between neighboring probes (Fig. 2). Preferably the fluorophore should be connected to the non-target complementary stem sequence to avoid interactions between the fluorophore and the target sequence (Fig. 3C). In this paper the fluorophore was attached to the target-complementary stem sequence, thus the results would probably improve by using the other configuration. We are currently successfully using molecular beacons of both configurations (F.Dahl et al., unpublished results). The extent of inter-molecular quenching between neighboring molecular beacons of the traditional design probably varies with the stability of the stem sequences, the distance between the molecular beacons, and the temperature during the polymerization reaction. One could probably find conditions under which traditional molecular beacons are not quenched when hybridizing to the RCA products. However, by using the present design, the inter-molecular quenching can be avoided completely, without optimizations.
Precise quantification and specific recognition are important for accurate measurements and comparisons of mRNA expression levels. Therefore, the combination of the specificity obtained using padlock probe circularization and the precise quantitative detection described herein should prove useful for quantification of cDNA or mRNA sequences. The coefficient of variation in signal (1.7%) in our experiments corresponds to a variation in circularized probes of 2.7% (range 0.45–5.2%). The quantitative precision was greatly increased by introducing an internal reference circle that reports in a separate color. Such an active reference can normalize for differences in reaction and detection conditions among reaction wells. We noticed further that the linear range of quantification increased with the use of an internal control. This effect was due to that at the highest concentrations of HEX probe circles, the relatively small increase of ΔmaxHEX signal was compensated by a decrease of the corresponding ΔmaxFAM signal, resulting in an enhanced increase of the ΔmaxHEX/ΔmaxFAM signal ratio (Fig. 4B). For quantitative studies of specific nucleic acid sequences, a reference circle can be added to a sample in a known amount for absolute measurements, or a control probe can be circularized by detecting a reference nucleic acid sequence for relative measurements.
As concentration differences of 15% can be reliably distinguished (Fig. 4), the precision in quantification compares advantageously with that of the Taqman assay, currently the most widely used quantitative RT–PCR method. Interestingly, the precision of quantification we report is similar to that of the recently described quantitative invasive cleavage assay (23), which also represents a linear amplification technique, in contrast to the exponential amplification obtained in the Taqman assay. Therefore, the limited precision of quantification in the Taqman assay may be a consequence of the exponential mode of amplification. The sensitivity and dynamic range of the Taqman assay is however superior to the method described here. We are currently developing means to greatly increase the sensitivity of the RCA with preserved quantification accuracy (F.Dahl et al., in preparation).
Acknowledgments
ACKNOWLEDGEMENTS
Dr Margarita Salas kindly provided Φ29 DNA polymerase. Eurogentec provided padlock probes and molecular beacons of excellent quality. Ulf Landegren contributed valuable comments to the paper. M.N. was supported by a long-term EMBO fellowship. M.N. and M.G. were further funded by the Beijer Foundation, the Foundation for Medical Research in Uppsala, and Polysaccharide Research AB in Uppsala. F.D. was supported by the Swedish Research Council.
REFERENCES
- 1.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S., and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 3.Li J.J., Geyer,R. and Tan,W. (2000) Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res., 28, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J.J., Fang,X., Schuster,S.M. and Tan,W. (2000) Molecular beacons: a novel approach to detect protein–DNA interactions. Angew. Chem. Int. Ed., 39, 1049–1052. [DOI] [PubMed] [Google Scholar]
- 5.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer B., Wiltshire,S., Lambert,J., O’Malley,S., Kukanskis,K., Zhu,Z., Kingsmore,S.F., Lizardi,P.M. and Ward,D.C. (2000) Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl Acad. Sci. USA, 97, 10113–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A., and Xu,S.-Q. (1995) Rolling replication of short DNA circles. Proc. Natl Acad. Sci. USA, 92, 4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu D., Daubendiek,S.L., Zillman,M.A., Ryan,K. and Kool,E.T. (1996) Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc., 118, 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banér J., Nilsson,M., Mendel-Hartvig,M. and Landegren,U. (1998) Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res., 22, 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson M., Malmgren,H., Samiotaki,M., Kwiatkowski,M., Chowdhary,B.P. and Landegren,U. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science, 265, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson M., Barbany,G., Antson,D.-O., Gertow,K. and Landegren,U. (2000) Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol., 18, 791–793. [DOI] [PubMed] [Google Scholar]
- 12.Antson D.-O., Isaksson,A., Landegren,U. and Nilsson,M. (2000) PCR-generated padlock probes detect single nucleotide variation in genomic DNA. Nucleic Acids Res., 28, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsson M., Antson,D.-O., Barbany,G. and Landegren,U. (2001) RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res., 29, 578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson M., Krejci,K., Koch,J., Kwiatkowski,M., Gustavsson,P. and Landegren,U. (1997) Padlock probes reveal single-nucleotide differences, parent of origin and in situ distribution of centromeric sequences in human chromosomes 13 and 21. Nature Genet., 16, 252–255. [DOI] [PubMed] [Google Scholar]
- 15.Christian A.T., Pattee,M.S., Attix,C.M., Reed,B.E., Sorensen,K.J. and Tucker,J.D. (2001) Detection of DNA point mutations and mRNA expression levels by rolling circle amplification in individual cells. Proc. Natl Acad. Sci. USA, 98, 14238–14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faruqi A.F., Hosono,S., Driscoll,M.D., Dean,F.B., Alsmadi,O., Bandaru,R., Kumar,G., Grimwade,B., Zong,Q., Sun,Z. et al. (2001) High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics, 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong X., Lizardi,P.M., Huang,X., Bray-Ward,P.L. and Ward,D.C. (2001) Visualization of oligonucleotide probes and point mutations in interphase nuclei and DNA fibers using rolling circle DNA amplification. Proc. Natl Acad. Sci. USA, 98, 3940–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallur G., Luo,C., Fang,L., Cooley,S., Dave,V., Lambert,J., Kukanskis,K., Kingsmore,S., Lasken,R. and Schweitzer,B. (2001) Signal amplification by rolling circle amplification on DNA microarrays. Nucleic Acids Res., 29, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi X., Bakht,S., Devos,K.M., Gale,M.D. and Osbourn,A. (2001) L-RCA (ligation-rolling circle amplification): a general method for genotyping of single nucleotide polymorphisms (SNPs). Nucleic Acids Res., 29, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sproat B.S., Lamond,A.I., Beijer,B., Neuner,P. and Ryder,U. (1989) Highly efficient chemical synthesis of 2′-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res., 17, 3373–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurata S., Kanagawa,T., Yamada,K., Torimura,M., Yokomaku,T., Kamagata,Y. and Kurane,R. (2001) Fluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY((R)) FL-labeled probe or primer. Nucleic Acids Res., 29, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.deVega M., Lazaro,J.M., Salas,M. and Blanco,L. (1998) Mutational analysis of phi29 DNA polymerase residues acting as ssDNA ligands for 3′–5′ exonucleolysis. J. Mol. Biol., 279, 807–822. [DOI] [PubMed] [Google Scholar]
- 23.Eis P.S., Olson,M.C., Takova,T., Curtis,M.L., Olson,S.M., Vener,T.I., Ip,H.S., Vedvik,K.L., Bartholomay,C.T., Allawi,H.T. et al. (2001) An invasive cleavage assay for direct quantitation of specific RNAs. Nat. Biotechnol., 19, 673–676. [DOI] [PubMed] [Google Scholar]