Abstract
Defects of mitochondrial DNA (mtDNA) are an important cause of disease and play a role in the ageing process. There are multiple copies of the mitochondrial genome in a single cell. In many patients with acquired or inherited mtDNA mutations, there exists a mixture of mutated and wild type genomes (termed heteroplasmy) within individual cells. As a biochemical and clinical defect is only observed when there are high levels of mutated mtDNA, a crucial investigation is to determine the level of heteroplasmic mutations within tissues and individual cells. We have developed an assay to determine the relative amount of deleted mtDNA using real-time fluorescence PCR. This assay detects the vast majority of deleted molecules, thus eliminating the need to develop specific probes. We have demonstrated an excellent correlation with other techniques (Southern blotting and three- primer competitive PCR), and have shown this technique to be sensitive to quantify the level of deleted mtDNA molecules in individual cells. Finally, we have used this assay to investigate patients with mitochondrial disease and shown in individual skeletal muscle fibres that there exist different patterns of abnormalities between patients with single or multiple mtDNA deletions. We believe that this technique has significant advantages over other methods to quantify deleted mtDNA and, employed alongside our method to sequence the mitochondrial genome from single cells, will further our understanding of the role of mtDNA mutations in human disease and ageing.
INTRODUCTION
Mitochondria contain the only non-chromosomal DNA in human cells. The mitochondrial genome is a small (16.5 kb) DNA molecule that is present in multiple copies in individual mitochondria. It encodes 13 essential polypeptides of the respiratory chain and the 24 RNAs required for intramitochondrial protein synthesis (1). Mutations in this genome are recognised as an important cause of disease (2,3) and over 200 different pathogenic defects (point mutations or large-scale rearrangements) have been identified (4,5). Pathogenic mitochondrial DNA (mtDNA) mutations are frequently heteroplasmic, with mutated and wild type mtDNA coexisting in the same cell (6). Single cell and tissue culture studies have shown that the ratio of wild type to mutated mtDNA is critical in determining the expression of the genetic defect, with mutant loads in excess of 60% required to cause respiratory chain dysfunction within an individual cell (7–9).
Whilst the role of mtDNA defects in disease is well established, the role of somatic mutations in human ageing remains controversial (10,11). There is an age-dependent increase in somatic mtDNA mutations presumably due in part to mtDNA lacking both protective histones and many of the DNA repair mechanisms of the nuclear genome (12–14). However, the level of an individual mtDNA mutation within a tissue is low (<1%) when compared with the level seen in patients with inherited disease (15–17). An important finding with increasing age is the accumulation of cytochrome c oxidase (COX)-deficient cells in human tissues (18,19). These COX-deficient cells are also present in patients with primary mtDNA defects and are known to contain high levels of mutated mtDNA (9,20). In ageing it has been shown that some of the COX-deficient cells contain high levels of deleted mtDNA (21).
We have speculated that the high level of mutated mtDNA in COX-deficient cells is due to clonal expansion of an individual deletion or point mutation. To test the hypothesis that clonal expansion of mtDNA defects is a common phenomenon in ageing, it is important to develop techniques that will allow analysis of mtDNA in individual cells. We have recently described a method to analyse the entire mtDNA sequence from individual cells to detect clonal expansion of individual point mutations (22) and there are well described methods to quantify the level of heteroplasmic point mutations within individual cells (23–25). The problem we address in this paper is the detection of deleted mtDNA in individual COX-deficient cells. We describe the development of a real-time PCR technique to quantify the level of deleted mtDNA in individual cells. We show the value of this technique both in the investigation of patients with mtDNA deletions and in studying the role of mtDNA mutations in the ageing process.
MATERIALS AND METHODS
Patients
For the initial studies, DNA extracted from blood of a control subject was used. Quadriceps muscle from 12 patients with known mitochondrial disease was studied. These patients had clinical features and muscle biopsy findings suggestive of mitochondrial disease, including the presence of COX-deficient fibres. In all patients, Southern blot and long-range PCR analysis was performed on total muscle DNA homogenates to establish the molecular defect as either a single deletion (patients S1–S6) or multiple deletions (patients M1–M6) (26). The size and site of each single deletion was mapped by amplifying across the deletion, followed by sequencing of the breakpoint (27).
Histochemical assessment
Cryostat sections (20 µm) were cut from transversely orientated muscle blocks. The blocks had been mounted previously for sectioning and frozen in isopentane cooled to –190°C in liquid nitrogen. Sequential assay of COX and succinate dehydrogenase (SDH) activities identified COX-deficient fibres (9). Type 1 (oxidative fibres) and type 2 (glycolytic fibres) were identified by the assay of COX activity or by reacting sections for pH-dependent ATPase activity.
Isolation of DNA from blood, skeletal muscle and single muscle fibres
DNA was extracted from blood and whole muscle homogenate using standard methods. Individual muscle fibres were micro-dissected from 20 µm transverse sections using fine glass capillaries. The fibres were transferred to sterile microfuge tubes, each containing 10 µl of cell lysis buffer (50 mM Tris–HCl pH 8.5, 1 mM EDTA, 0.5% Tween-20, 200 µg/ml proteinase K). The fibres were incubated at 55°C for 2 h and then at 95°C for 10 min to denature the proteinase K (28).
Real-time PCR
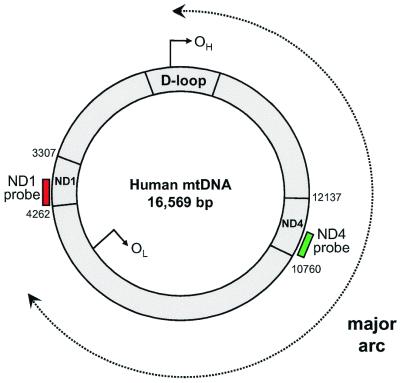
Our aim was to develop a real-time PCR protocol that could reliably quantify the amount of mtDNA in two different regions of the mitochondrial genome, one which is rarely deleted in patients, and another that is absent in the majority of patients with large-scale deletions (Fig. 1). The size and breakpoint positions of many mtDNA deletions have been carefully mapped in a large number of patients with single and multiple deletions (5). From these data, there is a region within the ND1 gene that is rarely deleted in patients (present in the genome remaining in 94% of cases with single deletions and 100% of those with multiple mtDNA deletions), and a region of the ND4 gene that is deleted in the majority of patients (82% of cases with a large-scale single deletion and 96% of cases with multiple mtDNA deletions). Having identified these regions we designed primers and probes using commercially available software (Primer Express). PCR primers and fluorogenic probes for regions of ND1 (forward primer, L3485–3504; reverse primer, H3532–3553; probe, L3506–3529) and ND4 (forward primer, L12087–12109; reverse primer, H12140–12170; probe, L12111–12138) were synthesised. Five microlitres of DNA lysate from a single muscle fibre was amplified separately with the ND1 and ND4 primer/probe combinations. To each sample 4.22 µl nanopure water and 12.5 µl TaqMan Universal PCR MasterMix [2.50 µl of 10× buffer A, 5.0 µl of 10 mM MgCl2, 0.5 µl of each dNTP (10 mM), 0.25 µl of 1 U/µl AmpErase uracil-N-glycosylase (UNG), 0.13 µl of 5 U/µl AmpliTaq Gold DNA polymerase and 2.62 µl of nanopure water] were added. The concentration of the PCR primers was 300 nM and the fluorogenic probes 100 nM. PCR and fluorescence analysis was performed using the Perkin Elmer GeneAmp 5700 sequence detection system. Amplification conditions were: 2 min at 50°C (for optimal AmpErase UNG activity), 10 min at 95°C (for deactivation of AmpErase UNG and activation of AmpliTaq Gold) then 40 cycles of 15 s at 95°C and 1 min at 60°C (for probe/primer hybridisation and DNA synthesis).
Figure 1.
Schematic representation of the mitochondrial genome. The sites of the probes used for the real-time PCR are shown. The majority of mtDNA deletions involve the major arc of the mitochondrial genome between the origin of heavy strand replication [OH (nucleotides 110–441)] and the origin of the light strand replication [OL (nucleotides 5721–5798)]. Thus, in many patients the ND4 gene is deleted whilst ND1 (which is outside this region) remains even in the deleted molecule.
Threshold cycle calculations
Threshold cycle number (Ct) was calculated with GeneAmp 5700 SDS software v1.7 (Applied Biosystems). The threshold was set at 10 times the standard deviation above the mean baseline emission value for the first 15 cycles.
Southern blotting and competitive three-primer PCR
For comparison with other methods of analysing the level of deleted mtDNA, we quantified the levels of the 4977 bp common deletion, in which ND4 is deleted, by Southern blot analysis and three-primer competitive PCR (9) using the conditions and primers described by our group previously (21).
RESULTS
Sensitivity of the PCR in relationship to mtDNA copy number
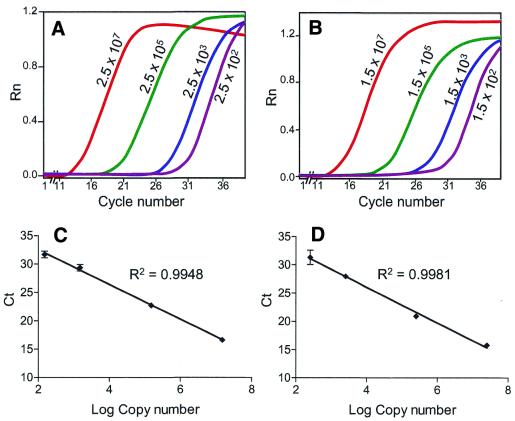
We wished to determine if it was possible to reliably detect and quantify the number of copies of ND1 and ND4 genes within single cells. For this we wished to determine the linearity of the PCR over a wide range of template concentrations. We generated templates for ND1 (2992–3640) and ND4 (11633–13595) by PCR. The concentration of each fragment was determined spectrophotometrically and real-time PCR amplifications over a copy number range of 2.5 × 102 to 2.5 × 107 and 1.5 × 102 to 1.5 × 107 for ND1 and ND4, respectively, were performed (Fig. 2A and B). The relationship between the Ct value and the logarithm of the initial copy number was linear, with correlation coefficients of 0.9948 for ND1 and 0.9981 for ND4 (Fig. 2C and D). We determined whether the technique was reproducible by determining the intra-assay variability for each template (three separate experiments with three independent observations) and the variability was <2.5% for both templates. We then determined the variability between assays by measuring Ct of the ND1 and ND4 templates in three separate experiments; the maximum variability for the ND1 template was 1.5% and for the ND4 template 5.2%. Using the standard curves generated from these templates, the mtDNA copy number in a single muscle fibre was calculated. The values for eight muscle fibres isolated from 20 µm sections ranged from 1.6 × 104 to 8.5 × 104. Thus, the amount of mtDNA in single muscle fibres is well within the linear range for quantification of the level of ND1 and ND4. We also determined the amount of mtDNA in 16 type 1 (oxidative) muscle fibres and 16 type 2 (glycolytic) muscle fibres from the same individual. As expected, the mean copy number was greater in the type 1 fibres (mean = 5.2 × 104) compared with type 2 fibres (mean = 2.6 × 104) (P < 0.05).
Figure 2.
Sensitivity of real-time PCR to mtDNA copy number. Fluorescent intensity at decreasing copy number concentrations for ND1 (A) and ND4 (B) are shown. There is an inverse linear relationship between the logarithm of the initial copy number and the threshold cycle (Ct) for ND1 (C) and ND4 (D). Each value represents the mean ± SD for four independent observations.
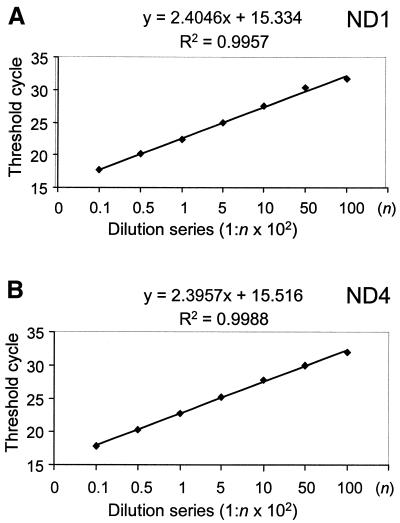
The above experiment confirmed the linearity of each template with copy number; we also wished to compare the efficiency of each reaction with whole mitochondrial genomes. DNA extracted from normal blood (500 ng/µl) was diluted serially, 1:10 to 1:10 000, and amplified using the ND1 and ND4 primers. As can be observed from Figure 3, there is an inverse linear relationship between the Ct values and the logarithm of the input blood DNA. In addition, as the gradient for amplification of ND1 and ND4 are the same, this shows that each template is amplified with the same degree of efficiency. Thus, the Ct values can be used as a measure of input DNA and to quantify the relative amount of ND1 to ND4 with the following equation: R = 2–ΔCt (29), where R is the calculated relative copy number and ΔCt is the CtND1 – CtND4.
Figure 3.
Relationship of DNA concentration to threshold cycle. Total blood DNA was serially diluted and the amplification of ND1 (A) and ND4 (B) compared at different concentrations of the original DNA (range of dilutions from 1 in 10 to 1 in 10 000).
Comparison of real-time PCR with other methods to determine the relative amount of deleted mtDNA
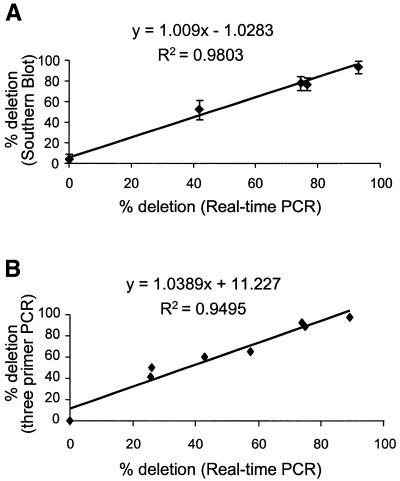
Southern blot analysis and three-primer competitive PCR methods are established methods for determining the amount of deleted mtDNA. Muscle DNA samples with varying concentrations of the 4977 bp common deletion were analysed by real-time PCR, Southern blot and three-primer competitive PCR. For real-time PCR, each sample was analysed in triplicate for both ND1 and ND4. There was a significant correlation of the level of deletion detected by real-time PCR, compared with Southern blotting and three-primer competitive PCR (Fig. 4). Furthermore, we confirmed reproducibility of this technique by repeating the comparative experiments with Southern blot on three separate occasions (Fig. 4).
Figure 4.
Comparison of the percentage of deleted mtDNA detected by real-time PCR to either Southern blot (A) or three-primer competitive PCR (B). DNA was extracted from patients and the different techniques compared. For the comparison between the real-time PCR and Southern blotting, the experiment was repeated on three separate occasions and the means ± SD are shown.
Application of real-time PCR to samples from patients with single or multiple mtDNA deletions
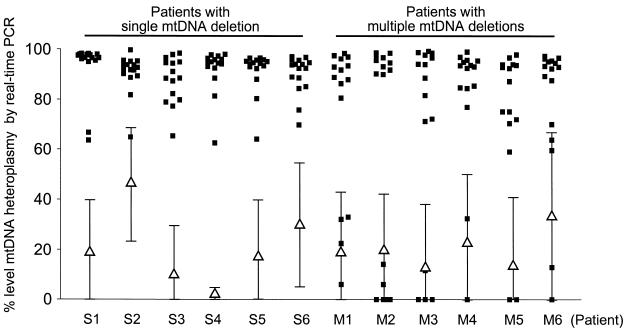
Fifteen fibres with normal COX activity and 15 COX-deficient fibres from each patient were isolated, lysed and amplified by real-time PCR. All COX-deficient fibres from patients with a known single mtDNA deletion had high ΔCt values, corresponding to high levels of deletion (>60%) (Fig. 5). This result was expected as in all cases ND4 was deleted. The COX-normal fibres also contained deleted mtDNA, but at a substantially lower level than the COX-deficient fibres [COX-deficient fibres, 90.7 ± 8.9% (mean ± SD); COX-normal fibres, 21.2 ± 25.1%]. In patients with multiple mtDNA deletions the COX-deficient fibres gave a different result to that seen in patients with a single deletion. The majority of fibres had high ΔCt values corresponding to high levels of deletion (>60%), but in all patients there were some COX-deficient fibres with normal ΔCt values, indicating the deletion in these fibres did not encompass ND4. Again there was a clear difference in the level of deletion between COX-deficient and COX-normal fibres [COX-deficient fibres, 72.1 ± 35.2% (mean ± SD); COX-normal fibres, 22.1 ± 27.8%].
Figure 5.
Real-time PCR of respiratory-deficient fibres from patients with mitochondrial myopathies. The figure shows the calculated percentage of deleted mtDNA (squares) in individual cells from COX-deficient fibres from patients with either single (S1–S6) or multiple (M1–M6) deletions. The values for 15 individual COX-deficient fibres for each patient are shown. The means ± SD for the COX-positive fibres are also shown for each patient (triangles).
DISCUSSION
Our studies have shown that real-time PCR is a valuable technique in determining the level of deleted mtDNA molecules in single cells. In our hands the technique is reproducible even using very low quantities of mtDNA. The technique will be of value not only in the diagnosis of patients with suspected mtDNA deletions, but also investigating the role of clonal expansion of mtDNA deletions in ageing. These studies have been performed using a real-time PCR machine that is capable of detecting the amplification of a single target template in each tube. A further increase in sensitivity will be possible using a real-time PCR machine capable of detecting two such reactions in the same tube by the use of different fluorescent dyes.
Real-time PCR is only one of several methods that can be used to quantify the level of rearranged mtDNA molecules, but we believe it has significant advantages over other techniques. Southern blotting is the standard method to measure the level of the deletion in DNA extracted from tissues. The major disadvantage of Southern blotting is the large amount of DNA required for analysis, and it certainly is not a technique that can be used to investigate mtDNA deletions in single cells. In contrast, three-primer competitive PCR is a semi-quantitative method used to determine the level of a deletion in a single cell. The major disadvantage of this technique is that it is specific to an individual deletion and fresh primers and conditions have to be devised for each deletion to be detected. This may be possible when a patient has a single deletion but it is not possible to use this technique for the investigation of patients with multiple deletions or in the investigation of mtDNA deletions in ageing cells. For both Southern blotting and three-primer competitive PCR, we obtained a good correlation with the results using real-time PCR. The identification of deleted molecules in individual cells is also possible using techniques such as long-range PCR (30,31) and primer-shift PCR (20,32). These techniques are not quantitative and the value of simply detecting a deletion is limited. Whilst other workers have previously described a real-time PCR method to detect a common mtDNA deletion in blood DNA (33), our specific aim was to apply this technique to the study of mtDNA rearrangements in individual cells, the level of the biochemical defect, and to confirm the validity of this approach using patient samples.
Although our original idea was merely to use the samples from patients to validate the technique of quantitative PCR, the technique has shown some very interesting differences between patients with single and multiple deletions. In all our patients with single deletions the COX-deficient cells had high levels of deletion removing ND4. In patients with multiple deletions this was not the case and all patients had COX-deficient cells that did not contain high levels of mitochondrial genome with a deletion removing ND4. Thus, this technique can rapidly differentiate between these two conditions. Whilst long-range PCR is also a very helpful technique for determining if there are single or multiple deletions, this technique can give misleading results when amplifying samples from older individuals due to the non-quantitative nature of the PCR.
Our findings in the patients with multiple mtDNA deletions were a little surprising. These are an interesting group of patients whose primary genetic defect is not in the mitochondrial genome. In these patients the defect is inherited in an autosomal manner and genetic defects have been identified in a number of genes involved in the maintenance of mtDNA, including ANT1 (34), POLG (35) and Twinkle (36), a mitochondrial helicase. In these patients, different deletions clonally expand within individual cells to high levels, and thus result in COX deficiency. It has been stated that in patients with multiple mtDNA deletions ∼96% of the deletions involve ND4; this was not found in our patients, which suggests that previous studies have underestimated the percentage of molecules in which the deletion involves other parts of the genome. Alternatively, as the primary defect often involves enzymes involved in mtDNA maintenance, it is possible that some of the other COX-deficient muscle fibres contain clonally expanded mtDNA point mutations. In none of the fibres without deleted mtDNA did we detect mtDNA depletion, as the mtDNA copy number was the same between fibres that contained deleted mtDNA species and those that did not.
There is a similarity between the situation in patients with multiple mtDNA deletions and the normal ageing process. In both groups there is the presence of COX-deficient cells. In ageing, somatic mutations occur, presumably due to a combination of DNA damage with inadequate repair. It is speculated that each COX-deficient cell should represent the clonal expansion of an individual mtDNA mutation (37). Our development of single cell sequencing of the mitochondrial genome will allow us to detect clonal expansion of mtDNA point mutations. We believe real-time PCR will provide a technique to investigate for the possibility of clonal expansion of partially deleted mtDNA molecules.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Fight for Sight, the Muscular Dystrophy Campaign and the Wellcome Trust for financial support.
REFERENCES
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1981) Sequence and organisation of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Chinnery P.F., and Turnbull,D.M. (1999) Mitochondrial DNA and disease. Lancet, 354 (Suppl. 1), S17–S21. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D.C., (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 4.Servidei S., (2001) Mitochondrial encephalomyopathies: gene mutation. Neuromusc. Disord., 11, 774–779. [DOI] [PubMed] [Google Scholar]
- 5.MITOMAP:, A Human Mitochondrial Genome Database (2001) Center for Molecular Medicine, Emory University, Atlanta, GA, USA. http://www.gen.emory.edu/mitomap.html.
- 6.Lightowlers R.N., Chinnery,P.F., Turnbull,D.M. and Howell,N. (1997) Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet., 13, 450–455. [DOI] [PubMed] [Google Scholar]
- 7.Chomyn A., Martinuzzi,A., Yoneda,M., Daga,A., Hurko,O., Johns,D., Lai,S.T., Nonaka,I., Angelini,C. and Attardi,G. (1992) MELAS mutation in mtDNA binding site for transcription termination factor causes defect in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl Acad. Sci. USA, 89, 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulet L., Karpati,G. and Shoubridge,E.A. (1992) Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet., 51, 1187–1200. [PMC free article] [PubMed] [Google Scholar]
- 9.Sciacco M., Bonilla,E., Schon,E.A., DiMauro,S. and Moraes,C.T. (1994) Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum. Mol. Genet., 3, 13–19. [DOI] [PubMed] [Google Scholar]
- 10.Linnane A.W., Marzuki,S., Ozawa,T. and Tanaka,M. (1989) Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet, 1, 642–645. [DOI] [PubMed] [Google Scholar]
- 11.Lightowlers R.N., Jacobs,H.T. and Kajander,O.A. (1999) Mitochondrial DNA—all things bad? Trends Genet., 15, 91–93. [DOI] [PubMed] [Google Scholar]
- 12.Clayton D.A., Doda,J.N. and Freiberg,E.C. (1974) The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA, 71, 2777–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter C., (1982) Do mitochondrial DNA fragments promote cancer and aging? FEBS Lett., 241, 1–5. [DOI] [PubMed] [Google Scholar]
- 14.Croteau D.L., Stierum,R.H. and Bohr,V.A. (1999) Mitochondrial DNA repair pathways. Mutat. Res., 434, 137–148. [DOI] [PubMed] [Google Scholar]
- 15.Cortopassi G., Shibata,D., Soong,N.W. and Arnheim,N. (1992) A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl Acad. Sci. USA, 89, 7370–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corral-Debrinski M., Horton,T., Lott,M.T., Shoffner,J.M., Beal,M.F. and Wallace,D.C. (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nature Genet., 2, 324–329. [DOI] [PubMed] [Google Scholar]
- 17.Melov S., Shoffner,J.M., Kaufman,A. and Wallace,D.C. (1995) Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res., 23, 4122–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller-Hocker J., (1990) Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age-related alteration. J. Neurol. Sci., 100, 14–21. [DOI] [PubMed] [Google Scholar]
- 19.Cottrell D.A., Blakely,E.L., Johnson,M.A., Ince,P.G., Borthwick,G.M. and Turnbull,D.M. (2001) Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Aging, 22, 265–272. [DOI] [PubMed] [Google Scholar]
- 20.Moslemi A.R., Melberg,A., Holme,E. and Oldfors,A. (1996) Clonal expansion of mitochondrial DNA with multiple deletions in autosomal dominant progressive external ophthalmoplegia. Ann. Neurol., 40, 693–694. [DOI] [PubMed] [Google Scholar]
- 21.Brierley E.J., Johnson,M.A., Lightowlers,R.N., James,O.F.W. and Turnbull,D.M. (1998) Role of mitochondrial DNA mutations in human ageing: implications for the central nervous system and muscle. Ann. Neurol., 43, 217–223. [DOI] [PubMed] [Google Scholar]
- 22.Taylor R.W., Taylor,G.A., Durham,S.E. and Turnbull,D.M. (2001) The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res., 29, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanno Y., Yoneda,M., Nonaka,I., Tanaka,K., Miyatake,T. and Tsuji,S. (1991) Quantitation of mitochondrial DNA carrying tRNALys mutation in MERRF patients. Biochem. Biophys. Res. Commun., 179, 880–885. [DOI] [PubMed] [Google Scholar]
- 24.Moraes C.T., Ricci,E., Bonilla,E., DiMauro,S. and Schon,E.A. (1992) The mitochondrial tRNALeu(UUR) mutation in mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am. J. Hum. Genet., 50, 934–939. [PMC free article] [PubMed] [Google Scholar]
- 25.Szuhai K., Ouweland,J., Dirks,R., Lemaitre,M., Truffert,J., Janssen,G., Tanke,H., Holme,E., Maassen,J. and Raap,A. (2001) Simultaneous A8344G heteroplasmy and mitochondrial DNA copy number quantification in myoclonus epilepsy and ragged-red fibers (MERRF) syndrome by a multiplex molecular beacon based real-time fluorescence PCR. Nucleic Acids Res., 29, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor R.W., Wardell,T.M., Blakely,E.L., Borthwick,G.M., Brierley,E.J. and Turnbull,D.M. (2000) Analysis of mitochondrial DNA mutations: deletions. In Barnett,Y.A. and Barnett,C.R. (eds), Aging Methods and Protocols. Methods in Molecular Medicine. Humana Press Inc., Totowa, NJ, vol. 38, pp. 245–264. [DOI] [PubMed]
- 27.Schon E.A., Rizzuto,R., Moraes,C.T., Nakase,H., Zeviani,M. and DiMauro,S. (1989) A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science, 244, 346–349. [DOI] [PubMed] [Google Scholar]
- 28.Zhou L., Chomyn,A., Attardi,G. and Miller,C.A. (1997) Myoclonic epilepsy and ragged red fibers (MERRF) syndrome: selective vulnerability of CNS neurons does not correlate with the level of mitochondrial tRNAlys mutation in individual neuronal isolates. J. Neurosci. 17, 7746–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 30.Fromenty B., Manfredi,G., Sadlock,J., Zhang,L., King,M.P. and Schon,E. (1996) Rapid mapping of identified partial duplications of human mitochondrial DNA by long PCR. Biochim. Biophys. Acta, 1308, 222–230. [DOI] [PubMed] [Google Scholar]
- 31.Reynier P., and Malthiery,Y. (1995) Accumulation of deletions in mtDNA during tissue aging: analysis by long PCR. Biochem. Biophys. Res. Commun., 217, 59–67. [DOI] [PubMed] [Google Scholar]
- 32.Santorelli F.M., Sciacco,M., Tanji,K., Shanske,S., Vu,T.H., Golzi,V., Griggs,R.C., Mendell,J.R., Hays,A.P., Bertorini,T.E., Pestronk,A., Bonilla,E. and DiMauro,S. (1996) Multiple mitochondrial DNA deletions in sporadic inclusion body myositis: a study in 56 patients. Ann. Neurol., 39, 789–795. [DOI] [PubMed] [Google Scholar]
- 33.Schinogl P., Müller,M. and Steinborn,R. (2001) Quantification of the 4977-bp deletion in human mitochondrial DNA using real-time PCR. Forensic Sci. Int., 122, 197–198. [DOI] [PubMed] [Google Scholar]
- 34.Kaukonen J., Juselius,J.K., Tiranti,V., Kyttala,A., Zeviani,M., Comi,G.P., Keranen,S., Peltonen,L. and Suomalainen,A. (2000) Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science, 289, 782–785. [DOI] [PubMed] [Google Scholar]
- 35.Van Goethem G., Dermaut,B., Lofgren,A., Martin,J.J. and Vam Broeckhaven,C. (2001) Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nature Genet., 28, 211–212. [DOI] [PubMed] [Google Scholar]
- 36.Spelbrink J.N., Li,F.Y., Tiranti,V., Nikali,K., Yuan,Q.P., Tatriq,M., Wanrooij,S., Garrid,N., Comi,G., Morandi,L., Santoro,L., Toscano,A., Fabrizi,G.M., Somer,H., Croxen,R., Beeson,D., Poulton,J., Suomalainen,A., Jacobs,H.T., Zeviani,M. and Larsson,C. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4 like protein localized in the mitochondria. Nature Genet., 28, 223–231. [DOI] [PubMed] [Google Scholar]
- 37.Elson J.L., Samuels,D.C., Turnbull,D.M. and Chinnery,P.F. (2001) Random intracellular drift explains the clonal expansion of the mitochondrial DNA mutations with age. Am. J. Hum. Genet., 68, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]